Abstract
Background
Prader-Willi syndrome (PWS) is a complex genetic disorder with errors in genomic imprinting, generally due to a paternal deletion of chromosome 15q11-q13 region. Maternal disomy 15 (both 15s from the mother) is the second most common form of PWS resulting from a trisomic zygote followed by trisomy rescue in early pregnancy and loss of the paternal chromosome 15. However, trisomy 15 or mosaicism for trisomy 15 may be present in the placenta possibly leading to placental abnormalities affecting gestational age and delivery.
Methods and Subjects
We examined growth and gestational data from 167 PWS infants (93 males and 74 females; 105 infants with 15q11-q13 deletion and 62 infants with maternal disomy 15) to determine if there are differences in gestation between the two genetic subtypes.
Results
No significant differences in growth data (birth weight, length, head circumference) or average gestational ages were found between the two genetic subgroups. However, post-term deliveries (> 42 weeks gestation) were more common in the maternal disomy group (i.e., 12 of 62 infants) compared with the deletion group (i.e., 7 of 105 infants) (chi-square test = 6.22; p < 0.02). The distribution of gestational ages in the 15q11-q13 deletion group was more bell-shaped or normal while the distribution in the maternal disomy group suggested a bimodal pattern.
Conclusions
Maternal disomy 15 in PWS may contribute to disturbances in gestational age and delivery by impacting on placental structure or function secondary to the abnormal chromosomal number in the placental cells or in mechanisms leading to the maternal disomy status in PWS infants.
Keywords: Prader-Willi syndrome, Maternal disomy 15, Trisomy rescue, Genomic imprinting, Abnormal gestation
Introduction
Prader-Willi syndrome (PWS) is a complex neurogenetic disorder that arises from lack of expression of paternally inherited genes located in the chromosome 15q11-q13 region [1]. PWS generally occurs sporadically and is characterized by infantile hypotonia, a poor suck, hypogenitalism/hypogonadism, mental deficiency and behavioral problems, hyperphagia leading to early childhood obesity, short stature due to growth hormone deficiency, small hands and feet and a characteristic facial appearance including a narrow bifrontal diameter, short upturned nose, thin upper lip and down turned corners of the mouth, almond-shaped eyes and sticky saliva with enamel hypoplasia. The hyperphagia or increased ingestion of food and early childhood obesity is extremely severe in some cases. PWS is considered the most common genetic syndrome leading to life-threatening obesity with an estimated frequency of 1 in 10,000 to 20,000 individuals. About 400,000 people with this disorder are affected worldwide and present in all ethnic groups but reported disproportionately more in Caucasians [2].
The course and early natural history of PWS can be divided into two distinct clinical stages. The first stage occurs during the neonatal period and early infancy and characterized by varying degrees of hypotonia, a weak cry, a narrow forehead, developmental delay, temperature instability, a poor suck reflex, sticky saliva, feeding difficulties sometimes requiring gastrostomy or stomach tube placement, hypogonadism, and underdevelopment of the sex organs. Failure-to-thrive is noted during this first stage [2–4].
The second stage usually begins around 2 years of age and characterized by continued developmental delay or psychomotor retardation and onset of hyperphagia leading to obesity. Other features noted during the second stage may include speech articulation problems, foraging for food, rumination, unmotivated sleepiness (found in greater than 50% of subjects), physical inactivity, decreased pain sensitivity, skin picking and other forms of self-injurious behavior, prolonged periods of hypothermia, strabismus, hypopigmentation, scoliosis, obstructive sleep apnea, and abnormal oral pathology (enamel hypoplasia, dental caries, malocclusion, and decreased saliva).
Decreased fetal movement is also noted by the mothers in nearly all PWS pregnancies. About one fourth of babies with PWS are delivered in breech presentation. Approximately one half of babies with PWS are born pre- or post-term (2 weeks earlier or later than the anticipated delivery date of 40 weeks). Mild prenatal growth retardation is noted, with an average birth weight of 2.8 kg. Low birth weight is seen in about 30% of deliveries [5].
Genetic evaluations and subtypes
There are several genetic disturbances that have been identified as causes of PWS. Approximately 70% of PWS cases are caused by a non-inherited (i.e., de novo) deletion in the paternally derived chromosome 15q11-q13 region. The typical 15q11-q13 deletion has been classified into two types, type I and type II, depending on the size and chromosome breakpoint position. Those with the larger typical type I deletion have more clinical problems such as obsessive-compulsive disorders, self-injury and poorer academic performance than those PWS subjects with the smaller typical type II deletion [6, 7]. Genetic subtypes are determined through genetic testing such as fluorescence in situ hybridization (FISH) studies using DNA probes from the 15q11-q13 region that will identify the typical deletion while the remaining cases have no obvious defect revealed by FISH. Approximately 25% of PWS cases result from maternal disomy 15 (i.e., two maternal chromosome 15s) and no paternal chromosome 15. About 3% of PWS cases have biparental or normal inheritance of chromosome 15 and results from defects (microdeletions or epimutations) of the imprinting center controlling the activity of imprinted genes in the 15q11-q13 region. Rarely, other chromosome 15q11-q13 rearrangements occur such as translocations, inversions or marker chromosomes [1, 4, 5].
Methylation DNA testing which measures the methylation status of the genes in the region can be used for laboratory diagnosis of PWS. Methylation testing is considered to be 99% accurate in the diagnosis of PWS but does not allow for identification of the specific genetic subtype [8, 9]. Additional testing besides FISH is required to identify maternal disomy 15 or imprinting defects.
Several genes or transcripts have been mapped to the 15q11-q13 region that are imprinted, with most having only paternal expression, including SNURF-SNRPN, small nucleolar RNAs (snoRNAs), necdin, MKRN3 and MAGEL2. Candidate genes for causing PWS should be paternally expressed and maternally silenced, located within the chromosome 15q11-q13 region and involved directly or indirectly in brain development and function. The promoter and first exon of SNURF-SNRPN are integral components of the imprinting center that controls the regulation of imprinting throughout the chromosome 15q11-q13 region. A disruption of this complex locus will cause loss of function of paternally expressed genes in this region, leading to PWS [1, 4, 10].
Maternal disomy 15 is the second most frequent finding in PWS due to four possible mechanisms proposed by Cassidy et al. [11]. These include: 1) a disomic egg with two maternal chromosome 15s plus a monosomic or normal sperm producing a trisomic zygote followed by subsequent loss of the paternal chromosome 15; 2) a disomic egg plus a nullisomic sperm without a chromosome 15 leading to a normal chromosome count; 3) a monosomic egg plus a nullisomic sperm producing a monosomic zygote followed by duplication of the maternal chromosome 15; and 4) postfertilization nondisjunction producing mosaicism for trisomic and monosomic cell lines with subsequent duplication in the monosomic line. Hence, during meiosis, the diploid set of human chromosomes (n = 46) is reduced to a haploid set (n = 23). Nondisjunction of the homologous chromosome 15s during female meiosis I or nondisjunction of the two sister (identical) chromatids during meiosis II results in an oocyte with two maternal chromosome 15s or an oocyte with no chromosome 15. Fertilization of an oocyte with two maternal chromosome 15s by a normal sperm with one chromosome 15 leads to a zygote trisomic for chromosome 15. This condition is not compatible with normal development and is a relatively common cause of early miscarriages, but through trisomy rescue of the fetus the pregnancy is salvaged and not spontaneously aborted. In two thirds of cases, one of the two maternal chromosome 15s will be lost from the trisomic cell. This results in a normal set of chromosomes. If, however, the paternal chromosome is lost, the cell is left with two maternal chromosome 15s. The fetus is delivered at term having PWS and normal cytogenetic findings but with maternal disomy 15.
Cassidy et al. [11] further reported a child with PWS who was born to a 43 year old mother with prenatal diagnosis by chorionic villus sampling (CVS) at 10 weeks gestation showing trisomy 15 in all cells. Amniocentesis studies at 16 weeks gestation were normal without evidence of trisomy 15. Trisomy 15 is one of the more common autosomal trisomies reported in a series of abortuses comprising about 8% [12]. The child was born with hypotonia, a poor suck and weak cry and features consistent with PWS. However, the chromosome count was normal but testing with DNA markers was consistent with maternal disomy 15. This case illustrates that maternal disomy 15 most likely occurs from nondisjunction during maternal meiosis followed by a postzygotic correction of the meiotic error. However, trisomy 15 or mosaicism for trisomy 15 may be present in the placenta possibly leading to placental abnormalities (structural and/or functional) affecting gestational age and delivery.
Maternal disomy may be initially detected as mosaic trisomy 15 during routine prenatal cytogenetic analysis as supported by a study of seven cases of trisomy 15 mosaicism [13] in which two cases were consistent with maternal disomy 15 supporting the theoretical expectation of one-third of the possible outcomes resulting in maternal disomy. Therefore, uniparental disomy testing should be offered in all cases of mosaic trisomy 15 encountered in CVS or amniocentesis studies.
To further investigate the trisomy rescue event and timing in abnormal cells from the early pregnancy, the X chromosome inactivation ratio using the polymorphic androgen receptor (AR) gene located at Xq11.2 can be used. X inactivation patterns can be assessed using the AR gene in females informative at the polymorphic CAG repeat region of exon 1 from the gene following DNA digestion with methyl-sensitive restriction enzymes (e.g., HpaII) and PCR amplification of the polymorphic AR gene. X chromosome skewness (i.e., one X chromosome may be more or less active compared with the second X chromosome in somatic cells) is assigned at an arbitrary ratio of highly skewed (e.g., > 80%:20%) or extremely skewed (e.g., > 90%:10%) [14–17]. In healthy females, X chromosome inactivation is considered to follow a bell-shaped distribution with highly skewed patterns being uncommon events [18]. The inactivation of the X chromosome in females is generally considered to be random with regards to which X is inactivated. In females each of the two X chromosomes becomes inactivated at random to allow for equal gene dosage for X-linked genes in normal females and males. In certain cases the X chromosome inactivation skewness is not random and skewness of the X inactivation ratio occurs such as in X-autosomal chromosome rearrangements, mutations of the gene controlling the X inactivation process (i.e., XIST), certain X-linked disorders (e.g., Rett) and monozygotic twinning [19, 20]. Since about 2% of pregnancies detected by chorionic villus sampling are associated with confined placental mosaicism (CPM) [21], it may be a significant contributor to both skewed X inactivation observed in some newborns and expression of X-linked recessive diseases in females.
Lau et al. [19] reported extreme skewness in 7 of 12 cases with a meiotic origin of the trisomy and none of 10 cases examined with a somatic origin of autosomal trisomic cases involving CPM but in only 1 of 27 control adult females. They also reported extreme X chromosome skewness in 3 of 10 informative females with maternal disomy 15 supporting trisomy rescue in a proportion of maternal disomy 15 cases associated with CPM. Furthermore, Butler et al. [22] reported results from a cohort of PWS females with the 15q deletion or maternal disomy 15 and female controls using the androgen receptor gene assay system in peripheral blood. A significantly larger number of PWS females with maternal disomy were found with extreme X chromosome skewness (95% vs 5%) compared with PWS deletion or control females. These results indicate a trisomy rescue event early in embryo development and a small number of cells survived with a selective advantage for cell proliferation due to the normal chromosome complement and maternal disomy 15 but with extreme X chromosome skewness. These females could also be at risk for X-linked recessive disorders along with PWS.
Results and discussion
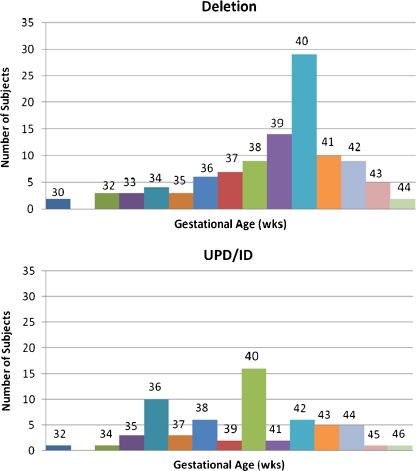
We recently examined growth measures at birth and gestational age data from 167 PWS infants (93 males and 74 females). The diagnosis of PWS was confirmed by molecular genetic testing and genetic subtypes determined [105 subjects with 15q11-q13 deletion; 62 subjects with maternal disomy 15/imprinting defects (UPD/ID)]. The gestational age, birth weight, birth length, birth head circumference and body mass index (BMI) data are shown in Table 1. No significant mean differences in growth or gestational age data were observed among the genetic subtype categories (independent t tests; p > 0.05). However, post-term deliveries (>42 weeks gestation) were more common in the UPD/ID group (i.e., 12 of 62 subjects) compared with those with the 15q11-q13 deletion (i.e., 7 of 105 subjects) (chi-square test = 6.22; p < 0.02). In addition, the distribution of gestational ages in the 15q11-q13 deletion group was more bell-shaped or normal in appearance but with skewing to the left. The modal number was 40 weeks gestation for each subject group while a second peak at 36 weeks gestation was present in the UPD/ID group suggesting a bimodal distribution pattern (Fig. 1).
Table 1.
Gestational and growth data in Prader-Willi syndrome genetic subtypes
| Variables | Deletion(n = 105) | Deletion Standard Deviation | UPD/ID (n = 62) | UPD/ID Standard Deviation | T-Test Values* |
|---|---|---|---|---|---|
| Mean Gestational Age (wks) | 38.78 | 2.97 | 39.52 | 3.08 | 0.13 |
| Mean Birth Weight (kg) | 2.87 | 0.56 | 2.82 | 0.57 | 0.62 |
| Mean Birth Length (cm) | 49.94 | 3.02 | 49.03 | 4.28 | 0.16 |
| Mean Head Circumference (cm) | 35.24 | 2.22 | 34.12 | 3.00 | 0.26 |
| Mean Body Mass Index (BMI) (kg/m2) | 11.41 | 1.85 | 11.83 | 2.42 | 0.27 |
* Independent t-test values (not significant; p > 0.05)
Fig. 1.
Histograms showing the gestational ages and number of subjects for the Prader-Willi syndrome subjects with the 15q11-q13 deletion (n = 105) and maternal disomy 15/imprinting defect (UPD/ID) subject (n = 62). Numbers at top of each bar represent the gestational age in weeks
Current medical practice and obstetrical care supports the induction of pregnancies by a specified gestational age (e.g., 42 weeks). Therefore, we will not know how post-term or mature the gestational age may become in future PWS pregnancies as extended pregnancies are now frequently induced at or before 42 weeks.
Our observations on gestational ages in PWS may stimulate additional studies on the effects of maternal disomy 15 on placental anatomy and function, specifically genetic factors, pathophysiology, growth factors, anatomical size and weight, and proteins regulating placental function and influencing gestational age and parturition. These investigations could include placental pathology studies in PWS fetuses, gene expression studies, animal modeling and skewness of X chromosome inactivation studies as a measure of early trisomy rescue of abnormal cells in the embryo. For example, skewed patterns of X inactivation in CPM cases are thought to result from a reduction in the size of the early embryonic cell pool because of either poor early growth or subsequent selection against trisomic cells. Extreme X chromosome skewness (>90%) in PWS females with maternal disomy 15 may indicate a small pool of embryonic progenitor cells originating from rescue of trisomy 15 and further suggest that those females with skewness could be at risk for X- linked recessive disorders along with PWS influencing clinical outcomes and pregnancies.
Additionally, maternal disomy 15 or uniparental disomy (UPD) causing PWS is of two types: heterodisomy or isodisomy; the disomic type may also impact on the pregnancy and clinical outcome. Most PWS subjects with UPD have the heterodisomic form. Maternal heterodisomy occurs when the baby inherits each of the mother’s chromosome 15s but with no chromosome 15 from the father. Maternal isodisomy results when two identical chromosome 15s are inherited from the mother only as a result of nondisjunction in meiosis II or from nondisjunction in meiosis I with crossing over and possibly a somatic recombination in early pregnancy producing a segmental or partial form of isodisomy but for only a region of chromosome 15.
Maternal isodisomy may also lead to other genetic disorders in the PWS patient. For example, if the mother is a carrier of an autosomal recessive gene mutation on chromosome 15, for example, Bloom syndrome gene (BLM) located at 15q26.1 [23] and this chromosome region (e.g., due to segmental isodisomy) or the entire chromosome shows isodisomy (e.g., nondisjunction in meiosis II producing identical chromatids), then the PWS offspring would also present with the recessive genetic disorder by inheriting both of the mother’s recessive alleles. As in other nondisjunction cases, the risk of UPD, or specifically maternal disomy 15, increases with maternal age.
Disturbances of growth factors influenced by maternal disomy 15 may contribute to abnormalities of placental growth or function in PWS and pregnancy outcome. There are no recognized growth factor genes in the proximal long arm of chromosome 15 that should affect placental growth or other imprinted genes in humans on chromosome 15. However, the insulin-like growth factor receptor 1 (IGF1R) gene is located on the distal long arm of chromosome 15 and involved with growth and development [24]. IGF1R is a member of a gene family of growth factors and receptors known to be imprinted in humans (e.g., IGF2) and transmits the biological effects of the major growth factors IGF1 and IGF2 during pre- and post-natal growth. In mice, the paternally expressed Igf2 gene and the maternally expressed Igf2r gene stimulate and inhibit embryonic growth, respectively. Abnormalities in these genes are known to cause Beckwith-Wiedemann syndrome, an overgrowth syndrome with abnormal placental growth and polyhyramnios due to imprinting disturbances on chromosome 11 including the IGF2 gene [25]. Another imprinted gene that could impact on placental growth and function is the GRB10 gene located on chromosome 7. This maternally expressed gene is a candidate for causing Silver-Russell syndrome (SRS), a clinically heterogenous growth retardation disorder with asymmetry due to maternal disomy 7 in a subset of affected individuals. In mice, loss of the maternal copy of the gene results in both fetal and placental overgrowth further demonstrating its role as a growth suppressor [26].
Generalized imprinting defects in some patients with imprinting disorders have been reported which could lead to disturbances of growth factor genes implicated in placental and fetal growth [27]. Therefore, one could speculate that differences in gestational age in PWS UPD subjects compared with those PWS subjects with the 15q11-q13 deletion could be triggered by maternal disomy 15 and/or trisomy 15 rescue events in early pregnancy. Imprinted gene products are critical regulators of growth and development and more research is needed to further address the observations of abnormal gestation in pregnancies with PWS and maternal disomy 15.
Acknowledgements
The authors would like to acknowledge the families with Prader-Willi syndrome. We thank Jasmin Tanaja and Cindy Yoo from University of California-Irvine for data retrieval. This study was partially supported by NIH rare disease grant (1U54RR019478).
References
- 1.Bittel DC, Butler MG. Prader-Willi syndrome: Clinical genetics, cytogenetics and molecular biology. Expert Rev Mol Med. 2005;7(14):1–20. [DOI] [PMC free article] [PubMed]
- 2.Butler MG, Hanchett J, Thompson T. Clinical findings and natural history of Prader-Willi syndrome. In: Butler MG, Lee PDK, Whitman BY, editors. Management of Prader-Willi Syndrome. 3rd ed. Springer-Verlag Publishers: New York; 2006. p. 3–48.
- 3.Goldstone AP, Holland AJ, Hauffa BP, Hokken-Koelega AC, Tauber M. Recommendations for the diagnosis and management of Prader-Willi syndrome. J Clin Endocrinol Metab. 2008;93(11):4183–97. [DOI] [PubMed]
- 4.Cassidy SB, Driscoll DJ. Prader-Willi syndrome. Eur J Hum Genet. 2009;17(1):3–13. [DOI] [PMC free article] [PubMed]
- 5.Butler MG, Thompson T. Prader-Willi syndrome: Clinical and genetic findings. The Endocrinologist. 2000;10:3S–16S. [DOI] [PMC free article] [PubMed]
- 6.Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004;113(3 Pt 1):565–73. [DOI] [PMC free article] [PubMed]
- 7.Bittel DC, Kibiryeva N, Butler MG. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in Prader-Willi syndrome. Pediatrics. 2006;118(4):e1276–1283. [DOI] [PMC free article] [PubMed]
- 8.Kubota T, Das S, Christian SL, Baylin SB, Herman JG, Ledbetter DH. Methylation-specific PCR simplifies imprinting analysis. Nat Genet. 1997;16(1):16–7. [DOI] [PubMed]
- 9.Glenn CC, Porter KA, Jong MT, Nicholls RD, Driscoll DJ. Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet. 1993;2(12):2001–5. [DOI] [PubMed]
- 10.Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–75. [DOI] [PubMed]
- 11.Cassidy SB, Lai LW, Erickson RP, Magnuson L, Thomas E, Gendron R, et al. Trisomy 15 with loss of the paternal 15 as a cause of Prader-Willi syndrome due to maternal disomy. Am J Hum Genet. 1992;51(4):701–8. [PMC free article] [PubMed]
- 12.Warburton D, Byrne J, Canki N. Trisomy. In: Warburton D, Byrne J, Canki N, editors. Chromosome anomalies and prenatal development: An atlas. New York: Oxford University Press; 1991. p. 57–62.
- 13.Christian SL, Smith AC, Macha M, Black SH, Elder FF, Johnson JM, et al. Prenatal diagnosis of uniparental disomy 15 following trisomy 15 mosaicism. Prenat Diagn. 1996;16(4):323–32. [DOI] [PubMed]
- 14.Harris A, Collins J, Vetrie D, Cole C, Bobrow M. X inactivation as a mechanism of selection against lethal alleles: further investigation of incontinentia pigmenti and X linked lymphoproliferative disease. J Med Genet. 1992;29(9):608–14. [DOI] [PMC free article] [PubMed]
- 15.Sangha KK, Stephenson MD, Brown CJ, Robinson WP. Extremely skewed X-chromosome inactivation is increased in women with recurrent spontaneous abortion. Am J Hum Genet. 1999;65(3):913–7. [DOI] [PMC free article] [PubMed]
- 16.Maier EM, Kammerer S, Muntau AC, Wichers M, Braun A, Roscher AA. Symptoms in carriers of adrenoleukodystrophy relate to skewed X inactivation. Ann Neurol. 2002;52(5):683–8. [DOI] [PubMed]
- 17.Talebizadeh Z, Bittel DC, Veatch OJ, Kibiryeva N, Butler MG. Brief report: Non-random X chromosome inactivation in females with autism. J Autism Dev Disord. 2005;35(5):675–81. [DOI] [PMC free article] [PubMed]
- 18.Migeon BR. Non-random X chromosome inactivation in mammalian cells. Cytogenet Cell Genet. 1998;80(1–4):142–8. [DOI] [PubMed]
- 19.Lau AW, Brown CJ, Penaherrera M, Langlois S, Kalousek DK, Robinson WP. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am J Hum Genet. 1997;61(6):1353–61. [DOI] [PMC free article] [PubMed]
- 20.Krepischi AC, Kok F, Otto PG. X chromosome-inactivation patterns in patients with Rett syndrome. Hum Genet. 1998;102(3):319–21. [DOI] [PubMed]
- 21.Ledbetter DH, Zachary JM, Simpson JL, Golbus MS, Pergament E, Jackson L et al. Cytogenetic results from the U.S. Collaborative Study on CVS. Prenat Diagn. 1992; 12(5):317–45. [DOI] [PubMed]
- 22.Butler MG, Theodoro MF, Bittel DC, Kuipers PJ, Driscoll DJ, Talebizadeh Z. X-chromosome inactivation patterns in females with Prader-Willi syndrome. Am J Med Genet A. 2007;143(5):469–75. [DOI] [PMC free article] [PubMed]
- 23.Woodage T, Prasad M, Dixon JW, Selby RE, Romain DR, Columbano-Green LM, et al. Bloom syndrome and maternal uniparental disomy for chromosome 15. Am J Hum Genet. 1994;55(1):74–80. [PMC free article] [PubMed]
- 24.Roback EW, Barakat AJ, Dev VG, Mbikay M, Chretien M, Butler MG. An infant with distal deletion of the long arm of chromosome 15 (q26.1-qter) and loss of insulin-like growth factor 1 receptor gene. Am J Med Genet. 1991; 38:74–9. [DOI] [PMC free article] [PubMed]
- 25.Eggermann T. Silver-Russell and Beckwith-Wiedemann syndromes: Opposite (epi)mutations in 11p15 result in opposite clinical pictures. Horm Res. 2009;71:30–5. [DOI] [PubMed]
- 26.Eggermann T, Eggermann K, Schonherr N. Growth retardation versus overgrowth: Silver-Russell syndrome is genetically opposite to Beckwith-Wiedemann syndrome. Trends Genet. 2008;24:195–204. [DOI] [PubMed]
- 27.Bliek J, Verde G, Callaway J, Maas SM, de Crescenzo A, Sparago A, et al. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2009;17:611–9. [DOI] [PMC free article] [PubMed]