Abstract
The acyl-AMP forming family of adenylating enzymes catalyze two-step reactions to activate a carboxylate with the chemical energy derived from ATP hydrolysis. X-ray crystal structures have been determined for multiple members of this family and, together with biochemical studies, provide insights into the active site and catalytic mechanisms used by these enzymes. These studies have shown that the enzymes use a domain rotation of 140° to reconfigure a single active site to catalyze the two partial reactions. We present here the crystal structure of a new medium chain acyl-CoA synthetase from Methanosarcina acetivorans. The binding pocket for the three substrates is analyzed, with many conserved residues present in the AMP binding pocket. The CoA binding pocket is compared to the pockets of both acetyl-CoA synthetase and 4-chlorobenzoate:CoA ligase. Most interestingly, the acyl binding pocket of the new structure is compared with other acyl- and aryl-CoA synthetases. A comparison of the acyl-binding pocket of the acyl-CoA synthetase from M. acetivorans with other structures identifies a shallow pocket that is used to bind the medium chain carboxylates. These insights emphasize the high sequence and structural diversity among this family in the area of the acyl binding pocket.
Keywords: Adenylate-forming enzyme, substrate specificity, X-ray crystallography
Enzyme ligases use the chemical energy derived from hydrolysis of the phosphate esters of ATP to generate chemical bonds. We have been studying a large family of acyl-AMP forming ligases that activate a substrate carboxylate with ATP to form an acyl- or aryladenylate. In a second enzymatic step, these enzymes catalyze the formation of a thioester with CoA or the CoA derived pantetheine cofactor of an acyl carrier protein. Work in our lab has recently demonstrated a novel mechanism that these enzymes use to segregate kinetically and conformationally the activities of these two half-reactions 1–4. In this model, the ping-pong catalytic mechanism uses domain rearrangements to isolate the two partial reactions.
The acyl-AMP forming superfamily of adenylating enzymes can be divided into three sub-families, acyl- and aryl-CoA synthetases, the adenylation domains of non-ribosomal peptide synthetases (NRPSs), and firefly luciferase enzymes 5–7. The first two sub-families are closely related and catalyze the formation of a thioester between the original carboxylate substrate and either CoA or the pantetheine cofactor of an NRPS acyl carrier protein (Scheme 1). The acyl- and aryl-CoA synthetase subfamily is broadly reactive with a diverse array of organic acids, ranging from small molecules like acetate and propionate to larger fatty acids and aromatic compounds. In the reaction catalyzed by firefly luciferase, the activated luciferyl-adenylate reacts in an oxidative decarboxylation reaction to produce a high energy intermediate that decomposes to yield a photon of light 8. Strengthening the ties between the different sub-families, the luciferase enzyme has been reported to catalyze the formation of luciferyl-CoA9.
Scheme 1.
A number of crystal structures have been determined for members of the adenylate-forming family of enzymes 1,2,10–22. These structures have demonstrated that the enzymes are composed of two domains, a larger N-terminal domain and a smaller C-terminal domain. Interestingly, these studies identified two catalytically relevant conformations, one for the first adenylation reaction, and a second conformation for the thioester-forming reaction. The two conformations differ by a ~140° domain rotation that occurs between the first and second half-reactions. A conserved residue, either an aspartic acid or less commonly a lysine, is present at the pivot point between the two domains. This hinge residue adopts alternate main chain torsion angles to accommodate the large conformational change. Our structural and functional studies of 4-chlorobenzoyl-CoA ligase have demonstrated that the rotation of the C-terminal domain serves to remove from the active site several residues that play catalytic roles in the initial adenylation reaction to thereby avoid steric hindrance of the thioester-forming reaction3. Accompanying this removal of these residues, the rotation of the C-terminal domain creates a binding tunnel for the CoA pantetheine moiety.
The structures of these enzymes have allowed the characterization and prediction of the binding pocket of different family members, most notably, the NRPS adenylation domains23,24. Using several well-conserved sequences within the N-terminal domain as “anchor residues”, it is possible to identify the regions of the protein that form the amino acyl-binding pocket in different family members. A more elaborate computational approach using support vector machines correlates well with these earlier methods and extends the predictions to additional NRPS adenylation domains25. These studies have allowed the prediction of enzyme specificity in certain instances26 yet the ultimate goal of engineering the complete synthesis of a novel peptide by altering the adenylation domain specificity remains elusive. An important determinant in predictions of NRPS adenylation domain activity is the presence of a conserved aspartic acid residue that interacts with the amino group of the substrate amino acids. Because of the fixed position of the carboxylate and the α-amino group of the amino acid substrate, the side chains of the L-amino acids are directed into a single pocket composed of enzyme residues that are tailored for the specific amino acid substrate.
Less well characterized are the binding pockets of acyl- and aryl-CoA synthetases and thus a protein sequence is not sufficient to predict with certainty the specific activity. To date, structures have been determined for short chain acyl-CoA synthetases2,15,20, several aryl-CoA synthetases (often called ligases)1,10, a medium chain acyl-CoA synthetase 16, and a longer chain fatty acyl-CoA synthetase14. These structures have provided insights for initial attempts at modulating substrate specificity27.
To provide further insight into this enzyme family, and to allow a better understanding of the relationship between acyl substrate specificity and the acyl binding pocket residues, Ingram-Smith et al. have recently cloned a putative acyl-CoA synthetase from the methanoarchaeon Methanosarcina acetivorans and characterized the catalytic preferences of this enzyme. This enzyme was analyzed for the formation of acyl-adenylates and acyl-CoA thioesters with a panel of substrates. This analysis showed that the enzyme is most active with propionate, butyrate, and the branched analogs, 2-methyl-propionate, butyrate, and pentanoate. The specific activity is weaker for smaller or larger acids. Interestingly, with some substrates, the enzyme appears to release the acyl-adenylate intermediate in the presence of CoA while the affinity of the enzyme for some substrates causes the acyl-adenylate to be retained until CoA binds. The full functional characterization of this protein will be presented elsewhere (C. Ingram-Smith, Y. Meng, L. L. Cooper, and K.S. Smith, submitted).
We present here the three-dimensional structure of this enzyme, solved by multi-wavelength anomalous dispersion phasing methods and refined against data to 2.1 Å resolution. This work identifies the active site residues of this newly identified family member and demonstrates the architecture for the acyl-binding pocket. The pocket of this acyl-CoA synthetase is shown to have a cavity volume comparable to the enzyme 4-chlorobenzoyl-CoA ligase. The pocket, however, is much more shallow in the current structure than previously observed acyl binding pockets within this family. The pocket of this enzyme is compared to the acyl binding pockets of previously characterized enzymes. The structural diversity of these active site residues are described and emphasize the need for structural information to enable the prediction of substrate specificity with the acyl-CoA synthetases.
Materials and Methods
Protein expression and purification
The AAE gene was amplified from genomic DNA from Methanosarcina acetivorans into pET19b plasmid using standard PCR protocols. The encoded sequence is represented in the Uniprot sequence database by accession number Q8TLW1_METAC. The sequence in the plasmid initiates at Met4 of the deposited sequence as this ATG codon likely represents the physiological start of the protein. The cDNA as ligated into the pET19b plasmid resulting in the production of a protein containing the His10 purification tag and the enterokinase protease site. The tag was left on for crystallization. The AAE protein was purified from E. coli RosettaBlue (DE3) cells requiring selection for both ampicillin and chloramphenicol resistance. Cells were grown to an OD600 of ~ 0.6 at 37 °C and protein production was induced overnight at 21°C by adding IPTG to a final concentration of 0.5 mM. Cells were harvested by centrifugation and frozen at −80 °C. Cells were thawed and lysed by sonication in lysis buffer containing 25 mM Tris-HCl (pH 7.5 at 4 °C), 10 % glycerol, and 0.2 mM TCEP prior to loading onto the column. His-tagged AAE was purified through IMAC chromatography using 5 ml Ni2+ HisTrap columns (GE Healthcare Life Sciences, Piscataway, NJ). The column was washed with lysis buffer containing 50 mM imidazole and the AAE protein was eluted using lysis buffer with 300 mM imidazole. AAE fractions were pooled and dialyzed overnight against 1L 25 mM Tris-HCl (pH 7.5 at 4 °C), 20 % glycerol, 0.5 mM EDTA, 0.2 mM TCEP and the protein was concentrated to 10 mg/ml and frozen by directly pipetting into liquid nitrogen28. The yield of a purification preparation of AAE was approximately 15 mg per L of cell culture.
SeMet-labeled protein was produced using the T7 Express Crystal Competent E. coli cell line(New England BioLabs, Beverly, MA). Briefly, a culture of cells was grown overnight in 100 mL LB media. The cells were pelleted by centrifugation and resuspended in 100 ml M9 minimal media containing 0.1 mg/mL ampicillin. The cells were used to inoculate two flasks containing 1 L of M9 media and grown at 37°C to an OD600 of ~0.6. An amino acid cocktail was added to a final concentration of 1 mg/mL each of lysine, threonine, and phenylalanine, and 0.5 mg/mL each of leucine, isoleucine, and valine. Cells were grown for 30 minutes and selenomethionine was added to a final concentration of 0.05 mg/mL. Protein expression was induced with 0.5 mM IPTG. Cell growth continued overnight at 16°C and cells were harvested by centrifugation. Purification of SeMet labeled protein was performed as for wild-type cells. Using the alternate cell line, a protein yield of ~50 mg per L of cell culture was concentrated to 6mg/mL and stored at − 80°C for crystallization.
Crystallization and data collection
Crystallization conditions for AAE were originally identified by batch method using the high throughput-screening robot at the Hauptman-Woodward Medical Research Institute29. Crystals of AAE were grown by vapor diffusion using the hanging-drop method. Wild-type AAE protein was incubated in a 1:1 ratio with mother liquor at room temperature using precipitant consisting of 80 % PEG 400, 100 mM Mg(NO3)2•6H2O, 50 mM HEPES (pH 7.5 at 4 °C). Selenomethionine labeled protein crystals of the same morphology were grown at 14 °C using a precipitant containing 50 % PEG 400, 100 mM Mg(NO3)2•6H2O, 50 mM HEPES (pH 7.5 at 4 °C). The crystals were transferred to the mother liquor which contained 80 % PEG 400 as cryoprotectant and were cryo-cooled directly in the stream of N2 gas at −160 °C. Crystallographic data for native and selenomethionine protein crystals were collected remotely at beamlines 11-1 and 9-1, respectively, at the Stanford Synchrotron Radiation Laboratory (SSRL)30. Native crystals diffracted to 2.1 Å and the space group determined was P212121.
Three-wavelength MAD data were collected. The detector was set at a distance of 370 mm and 100 frames of data at 1° oscillation width and 5 s per frame were collected at each wavelength. Data were integrated and scaled using HKL200031. Final data collection statistics are shown for the MAD and native data in Table 1
Table 1.
Data collection statistics for the three wavelength SeMet and the native crystals.
| Peak | Inflection | Remote | Native | |
|---|---|---|---|---|
| Wavelength (Å) | 0.97894 | 0.979 | 0.99 | 0.988 |
| Resolution (Å) | 2.4 | 2.4 | 2.4 | 2.1 |
| Unit-cell parameters | ||||
| a (Å) | 90.4 | 91.7 | ||
| b (Å) | 96.2 | 96.4 | ||
| c (Å) | 140.5 | 141.5 | ||
| Rmerge (%)a | 10.3 | 10.1 | 9.9 | 8.2 |
| (48.7) | ||||
| Completeness (%)a | 98.8 | 97.7 | 97.5 | 99.9 |
| (99.7) | ||||
| I/σa | 10.6 | 10.1 | 10.1 | 9.4 |
| (2.0) | ||||
| No. of observations | 184432 | 180,400 | 180,057 | 334108 |
| No. of unique reflections | 47673 | 47,462 | 47,766 | 72781 |
Value in parenthesis is for the highest resolution shell (2.2-2.1 Å).
Structure determination and refinement
Attempts to solve the structure of AAE by molecular replacement were unsuccessful, despite the use of multiple software packages and multiple search models, including Acs (1PG4), CBAL (1TS5), fAcs (1V26), and the NRPS adenylation domains (1AMU and 1MDB). Where possible, we tested protein models in both the adenylate- and thioester-forming conformations, as well as a model consisting of only the larger N-terminal domain. This difficulty with molecular replacement has been observed by us and others with previous members of the family and reflects the low sequence identity, often in the range of 20–25 %, as well as the structural variability between different family members.
The structure of AAE was therefore determined by multi-wavelength anomalous dispersion using the selenomethionine-labeled protein. The protein crystallized in the orthorhombic space group P212121. Matthews coefficient analysis suggested there were two AAE molecules in the asymmetric unit (VM = 2.4). The positions of the selenium heavy atoms were determined using BnP program32. The SnB algorithm was able to identify twenty-six selenium atoms out of twenty-eight and BnP was further used to improve the final phases. The final phases from BnP were imported into RESOLVE 33 for additional solvent flattening and automated chain tracing. RESOLVE was able to identify a total of 680 residues (out of 1114 in the asymmetric unit) however only 63 were properly assigned within the sequence. The molecular model was built through extensive manual model building with COOT34. Once the same stretch of residues could be identified in both chains, residues that were built into one model were rotated into the other chain for assistance in building. An initial model was built into the experimental map that contained 436 residues in each chain. In this model, 337 residues were placed with side chains, while 99 were included as polyalanine. This initial model was submitted to REFMAC535,36 using data to 2.4 Å and refined to an R-factor of 41.2% (R-free of 46%). Continued iterative model building and refinement allowed extension to include the higher resolution data and completion of the model. The final model contains residues 3-536 of chain A and 3-539 of chain B. Water molecules were placed using the find water option of COOT and confirmed manually. In the final stages of refinement, each N-and C-terminal domain was assigned as a group for TLS anisotropic refinement37, which further reduced the Rfree.
Calculation and comparison of cavity volume
The cavity volumes within the acyl-binding pocket of Acs2 (1PG4), fAcs14 (1V26), and CBAL3 (3CW9) were compared to the acyl-binding pocket of AAE in thioester forming conformation. The cavity size was analyzed using VOIDOO38 with the probe radius of 1.2 Å39, a primary grid spacing of 0.5 Å, and remaining values as defaults. The cavities were generated using the Probe-Occupied option, which reports the volume of the cavity occupied by a rolling probe. The resulting cavities were observed via PyMOL40. If present, the acyl ligand was removed from the file prior to cavity calculation however the remaining ligands, CoA and AMP, were left intact. To determine the size of the pocket for AAE, AMP and CoA were modeled into their respective binding pockets from a superposition of Acs onto AAE to allow determination of the size of acyl pocket alone and not the size of the combined acyl-adenylate or acyl thioester pockets. Two additional water molecules were added into the structure of AAE for this calculation to avoid what has been referred to as the “leaking effect”41 where the cavity recognized by VOIDOO merges into adjacent areas.
Nano-LC/MS protein sequencing
The protocol for sequencing the AAE protein by high resolution nano-LC coupled with mass spectrometry is described in detail in the supplemental material. Briefly, the AAE protein was incubated with 1 mM TCEP-HCl and 50 mM iodoacetamide and subsequently digested with trypsin or the Glu-C endoproteinase. The proteolytic peptides were separated by nano-liquid chromatography on a reversed phase C18 nano-column and identified using a Thermofisher LTQ/Orbitrap high-resolution mass spectrometer. Additional experimental details, as well as sequence coverage (Figure S1) and the spectra of the relevant peptides (Figure S2), are included in the supplemental material.
Sequence Alignment and superposition of structures
The sequence of the N-terminal domain of AAE was aligned with the sequence of the same region from Acs, fAcs, BzCL, and CBAL to compare the acyl binding pockets. A multiple sequence alignment was prepared initially using ClustalW 42,43. As many alignments based on sequence alone are challenged by the proper placement of loops and insertions, the protein structures were then manually inspected and the alignment was modified to present a proper structural alignment. The coordinates of Acs, CBAL, fAcs, and AAE were superposed using LSQKAB 44 within CCP4 suite 35 for structure comparison. Figures 1, 2, 3, 4, and 6 were made with PYMOL 40.
Figure 1.
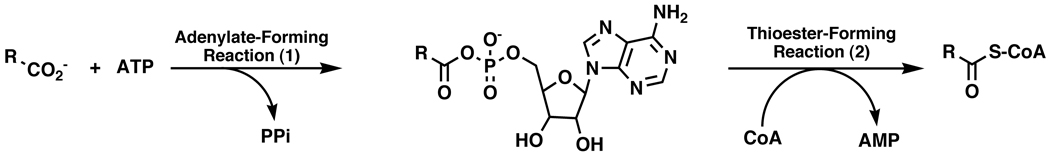
Structure determination of AAE. A. Stereorepresentative electron density is shown for the experimental, solvent-flattened phases. A region of the final N-terminal domain is shown superimposed on map calculated with coefficients and phases derived from RESOLVE. Carbon atoms are shown yellow, oxygens are shown red, nitrogens are blue, and sulfur atoms are shown orange. The map is contoured at 1 σ. Because of slight differences in the native and MAD crystal packing, the final model was superimposed on the original coordinate file with LSQKAB44. B. Refined electron density map generated with final calculated phases and coefficients of the form 2Fo-Fc and contoured at 1.25 σ. Superimposed on the map is the final refined model colored as in panel A. The atoms that form the observed linkage between Lys256 and Cys298 are colored green indicating that their exact composition is unknown. C. Potential chemical structures of the linkage between Lys256 and Cys298 observed in the native structure.
Figure 2.
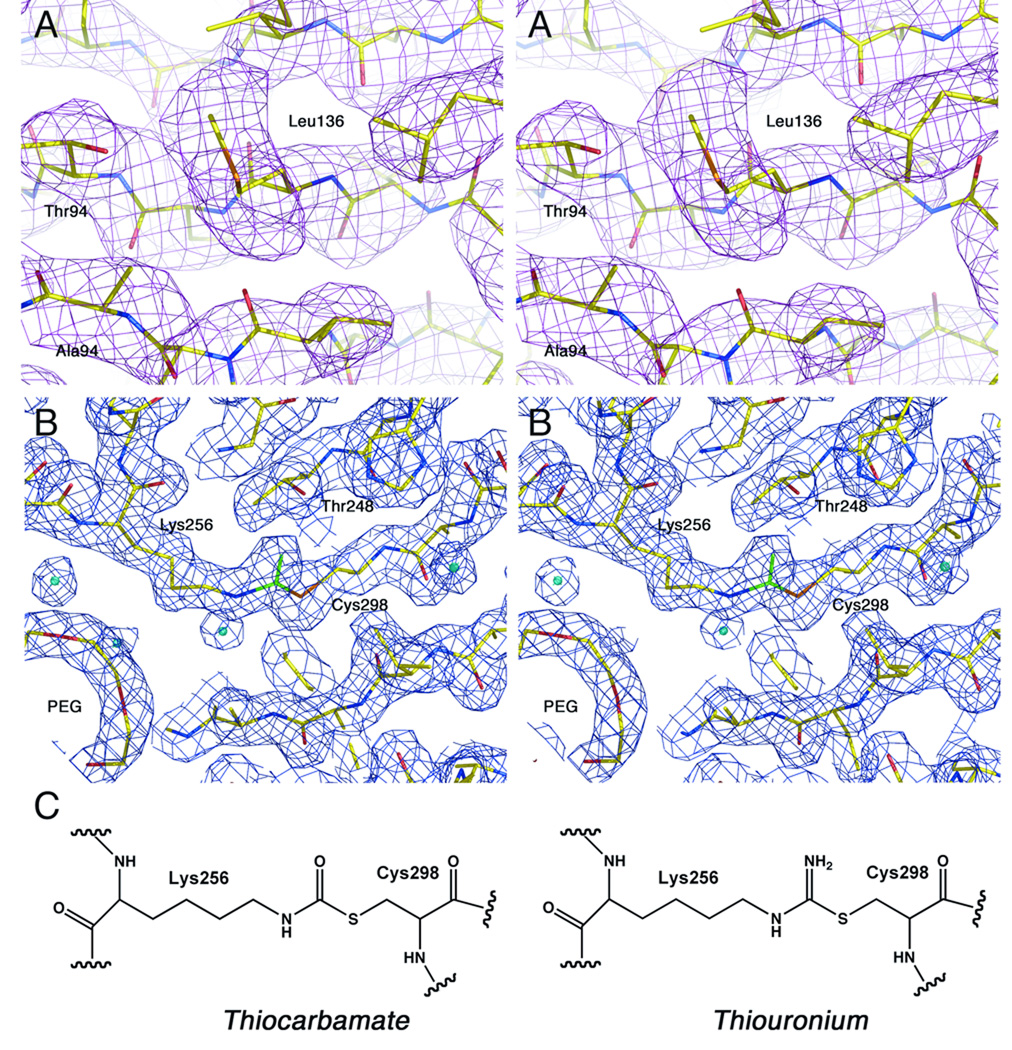
Ribbon diagram representing the AAE crystal structure. The large, N-terminal domain is shown with yellow helices. The two central β-sheets that form the core of this domain are shown in dark blue and light blue, while the distorted β-sheet at the end of this domain is shown in magenta. The C-terminal domain is shown in red. The active site, positioned between the two domains, is indicated with an arrow.
Figure 3.
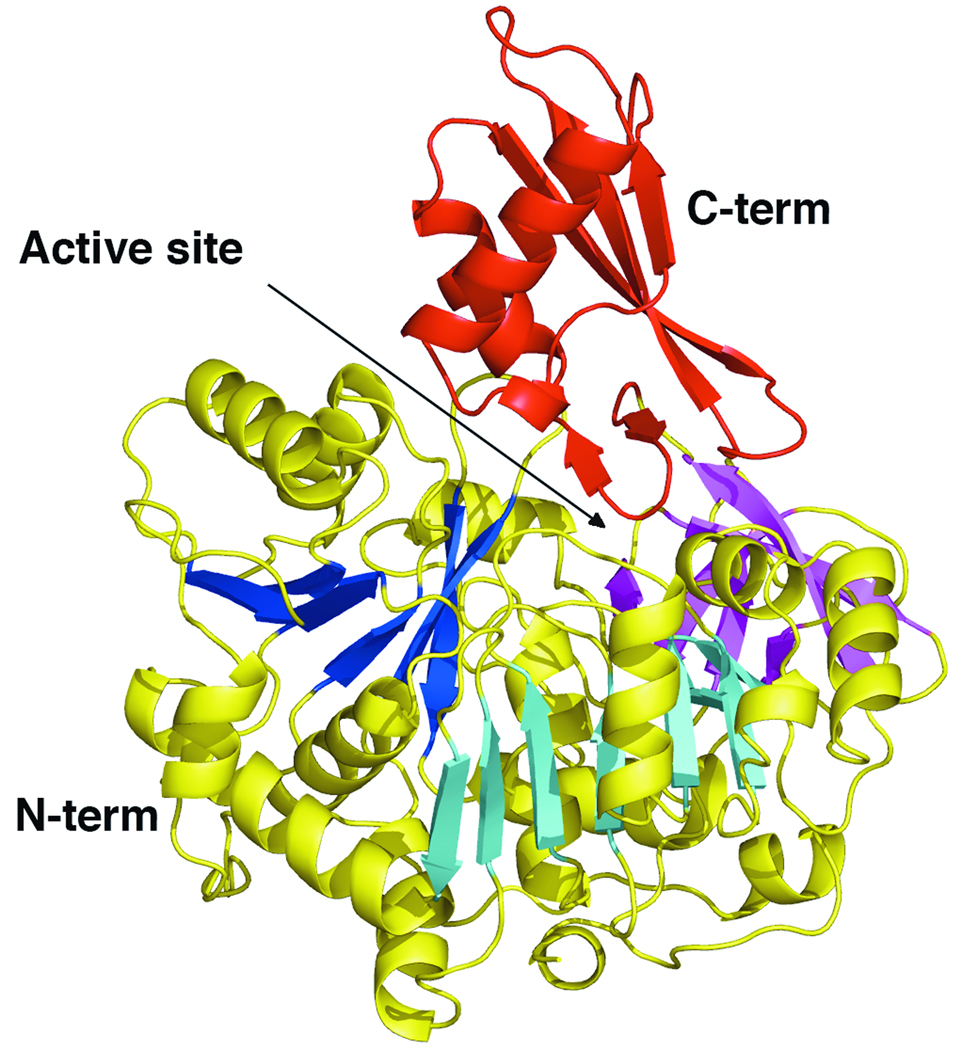
Active Site Superpositions. A. The adenylate binding pocket of AAE is shown in a stereorepresentation superimposed on the binding pocket residues of CBAL (PDB 3CW9), along with the AMP molecule co-crystallized in the CBAL structure. AAE residues that form the AMP binding pocket are shown in stick representation and are properly positioned to interact with a nucleotide. Atoms are colored grey (carbons), red (oxygens), and blue (nitrogens) for the AAE protein. Atoms from the AMP from the CBAL structure are colored similarly, except the carbon atoms are shown green and the phosphate atom is shown orange. The homologous CBAL residues, notably the P-loop residues Tyr160-Thr164, Ala280-Ala282, Tyr304, Thr306, Thr307, Asp385, Arg400, and Asp402, are shown with a thin green line. B. Overlay of the CoA binding pocket from CBAL with AAE demonstrating the presence of the pantetheine tunnel and hydrophobic residues that are may bind to adenine of CoA. The CoA molecule derived from the CBAL structure is colored as the AMP ligand in panel A. Superimposed in green are CBAL residues Ser407, Trp440, and Phe473. Also present in the refined model of AAE is a molecule of HEPES (with carbon atoms displayed in pale yellow) and PEG (all atoms shown in yellow). Coloring of atoms is the same as in panel A. Residue numbers reflect the numbering in AAE.
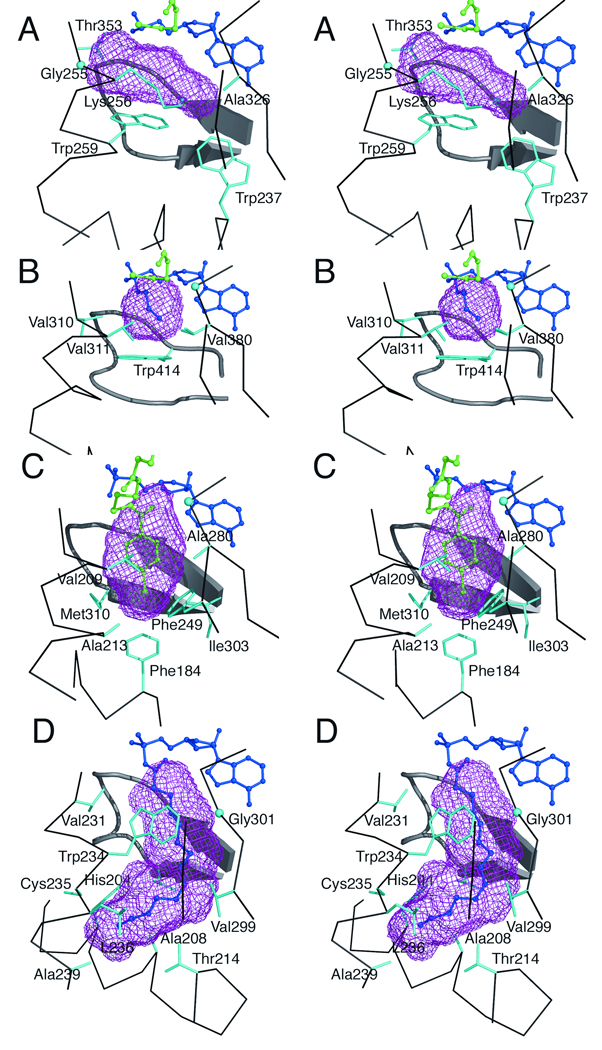
Figure 4.
The acyl- and aryl-binding pockets, calculated with VOIDOO, of several adenylate-forming enzymes. Stereorepresentations of the binding pockets are shown for A. AAE B. Acs C. CBAL and D. fAcs. The protein backbone surrounding the binding pocket is shown, as are residue side chains that contact the binding pocket. The ligands are shown in ball-in-stick representation with AMP or the acyl-adenylate shown in dark blue and regions of the pantetheine of CoA shown in green. In panel A, the AMP and CoA ligands were modeled from PDB 1PG4 as described in the methods. In panel C, the 4-chlorophenacyl-CoA molecule is shown in green. Glycine residues, lacking a side chain, are shown as spheres denoting the Cα position: Gly255 and Gly351 of AAE, Gly387 of Acs, Gly281of CBAL and Gly301 of fACS. The binding pockets were identified with VOIDOO as described in the text.
Results and Discussion
Overall Structure Description
The three-dimensional crystal structure of the 557 residue AAE protein was solved by MAD phasing of selenomethionine labeled protein. The heavy atom substructure was determined and experimental phases were used for model building. The protein crystallized in the orthorhombic space group P212121 with two molecules of AAE in the asymmetric unit. The AAE protein runs on a size exclusion chromatography column as a monomer and the two molecules in the asymmetric unit do not interact with a significant interface.
The final model contains residues 3-536 in molecule A and 3-539 in molecule B. The C-terminal helix observed in some members of the adenylate forming family of enzymes was disordered in the AAE structure. Weak density was observed for several residues of this C-terminal region of chain B however it could not be placed within the sequence and, when refined as polyalanine, the B-factors were significantly higher than observed in other regions of the protein. As such, this region was left out of the final model. Iterative cycles of refinement were performed against the 2.1 Å data obtained from crystals of wild-type AAE protein. The final model contains 1079 protein residues, 407 water molecules, three molecules of the HEPES buffer, five molecules of PEG, two solvated Mg2+ ions, two NO3 ions and six molecules of glycerol. The ordered PEG molecules were modeled as three molecules containing three ethylene glycol units and two molecules containing five ethylene glycol units. These are likely part of longer PEG molecules with disordered ends. Interestingly, one of the PEG molecules partially fills the active site of the enzyme passing from the pantetheine channel into the acyl binding pocket and into the nucleotide pocket in both molecules. Crystallographic data and final refinement statistics against the high-resolution synchrotron data are presented in Table 1 and Table 2. Representative experimental electron density is shown in Figure 1. Before we describe the AAE protein, we note an interesting feature in the electron density that we characterized further. In both the MAD and native refinement maps, continuous density was observed between the side chains of Cys298 and Lys256 (Figure 1B); attempts to model this density as cysteine sulfenic acid (amino acid side chain -CH2-S-OH), mercaptocysteine (-CH2-S-SH), or as a metal ion did not refine satisfactorily. Inspection of the electron density and trial refinements suggested that the density was most consistent with a covalent cross-link that was formed by a formylated Lys256 side chain that was then attacked by the Cys298 thiol to form either a thiocarbamate or thiouronium linkage (Figure 1C). To our knowledge, no such cross-link has been observed between protein side chains in a protein crystal structure, however a chemical precedent for the thiouronium linkage exists in the pentein superfamily of enzymes. Members of this family45,46 use a nucleophilic active site cysteine to attack the guanidino group of the arginine substrate to form a thiouronium linkage. This intermediate then reacts further to catalyze a variety of steps in the arginine degradative pathway.
Table 2.
Refinement Statistics
| Resolution Range | 40.0-2.1 Å |
| Rcryst | 17.5 % |
| Rcryst (highest resolution shell) | 19.9 % |
| Rfree | 20.7 % |
| Rfree (highest resolution shell) | 24.1 % |
| Wilson B factor (Å3) | 28.7 |
| Average B factora, chain A (Å3), number of atoms | 28.4, 4191 |
| Average B factora, chain B (Å3), number of atoms | 26.7, 4226 |
| Average B factor, solvent (Å3), number of atoms | 36.9, 430 |
| Average B factor, additives (Å3), number of atoms | 44.9, 149 |
| Average B factor, ions (Å3), number of atoms | 40.2, 10 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Angles (°) | 1.2 |
B-factors were calculated prior to the inclusion of TLS refinement as refinement was nearly complete.
We investigated whether this cross-link exists in the native protein prior to crystallization or whether this was a potential consequence of the diffraction experiment. The native AAE protein was subjected to a nano-LC/MS based protein-sequencing strategy. Stringent filtering approaches in conjunction with high mass accuracy (approximately 2 ppm) achieved by Orbitrap, resulted in highly confident identification. By combining the identified proteolytic peptides from trypsin and Glu-C proteases, 94% sequence coverage was achieved (Figure S1). The proteolytic peptides containing Lys256 and Cys298 were identified with high confidence. Identification spectra are shown in Figure S2. We were concerned that the reductive alkylation of the sample necessary to prevent disulfide formation and to achieve high coverage of proteolytic fragments could cleave the putative linkage. The thiouronium linkage has however been shown to be stable to reductive alkylation45. In arginine deiminase, the covalent substrate-enzyme intermediate was identified through MS/MS peptide sequencing that showed the thiouronium species present after alkylation. Indeed, in one peptide, the thiouronium adduct and an alkylated cysteine are both clearly identified on two neighboring cysteine residues45. Additionally, it has been shown that formation of the thiouronium linkage in dimethylarginase prevents inactivation by iodoacetamide demonstrating that iodoacetamide cannot displace the thiouronium47.
The fact that the “free” proteolytic peptides (i.e., those containing only one amino acid backbone) containing these residues were identified suggests that the covalent linkage observed in the electron density does not exist in the native protein preparation and that the cross-link was a radiation induced event and is not an intrinsic part of the native protein. We cannot however rule out the possibility that the native protein preparation is a mixture of cross-linked protein and protein containing free side chains and that the chemical cross-link was not identified in the mass spectrometry experiment. Additionally, it is possible that the covalent cross-link formed during the several weeks that the protein was incubated in the crystallization experiment. We also note that we compared the sequences of related proteins in PFAM 0050148 for the presence of a lysine and cysteine residue at equivalent positions. Of 400 diversely representative sequences, there were only three other proteins containing both the lysine and cysteine residues, including a murine medium chain acyl-CoA synthetase, and putative acyl-CoA synthetases from Archaeoglobus fulgidus and Streptomyces coelicolor.
The crystal structure of AAE (Figure 2) illustrates a large N-terminal domain that is formed by ten α-helices and two 8-stranded β-sheets followed by a distorted four-stranded sheet. The final 106 residues form a C-terminal domain that begins with the anti-parallel loop at residue 453, followed by two α-helices and the final sheet of the C-terminal domain. The final 21 (chain A) or 18 (chain B) residues are disordered. The two molecules superimpose with LSQKAB44 with an rms displacement between Cα positions of 0.4 Å. The two molecules show no significant differences; for subsequent analysis, we will consider and discuss molecule A.
As noted above, enzymes within the adenylate-forming family have been shown to adopt two different conformations. Biochemical and structural data3,4,20 support the hypothesis that these enzymes catalyze the first adenylation half-reaction and then rotate the C-terminal domain by ~140° to a second conformation that is used for CoA binding and catalysis of the thioester-forming reaction. For purposes of discussion, the adenylate-forming conformation is referred to as “conformation 1” while the thioester-forming conformation is referred to as “conformation 2”.
Interestingly, the structure of AAE was determined in the second, thioester-forming conformation in the absence of ligands. The structure is similar to prior adenylate-forming enzymes determined in this conformation and a superposition was performed with 4-chlorobenzoyl-CoA ligase (CBAL, 3CW9), acetyl-CoA synthetase (Acs, 1PG4) and the thermophilic fatty-acyl-CoA synthetase (fAcs, 1V26). The Cα positions of AAE and CBAL superimpose with an rms displacement of 1.9 Å over 346 residues, of AAE and Acs with an rms displacement of 1.5 Å over 292 amino acids, and of AAE and fAcs with an rms displacement of 1.8 Å over 294 residues.
We had originally proposed1,2 that the binding of CoA was the trigger that caused the C-terminal domain to rotation from the adenylate-forming conformation to the thioester-forming conformation. This hypothesis was based on the fact that the only structures determined in the thioester-forming conformation contained CoA bound reactions2,3 or were crystallized with CoA present in solution, although not apparent in the density14. Recently, the structure of DltA, a D-alanine:D-alanyl-carrier protein ligase that is involved in lipoteichoic acid biosynthesis, was determined in the thioester-forming conformation while bound only to AMP22. Additionally, a human medium chain acyl-CoA synthetase (mAcs) has been structurally characterized in multiple states16. This protein was characterized in the thioester-forming conformation in both in the unliganded state as well as bound to either AMP or CoA.
Thus it appears that some members of the adenylate-forming family, including the AAE enzyme described here, adopt conformation 2 in the absence of CoA. Members of this family likely exist in an equilibrium between these two states and the binding of ligands will drive them into the conformation necessary for activity. The factors that influence the crystallization of an unliganded protein in one conformation or the other are likely to be complex. We have tried to correlate the sequences and structures of multiple enzymes with the crystallographic conformation that was observed but, with the exception of the presence of a ligand, have been unable to identify additional factors.
Analysis of AAE residues in the AMP binding pocket
The active site of the adenylate-forming enzymes is located at the interface between the two domains. The acyl binding pocket is located entirely within the N-terminal domain. The AMP binding pocket is positioned at the interface with more residues from the N-terminal domain contributing to binding. The CoA substrate binding site, however, spans the two domains with the CoA nucleotide positioned on the surface of the C-terminal domain and the pantetheine moiety passing between the two domains to the acyl binding pocket. Recent biochemical4,49 and structural3,16 analyses provide insights into the catalytic mechanisms of this enzyme family.
To characterize and compare the amino acid residues lining the active site in AAE with those shown to be important for enzymatic activity of other family members, the structure of AAE was superimposed on the structure of CBAL3 bound to the inhibitor 4-chlorophenacyl-CoA and AMP. We studied the amino acid residues that surround the AMP nucleotide-binding site (Figure 3A). The well conserved Asp385 and Arg400 residues of CBAL that interact with the ribose 2’ and 3’ hydroxyls of AMP were found to be present in the AAE structure at Asp435 and Arg450 preceding the hinge at Asp452. These residues have been demonstrated to be important for AMP binding in CBAL where a mutation of either to alanine impairs the reaction by reducing the activity several 100-fold4. Residues Gly281, Ala282, Thr283, and Tyr304 in CBAL, which surround the adenine ring, are found to be substituted in the AAE structure with Gly327, Glu328, Pro329 and the aromatic side chain Phe350, which is positioned to stack against the adenine ring in the similar manner as in Tyr304 in CBAL. Thr353 and Ser210, homologs of CBAL residues Thr307 and Thr161, are also appropriately positioned to interact with the nucleotide. Ser210 is a part of a highly conserved glycine- and serine/threonine-rich sequence that we and others have compared to the P-loop of other ATP binding proteins2,22. The ATP binding pocket of AAE thus seems well conserved with other members of the adenylate-forming family. These residues, which are conserved at the level of structure, are used below to perform the structural alignment.
Two recent structural studies mentioned above provide exciting new information about the ATP binding pocket and the interaction of the P-loop with ATP. New structures of the medium chain acyl-CoA synthetase16 and of DltA19 were determined bound to the ATP in a productive interaction. These structures provide insights into the interactions between the conserved residues of the P-loop with ATP and also suggest that the universally conserved catalytic lysine of the C-terminal domain appears to interact with multiple negatively charged groups of the reactants and intermediates during the catalytic cycle. Additionally, these structures raise the intriguing possibility of a preliminary binding mode for ATP19.
Analysis of AAE residues in the CoA binding pocket
The crystal structures of Acs, mAcs, and CBAL have all been determined bound to CoA or a CoA-adduct2,3,16. The CoA binding site is composed of a nucleotide binding pocket on the surface of the protein and a “pantetheine tunnel” that passes between the two domains and enters the mostly buried active site. Interestingly, the three proteins exhibit distinct CoA nucleotide binding pockets with the nucleotide interacting mostly with the N-terminal domain in Acs and mAcs, and primarily with the C-terminal domain in CBAL. The positions of the CoA nucleotide in Acs and mAcs differ by ~8–10Å.
The pantetheine tunnel of AAE is well conserved in structure, passing between the loops consisting of Ala250-Lys256, Ala299-Ile303, and Ser458-Lys461 (Figure 3B). At the level of sequence identity, however, there is limited conservation around this region of the protein. A stringently conserved glycine residue (Gly 459) is present on the loop that follows the hinge (Asp452) for the conformational change in the C-terminal domain. This glycine is present in all members of the adenylate-forming family. Site-directed mutagenesis of this glycine (Gly524 in Acs) to leucine20 compromises the thioester forming reaction specifically. Interestingly, the lower region of the pantetheine tunnel in AAE contains a PEG molecule that approximates the position of the cysteamine moiety of the CoA ligand. The oxygens of the PEG molecule (Figure 3B) do not appear to hydrogen bond with the protein atoms in any of the interactions that have been observed in the structures of Acs and CBAL. The PEG spans the different binding pockets and does not appear to serve as a functional mimic of the pantetheine or any of other ligands.
The structure of AAE was compared to the homologous enzymes to characterize how the CoA nucleotide would bind in AAE (Figure 3B). In the structure of CBAL3, two aromatic residues from the C-terminal domain form stacking interactions with the adenine moiety of CoA. The Phe473 residue of CBAL is conserved in AAE with Tyr525, while Trp440 of CBAL is replaced with Arg490. Interestingly, an aromatic residue from an adjacent loop, Tyr460, is positioned near the modeled CoA nucleotide of the superimposed structure and thus may substitute for Trp440. AAE may bind CoA in a binding mode similar to CBAL however additional structural and kinetic experiments will be necessary to identify the true binding interactions.
Conformational change and catalysis
Extensive kinetic analysis4,49 of CBAL strongly supports the domain alternation hypothesis, namely that the adenylation partial reaction is carried out using conformation 1 and the thioesterifcation partial reaction is catalyzed using conformation 2. Additional kinetic evidence exists for Acs20 and luciferase50–52. It has however been suggested that other members of this family can catalyze both partial reactions in either of the alternate conformations14,53.
As we3,4 and others16 have noted, conformation 2 is unable to bind to ATP in a productive manner because of a steric clash between a C-terminal region of the protein and the triphosphate moiety of the nucleotide. This demonstrates that PPi must be released prior to, or coincident with, the domain rotation from conformation 1 to conformation 2. Similarly, a conserved aromatic residue (Trp254 in AAE, His207 in CBAL) has been shown to rotate between two positions in the alternate conformations. In conformation 1 it is positioned near the substrate carboxylate where it prevents access of the CoA thiol. Thus, CoA is unable to bind productively to conformation 1, also supporting the structural segregation of the two partial reactions.
We believe that the cumulative structural and biochemical evidence supports the domain alternation hypothesis and believe it is likely to be valid for the full enzyme family, including AAE. The strong conservation of residues on both faces of the mobile C-terminal domain therefore seems to support the ability of the two known conformations to catalyze only one of the two specific partial reactions. All members of the family contain a C-terminal lysine (Lys544 in AAE) that is responsible4,20,51,54,55 for the adenylation partial reaction as well as a universally conserved glycine residue (Gly459 in AAE) that is necessary to open the pantetheine tunnel in the thioester-forming conformation3,20. Additionally, we have recently demonstrated that the mutation of the hinge residue of CBAL to a proline severely compromises its ability to rotate into the conformation necessary for the thioester-forming reaction. The microscopic rate constant for the adenylate-forming partial reaction is reduced by a factor of three, while the rate of thioesterification reaction is reduced by four orders of magnitude49.
It is of course possible that the domain alternation strategy will not be used by some members of the enzyme superfamily. Examination of alternate proposals will require that other family members are tested with both structural and functional studies. Additional mutagenesis studies are underway.
Analysis of acyl substrate binding pocket in AAE
An acyl substrate-binding pocket is present in AAE and, as noted above, is partially filled with a PEG molecule that is shared with the adjacent AMP and pantetheine binding sites. Superposition of AAE, Acs, and CBAL allowed the identification of residues in AAE that form the acyl substrate-binding pocket. Interestingly, Trp259 of AAE truncates the acyl substrate-binding pocket (Figure 3B, Figure 4A) and forms a “floor” that prevents the AAE enzyme from binding to larger acyl substrates. This residue is positioned similarly to Trp414 of Acs (Figure 4B), although the two residues derive from opposite sides of the active site. Additional residues that form the pocket in AAE include Gly255-Lys256, Ala326-Gly327, Gly351, and the main chain backbone of Gln352 and Thr353. Because Trp259 of AAE and Trp414 of Acs form a similar base of the acyl substrate pocket, we looked for amino acid substitutions that would allow AAE to accommodate the larger acyl substrates. Of particular note, two residues in Acs that form the acetate pocket, Val310 and Val386, are both replaced with smaller residues, Gly255 and Ala326 in AAE. Additionally, Thr311 of Acs is replaced with Lys256 in AAE, a larger residue. It is difficult to identify conclusively the role of this residue in the active site as the Lys256 side chain is pulled away from the acyl-binding pocket by its interaction with the side chain of Cys298. The replacement of several residues from Acs with smaller residues in AAE likely accounts for the preference for larger acyl substrates in AAE.
To analyze more rigorously the acyl substrate-binding pocket of AAE, we used VOIDOO38 to measure the volume of the acyl substrate cavity and compared this to the values obtained from the structures of CBAL, Acs, and fAcs.
We compared AAE to the structures of Acs (1PG4), fAcs (1V26), and CBAL (3CW9). If necessary, the bound carboxylate substrate was removed from the active site. The observed pocket sizes for Acs, CBAL, and fAcs are 72 Å3, 147 Å3, and 365 Å3, respectively (Figure 4). The volume of the pocket for the AAE acyl-binding pocket is 138 Å3, approximately the size of the binding pocket in CBAL. The binding pocket for AAE is not as deep as that observed in CBAL but rather is wider, a result of the presence of Trp259 that truncates the binding pocket and the replacement of several side chains with Gly255 and Ala326, as described above.
The residues identified in VOIDOO as forming the binding pocket in AAE are Trp237, Gly255, Lys256, Trp259, Ala326, Thr353, and the main chain backbone of Gly349-Gln352. These new binding residues are distinct from the pockets identified previously, and provide more insights into the acyl- and aryl-binding pockets of members of the adenylate-forming family.
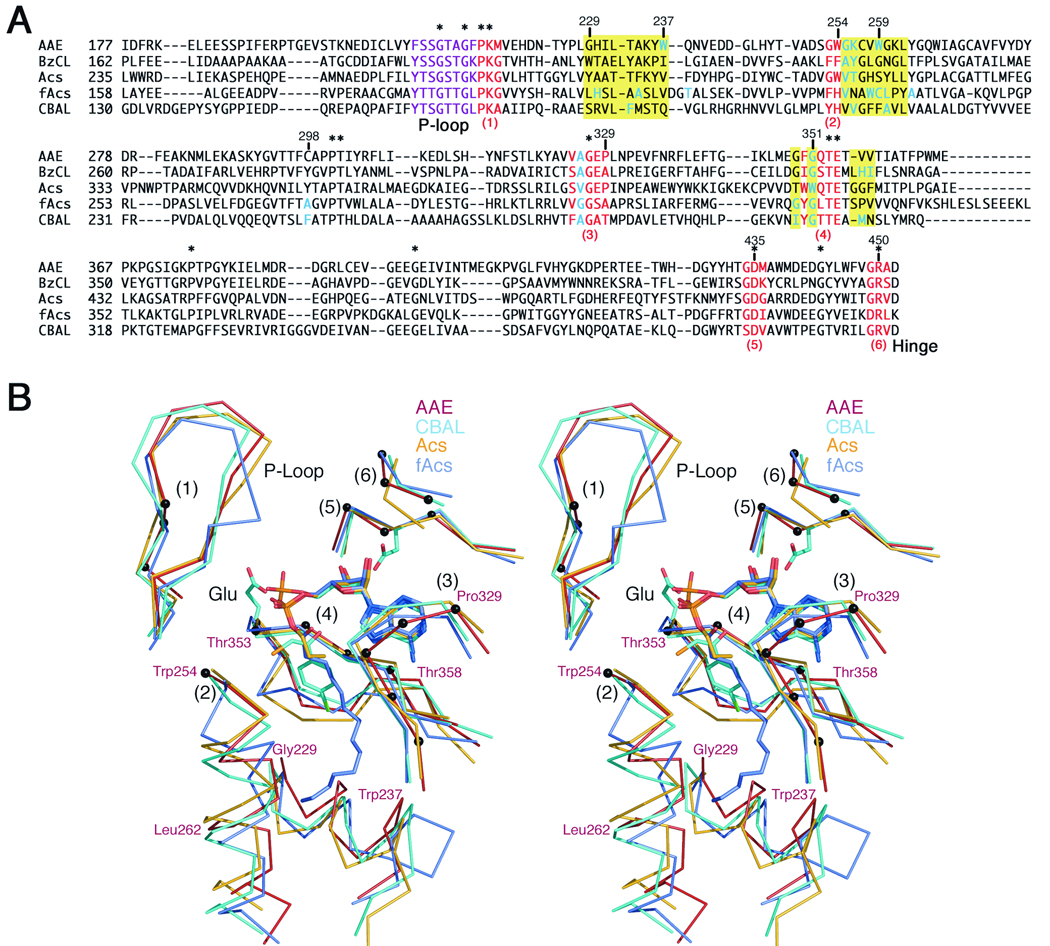
We then created a structure based alignment of the core of the N-terminal domains to analyze the residues that form the pockets in the five structurally characterized acyl- and aryl-CoA synthetases: acetyl-CoA synthetase, Acs2; 4-chlorobenzoyl-CoA ligase, CBAL1,3; benzoyl-CoA ligase, BzCL10; and the fatty acyl-CoA synthetase from Thermus thermophilus HB8, fAcs14. We used LSQKAB44 with six regions of the N-terminal domain that are structurally conserved in the entire family. These regions of the proteins interact with the ATP in a highly conserved manner. The residues used in the alignment are shown in red in Figure 5A and include: 1. Pro217-Met219, located at the end of the P-loop region that binds the ATP phosphates. 2. Gly253-Trp254. The position of Trp254 is always conserved as an aromatic side chain and we have noted3,4 that it exhibits a side chain torsional rotation around the χ1 bond between the two conformational states. 3. Gly327-Pro329, three residues that form a planar lid that stacks against the adenine ring. 4. Phe350-Glu354, a conserved motif of ϕGxTE, where ϕ is an aromatic residue that stacks against the adenine ring and the Glu has been shown to coordinate the Mg2+ ion that binds with ATP12. 5. Gly434-Met436, residues that include the universally conserved aspartate that interacts with the ribose hydroxyls, and 6. Gly449-Ala451, a final motif that contains a universally conserved Arg residue that interacts with the ribose hydroxyls as well. These six regions, in particular regions 2 and 4, will be discussed in greater detail below.
Figure 5.
Alignments of Superfamily Members. A. Multiple sequence alignment of AAE from Methanosarcina acetivorans, BzCL from Burkholderia xenovorans LB400 (2V7B), Acs from Salmonella enterica (1PG4), fAcs from T. thermophilus (1v25), and CBAL from Alcaligenes sp. (1CW9), showing the amino acid residues lining the acyl binding pocket region. The alignment was manually adjusted to preserve the structural basis for the alignment. Residues that are conserved in all five proteins are highlighted with an asterisk. The six regions in red are well-conserved structurally and served as constant regions to define the alignment with LSQKAB. The phosphate binding loop is highlighted in purple. The three regions that are defined in yellow form the acyl- and aryl-binding pockets of these enzymes. Residues that contribute side chains to the pockets are shown in blue. B. Stereorepresentation of the acyl- and aryl-binding pockets of the acyl-CoA synthetases. AAE is depicted in red, CBAL in light blue, Acs in orange, and fAcs is shown in slate grey. The six structurally conserved regions noted in red in the sequence alignment and discussed in the text are indicated with (1)–(6). The Cα positions of the AAE enzyme from these six residues are shown with black spheres. The active site shows the AMP or adenylate from the three liganded structures. The structural deviation of the three regions that form the acyl pockets are labeled using the AAE numbering: Gly229-Trp237, Trp254-Leu262, and Thr353-Thr358.
Using these well-conserved residues to anchor the alignment, we then modified the strict sequence alignment generated with CLUSTALW42,43 to produce an accurate structure based alignment (Figure 5A). This sequence alignment and the structural comparison (Figure 5B) were carefully analyzed to note the distinctions between the acyl- and aryl-binding pockets of different family members.
The structure-based sequence alignment illustrates the extremely limited sequence conservation within this region of the protein. This stretch of ~250 residues that forms the core of the large N-terminal domain contains only 14 residues (5%) that are present in all five proteins. Loosening the criteria to search for residues that are conserved in four of the five proteins, an additional 18 residues are identified, bringing the total conserved residues to 32 (11.5%). This demonstrates the extreme divergence between the sequences of different family members. This has greatly hindered efforts at deriving sequence-based predictions of substrate specificity and also of determining structures by molecular replacement, a more practical consideration.
The regions that form the acyl-binding pocket are located on three regions of the protein that are highlighted in yellow in the sequence alignment (Figure 5A). The first is a helix that follows the P-loop and is located in AAE at residues Gly229-Trp237. The second region is a mostly helical stretch that is located after the aromatic residue at position Trp254 in AAE. The final stretch of residues that form the acyl binding pocket are on the two strands that surround the conserved Mg2+ binding loop at position Thr353 and Glu354 in AAE.
Analysis of the primary sequence of these three regions shows that there is not a single uniformly conserved residue within these three regions of the protein. This divergence makes it difficult to properly align the different regions. Surprisingly, comparing the sequences from bacterial Acs2 and the Acs enzyme from yeast15 shows that only 10 of the 23 residues in these regions are conserved (not shown). Also apparent from the alignment is that these regions of the protein contain several interruptions and deviations from standard secondary structural elements. For example, the fatty acyl-CoA synthetase (fAcs) begins the helix at region 2 with five residues (Val231-Cys235) that adopt a single turn of a left-handed α-helix before adopting a standard helix beginning at residue Leu236. The helices of the other proteins, although all initiating at the aromatic residue of region 2, differ in orientation by as much as 20° resulting in an overall increase or decrease in the size of the cavity. Additionally, in three of the proteins, AAE, Acs, and BzCL, contain altered backbone torsion angles at several positions in the helix. The presence of Pro247 in BzCL, for example, disrupts the angles between Gly243 and Leu244 one turn higher on the helix. A similar effect is seen in Acs where the proline at position 320 alters the torsion angles at Tyr315 and Leu316. In AAE, Gln265 inserts into the axis of the helix to disrupt the hydrogen bonding and torsion angles at Gly260 and Lys261.
An insertion is also apparent at the strand that follows the region of the protein that coordinates the Mg2+ ion (region 4). BzCL, Acs, and fAcs all contain an extra residue (Figure 5) compared to AAE or CBAL. This insertion does not appear to impact the small acyl binding pocket of Acs, however it repositions His339 of BzCL, which forms the base of the aryl-binding pocket, and Pro331 and Val332 of fAcs, which also contribute to the pocket. It is noteworthy that, because of the sequence divergence, there is no clear way to predict whether the insertion will be present or not, and therefore it is difficult to identify the residues from this region that form the acyl pocket.
A comparison of the binding pockets of AAE and Acs (Figure 4) shows an interesting feature. Both proteins contain a tryptophan residue that forms the “floor” of the binding pocket. In Acs this residue is Trp414, a position that is almost universally conserved as a Gly, as we have noted before1,2. Mutation of this Trp residue in the Acs enzyme from M. thermoautotrophicus to an alanine results in a larger pocket that is able to accommodate acids as large as heptanoate27. The glycine residue in the proteins with larger substrates opens the acyl-binding pocket to allow it to project more deeply into the core of the N-terminal domain. AAE contains a glycine at the position of Trp414 of Acs and would therefore be expected to contain a deeper binding pocket. In contrast, as we have noted above, the side chain of Trp259 is directed from the opposite face of the active site to perform a similar role of truncating the carboxylate pocket. Thus, it appears that two different family members, Acs and AAE, use the indole ring of a Trp from opposite sides of the active site to form the base of the binding pocket, a structural feature that would have been difficult to predict from sequence alignments alone.
As noted above, the acyl binding pockets of the adenylation domains of the NRPS proteins have been carefully analyzed and the homologous features in these pockets have enabled the prediction of substrate specificity on the basis of a series of residues that form a specificity determining code23,24. These studies were aided by the relative uniformity of the binding pockets of the NRPS adenylation domains. The amino acyl substrates for these domains all contain the carboxylate, which is oriented towards the α-phosphate of ATP, and the amino group, which interacts with a conserved Asp residue. The conserved positioning of these two groups, bound to the tetrahedral α-carbon, directs the amino acid side chain towards the base of this acyl pocket in all NRPS adenylation domains. The chemical properties of the amino acid side chain then are complemented by the protein residues that form this pocket. In a more limited analysis of aryl acid adenylating enzymes, the structure of DhbE, a self-standing adenylation domain that activates 2,3-dihydroxybenzoate, provided a modified specificity determining code through a comparison with other adenylation domains that activate salicylic acid17.
In the acyl- and aryl-CoA synthetases, the lack of consistent chemical features on the chemically diverse alkyl and aryl ligands may make the identification of a similar specificity determining code more difficult. Our study with AAE has demonstrated that enzymes in the broader adenylate-forming family have more diverse acyl binding pockets and an analysis based primarily on sequence alignment may not prove as useful in the prediction of function of uncharacterized members of the family. Additional structures of proteins of this family and correlations with functional analyses will prove useful in the prediction of substrate specificity.
Supplementary Material
The protocol for sequencing the AAE protein by high resolution nano-LC coupled with mass spectrometry is described in detail in the supplemental material.
Abbreviations
- Acs
Acetyl-CoA synthetase
- NRPS
Non-ribosomal peptide synthetase
- AAE
Acyl-adenylating enzyme from Methanosarcina acetivorans
- CBAL
4-chlorobenzoic acid: CoA ligase
- fAcs
long chain fatty acyl-CoA synthetase
- mAcs
medium chain acyl-CoA synthetase
- DhbE
2,3-dihydroxybenzoate-activating E domain
- HEPES
N-(2-hydroxyethyl)piperzine-N’-2-ethanesulfonate
- AMP
adenosine-5’-monophosphate
- ATP
adenosine-5-triphosphate
- IPTG, SeMet
selenomethionine; isopropyl-β–D-1-thiogalactopyranoside
- TCEP-HCl
Tris(2-carboxyethyl)phosphine hydrochloride
- IMAC
immobilized metal-affinity chromatography
- MAD
Multiwavelength Anomalous Dispersion
- PEG
polyethylene glycol
- rms
root mean square.
Footnotes
This research was supported by funds from grant NIH-GM068440 (A. M. G.) and NIH-GM069374 (K. S. S.), the South Carolina Experiment Station Project SC-1700198 (K.S.S), and Technical Contribution No. 5551 of the Clemson University Experiment Station. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The LC-MS/MS system was obtained by a Shared Instrument Grant (S10-RR14592) from the National Center for Research Resources, National Institutes of Health.
The X-ray coordinates and structure factors for the AAE enzyme have been deposited in the Brookhaven Protein Data Bank (Accession number 3ETC).
References
- 1.Gulick AM, Lu X, Dunaway-Mariano D. Crystal structure of 4-chlorobenzoate:CoA ligase/synthetase in the unliganded and aryl substrate-bound states. Biochemistry. 2004;43(27):8670–8679. doi: 10.1021/bi049384m. [DOI] [PubMed] [Google Scholar]
- 2.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry. 2003;42(10):2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 3.Reger AS, Wu R, Dunaway-Mariano D, Gulick AM. Structural characterization of a 140° domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase. Biochemistry. 2008;47(31):8016–8025. doi: 10.1021/bi800696y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu R, Cao J, Lu X, Reger AS, Gulick AM, Dunaway-Mariano D. Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis. Biochemistry. 2008;47(31):8026–8039. doi: 10.1021/bi800698m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, Chang KH, Liang PH, Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31(24):5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- 6.Scholten JD, Chang KH, Babbitt PC, Charest H, Sylvestre M, Dunaway-Mariano D. Novel enzymic hydrolytic dehalogenation of a chlorinated aromatic. Science. 1991;253(5016):182–185. doi: 10.1126/science.1853203. [DOI] [PubMed] [Google Scholar]
- 7.Turgay K, Krause M, Marahiel MA. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6(18):529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 8.White HE, Miano JD, Umbreit M. Letter: on the mechanism of firefly luciferin luminescence. J Am Chem Soc. 1975;97(1):198–200. doi: 10.1021/ja00834a040. [DOI] [PubMed] [Google Scholar]
- 9.Fraga H, Fernandes D, Fontes R, Esteves da Silva JC. Coenzyme A affects firefly luciferase luminescence because it acts as a substrate and not as an allosteric effector. FEBS J. 2005;272(20):5206–5216. doi: 10.1111/j.1742-4658.2005.04895.x. [DOI] [PubMed] [Google Scholar]
- 10.Bains J, Boulanger MJ. Biochemical and structural characterization of the paralogous benzoate CoA ligases from Burkholderia xenovorans LB400: defining the entry point into the novel benzoate oxidation (box) pathway. J Mol Biol. 2007;373(4):965–977. doi: 10.1016/j.jmb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4(3):287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 12.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non- ribosomal biosynthesis of gramicidin S. Embo J. 1997;16(14):4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du L, He Y, Luo Y. Crystal structure and enantiomer selection by D-alanyl carrier protein ligase DltA from Bacillus cereus. Biochemistry. 2008;47(44):11473–11480. doi: 10.1021/bi801363b. [DOI] [PubMed] [Google Scholar]
- 14.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J Biol Chem. 2004;279(30):31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 15.Jogl G, Tong L. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry. 2004;43:1425–1431. doi: 10.1021/bi035911a. [DOI] [PubMed] [Google Scholar]
- 16.Kochan G, Pilka ES, von Delft F, Oppermann U, Yue WW. Structural Snapshots for the Conformation-dependent Catalysis by Human Medium-chain Acyl-coenzyme A Synthetase ACSM2A. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.03.064. in press. [DOI] [PubMed] [Google Scholar]
- 17.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440(7082):372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- 19.Osman KT, Du L, He Y, Luo Y. Crystal structure of Bacillus cereus D-alanyl carrier protein ligase (DltA) in complex with ATP. J Mol Biol. 2009;388(2):345–355. doi: 10.1016/j.jmb.2009.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Reger AS, Carney JM, Gulick AM. Biochemical and Crystallographic Analysis of Substrate Binding and Conformational Changes in Acetyl-CoA Synthetase. Biochemistry. 2007;46(22):6536–6546. doi: 10.1021/bi6026506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu R, Reger AS, Cao J, Gulick AM, Dunaway-Mariano D. Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range(,)(,) Biochemistry. 2007;46(50):14487–14499. doi: 10.1021/bi701609w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yonus H, Neumann P, Zimmermann S, May JJ, Marahiel MA, Stubbs MT. Crystal structure of DltA. Implications for the reaction mechanism of nonribosomal peptide synthetase adenylation domains. J Biol Chem. 2008;283(47):32484–32491. doi: 10.1074/jbc.M800557200. [DOI] [PubMed] [Google Scholar]
- 23.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7(3):211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 24.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6(8):493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 25.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33(18):5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1(5):265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 27.Ingram-Smith C, Woods BI, Smith KS. Characterization of the acyl substrate binding pocket of acetyl-CoA synthetase. Biochemistry. 2006;45(38):11482–11490. doi: 10.1021/bi061023e. [DOI] [PubMed] [Google Scholar]
- 28.Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WG, Mehlin C. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 1):203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 29.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142(1):170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 30.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9(Pt 6):401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski ZM. W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Weeks CM, Blessing RH, Miller R, Mungee R, Potter SA, Rappleye J, Smith GD, Xu H, Furey W. Towards automated protein structure determination: BnP, the SnB-PHASES interface. Z Kristallogr. 2002;217:686–693. [Google Scholar]
- 33.Terwilliger TC. Maximum-likelihood density modification with pattern recognition of structural motifs. Acta Crystallogr D Biol Crystallogr. 2001;57:1755–1762. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 1):122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 38.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50(pt 2):178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 39.Hubbard SJ, Argos P. Detection of internal cavities in globular proteins. Protein Eng. 1995;8(10):1011–1015. doi: 10.1093/protein/8.10.1011. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The Pymol Molecular Graphics System. DeLano Scientific. 2002 [Google Scholar]
- 41.Cuff AL, Martin AC. Analysis of void volumes in proteins and application to stability of the p53 tumour suppressor protein. J Mol Biol. 2004;344(5):1199–1209. doi: 10.1016/j.jmb.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 43.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. A Solution for the Best Rotation to Relate Two Sets of Vectors. Acta Crystallogr A. 1976;32:922–923. [Google Scholar]
- 45.Galkin A, Lu X, Dunaway-Mariano D, Herzberg O. Crystal structures representing the Michaelis complex and the thiouronium reaction intermediate of Pseudomonas aeruginosa arginine deiminase. J Biol Chem. 2005;280(40):34080–34087. doi: 10.1074/jbc.M505471200. [DOI] [PubMed] [Google Scholar]
- 46.Linsky TW, Monzingo AF, Stone EM, Robertus JD, Fast W. Promiscuous partitioning of a covalent intermediate common in the pentein superfamily. Chem Biol. 2008;15(5):467–475. doi: 10.1016/j.chembiol.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006;45(17):5618–5630. doi: 10.1021/bi052595m. [DOI] [PubMed] [Google Scholar]
- 48.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu R, Reger AS, Lu X, Gulick AM, Dunaway-Mariano D. The Mechanism of Domain Alternation in the Acyl-Adenylate Forming Ligase Superfamily Member 4-Chlorobenzoate: Coenzyme A Ligase. Biochemistry. 2009 doi: 10.1021/bi9002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branchini BR, Magyar RA, Murtiashaw MH, Anderson SM, Zimmer M. Site-directed mutagenesis of histidine 245 in firefly luciferase: a proposed model of the active site. Biochemistry. 1998;37(44):15311–15319. doi: 10.1021/bi981150d. [DOI] [PubMed] [Google Scholar]
- 51.Branchini BR, Murtiashaw MH, Magyar RA, Anderson SM. The role of lysine 529, a conserved residue of the acyl-adenylate- forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39(18):5433–5440. doi: 10.1021/bi9928804. [DOI] [PubMed] [Google Scholar]
- 52.Branchini BR, Southworth TL, Murtiashaw MH, Wilkinson SR, Khattak NF, Rosenberg JC, Zimmer M. Mutagenesis evidence that the partial reactions of firefly bioluminescence are catalyzed by different conformations of the luciferase C-terminal domain. Biochemistry. 2005;44(5):1385–1393. doi: 10.1021/bi047903f. [DOI] [PubMed] [Google Scholar]
- 53.Tian Y, Suk DH, Cai F, Crich D, Mesecar AD. Bacillus anthracis o-Succinylbenzoyl-CoA Synthetase: Reaction Kinetics and a Novel Inhibitor Mimicking Its Reaction Intermediate. Biochemistry. 2008 doi: 10.1021/bi801311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gocht M, Marahiel MA. Analysis of core sequences in the D-Phe activating domain of the multifunctional peptide synthetase TycA by site-directed mutagenesis. J Bacteriol. 1994;176(9):2654–2662. doi: 10.1128/jb.176.9.2654-2662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horswill AR, Escalante-Semerena JC. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry. 2002;41(7):2379–2387. doi: 10.1021/bi015647q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The protocol for sequencing the AAE protein by high resolution nano-LC coupled with mass spectrometry is described in detail in the supplemental material.