Abstract
The tetrapeptide, FMRFamide, was first discovered in 1977 in the molluscan nervous system and was found to affect the contractile force of molluscan cardiac muscle and other muscles [1]. Since then, numerous FMRFamide-related peptides (FaRPs) have been reported in both invertebrate and vertebrate species [2-9]. We have previously reported the detection and identification of numerous FaRPs in Cancer borealis pericardial organs (POs), one of the major neurosecretory structures in the crustaceans [2-3]. Here, we have developed two immunoaffinity-based methods, immunoprecipitation (IP) and immuno-dot blot screening assay, for the enrichment of FaRPs in C. borealis POs. A combined mass spectrometry (MS)-based approach involving both matrix-assisted laser desorption/ionization Fourier transform mass spectrometry (MALDI-FTMS) and nanoscale liquid chromatography coupled to electrospray ionization quadrupole time-of-flight tandem mass spectrometry (nanoLC-ESI-QTOF MS/MS) is used for a more comprehensive characterization of the FaRP family by utilizing high mass accuracy measurement and efficient peptide sequencing. Overall, 17 FMRFamide-related peptides were identified using these two complementary immuno-based approaches. Among them, three novel peptides were reported for the first time in this study.
Keywords: FMRFamide-related peptide, neuropeptides, immuno-dot blot screening assay, immunoprecipitation, MALDI-FTMS, ESI-QTOF
Introduction
The tetrapeptide FMRFamide was first isolated and identified in the ganglia of the venus clam Macrocallista nimbosa [1]. Since then, numerous peptides with the C-terminal motif Arg-Phe-NH2 have been discovered and characterized in a wide variety of invertebrate and vertebrate species [4-10]. Physiological studies of FMRFamide-related peptides (FaRPs) in invertebrates has revealed their highly varied biological functions including cardioexcitatory activities [1, 11-13], control of muscle contraction [13-16], neuromodulation [17, 18], and control of locomotor activity [13, 19].
Pericardial organs (POs) are one of the major neurosecretory structures in crustaceans, releasing numerous hormones that regulate a variety of physiological processes. Previous studies have reported the identification of numerous FaRPs in the Cancer borealis POs by mass spectrometric methods [2, 3]. However, a complete biochemical profile of FaRPs in POs is lacking due to the system's chemical complexity, FaRPs' low concentration, and signal suppression effects caused by the sample matrix. Therefore, a critical first step to elucidating the structure and functional relationship of this neuropeptide family is the development of methods that can enhance the detection of these low abundance peptides in complex samples.
Immuno-based methodologies have been widely employed in specific peptide/protein family detection or enrichment by antibody recognition of the specific consensus motif. These antibody based methods can facilitate highly specific targeting of single neuropeptide families. This study employs both immunoprecipitation (IP) reactions and immuno-dot blot screening assays to identify FaRPs in crude PO extracts and to screen for FaRPs in HPLC fractions, respectively. FaRP rich samples are analyzed by a combined mass spectrometric approach that harnesses the high mass accuracy/mass resolution properties of a MALDI-FTMS and the efficient de novo capabilities of nanoESI-QTOF MSMS in order to increase the coverage of FaRPs identified in Cancer borealis POs.
Materials and Methods
Animals, tissue collection, and tissue extraction
Jonah crabs (Cancer borealis) were obtained from commercial suppliers (Commercial Lobster and Seafood, Boston, MA; Marine Biological Laboratory, Woods Hole, MA) and housed in tanks containing recirculating, aerated artificial seawater (10 °C). Crabs were cold-anesthetized by packing on ice for at least 30 min prior to dissection. Dissection of the pericardial organs (POs) was performed in chilled (∼4 °C) physiological saline (composition: 440 mM NaCl; 11mM KCl; 13 mM CaCl2; 26 mM MgCl2; 10 mM HEPES acid; pH 7.4-7.5 [adjusted with 1 M NaOH]). The details of the dissection were described previously [19].
C. borealis POs were stored in a minimal volume (fully submerge tissue) of ice-cold acidified methanol (90% methanol [Fisher Scientific], 9% glacial acetic acid Fisher Scientific], and 1% deionized water) and stored at -80 °C until use. Twenty POs (10 crabs) were placed in a 0.1 mL glass tissue grinder (Wheaton), submerged in 100 μL of ice-cold acidified methanol, and mechanically homogenized. The homogenate was then transferred to a 0.6 mL low retention microcentrifuge tube (Fisher Scientific) and centrifuged at 16000 × g for 10 min. The supernatant fraction was transferred to a clean vial, and the pellet was re-extracted using ice-cold acidified methanol. The supernatant fractions were combined and concentrated to dryness using a Savant SVC 100 SpeedVac (Thermo Electron) concentrator at medium heat. Finally, 100 μL of 0.1 % formic acid(aq) (v/v, Fluka) was added to the dried extract. This resuspended extract was then vortexed and centrifuged. The supernatant was transferred to a new microcentrifuge tube and was used in future immuno-based experiments.
Immunoprecipitation
A seize X protein G immunoprecipitation kit (Pierce, IL) was used in this experiment. An aliquot of 30 μL of the protein G beads slurry (Pierce, IL) was combined with 300 μL binding/wash buffer (Pierce, IL) and added to a Handee spin cup (Peirce, IL). The beads were washed by centrifugation at 3800 × g for 2 min. The flow through was discarded and the wash step was repeated twice more. The protein G beads were allowed to interact with 2 μL of 1:100 rabbit anti-mouse polyclonal FMRFamide antibody (Abcam Inc., MA) in 60 μL binding/wash buffer with gentle rocking for 2 hrs at room temperature. After incubation, unbound FMRFamide antibody was removed by washing the beads three times with 300 μL binding/wash buffer. Either 100 μL of a standard peptide solution or 100 μL PO extract was added to the bead slurry and allowed to incubate for 3 hrs at room temperature with gentle rocking. The pH of the PO extract was adjusted to neutral by using 0.1 M ammonium bicarbonate prior to the incubation. The sequences and concentrations of the four peptide standards were: SDRNFLRFa (FMRFamide-like peptide I, lobster, m/z 1053.5588), 1.0 μM; RPPGFSPFR (Bradykinin, m/z 1060.5687), 1.0 μM; TNRNFLRFa (FMRFamide-like peptide II, lobster, m/z 1066.5905), 1.0 μM; and RPKPQQFFGLMa (Substance P, m/z 1347.7354), 1.0 μM. After incubation, the beads were washed three times with 300 μL binding/wash buffer and resuspended in 20 μL of 2% formic acid(aq) (v/v). After 15 min of incubation with gentle rocking, peptides and antibodies were eluted from the protein G beads. The eluate was then analyzed by MALDI-TOF/TOF MS, MALDI FTMS, and/or nanoESI-QTOF MS/MS.
Offline HPLC fractionation
Offline HPLC fractionation was performed using a Rainin Dynamax HPLC system equipped with a Dynamax UV-D II absorbance detector (Model SD-200, Rainin Instrument Inc., Woburn, MA). The mobile phases used were: (A) deionized water with 0.1% formic acid; (B) acetonitrile (HPLC grade, Fisher Scientific) with 0.1% formic acid. 20 μL of concentrated sample was injected via a Rainin HPLC onto a Macrosphere C18 reverse phase column (2.1 mm i.d. × 250 mm length, 5 μm particle size; Alltech Assoc. Inc., Deerfield, IL) at a flow rate of 1 mL/min. The peptides were eluted with a linear mobile phase B gradient of 5 – 95% over 120 min. Fractions were collected every minute for 120 min using a Rainin Dynamax FC-4 fraction collector (120 total fractions).
Immuno-dot blot screening assay
The HPLC fractions were assayed for immuno-reactivity to a rabbit anti-mouse polyclonal FMRFamide antibody. Briefly, 30 μL of each fraction was combined with 6 μL of 1 mg/mL bovine serum albumin (BSA, RIA grade, Fluka, Switzerland) in phosphate-buffered saline (PBS) and concentrated to dryness in a SpeedVac. Concentrated fractions were then resuspended in 6 μL of deionized water. 1 μL of each fraction was then spotted onto a nitrocellulose membrane (Schleicher & Schuell Bioscience Inc., Keene, NH) and allowed to dry in a 60 °C oven for 1 hr. The membrane was then fixed for 16 hrs at 37 °C with 2% glutaraldehyde (EM grade, Ted Pella Inc., Redding, CA) vapor in a sealed container. After fixation, a 2% glutaraldehyde solution in PBS was prepared and placed on the membrane and incubated for 1 hr to complete the fixing of the peptides. Each blot was washed six times for 15 min in PBS, blocked with blotto (5% non-fat dry milk, 0.1% Triton X-100, 0.001% Thimerosal in PBS) for 30 min, and then incubated with the FMRFamide antiserum (diluted 1:35,000 in blotto) for 16 hours at 4 °C. The blots were washed three times for 15 min in a 10% blotto solution to rinse away unbound antibody. The blot was then incubated for 1 hr with a secondary goat anti-rabbit antiserum coupled to horseradish peroxidase at a dilution of 1:500 in blotto. Unbound antibodies were removed by washing each blot three times for 15 min with 10% blotto. The bound antibodies were visualized by placing the blot in a solution of 0.006% H2O2, 0.03% 3,3′-diaminobenzidine 4-HCl, 0.5% CoCl2 in PBS (v/v/v) for 4-5 min. The membrane was washed thoroughly with deionized water and dried at room temperature. Immunoreactive fractions were identified by a reddish-brown staining in which the fraction had been added to the membrane [20-22]. The immunoreactive fractions were further analyzed by MALDI-FTMS.
MALDI-TOF/TOF MS
A model 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for peptide detection in control experiments. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). Mass spectra were obtained by averaging 625 laser shots covering mass range m/z 1000-1400. A saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA; Sigma-Aldrich, St. Louis, MO) in 50:50 acetonitrile: 0.1% trifluoroacetic acid(aq) (v/v) was used as matrix. For sample spotting, 0.35 μL of IP eluate was combined with 0.35 μL of matrix on a MALDI plate and allowed to dry prior to analysis.
MALDI-FTMS
Matrix-assisted laser desorption/ionization Fourier transform mass spectrometry (MALDI-FTMS) experiments were performed on an IonSpec ProMALDI Fourier transform mass spectrometer (Lake Forest, CA) equipped with a 7.0 Tesla actively-shielded superconducting magnet. The FTMS instrument contains a high pressure MALDI source where the ions from multiple laser shots can be accumulated in the external hexapole storage trap before the ions are transferred to the ion cyclotron resonance (ICR) cell via a quadrupole ion guide. A 337 nm nitrogen laser (Laser Science, Inc., Franklin, MA) was used for ionization/desorption. The ions were excited prior to detection with an rf sweep beginning at 7050 ms with a width of 4 ms and amplitude of 150 V base to peak. The filament and quadrupole trapping plates were initialized to 15 V, and both were ramped to 1 V from 6500 to 7000 ms to reduce baseline distortion of peaks. Detection was performed in broadband mode from m/z 108.00 to 4500.00. For sample spotting, 0.35 μL of sample was combined with 0.35 μL of matrix on a MALDI plate and allowed to dry prior to analysis.
NanoLC-ESI-QTOF MS/MS
NanoLC-ESI-QTOF MS/MS analysis of immuno-purified tissue extracts was performed using a Waters capillary LC system coupled to a QTOF Micromass spectrometer (Waters Corp., Milford, MA, USA). Chromatographic separations were performed on a reverse phase C18 capillary column (75 μm i.d. × 150 mm length, 3 μm particle size; Micro-Tech Scientific Inc.). The mobile phases used were: A, deionized H2O with 5% acetonitrile and 0.1% formic acid; B, acetonitrile with 5% deionized H2O and 0.1% formic acid; C, deionized H2O with 0.1% formic acid. An aliquot of C. borealis PO immunoprecipitation elution solution was injected and loaded onto the trap column (PepMap C18; 300 μm i.d. × 1 mm length, 5 μm particle size, 100 Å pore size; LC Packings, Sunnyvale, CA, USA) using mobile phase C at a flow rate of 30 μL/min for 3 min. Following loading, the stream select module was switched in line with the analytical capillary column, and a linear gradient of mobile phases A and B was carried out progressing from 5% to 45% B over 60 minutes. A flow splitter was added between the mobile phase mixer and the stream select module to reduce the flow rate from 12 μL/min to 200 nL/min.
The nanoflow electrospray source conditions were set as follows: capillary voltage 3800 V, sample cone voltage 40 V, extraction cone voltage 1 V, source temperature 120 °C, cone gas (N2) 13 L/hr. Data dependent acquisition was employed for the MS survey scan, the selection of precursor ions, and subsequent MS/MS of the selected parent ions. The MS scan range was m/z 100 to 2000 and the MS/MS scan was m/z 50 to 2000. De novo sequencing was performed using the PepSeq peptide sequencer of MassLynx 4.0 software (Waters) in combination with manual sequencing.
Results and Discussion
A commercial rabbit anti-mouse polyclonal FMRFamide antibody was used in both immuno-based FaRP enrichment strategies. The efficacy of this antibody for FaRPs was first determined by performing IP experiments using a standard solution of neuropeptides. This solution contained two positive control peptides (SDRNFLRFa [FMRF amide-like peptide I, lobster, m/z 1053.5588], 1.0 μM; TNRNFLRFa [FMRF amide-like peptide II, lobster, m/z 1066.5905], 1.0 μM) and two negative control neuropeptides (RPPGFSPFR [Bradykinin, m/z 1060.5687], 1.0 μM; RPKPQQFFGLMa [Substance P, m/z 1347.7354], 1.0 μM). It was hypothesized that the polyclonal FMRFamide antibody would have cross-reactivity with FaRPs, but minimal cross-reactivity with peptides not containing an FxRFamide C-terminus. Figure 1 shows the mass spectra of the peptide standard solution prior to IP (top spectrum) and after IP (bottom spectrum) acquired on an ABI 4800 MALDI-TOF/TOF mass spectrometer. The bottom spectrum shows that the polyclonal FMRFamide antibody reacts with FaRPs (FMRFamide-like peptide I, lobster, and FMRFamide-like peptide II, lobster), thus enabling selective detection of these two peptides containing an FLRFamide C-terminus. It should be pointed out that the two FaRPs present in the control sample had different reaction efficiencies. This may be attributed to the polyclonal nature of the antibody in that one FaRP may bind to the antibody with greater affinity than a different FaRP. Therefore, FaRP immuno-based strategies using this antibody will have greater efficacy for some FaRPs and not others. Regardless of the antibody's differential reactivity with FaRPs, the data suggests that the antibody has strong specificity for FaRPs and may be useful for pulling down peptides of this family in more complex samples (e.g. crustacean PO).
Figure 1.
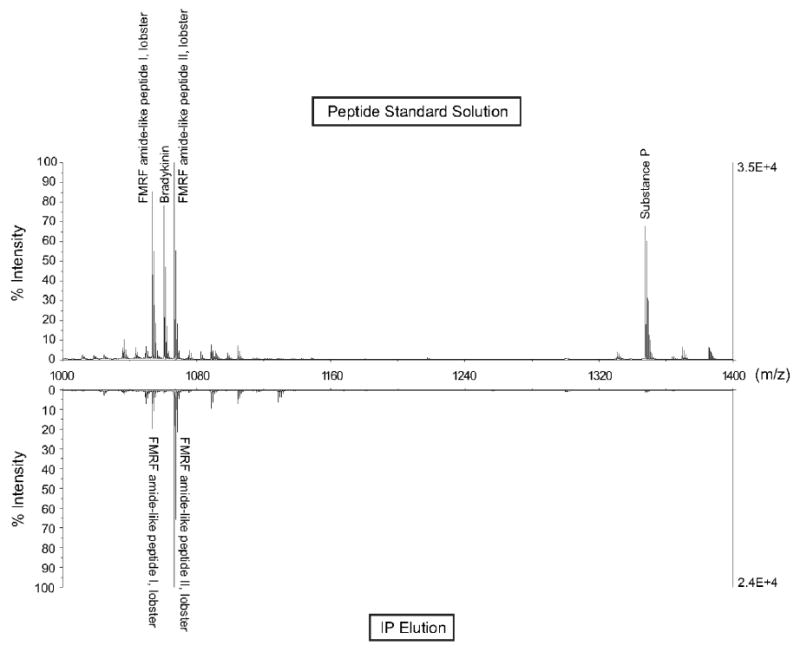
Comparison of peptide standard solution before (top) and after (bottom) FaRP immunoprecipitation. The peptide standard solution prepared in binding/wash buffer contained 10-6 M of the following peptides: FMRF amide-like peptide I, lobster (m/z 1053.5588, SDRNFLRFa), bradykinin (m/z 1060.5687, RPPGFSPFR), FMRF amide-like peptide II, lobster (m/z 1066.5905, TNRNFLRFa), and substance P (m/z 1347.7354, RPKPQQFFGLMa). IP elution reveals selective enrichment and detection of two FaRP peptides.
FaRP IP of C. borealis PO extract
Figure 2 shows the neuropeptide profile within PO extract before (a) and after (b) IP from m/z 825 – 1250. Figure 2a shows that both RYamide and FaRP families are observed in this m/z range prior to IP, with the two RYamide peptides (m/z 976.46 and 1030.47) having the greatest ion intensities. The high ionization efficiency of the two RYamide peptides suppresses the peak intensities of other ion species in this range (e.g. FaRPs). As shown in Figure 2a, eight FaRPs were identified in the PO extract. Figure 2b shows the mass spectrum of the same PO extract post IP. The two RYamide peptides (pEGFYSQRYamide [m/z 1030.47] and SGFYANRYamide [m/z 976.46]) are removed from the sample solution during wash steps because they do not bind to the FMRFamide antibody. As a result, detection of additional FaRPs is likely due to the removal of the highly abundant peptides, enhancing the detection of lower abundance peptides. It was also noted that antibody non-specific binding was minimal, with the FaRPs dominating the spectrum. Only a few unidentified ion peaks were observed in the IP enriched spectrum such as m/z 1005.57, 1101.60, 1041.78, 1057.77, 1204.62, and 1217.89. These ion peaks may be putative FaRPs or peptides pulled down by non-specific binding. Molecular ions at m/z 1005.57 and 1101.60 were further characterized and revealed as novel FaRPs via nanoESI-QTOF MS/MS de novo sequencing. Therefore, this immunoaffinity-based technique is an effective isolation method for the enrichment of FaRPs.
Figure 2.

MALDI-FTMS mass spectra of C. borealis PO extract before (a) and after (b) FaRP immunoprecipitation. In contrast to panel a) where RYamide peptides dominating the mass spectrum, panel b) reveals enriched RFamide-related peptide profiles from the same extract upon immunoprecipitation reaction. Black stars represent putative novel FaRPs.
Further characterization of the FaRP immunoaffinity-enriched PO sample with nanoLC-ESI-QTOF MS/MS identified 13 FaRPs including three novel isoforms: GPRNFLRFamide (m/z 1005.57); YNRSFLRFamide (m/z 1101.60); and GPKNFLRFamide (m/z 977.57) (Figure 3). Among them, m/z 977.57 was not detected by MALDI-FTMS. The other two peaks m/z 1005.57 and 1101.60 were also present in the MALDI-FTMS spectrum with quite low abundance (Figure 2b), which could be the reason that they were not detected in the extract due to ion suppression effect. Ten previously known C. borealis FaRPs were identified by both MALDI-FTMS and nanoESI-QTOF MS/MS in this study except peptide at m/z 926.52, which was only detected by nanoESI-QTOF-MS/MS.
Figure 3.

NanoESI-QTOF MS/MS collision-induced dissociation spectra of three novel de novo sequenced FaRPs. NanoESI-QTOF MS/MS sequences of peptides (a) GPRNFLRFamide (503.282+), (b) YNRSFLRFamide (551.302+), and (c) GPKNFLRFamide (489.302+).
FaRP immuno-dot blot screening assay of C. borealis PO extract
An immuno-dot blot screening assay on C. borealis PO extract HPLC fractions was also performed and identified 46 immunoreactive fractions among a total of 120 fractions. MALDI-FTMS detection was performed as a fast screening strategy for identifying putative FaRPs in the immunoreactive fractions. Of the 46 fractions analyzed, 16 FaRPs were identified including two novel FaRPs m/z 977.63 and 1005.59. The immuno-dot blot screening assay is highly sensitive and specific, narrowing down the target fractions for the MALDI-FTMS detection and providing the snapshot of the FaRPs in each fraction. Further characterization of the immunoreactive HPLC fractions with nanoLC-ESI-QTOF tandem MS revealed 30 FaRPs with 3 additional novel peptides, which was previously published [3].
Comparison of immuno-based methods
The FaRP immunopreciptiation and FaRP immuno-dot blot screening assay provide complementary information of neuropeptide content in C. borealis PO extract. The IP allows a user to isolate FaRPs from whole cell or tissue extracts while the immuno-dot blot screening assay provides qualitative and sensitive identification of FaRP-rich fractions post off-line HPLC fractionation. Additionally, studying FaRP expression by IP simplifies complex sample matrices by removing highly abundant ion species in a flow-through step. As a result, the dynamic range of FaRPs in mass spectra is expanded, facilitating the detection and identification of novel FaRPs. Taken together, the two immuno-based methods provide complementary information regarding neuropeptide expression in PO tissue (Table 1). For example, peptides RSFLRFamide (m/z 824.62), pyrRNFLRFamide (m/z 962.72) and RDRNFLRFamide (m/z 1122.63) were observed only in immuno-dot blot screening assays and not by IP experiments. On the other hand, the novel peptide YNRSFLRFamide (m/z 1101.61) was only observed in IP experiments. It is also possible to apply IP approach to the immunoreactive HPLC fractions to further expand the dynamic range for FaRPs detection. Overall, by using both immuno-based methods for neuropeptide detection we were able to identify the largest number of FaRPs, including those previously reported FaRPs as well as three novel ones reported for the first time here, in C. borealis PO to date. The developed methodologies presented here could also be used to screen for other neuropeptide families present in the same or different neurosecretory organs, enabling fast and targeted neuropeptide identification and discovery.
Table 1.
Identified FaRPs by two immuno-based methodologies
| [M+H]+ | Sequence | Immuno-dot-blot | Immunoprecipitation |
|---|---|---|---|
| 824.49 | RSFLRFamide | + | - |
| 851.50 | RNFLRFamide | + | + |
| 926.52 | SKNYLRFamide | + | + |
| 938.53 | NRSFLRFamide | + | - |
| 962.53 | pyrRNFLRFamide | + | - |
| 965.54 | NRNFLRFamide | + | + |
| 966.53 | DRNFLRFamide | + | + |
| 1022.56 | GNRNFLRFamide | + | + |
| 1104.61 | GAHKNYLRFamide | + | + |
| 1122.63 | RDRNFLRFamide | + | - |
| 1146.61 | GYSKNYLRFamide | + | + |
| 1147.65 | APQRNFLRFamide | + | + |
| 1172.63 | AYNRSFLRFamide | + | + |
| 1181.62 | SENRNFLRFamide | + | + |
| 977.63 | GPKNFLRFamide | + | + |
| 1005.59 | GPRNFLRFamide | + | + |
| 1101.61 | YNRSFLRFamide | - | + |
Bold type indicates novel peptides detected in our study
+ indicates the presence of a peptide and − indicates absence of peptide signal.
Conclusions
Two immuno-based methods (immunoprecipitation and immuno-dot blot screening assay) were developed and utilized for the characterization of FaRPs in C. borealis PO. In total, 17 FaRPs were identified using the two immuno-based methods. Of the 17 FaRPs, three novel FaRPs were de novo sequenced for the first time in this study. The presented methodology can be utilized to characterize any peptide family of interest depending on the availability of an antibody. These immuno-based methods can also be used to search for peptide families that have never been identified in an organism. In addition, multiple antibodies may be employed to a single tissue extract in order to pull out specific families of interest within a single experiment. When such targeted isolation approaches are combined with high resolution and highly specific mass spectrometric tools, increased throughput and coverage of peptide family discovery can be achieved.
Acknowledgments
The authors thank the University of Wisconsin (UW) School of Pharmacy Analytical Instrumentation Center for access to the MALDI-FTMS instrument and the UW Biotechnology Center for access to the MALDI-TOF/TOF instrument. This work was supported in part by the School of Pharmacy and Wisconsin Alumni Research Foundation at the University of Wisconsin-Madison, a National Science Foundation CAREER Award (CHE-0449991), and the National Institutes of Health through grant 1RO1DK071801 (to L.L.). R.M.S. acknowledges the NIHsupported Clinical Neuroengineering Training Program Predoctoral Fellowship (NIH T90 DK070079). L.L. acknowledges an Alfred P. Sloan Research Fellowship.
Abbreviations
- FaRP
FMRFamide-related peptide
- PO
pericardial organ
- IP
immunoprecipitation
- i.d.
inner diameter
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- MALDI-FTMS
matrix-assisted laser desorption/ionization Fourier transform mass spectrometry
- MALDI TOF/TOF
matrix-assisted laser desorption/ionization time-of-flight/time-of-flight
- nanoLC-ESI-QTOF
nanoscale liquid chromatography coupled to electrospray ionization quadrupole
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. 1977;197:670–671. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- 2.Fu Q, Li L. De Novo sequencing of neuropeptides using reductive isotopic methylation and investigation of ESI QTOF MS/MS fragmentation pattern of neuropeptides with N-terminal dimethylation. Anal Chem. 2005;77:7783–7795. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- 3.Ma M, Wang J, Chen R, Li L. Expanding the crustacean neuropeptidome using a multifaceted mass spectrometric approach. J Proteome Res. 2009;8:2426–2437. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baratte B, Gras-Masse H, Ricart G, Bulet P, Dhainaut-Courtois N. Isolation and characterization of authentic Phe-Met-Arg-Phe-NH2 and the novel Phe-Thr-Arg-Phe-NH2 peptide from Nereis diversicolor. Eur J Biochem. 1991;198:627–633. doi: 10.1111/j.1432-1033.1991.tb16060.x. [DOI] [PubMed] [Google Scholar]
- 5.Price DA, Lesser W, Lee TD, Doble KE, Greenberg MJ. Seven FMRFamide-related and two SCP-related cardioactive peptides from Helix. J Exp Biol. 1990;154:421–437. doi: 10.1242/jeb.154.1.421. [DOI] [PubMed] [Google Scholar]
- 6.de With ND, van der Schors RC. SKPYMRFamide, a novel FMRFamide-related peptide in the snail Lymnaea stagnalis. Neuroreport. 1992;3:612–614. doi: 10.1097/00001756-199207000-00017. [DOI] [PubMed] [Google Scholar]
- 7.Dockray GJ, Reeve JRJ, Shively J, Gayton RJ, Barnard CS. A novel active pentapeptide from chicken brain identified by antibodies to FMRFamide. Nature. 1983;305:328–330. doi: 10.1038/305328a0. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 1999;897:239–252. doi: 10.1111/j.1749-6632.1999.tb07895.x. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Kim K, Nelson LS. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res. 1999;848:26–34. doi: 10.1016/s0006-8993(99)01972-1. [DOI] [PubMed] [Google Scholar]
- 10.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptidelike protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci USA. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesser W, Greenberg MJ. Cardiac regulation by endogenous small cardioactive peptides and FMRFamide-related peptides in the snail Helix aspersa. J Exp Biol. 1993;178:205–230. doi: 10.1242/jeb.178.1.205. [DOI] [PubMed] [Google Scholar]
- 12.Nichols R. FMRFamide-related peptides and serotonin regulate Drosophila melanogaster heart rate: Mechanisms and structure requirements. Peptides. 2006;27:1130–1137. doi: 10.1016/j.peptides.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Mercier AJ, Friedrich R, Boldt M. Physiological functions of FMRFamide-like peptide (FLPs) in crustaceans. Microsc Res Tech. 2003;60:313–324. doi: 10.1002/jemt.10270. [DOI] [PubMed] [Google Scholar]
- 14.Anctil M, Grimmelikhuijzen CJ. Excitatory action of the native neuropeptide antho-RFamide on muscles in the pennatulid Renilla kollikeri. Gen Pharmacol. 1989;20:381–384. doi: 10.1016/0306-3623(89)90277-2. [DOI] [PubMed] [Google Scholar]
- 15.Franks CJ, Holden-Dye L, Williams RG, Pang FY, Walker RJ. A nematode FMRFamide-like peptide, SDPNFLRFamide (PF1), relaxes the dorsal muscle strip preparation of Ascaris suum. Parasitology. 1994;108:229–236. doi: 10.1017/s0031182000068335. [DOI] [PubMed] [Google Scholar]
- 16.Moulis A, Huddart H. RFamide neuropeptide actions on molluscan proboscis smooth muscle: interactions with primary neurotransmitters. J Comp Physiol [B] 2004;174:363–370. doi: 10.1007/s00360-004-0422-8. [DOI] [PubMed] [Google Scholar]
- 17.Weiss S, Goldberg JI, Chohan KS, Stell WK, Drummond GI, Lukowiak K. Evidence for FMRF-amide as a neurotransmitter in the gill of Aplysia californica. J Neurosci. 1984;4:1994–2000. doi: 10.1523/JNEUROSCI.04-08-01994.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson ID, Huddart H. Neuromodulation in molluscan smooth muscle: the action of 5-HT, FMRFamide and purine compounds. Gen Pharmacol. 1994;25:539–552. doi: 10.1016/0306-3623(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 19.Raffa RB. Supraspinal FMRFamide antagonizes morphine-induced horizontal, but not vertical, locomotor activity. Peptides. 1989;10:403–406. doi: 10.1016/0196-9781(89)90050-8. [DOI] [PubMed] [Google Scholar]
- 20.Sithigorngul P, Cho Nando J, Stretton AOW. A strategy for isolating rare peptides: isolation and sequencing of a large peptide present in a single neuron of the nematode Ascaris suum. Peptides. 2003;24:1025–1033. doi: 10.1016/s0196-9781(03)00184-0. [DOI] [PubMed] [Google Scholar]
- 21.Sithigornul P, Stretton AO, Cowden CA. A versatile dot-ELISA method with femtomole sensitivity for detecting small peptides. J Immunol Methods. 1991;141:23–32. doi: 10.1016/0022-1759(91)90206-u. [DOI] [PubMed] [Google Scholar]
- 22.Saideman SR, Ma M, Kutz-Naber KK, Cook A, Torfs P, Schoofs L, Li L, Nusbaum MP. Modulation of rhythmic motor activity by pyrokinin peptides. J Neurophysiol. 2007;97:579–595. doi: 10.1152/jn.00772.2006. [DOI] [PubMed] [Google Scholar]


