Abstract
The non-heme iron(II) dioxygenase family of enzymes contain a common 2-His–1-carboxylate iron-binding motif. These enzymes catalyze a wide variety of oxidative reactions, such as the hydroxylation of aliphatic C–H bonds. Prolyl 4-hydroxylase (P4H) is an α-ketoglutarate-dependent iron(II) dioxygenase that catalyzes the post-translational hydroxylation of proline residues in protocollagen strands, stabilizing the ensuing triple helix. Human P4H residues His412, Asp414, and His483 have been identified as an iron-coordinating 2-His–1-carboxylate motif. Enzymes that catalyze oxidative halogenation do so by a mechanism similar to that of P4H. These halogenases retain the active-site histidine residues, but the carboxylate ligand is replaced with a halide ion. We replaced Asp414 of P4H with alanine (to mimic the active site of a halogenase) and with glycine. These substitutions do not, however, convert P4H into a halogenase. Moreover, the hydroxylase activity of D414A P4H cannot be rescued with small molecules. In addition, rearranging the two His and one Asp residues in the active site eliminates hydroxylase activity. Our results demonstrate a high stringency for the iron-binding residues in the P4H active site. We conclude that P4H, which catalyzes an especially demanding chemical transformation, is recalcitrant to change.
Introduction
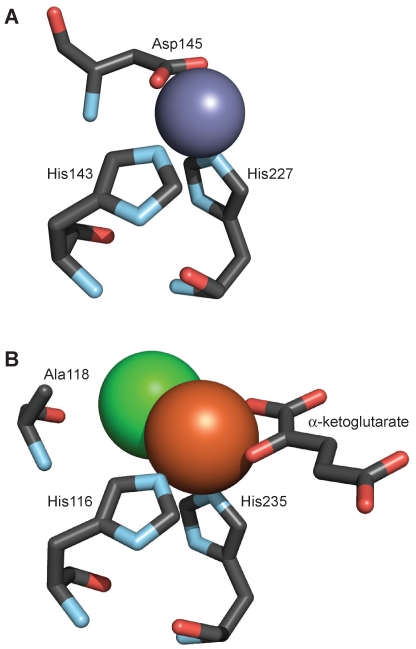
Iron is a common cofactor in enzymes that employ molecular oxygen as an oxidant. In addition to those iron-dependent enzymes that rely on heme, there is a class of non-heme, mononuclear iron(II) enzymes that catalyze the hydroxylation of aliphatic C–H bonds, dihydroxylation of arene double bonds, epoxidation of C–C double bonds, heterocyclic ring formation, and oxidative aromatic ring opening. The iron in these non-heme dioxygenases is commonly coordinated by a 2-His–1-carboxylate motif [1]. Two histidine (His) and either one aspartic acid (Asp) or one glutamic acid (Glu) residue bind the iron at the vertices of one triangular face of the octahedral metal center, forming a 2-His–1-carboxylate triad of facial ligands (Figure 1A). This arrangement leaves three coordination sites on the iron open for the substrate, oxygen, and other co-substrates.
Figure 1. Active sites of α-ketoglutarate-dependent iron(II) dioxygenases.
(A) The Chlamydomonas reinhardtii prolyl 4-hydroxylase contains a 2-His–1-carboxylate facial triad composed of His143, His227, and Asp145. A zinc ion (gray) replaced iron in the crystal structure [28]. (B) The SyrB2 halogenase has two histidine residues (His116, His235) that coordinate to the iron ion (orange). α-Ketoglutarate is also bound to the iron. Alanine (Ala118) is found in place of the carboxylate-containing residue of hydroxylases, allowing space for the chloride ion (green) [12].
A subset of non-heme iron(II) enzymes uses α-ketoglutarate as a co-substrate. These dioxygenases also contain the 2-His–1-carboxylate iron-binding motif. The substrate binds near the active site, while α-ketoglutarate binds directly to the iron in a bidentate manner. The reaction mechanism involves oxidative decarboxylation of α-ketoglutarate to produce succinate, CO2, and a high energy Fe(IV)-oxo intermediate [2]. In a hydroxylation reaction, one atom of oxygen is incorporated into the product, and the other into succinate.
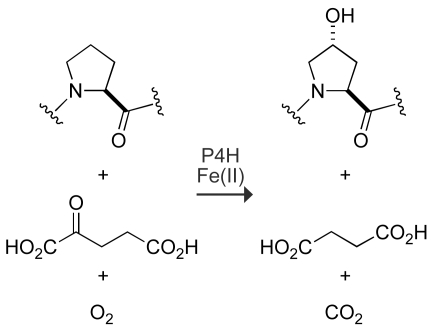
Prolyl 4-hydroxylase (P4H) is an α-ketoglutarate-dependent, iron(II) dioxygenase, and catalyzes the post-translational hydroxylation of proline residues (Pro) during collagen biosynthesis [3]. This hydroxylation reaction forms (2S,4R)-4-hydroxyproline (Hyp; Figure 2), which is necessary for the proper folding of the collagen triple helix [4]. Accordingly, P4H is an essential enzyme for animals [5]–[7].
Figure 2. Reaction catalyzed by P4H.
The active site of P4H is within the α subunit of the α2β2 enzyme tetramer. The β subunits (protein disulfide isomerase) keep the α subunits soluble and retained within the endoplasmic reticulum [8], [9]. The 2-His–1-carboxylate iron-binding residues of P4Hα, identified by site-directed mutagenesis, comprise His412, Asp414, and His483 [10], [11].
A recently discovered subclass of α-ketoglutarate-dependent, iron(II) dioxygenases catalyzes halogenation reactions in natural product biosynthesis. The halogenase SyrB2, found in Pseudomonas syringae, catalyzes the conversion of threonine to 4-chlorothreonine during the biosynthesis of syringomycin E. The three-dimensional structure of SyrB2 revealed a chloride ion present in place of a carboxylate ligand in the facial triad [12]. The Asp or Glu present in other mononuclear iron enzymes is replaced by alanine (Ala) in SyrB2, creating space in the active site for the binding of a chloride ligand to the iron (Figure 1B). The halogenase CytC3 likewise contains a 2-His motif rather than the 2-His–1-carboxylate facial triad [13].
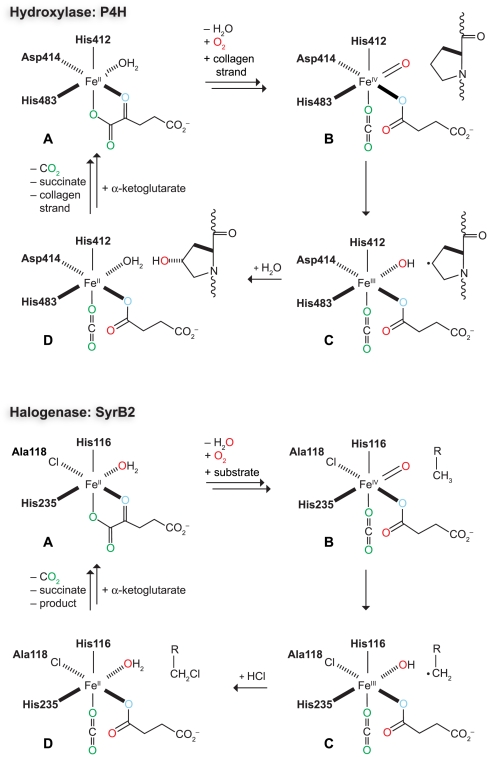
Despite the difference in iron-binding residues, halogenation is thought to follow a mechanism similar to that of P4H and other α-ketoglutarate-dependent iron(II) dioxygenases (Figure 3). Decarboxylation of α-ketoglutarate generates a reactive Fe(IV)-oxo intermediate, which abstracts a hydrogen atom from a C–H bond of the substrate [14], [15]. This reaction yields a substrate radical and a Fe(III)-OH complex. Hydroxylated products result from the recombination of the substrate radical with the iron-coordinated hydroxyl group. In contrast, the radical substrate intermediate of halogenases combines with the chlorine atom coordinated to the iron, instead of the hydroxyl group, to produce a chlorinated product. This discovery prompts the question of how the enzyme controls product formation.
Figure 3. Proposed reaction mechanisms for the hydroxylase P4H (top) and halogenase SyrB2 (bottom).
In both mechanisms, the intermediates are labeled A–D. A Iron(II) in the active site is bound by a 2-His–1-Asp motif in P4H, but a 2-His–1-chloride in SyrB2. The configuration of these residues is not known for human P4H, but is drawn in analogy to SyrB2 (Figure 2B). B The reactive iron(IV)-oxo species is formed upon decarboxylation of α-ketoglutarate. C The iron(IV)-oxo species abstracts a hydrogen atom from the substrate producing a radical intermediate. D In the hydroxylase reaction, the substrate radical recombines with the hydroxyl group. In the halogenase reaction, the substrate radical reacts with the chloride ligand.
Herein, we test the stringency of the iron-binding ligands in P4H. We determine whether altering the P4H active site so as to mimic that of a halogenase can convert its hydroxylase activity into halogenase activity. In addition, we characterize the spatial relationships within the active site of P4H by changing the relative positions of the residues that constitute the 2-His–1-carboxylate facial triad. The results provide insight on enzymes that catalyze difficult but important chemical transformations.
Results
Difference between Hydroxylases and Halogenases
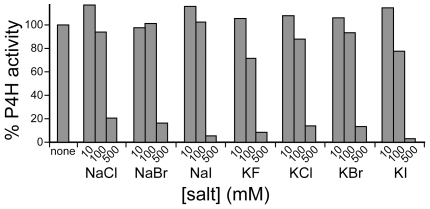
P4H, an α-ketoglutarate-dependent iron(II) dioxygenase, contains a 2-His–1-carboxylate motif. In related enzymes that catalyze halogenation reactions, instead of hydroxylations, the carboxylate-containing residue is replaced with Ala, providing space for a halide ion to bind to the iron. If the only difference between hydroxylation and halogenation is an aspartate versus an alanine, then exchanging these residues should interconvert the activities. The halogenation reaction requires the presence of halide ions. Accordingly, we first investigated the activity of P4H in the presence of salts. Up to 100 mM KF, NaCl, NaBr, NaI, KF, KCl, KBr, or KI had little effect on P4H activity (Figure 4). Higher salt concentrations (500 mM) decreased P4H activity by ∼80%.
Figure 4. Tolerance of P4H activity to salts.
P4H activity is shown in the absence and presence of increasing concentrations of sodium chloride (NaCl), sodium bromide (NaBr), sodium iodide (NaI), potassium fluoride (KF), potassium chloride (KCl), potassium bromide (KBr), and potassium iodide (KI). Reaction mixtures are described in the Materials and Methods section.
The D414A variant of P4H, which has an active site analogous to that of SyrB2, was produced, and, as expected [16], lost its hydroxylase activity. Still, no halogenated product from reaction mixtures containing D414A P4H was detected by high-performance liquid chromatography (HPLC) or mass spectrometry, even in the presence of 100 mM sodium fluoride, chloride, or bromide (Table 1). Subsequently, a D414G variant was made to provide even more space for a halide ion within the active site. This variant likewise had no detectable halogenase or hydroxylase activity.
Table 1. Biochemical attributes of P4H variants.
| P4H variant | Poly(proline) affinity | Hydroxylase activity | Halogenase activity |
| Wild-type | + | + | − |
| D414A | + | − | − |
| D414G | + | − | − |
| D414H | + | − | nd |
| D414H/H412D | + | − | nd |
| D414H/H438D | + | − | nd |
nd, Not determined.
Small Molecules Cannot Replace the Carboxylate in the P4H Active Site
Functional groups missing from amino-acid side chains can be restored by small molecules that rescue protein function [17]. In the D414A and D414G variants of P4H, the carboxylate group is absent. The sodium salt of azide, formate, or acetate (100 mM) was added to D414A P4H reaction mixtures, but did not lead to the formation of hydroxylated product. Nor was the activity of D414G rescued by nitrite or nitrate.
A 3-His Active Site Is Not Functional in P4H
A 3-His active site is found in cysteine dioxygenase, a non-heme, iron(II) dioxygenase related to P4H that does not utilize α-ketoglutarate. To mimic that enzyme, Asp414 was replaced with a histidine residue. The D414H variant was, however, unable to form a hydroxylated product detectable by either the HPLC-based enzyme assay or mass spectrometry.
Spatial Orientation of the 2-His–1-Asp Triad Is Critical for Activity
The 2-His–1-Asp triad occupies adjacent coordination sites on the iron. The relative position of these residues varies with respect to the sites occupied by α-ketoglutarate and oxygen [18]. The locations of the two histidine and aspartate residues were shuffled in the H412D/D414H and D414H/H483D variants. No hydroxylated product was detected in reactions with either of these P4H variants by either HPLC or mass spectrometry. Apparently, the relative positions of the components of the 2-His–1-carboxylate triad are critical for the hydroxylation activity of P4H.
Discussion
The 2-His–1-carboxylate motif is common to many α-ketoglutarate-dependent iron(II) dioxygenases. The related halogenases differ in that the iron is coordinated by only two His residues. In the halogenases, Ala is located in place of the Asp/Glu, and a chloride ion binds to the iron in place of the carboxylate ligand. In an attempt to coerce P4H to perform halogenation reactions, we created the D414A and D414G variants of P4H. Although these variants lose their hydroxylase activity, they do not gain detectable halogenase activity. Similar experiments have been conducted in other dioxygenases to explore the conversion of hydroxylases to halogenases, and vice versa. Replacing alanine with aspartate or glutamate in the halogenase SyrB2 abolished halogenase activity with no observable hydroxylase activity [12]. Substitution of aspartate for alanine in hydroxylase taurine dioxygenase (TauD) abolished hydroxylase activity with no gain in halogenase activity [19]. The D101A variant of TauD did bind iron, though not as efficiently as did the wild-type enzyme, but it was not known if sufficient chloride ion was binding in the active site. The affinity of D414A P4H and D414G P4H for iron was not determined, as even the wild-type P4H does not bind iron with high affinity (K M = 5 µM [20]). These P4H variants were able to be produced like the wild-type enzyme and had a similar affinity for poly(proline) (Table 1), as assessed by their retention during poly(proline)-affinity chromatography. Accordingly, we conclude that global protein conformation was unaffected by the amino-acid substitutions.
All data thus far shows that the difference between Fe(II) and α-ketoglutarate-dependent hydroxylases and halogenases extends beyond the presence or absence of an enzymic carboxyl group. Analysis of structural data of the halogenases CytC3 and SyrB2 suggests that residues that do not make a direct contact with the active-site iron are essential for binding α-ketoglutarate and a chloride ion [12], [13]. Residues that surround the active site form a network of hydrogen bonds, which appear to contribute to chloride binding in CytC3 and SyrB2, and could fail to do so in the P4H variants.
The hydroxylase activity could not be rescued by a number of small molecules. Chemical rescue of the residues within the 2-His–1-carboxylate motif of α-ketoglutarate-dependent iron(II) dioxygenases has been demonstrated with other enzymes. The activity of the H174A variant of phytanoyl-CoA 2-hydroxylase was rescued with the addition of imidazole [21]. Interestingly, the same substitution at the other iron-binding His residue was inert to rescue. Deleterious substitutions of the carboxylate ligand have also been rescued by small molecules, as demonstrated by the rescue of TauD D101A by formate [19]. These data provide clues about the accessibility of the enzymic active sites, as well as the tolerance to deviation in ligand position.
Although the 2-His–1-carboxylate motif is common to many iron(II) α-ketoglutarate-dependent dioxygenases, there are variations of the metal coordination motif [22]. Whereas the 2-His–1-carboxylate dioxygenases are quite selective for iron, enzymes containing three histidines and one glutamate (3-His–1-Glu) coordinate additional metals. Moreover, some dioxygenases do not utilize a carboxylate ligand at all. The halogenases, discussed previously, contribute just two histidine residues for iron binding. Enzymes with four histidine (4-His) and three histidine (3-His) ligands have also been identified. A comparison of the 3-His and 2-His–1-carboxylate metal sites revealed an overall structural similarity with slight differences in metal-ligand distances [23]. Conversion of a 2-His–1-carboxylate to a 3-His metal center was accomplished in tyrosine hydroxylase [24]. In P4H, the 3-His motif (that is, D414H P4H) did not have detectable enzymatic activity. Thus far, glutamate is the only amino acid that can replace Asp414 in P4H and maintain activity [11].
Instead of removing the carboxylate ligand, a second Asp/Glu could be added to the facial triad or the carboxylate could be relocated. His675 in aspartyl (asparaginyl) β-hydroxylase was replaced with Asp or Glu, and the enzyme retained 20 or 12% of its activity, respectively [25]. Gln or Glu could be substituted for His255 in TauD with 81 or 33% retention of its activity, respectively [19]. In P4H, however, no activity was found in the H412E or H483E variant [11]. Rearrangement of the His and Asp ligands in the P4H active site (as in the H412D/D414H and D414H/H483D variants) did not result in enzymatic activity, demonstrating that the native orientation of the 2-His–1-carboxylate facial triad is critical for P4H catalytic ability.
In conclusion, the 2-His–1-Asp facial triad in the active site of P4H is resistant to variation, including alternative iron-binding residues and the spatial positioning of the residues. Mutational analysis of the iron-binding residues within non-heme, iron(II) dioxygenases shows some flexibility at these locations, though no pattern has emerged as to which amino acids can endow function at a particular position. Although focus has been placed on the common 2-His–1-carboxylate iron-binding motif, differences among the non-heme iron(II) dioxygenases point toward the importance of residues that do not bind the iron directly. This secondary coordination sphere presumably alters the steric and electrostatic environments of the facial triad and influences the reaction mechanism. Three-dimensional structural information for human P4H, which has thus far eluded crystallization, will identify these additional residues, and could provide information that enables the design of P4H variants with transformed activity.
Materials and Methods
Materials
Escherichia coli strain Origami B (DE3) were obtained from Novagen (Madison, WI). Enzymes used for DNA manipulation were from Promega (Madison, WI) and DNA oligonucleotides for mutagenesis and sequencing were from Integrated DNA Technologies (Coralville, IA). DNA sequences were elucidated by capillary arrays on an Applied Bioscience automated sequencing instrument at the University of Wisconsin–Madison Biotechnology Center. Poly(proline) was from Sigma Chemical (St. Louis, MO). Luria–Bertani (LB) medium contained tryptone (10 g), yeast extract (5 g), and NaCl (10 g). Terrific broth (TB) medium contained tryptone (12 g), yeast extract (24 g), K2HPO4 (72 mM), KH2PO4 (17 mM), and glycerol (4 mL). All media were prepared in deionized, distilled water, and autoclaved.
Instrumentation
UV absorbance measurements were made with a Cary 50 spectrophotometer from Varian (Palo Alto, CA). Fast protein liquid chromatography (FPLC) was performed with an AKTA system from Amersham–Pharmacia (Piscataway, NJ) and the results were analyzed with the UNICORN Control System. High-performance liquid chromatography (HPLC) was carried out with a system from Waters (Milford, MA) that was controlled with the manufacturer's Millennium32 (Version 3.20) software. Mass spectrometry was performed with a Perkin–Elmer (Wellesley, MA) Voyager MALDI–TOF mass spectrometer at the University of Wisconsin–Madison Biophysics Instrumentation Facility.
Production and Purification of P4H Active–Site Variants
The pBK1.PDI1.P4H7 plasmid that directs the expression of the α and β subunits of human P4H served as a template for mutagenesis [26]. The QuikChange site-directed mutagenesis kit (Stratagene) was used to make point mutations in the gene encoding the α subunit. The P4H variants were produced and purified by using procedures reported previously [26]. Briefly, P4H cDNA expression in E. coli Origami B (DE3) cells was induced at 18°C for 18 h. Cells were harvested, lysed by sonication, and fractionated by precipitation with ammonium sulfate. P4H variants were purified by chromatography on a poly(proline)-affinity column, eluting with free poly(proline). Fractions were purified further by anion-exchange chromatography, followed by gel-filtration chromatography.
HPLC-Based Enzyme Activity Assay
The ability of P4H to catalyze the hydroxylation or halogenation of a proline residue was monitored with an HPLC-based assay described previously [16], [26], [27]. Assays were conducted for 20 min at 30°C in 100 µL of 50 mM Tris–HCl buffer, pH 7.8, containing dithiothreitol (100 µM), catalase (100 µg/mL), ascorbate (2 mM), FeSO4 (50–300 µM), α-ketoglutarate (0.5–25 mM), bovine serum albumin (1.0 mg/mL), and P4H (0.09–1.5 µM). The synthetic tetrapeptide substrate (dansyl-Gly–Phe–Pro–GlyOEt) was added to initiate the reaction. A reversed-phase analytical C18 column was used to separate the substrate and product peptides. Enzymic reaction mixtures were also analyzed by mass spectrometry. To assess the effect of salt on catalysis, KF, NaCl, NaBr, NaI, KF, KCl, KBr, or KI was added to reaction mixtures to 10, 100 or 500 mM. To attempt to rescue the enzymatic activity of D414A P4H and D414G P4H, the sodium salt of azide, nitrite, nitrate, formate, or acetate was added to reaction mixtures to 100 mM.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Grant R01 AR044276 (NIH). K.L.G. was supported by Chemistry-Biology Interface Training Grant T32 BM008505 (NIH). K.H.P. was supported by an A*STAR National Science Scholarship (Singapore) and a Hilldale Undergraduate/Faculty Research Fellowship (University of Wisconsin-Madison). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Koehntop KD, Emerson JP, Que L,, Jr. The 2-His–1-carboxylate facial triad: A versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 2.Costas M, Mehn MP, Jensen MP, Que L,, Jr. Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 3.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 4.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter AD, Page AP. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol Cell Biol. 2000;20:4084–4093. doi: 10.1128/mcb.20.11.4084-4093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, et al. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, et al. Loss of assembly of the main basement membrane collagen, Type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- 8.Vuori K, Pihlajaniemi T, Myllyla R, Kivirikko KI. Site-directed mutagenesis of human protein disulphide isomerase: Effect on the assembly, activity and endoplasmic reticulum retention of human prolyl 4-hydroxylase in Spodoptera frugiperda insect cells. EMBO J. 1992;11:4213–4217. doi: 10.1002/j.1460-2075.1992.tb05515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vuori K, Pihlajaniemi T, Marttila M, Kivirikko KI. Characterization of the human prolyl 4-hydroxylase tetramer and its multifunctional protein disulfide-isomerase subunit synthesized in a baculovirus expression system. Proc Natl Acad Sci USA. 1992;89:7467–7470. doi: 10.1073/pnas.89.16.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamberg A, Helaakoski T, Myllyharju J, Peltonen S, Notbohm H, et al. Characterization of human type III collagen expressed in a baculovirus system. J Biol Chem. 1996;271:11988–11995. doi: 10.1074/jbc.271.20.11988. [DOI] [PubMed] [Google Scholar]
- 11.Myllyharju J, Kivirikko KI. Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 13.Wong C, Fujimori DG, Walsh CT, Drennan CL. Structural analysis of an open active site conformation of nonheme iron halogenase CytC3. J Am Chem Soc. 2009;131:4872–4879. doi: 10.1021/ja8097355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffart LM, Barr EW, Guyer RB, Bollinger JM,, Jr., Krebs C. Direct spectroscopic detection of a C–H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galonic DP, Barr EW, Walsh CT, Bollinger J, J.M., Krebs C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat Chem Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- 16.Gorres KL, Raines RT. Direct and continuous assay for prolyl 4-hydroxylase. Anal Biochem. 2009;386:181–185. doi: 10.1016/j.ab.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 18.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J Inorg Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Grzyska PK, Muller TA, Campbell MG, Hausinger RP. Metal ligand substitution and evidence for quinone formation in taurine/α-ketoglutarate dioxygenase. J Inorg Biochem. 2007;101:797–808. doi: 10.1016/j.jinorgbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Tuderman L, Myllyla R, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 1. Role of co-substrates. Eur J Biochem. 1977;80:341–348. doi: 10.1111/j.1432-1033.1977.tb11888.x. [DOI] [PubMed] [Google Scholar]
- 21.Searls T, Butler D, Chien W, Mukherji M, Lloyd MD, et al. Studies on the specificity of unprocessed and mature forms of phytanoyl-CoA 2-hydroxylase and mutation of the iron binding ligands. J Lipid Res. 2005;46:1660–1667. doi: 10.1194/jlr.M500034-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Straganz GD, Nidetzky B. Variations of the 2-His–1-carboxylate theme in mononuclear non-heme FeII oxygenases. ChemBioChem. 2006;7:1536–1548. doi: 10.1002/cbic.200600152. [DOI] [PubMed] [Google Scholar]
- 23.Leitgeb S, Nidetzky B. Structural and functional comparison of 2-His–1-carboxylate and 3-His metallocentres in non-haem iron(II)-dependent enzymes. Biochem Soc Trans. 2008;36:1180–1186. doi: 10.1042/BST0361180. [DOI] [PubMed] [Google Scholar]
- 24.Fitzpatrick PF, Ralph EC, Ellis HR, Willmon OJ, Daubner SC. Characterization of metal ligand mutants of tyrosine hydroxylase: Insights into the plasticity of a 2-histidine-1-carboxylate triad. Biochemistry. 2003;42:2081–2088. doi: 10.1021/bi0271493. [DOI] [PubMed] [Google Scholar]
- 25.McGinnis K, Ku GM, VanDusen WJ, Fu J, Garsky V, et al. Site-directed mutagenesis of residues in a conserved region of bovine aspartyl (asparaginyl) β-hydroxylase: Evidence that histidine 675 has a role in binding Fe2+. Biochemistry. 1996;35:3957–3962. doi: 10.1021/bi951520n. [DOI] [PubMed] [Google Scholar]
- 26.Kersteen EA, Higgin JJ, Raines RT. Production of human prolyl 4-hydroxylase in Escherichia coli. Protein Exp Purif. 2004;38:279–291. doi: 10.1016/j.pep.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Gorres KL, Edupuganti R, Krow GR, Raines RT. Conformational preferences of substrates for human prolyl 4-hydroxylase. Biochemistry. 2008;47:9447–9455. doi: 10.1021/bi8009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koski MK, Hieta R, Hirsila M, Ronka A, Myllyharju J, et al. The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J Biol Chem. 2009;284:25290–25301. doi: 10.1074/jbc.M109.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]