Abstract
Protein tyrosine O-sulfation, a widespread post-translational modification, is mediated by two Golgi enzymes, tyrosylprotein sulfotransferase-1 and -2. These enzymes catalyze the transfer of sulfate from the universal sulfate donor PAPS to the hydroxyl group of tyrosine residues to form tyrosine O-sulfate ester and PAP. More than 60 proteins have been identified to be tyrosine sulfated including several G protein-coupled receptors, such as CC-chemokine receptor 8 (CCR8) that is implicated in allergic inflammation, asthma, and atherogenesis. However, the kinetic properties of purified TPST-1 and -2 have not been previously reported. Moreover, currently there is no available quantitative TPST assay that can directly monitor individual sulfation of a series of tyrosine residues which is present in most known substrates. We chose an MS-approach to address this limitation. In this study, an LC/ESI-MS-based TPST assay was developed to determine the kinetic parameters of individual TPSTs and a mixture of both isozymes using CCR8 peptides as substrates that have three tyrosine residues in series. Our method can differentiate between mono- and disulfated products and our results show that the Km,app for the monosulfated substrate was five-fold less than the nonsulfated substrate. The development of this method is the initial step in the investigation of kinetic parameters of the sequential tyrosine sulfation of chemokine receptors by TPSTs and in determining its catalytic mechanism.
Keywords: Tyrosylprotein sulfotransferase, tyrosine O-sulfation, LC/ESI-MS, kinetic measurements, chemokine receptor 8 (CCR8), chemokine receptor, TPST assay
Introduction
Protein tyrosine O-sulfation is a widespread post-translational modification that was suggested to affect an average of 1% of all tyrosine residues in all proteins, secreted and membrane-bound, in higher order eukaryotes [1–6]. This common modification is implicated in a wide variety of cellular processes such as adhesion, regulation of intracellular communication, and activation of signal transduction pathways and cellular responses, and these processes are involved in a broad list of pathophysiological functions [6–8]. Among the more than 60 tyrosine sulfated proteins identified to date, a great deal of interest is focused on the role of tyrosine sulfation in the binding of the seven transmembrane G protein-coupled receptors (GPCRs). Equally important has been the investigation of sulfated tyrosine as regards to chemokine receptor binding to their cognate ligands that were found to be relevant in several biological processes such as inflammatory responses, dendritic cell maturation, B and T cells development, Th1 and Th2 responses, infections, angiogenesis, and tumor biology [9–14]. Furthermore, chemokine receptors were found to act as co-receptors for HIV infection with tyrosine sulfation being crucial in HIV entry into cells [15–19].
Tyrosylprotein sulfotransferases (TPSTs) are ~50 kDa glycoproteins found in the trans-Golgi network that catalyze the transfer of sulfate from the universal sulfate donor 3'-phosphoadenosine-5'-phosphosulfate (PAPS) to the hydroxyl group of tyrosine residues within highly acidic motifs in proteins or polypeptides [20–25]. Two human TPST isozymes, TPST-1 and TPST-2 (EC 2.8.2.20) have been identified as having ~65% sequence homology and both are composed of 3 functional domains. These consist of a short N-terminal cytoplasmic domain, a single 17-residue transmembrane domain, and a luminal catalytic domain with a poorly conserved ~40 amino acid portion close to the membrane, the latter being unnecessary for enzyme catalysis [23–25]. Both TPST isozymes are broadly expressed in human and mouse tissues.
Although these two isozymes have similar domain structures, previous studies have indicated differences in their biological roles and substrate specificities as shown by several in vivo and in vitro experiments on the individual isozymes. In vivo experiments on the targeted disruption of the respective TPST genes showed distinct phenotypic differences between TPST-1 deficient and TPST-2 deficient mice [26–28]. In vitro substrate specificity studies using synthetic peptides indicated several differences in substrate preferences of the individual isozymes [24; 29; 30]. Seibert et.al. showed that TPST-1 preferentially sulfated Y14 of CC-chemokine receptor 5 (CCR5, an HIV co-receptor) 2–18 peptide, over Y15 as the initial sulfation site, while TPST-2 preferred Y15 over Y14 [29]. Our group presented results on the sulfation of CCR8 12–20 peptide (VTDYYYPDI) that showed the same preferences of TPST-1 and TPST-2 in the sulfation order of Y15 and Y17 (Y16 is not sulfated), but differences in the distribution of the sulfated products given the same experimental conditions [30].
Furthermore, experimental evidence has shown that tyrosylprotein sulfation plays an important biological role, which makes the characterization of TPST enzymes more particularly necessary to further the understanding of this process. Although progress has been made in recent years in understanding the enzymology and biology of tyrosine sulfation, the pace of research has been hampered by two factors, the lack of homogenous protein samples, and a fast and accurate assay for quantitative kinetics analysis. Most previous reports on kinetic properties of TPST have employed crude enzyme, such as microsomal or enriched Golgi membrane extracts [31–33]. One study of highly enriched TPST from adrenal medulla has been reported [34]. However, the purity of the enzyme was uncertain as was whether the material contained one or both TPST isozymes. In most of these studies, radioactive labeling assays using [35S]PAPS was used to monitor the formation of sulfated tyrosine products. Either TLC or paper chromatography was employed in analyzing the end-point [35S] signal. Accuracy of these methods can vary greatly thus compromising the accuracy of the kinetic parameters reported. More importantly, said methodology only allows the monitoring and quantification of total sulfation of tyrosine residues in products and could not differentiate between mono-, di-, trisulfated products which are present in most of the natural substrates such as CCR5, CCR8, PSGL-1, etc.
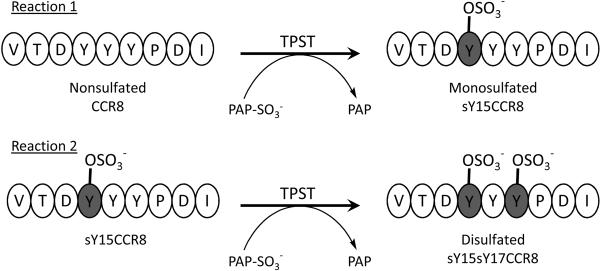
Here we report a new LC-MS-based enzyme assay adapted and modified from our previous direct infusion MS kinetic assay [35–39]. We have analyzed the formation of the mono-and disulfated products using reverse-phase (RP) HPLC coupled directly to a linear ion trap mass spectrometer (MS) equipped with an electrospray ionization source (ESI-MS). Using this method, the kinetic parameters were obtained for purified recombinant human TPST-1, TPST-2, and a 1:1 mixture of each isozyme with non- and monosulfated Y15 CCR8 12–20 synthetic peptides. This current methodology allows for the analysis and quantification of the monosulfated Y15 CCR8 and the disulfated Y15Y17 CCR8 products (Figure 1) (For clarity, “s” is used prior to Y to indicate sulfation in that particular tyrosine residue).
Figure 1.
Shown above are the tyrosine sulfation reactions of CCR8 12-20 peptides with the two TPST isozymes and a 1:1 mixture of TPST-1 and TPST-2. Reaction 1 shows the monosulfation of nonsulfated CCR8 to product sY15CCR8 while reaction 2 shows the formation of the disulfated product sY15sY17CCR8 from monosulfated sY15CCR8 as substrate.
By using this MS-based assay, we found that the affinity of the nonsulfated CCR8 to TPST-1 and TPST-2 are quite similar. We also determined that monosulfated Y15 CCR8 has approximately five times higher affinity than nonsulfated CCR8 for the enzymes. We chose to investigate the pure isozymes as well as a mixture of both in order to address the possibility that the two isozymes work in synergy within the Golgi. Our results indicate a detectable difference in apparent Km (Km,app) when the isozymes are combined, as compared to TPST-1 alone. This trend holds for the other kinetic constants as well. These results represent the first detailed analysis and comparison of the kinetic parameters of pure TPST-1 and -2, and its mixture. It also addresses the analysis of individual sulfation of multiple tyrosine residues of a peptide by pure isozymes and the mixture. This LC-MS-based TPST assay can be used to study other chemokine receptors as substrates, to test potential inhibitors of TPST, and to determine the mechanisms of the two TPST isozymes.
Experimentals
Materials
Recombinant human TPST-1 and TPST-2 were expressed and purified as previously reported [23; 24]. All CCR8 nonapeptides (amino acids 12–20), VTDYYYPDI (nonCCR8), VTDsYYYPDI (sY15CCR8), VTDsYYsYPDI (sY15sY17CCR8), and the sY15CCR8 I20G mutant (internal standard (IS), VTDsYYYPDG), were synthesized and HPLC-purified to ≥95% purity by Quality Controlled Biochemicals (QCB, Hopkinton, MA). PAPS (≥94% purity) was obtained from Prof. Sanford Singer (University of Dayton, OH) without additional purification [40]. All reagents used were of analytical grade and were purchased from Sigma-Aldrich (St. Louis, MO) while HPLC-grade solvents used were from Fisher Scientific (Fairborn, NJ).
Mass spectrometry
All mass spectra were acquired on an LTQ linear ion trap mass spectrometer with a Surveyor HPLC system coupled to an electrospray ionization source (Thermo Electron, San Jose, CA). Analytical samples were separated on a Zorbax Eclipse XDB C8 column (3μm, 80Å) (Agilent Technologies, Palo Alto, CA) using a gradient from 2 to 100% eluant B over 30 minutes at a flow rate of 75μL/min (eluant A, 20mM ammonium acetate, pH 7, in water; eluant B, 20mM ammonium acetate, pH 7, in 80% acetonitrile). The first 5 minutes of the analytical run was directed to waste for desalting purposes and then redirected to the source for MS analysis in the negative ionization mode. The ion transfer capillary was heated to 200°C and the spray voltage was held at a potential of 3.6 kV. Selected ion monitoring (SIM) mode was used for quantification, focusing on the total ion count (TIC) peak areas representing the IS and the product. Twenty scans for each of the IS and product chromatographic peaks were averaged and were used to determine their peak intensity ratio (IP/IIS).
Enzyme assay
The kinetic parameters of TPST-1, TPST-2, and a 1:1 TPST-1/TPST-2 mixture were measured by monitoring the sulfation of the tyrosine residues in the CCR8 peptides. All three were reacted with nonCCR8 (Reaction 1) and sY15CCR8 (Reaction 2) (Figure 1) to determine their respective apparent Km (Km,app) and Vmax (Vmax,app) values. All reactions were carried out in 20 mM MOPS, pH 7.5, 100 mM NaCl, and 10% glycerol at 30°C. Reaction conditions optimization experiments were performed using enzyme concentrations from 0.4 to 4 μM with quench times from 3–30 minutes to determine linearity (Figure 2, Supplemental Material 1 and 2). The optimal concentrations of TPST-2, TPST-1, and 1:1 TPST-1/TPST-2 mixture in the reactions were determined to be 1.5, 3.0, and 1.5 μM, respectively, when reacted to varying concentrations of the CCR8 peptides while having constant and saturating PAPS concentration of 1 mM. When PAPS was varied and the CCR8 peptide was held constant at 300 uM, TPST-2, TPST-1, and 1:1 mixture of TPST-1 & -2 concentrations used were 375, 800, and 375 nM, respectively. All reactions were initiated by the addition of enzyme in a final volume of 50 μL. Reaction mixtures were incubated at 30°C at their optimal reaction times: 10 min (TPST-2), 15 min (TPST-1), and 10 min (TPST-1 and -2) and terminated by transferring 10 μL aliquots of the reaction mixtures into 40 μL methanol containing the IS. Under these conditions, all initial velocity reactions utilized less than 10% of the substrates as evidenced by product formation. The IS, although similar structurally to sY15CCR8, did not interfere with the reaction. The quenched reaction mixtures were then diluted with 20 mM ammonium acetate pH 7 buffer in preparation for online LC-MS analysis.
Figure 2.
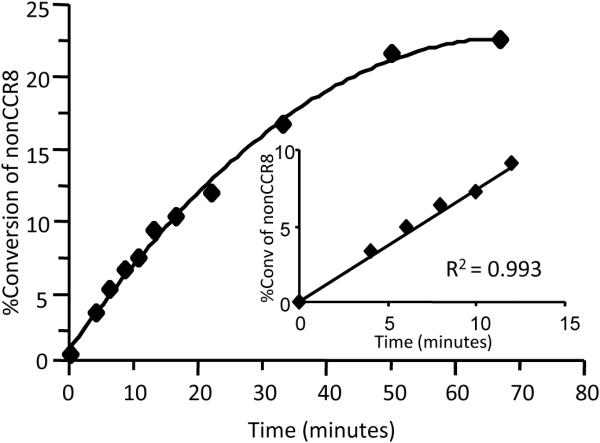
Progress curve for TPST-2 reaction time with insert showing the linear range. Optimal reaction time with linear response of substrate conversion to product can be reached when reaction time is ≤ minutes. TPST-2 concentration was set at 1.5 μM while PAPS at 1 mM and nonsulfated CCR8 at 20 μM.
Quantification and Data Analysis
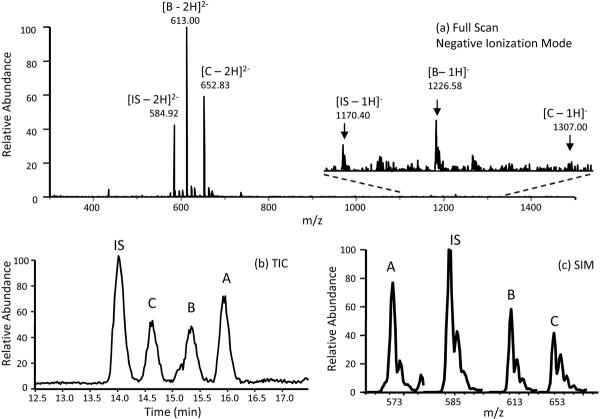
VTDsYYYPDG peptide was used as IS because its molecular mass and ionization efficiency are very similar to the sulfated products. All samples contained 0.5 μM IS prior to LC-MS analysis. Since proteins and peptides were observed to be multiply-charged in ESI [41], in this study, the doubly-charged species of each of the peptides were observed predominantly as shown in Figure 3. For quantification purposes, both the singly-charged and the doubly-charged species of each peptide were monitored in SIM and their intensities, I, were summed to represent their respective total intensities, ITOT.
Figure 3.
RP-HPLC/ESI-MS analysis of sulfated and nonsulfated CCR8 12–20 standard peptides. (a) Full scan analysis of the IS, monosulfated product B, and disulfated product C showing their respective singly-charged (insert) and doubly-charged species. (b) Total ion chromatogram (TIC) showing the separation and order of elution of the 4 CCR8 peptides. (c) Mass spectrum in the selected ion monitoring (SIM) mode showing the most abundant signals for the doubly-charged ions of the 4 CCR8 peptides. A = nonsulfated CCR8, B = sY15CCR8, C = sY15sY17CCR8
| (Equation 1) |
A seven-point calibration curve was used for product quantification where the samples contained varying concentrations of the monitored product while keeping the IS concentration constant. Each of the calibration standards was analyzed in SIM mode to obtain the ratio of the intensity of the product (IP) and the IS (IIS), IP/IIS. Calibration curves were generated from IP/IIS versus [product]/[IS] plots and the slope of the line, m, was calculated such that [P] from each subsequent reaction could be determined from Equaiton 2.
| (Equation 2) |
Once [P] is calculated, it can be used to calculate the velocity of the reaction, υ, at any given substrate concentration and quench time, Tq, of the reaction by using equation 3:
| (Equation 3) |
This equation is valid only when the reaction is halted in the linear region of the reaction; i.e. when the percent conversion of substrate to product is ≤ 10%.
Initial velocity kinetics of reactions 1 and 2 were analyzed by plots of υ versus [substrate] at fixed cosubstrate concentrations. The data for each plot were fitted to the Michaelis-Menten equation to obtain Km,app and Vmax,app values for the varied [substrate]. The same data were analyzed by double reciprocal plots. Grafit v.5 (Erithacus Software Corp., Surrey, UK) was used for all curve fits. Kinetic constants are reported as the mean determined from triplicate experiments (n=3).
Validation of Method
The accuracy and precision of the method for quantification were internally validated by running replicates of standard samples of 0.5 μM monosulfated sY15CCR8 and disulfated sY15sY17CCR8 throughout the analytical batches. Average matrix spike percent recoveries were calculated to be 103% ± 7 for sY15CCR8, and 101% ± 12 for sY15sY17CCR8 (n = 19).
Results
LC/ESI-MS of Sulfation Products
All CCR8 peptides in standards, quality control samples, and reaction mixtures were separated by RP-HPLC and directly analyzed by ESI-Ion trap MS for peak identification and quantification. MS analysis was performed in the negative ion mode instead of the positive ion mode since the sulfoester bond is labile in the positive ion mode [42; 43].
To determine the order of elution of the CCR8 peptides in the reaction mixtures, a standard mix of the IS, sY15CCR8, and sY15sY17CCR8 were analyzed using the full scan mode (Figure 3a). Each peptide was represented by their respective singly-charged and doubly-charged species and their m/z values were noted for selected ion monitoring analysis. The total ion count (TIC) in Figure 3b shows the order of elution of the peptides with their doubly charged m/z signals chosen for SIM as shown in the mass spectrum (Figure 3c).
Kinetic constants for nonsulfated CCR8 (nonCCR8)
The activities of the TPSTs when reacted with nonCCR8 were characterized by monitoring the formation of the monosulfated product, sY15CCR8. Peak areas of the sY15CCR8's singly-charged ion, m/z 1226, and doubly-charged ion, m/z 613, were added and quantified using the internal standard peak area (m/z 1170 and m/z 585) and Equation 2.
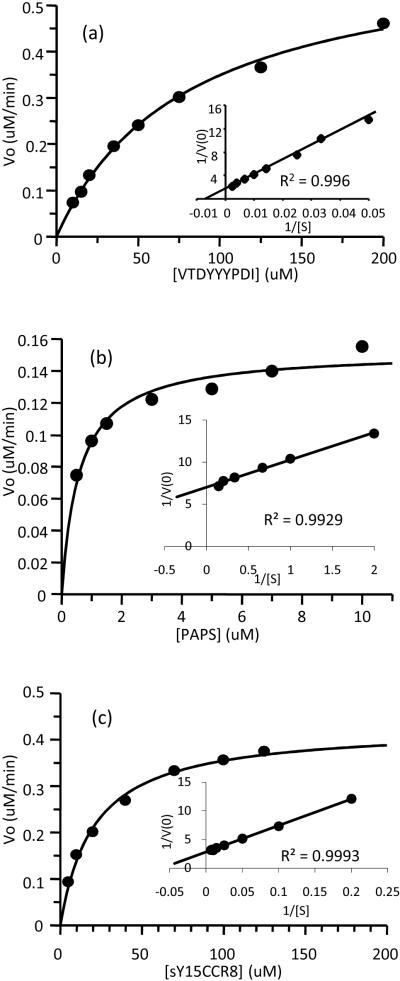
The nonCCR8 concentration range of 10–200 μM was used to determine its Km,app while [PAPS] was held constant at 1 mM. A preliminary concentration range of 0.5–10 μM for PAPS was then used while keeping [nonCCR8] constant at 300 μM. Enzyme inhibition was observed when [nonCCR8] ≥ 250 μM and when [PAPS] ≥ 50 μM. The final concentration ranges were used to determine all kinetic constants for nonCCR8 and PAPS when reacted with TPST-1, TPST-2, and with 1:1 mixture of both isozymes. Table 1 shows all kinetic constants measured for Reaction 1. Typical Michaelis-Menten hyperbolic saturation plots were observed for both nonCCR8 (Figure 4a, Supplemental Material 3a and 4a) and PAPS (Figure 4b, Supplemental Material 3c and 4c). The corresponding double-reciprocal plots (inserts) showed excellent linearity. The Km,app values for nonCCR8 with either of the isozymes or mixture were close to 0.1 mM; however, the Km,app using 1:1 mixture of TPST-1 & -2 was somewhat lower which suggests possible synergistic interaction between the 2 isozymes when binding to these particular substrates. The Km,app values for nonCCR8 and PAPS also indicated that the sulfate donor PAPS has a much higher binding affinity for the TPSTs than the sulfate acceptor nonCCR8 peptide. TPST-2 is also 10 times faster in converting nonCCR8 to sY15 product than TPST-1 as evident in the kcat values. In terms of catalytic efficiencies, the kcat/Km,app values of TPST-1 and TPST-2 clearly showed that TPST-2 is 10-fold more catalytically efficient than TPST-1 under these conditions (Table 1).
Table 1.
Comparison of kinetic constants of TPST-1, TPST-2, and 1:1 TPST-1 and -2 mixture
| Varied Substrate | Km,appaμM | kcatbmin-1 | kcat/Km,appc μM-1 s-1 | |
|---|---|---|---|---|
| Reaction 1: Monosulfation of nonCCR8 | ||||
| TPST-2 | nonCCR8 | 1.2 × 102 ± 10 | 0.50 ± 0.03 | 0.075 ± 0.010 |
| PAPS | 0.59 ± 0.10 | 14 ± 1 | ||
| TPST-1 | nonCCR8 | 99 ± 5 | 0.045 ± 0.007 | 0.0076 ± 0.0004 |
| PAPS | 0.50 ± 0.09 | 1.5 ± 0.1 | ||
| TPST-1 & -2, 1:1 mixture | nonCCR8 | 75 ± 4 | 0.43 ± 0.10 | 0.096 ± 0.005 |
| PAPS | 0.54 ± 0.09 | 13 ± 1 | ||
| Reaction 2: Disulfation of sY15CCR8 | ||||
| TPST-2 | sY15CCR8 | 23 ± 2.6 | 0.30 ± 0.01 | 0.21 ± 0.02 |
| TPST-1 | sY15CCR8 | 21 ± 0.1 | 0.02 ± 0.00 | 0.014 ± 0.001 |
| TPST-1 & -2, 1:1 mixture | sY15CCR8 | 17 ± 0.5 | 0.29 ± 0.01 | 0.29 ± 0.01 |
Km,app is the apparent Michaelis constant at a particular substrate at saturating co-substrate concentration;
kcat is the parameter that measures how fast the enzyme can turnover a substrate to product given [E]total, kcat = Vmax/[E]total;
Kcat/Km,app measures how efficient an enzyme in catalyzing the reaction.
For Reaction 1, experiments when nonCCR8 and PAPS are varied were performed while keeping the cosubstrate (PAPS and nonCCR8, respectively) at constant and saturating concentrations. For Reaction 2, sY15CCR8 concentrations were varied while keeping PAPS constant at 1 mM. The experimental details and data analyses are as described under “Experimentals”. All experiments were performed 3 trials each (n=3).
Figure 4.
Saturation plots and double reciprocal plots (inserts) of the substrates of TPST-2 (TPST-1 and TPST-1 and -2 similar plots not shown): (a) nonsulfated CCR8 as varied substrate while PAPS was held constant at 1mM; (b) PAPS as varied substrate while nonsulfated CCR8 was constant at 300μM; and (c) sY15CCR8 as varied substrate with PAPS constant at 1mM. TPST-2 concentration was constant at 1.5μM for (a) and (c), and was lowered to 375nM for (b). All experiments exhibited double reciprocal plots with excellent linearity.
Kinetic constants for monosulfated sY15CCR8
Km,app and Vmax,app values for sY15CCR8 were determined by monitoring the formation of the disulfated product, sY15sY17 CCR8 (Figure 1, Reaction 2), which is represented by m/z 1306 and 653, the singly-charged and doubly-charged ions, respectively. Their peak areas were added and quantified with respect to the IS.
An sY15CCR8 concentration range of 5–150 μM was used to determine its Km,app and Vmax,app while keeping [PAPS] constant at 1mM (Figure 4c, Supplemental Material 3b and 4b). Beyond 150 μM sY15CCR8 concentration, enzyme inhibition was observed. These concentrations were used for all kinetic constant determinations for TPST-1, TPST-2, and 1:1 mixture of TPST-1 and TPST-2. As observed with the nonCCR8 kinetic experiments, sY15CCR8 kinetic constants in Table 1, Reaction 2 show the Km,app for the two isozymes to be very similar while the 1:1 TPST-1 and -2 mixture is slightly lower. TPST-2 is more reactive than TPST-1 as can be observed again with at least a 10-fold difference in catalytic efficiency, kcat/Km,app.
Discussion
Previous kinetic studies on tyrosylprotein sulfotransferases involved the use of lysates from cultured cells or tissues as the enzyme source that almost certainly contained a mixture of the two isozymes [20; 32; 44–50]. Individual kinetic analysis of the two TPST isozymes became possible after the purification and cloning of TPST-1 and TPST-2 10 years ago. In most of the previous studies, a radioactive labeling assay using [35S] PAPS was used to monitor the formation of [35S] tyrosine sulfated products. This method only provides information on the total sulfation of peptides with multiple tyrosine sulfation sites and cannot distinguish among mono-, di- or trisulfated [35S]- labelled products. The ability of an assay to distinguish tyrosine sulfated products becomes essential in studying the kinetic parameters of the nonrandom, sequential sulfation of tyrosine residues of peptides. Several investigators have reported the importance of tyrosine sulfation in the binding of chemokine receptors to their respective chemokines [12–14; 16; 17]. As such, the cooperative sulfation of tyrosine clusters in peptides by TPSTs requires the monitoring of sulfation of unmutated peptides. Therefore, an efficient kinetic assay capable of characterizing the parameters for each isozyme with unmutated peptides as substrates is important for understanding the catalytic properties of tyrosylprotein sulfotransferases.
The LC/ESI-MS assay described herein was used to obtain the kinetic constants of TPST-1 and TPST-2, individually, and its 1:1 mixture when reacted with peptides modeled after the N-terminal extracellular domain of CCR8. The order of sulfation of nonCCR8 12–20 was first determined using a previously published method [30], at which point it was determined that Y15 is sulfated first followed by the disulfated sY15sY17CCR8 species (Figure 1). There were no trisulfated CCR8 products observed using the optimized reaction conditions in this study. For nonCCR8, both TPST-1 and -2 showed very similar Km,app for both substrate. However, the kcat values are 10 times different. A comparison of kcat/Km,app values of the two isozymes and their corresponding CCR8 peptides in Table 1 shows that TPST-2 is more catalytically efficient than TPST-1 in sulfating CCR8 substrates. This implies that CCR8 12–20 non- and monosulfated peptides are more suitable substrates of TPST-2 than TPST-1. In other studies, it was reported that PSGL-1 peptide is efficiently sulfated by TPST-1 and TPST-2 equally, while heparin cofactor II and C4α peptides are more effectively sulfated by TPST-1 than by TPST-2 [23; 24]. Differential substrate preferences of the isozymes was also evident during in vivo gene knock-out studies where TPST-1-deficient mice showed reduced body mass and increased post-implantation fetal death, whereas TPST-2-deficiency caused male mice infertility and primary hypothyroidism [26–28]. Overall, the kcat/Km,app values from this study agree with the postulation that TPST-1 and TPST-2 have overlapping but not identical substrate specificity in vitro and in vivo.
The kinetic data in Table 1 show that both isozymes have similar Km,app values when reacted with the same substrates. Interestingly, the Km,app for the equal mixture of TPST-1 and -2 for nonCCR8 and for sY15CCR8 are slightly lower than the individual isozymes. This may suggest some synergistic interaction between the 2 isozymes in sulfating the tyrosines, however, further experiments are needed to explore this possibility.
Few studies have focused on determining the order of sulfation of multiple tyrosine sulfation sites on peptides [29; 30]. General considerations indicate that acidic residues within ±5 residues of the tyrosines to be sulfated are necessary. It would appear that clusters of tyrosine residues are sulfated cooperatively; i.e. sulfation of one tyrosine is dependent on the prior sulfation of nearby tyrosines. In the case of nonCCR8, Y15 is predominantly sulfated first: interestingly, it has an aspartic acid residue immediately N-terminal to Y15. After initial formation of sY15CCR8, Y17 of sY15CCR8 is sulfated to form the disulfated sY15sY17CCR8 product [30]. The comparison of Km,app values of nonCCR8 and sY15CCR8 in Table 1 clearly suggests that sY15CCR8 has a higher binding affinity than nonCCR8 for both isozymes. Similarly, sY15CCR8 also showed higher kcat/Km,app than nonCCR8 for both isozymes, as well as for the mixture, which suggests that the two isozymes are more catalytically efficient in the formation of the disulfated sY15sY17CCR8 product. These differences in the kinetic parameters of the two substrates may suggest important interactions between the negative charges on the sulfated Y15 and the binding site on TPST, and on general TPST catalysis.
In summary, this study presents a detailed kinetic analysis of the monosulfation and disulfation of a peptide substrate of TPST-1 and TPST-2. The MS-based assay described is useful for the comparison of the catalytic properties of the two TPST isozymes. In particular, it can be used to observe the differences in kinetic constants of sulfation reactions in peptides with multiple tyrosine sulfation sites. Aside from the facile kinetic analysis for substrate specificity studies, the LC/ESI-MS-based assay can be used to test potential inhibitors of these isozymes, and thus be further extended to determine the catalytic mechanism of TPST-1 and TPST-2. These latter two investigations will be the focus of future studies.
Supplementary Material
Acknowledgements
We thank the NIH (Grant No. GM063581 and Grant No. HL074015) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huttner WB. Sulphation of Tyrosine Residues-a Widespread Modification of Proteins. Nature. 1982;299:273–276. doi: 10.1038/299273a0. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle PA, Huttner WB. Tyrosine Sulfation of Yolk Proteins 1, 2, and 3 in Drosophila Melanogaster. J Biol Chem. 1985;260:6434–6439. [PubMed] [Google Scholar]
- 3.Hille A, Rosa P, Huttner WB. Tyrosine Sulfation: A Post-Translational Modification of Proteins Destined for Secretion? FEBS Lett. 1984;177:129–134. doi: 10.1016/0014-5793(84)80996-5. [DOI] [PubMed] [Google Scholar]
- 4.Hille A, Braulke T, von Figura K, Huttner WB. Occurrence of Tyrosine Sulfate in Proteins--a Balance Sheet. 1. Secretory and Lysosomal Proteins. Eur J Biochem. 1990;188:577–586. doi: 10.1111/j.1432-1033.1990.tb15438.x. [DOI] [PubMed] [Google Scholar]
- 5.Hille A, Huttner WB. Occurrence of Tyrosine Sulfate in Proteins--a Balance Sheet. 2. Membrane Proteins. Eur J Biochem. 1990;188:587–596. doi: 10.1111/j.1432-1033.1990.tb15439.x. [DOI] [PubMed] [Google Scholar]
- 6.Moore KL. The Biology and Enzymology of Protein Tyrosine O-Sulfation. J Biol Chem. 2003;278:24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 7.Huttner WB. Tyrosine Sulfation and the Secretory Pathway. Annu Rev Physiol. 1988;50:363–376. doi: 10.1146/annurev.ph.50.030188.002051. [DOI] [PubMed] [Google Scholar]
- 8.Walsh G, Jefferis R. Post-Translational Modifications in the Context of Therapeutic Proteins. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 9.Kehoe JW, Bertozzi CR. Tyrosine Sulfation: A Modulator of Extracellular Protein-Protein Interactions. Chem Biol. 2000;7:R57–61. doi: 10.1016/s1074-5521(00)00093-4. [DOI] [PubMed] [Google Scholar]
- 10.Rossi D, Zlotnik A. The Biology of Chemokines and Their Receptors. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 11.Monigatti F, Hekking B, Steen H. Protein Sulfation Analysis--a Primer. Biochim Biophys Acta. 2006;1764:1904–1913. doi: 10.1016/j.bbapap.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Preobrazhensky AA, Dragan S, Kawano T, Gavrilin MA, Gulina IV, Chakravarty L, Kolattukudy PE. Monocyte Chemotactic Protein-1 Receptor Ccr2b Is a Glycoprotein That Has Tyrosine Sulfation in a Conserved Extracellular N-Terminal Region. J Immunol. 2000;165:5295–5303. doi: 10.4049/jimmunol.165.9.5295. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez J, Kremer L, Zaballos A, Goya I, Martinez AC, Marquez G. Analysis of Post-Translational Ccr8 Modifications and Their Influence on Receptor Activity. J Biol Chem. 2004;279:14726–14733. doi: 10.1074/jbc.M309689200. [DOI] [PubMed] [Google Scholar]
- 14.Colvin RA, Campanella GS, Manice LA, Luster AD. Cxcr3 Requires Tyrosine Sulfation for Ligand Binding and a Second Extracellular Loop Arginine Residue for Ligand-Induced Chemotaxis. Mol Cell Biol. 2006;26:5838–5849. doi: 10.1128/MCB.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Broder CC, Kennedy PE, Berger EA. Hiv-1 Entry Cofactor: Functional Cdna Cloning of a Seven-Transmembrane, G Protein-Coupled Receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine Sulfation of the Amino Terminus of Ccr5 Facilitates Hiv-1 Entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 17.Farzan M, Babcock GJ, Vasilieva N, Wright PL, Kiprilov E, Mirzabekov T, Choe H. The Role of Post-Translational Modifications of the Cxcr4 Amino Terminus in Stromal-Derived Factor 1 Alpha Association and Hiv-1 Entry. J Biol Chem. 2002;277:29484–29489. doi: 10.1074/jbc.M203361200. [DOI] [PubMed] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a Major Co-Receptor for Primary Isolates of Hiv-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Lusso P. Hiv and the Chemokine System: 10 Years Later. EMBO J. 2006;25:447–456. doi: 10.1038/sj.emboj.7600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee RW, Huttner WB. Tyrosine-O-Sulfated Proteins of Pc12 Pheochromocytoma Cells and Their Sulfation by a Tyrosylprotein Sulfotransferase. J Biol Chem. 1983;258:11326–11334. [PubMed] [Google Scholar]
- 21.Baeuerle PA, Huttner WB. Tyrosine Sulfation Is a Trans-Golgi-Specific Protein Modification. J Cell Biol. 1987;105:2655–2664. doi: 10.1083/jcb.105.6.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bundgaard JR, Vuust J, Rehfeld JF. New Consensus Features for Tyrosine O-Sulfation Determined by Mutational Analysis. J Biol Chem. 1997;272:21700–21705. doi: 10.1074/jbc.272.35.21700. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang Y, Lane WS, Moore KL. Tyrosylprotein Sulfotransferase: Purification and Molecular Cloning of an Enzyme That Catalyzes Tyrosine O-Sulfation, a Common Posttranslational Modification of Eukaryotic Proteins. Proc Natl Acad Sci U S A. 1998;95:2896–2901. doi: 10.1073/pnas.95.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang YB, Moore KL. Molecular Cloning and Expression of Human and Mouse Tyrosylprotein Sulfotransferase-2 and a Tyrosylprotein Sulfotransferase Homologue in Caenorhabditis Elegans. J Biol Chem. 1998;273:24770–24774. doi: 10.1074/jbc.273.38.24770. [DOI] [PubMed] [Google Scholar]
- 25.Beisswanger R, Corbeil D, Vannier C, Thiele C, Dohrmann U, Kellner R, Ashman K, Niehrs C, Huttner WB. Existence of Distinct Tyrosylprotein Sulfotransferase Genes: Molecular Characterization of Tyrosylprotein Sulfotransferase-2. Proc Natl Acad Sci U S A. 1998;95:11134–11139. doi: 10.1073/pnas.95.19.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang YB, Crawley JT, Aston CE, Moore KL. Reduced Body Weight and Increased Postimplantation Fetal Death in Tyrosylprotein Sulfotransferase-1-Deficient Mice. J Biol Chem. 2002;277:23781–23787. doi: 10.1074/jbc.M202420200. [DOI] [PubMed] [Google Scholar]
- 27.Borghei A, Ouyang YB, Westmuckett AD, Marcello MR, Landel CP, Evans JP, Moore KL. Targeted Disruption of Tyrosylprotein Sulfotransferase-2, an Enzyme That Catalyzes Post-Translational Protein Tyrosine O-Sulfation, Causes Male Infertility. J Biol Chem. 2006;281:9423–9431. doi: 10.1074/jbc.M513768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westmuckett AD, Hoffhines AJ, Borghei A, Moore KL. Early Postnatal Pulmonary Failure and Primary Hypothyroidism in Mice with Combined Tpst-1 and Tpst-2 Deficiency. Gen Comp Endocrinol. 2008;156:145–153. doi: 10.1016/j.ygcen.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seibert C, Cadene M, Sanfiz A, Chait BT, Sakmar TP. Tyrosine Sulfation of Ccr5 N-Terminal Peptide by Tyrosylprotein Sulfotransferases 1 and 2 Follows a Discrete Pattern and Temporal Sequence. Proc Natl Acad Sci U S A. 2002;99:11031–11036. doi: 10.1073/pnas.172380899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y, Hoffhines AJ, Moore KL, Leary JA. Determination of the Sites of Tyrosine O-Sulfation in Peptides and Proteins. Nat Methods. 2007;4:583–588. doi: 10.1038/nmeth1056. [DOI] [PubMed] [Google Scholar]
- 31.Mishiro E, Sakakibara Y, Liu MC, Suiko M. Differential Enzymatic Characteristics and Tissue-Specific Expression of Human Tpst-1 and Tpst-2. J Biochem (Tokyo) 2006;140:731–737. doi: 10.1093/jb/mvj206. [DOI] [PubMed] [Google Scholar]
- 32.Lee RW, Huttner WB. (Glu62, Ala30, Tyr8)N Serves as High-Affinity Substrate for Tyrosylprotein Sulfotransferase: A Golgi Enzyme. Proc Natl Acad Sci U S A. 1985;82:6143–6147. doi: 10.1073/pnas.82.18.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niehrs C, Kraft M, Lee RW, Huttner WB. Analysis of the Substrate Specificity of Tyrosylprotein Sulfotransferase Using Synthetic Peptides. J Biol Chem. 1990;265:8525–8532. [PubMed] [Google Scholar]
- 34.Niehrs C, Huttner WB. Purification and Characterization of Tyrosylprotein Sulfotransferase. Embo J. 1990;9:35–42. doi: 10.1002/j.1460-2075.1990.tb08077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge X, Sirich TL, Beyer MK, Desaire H, Leary JA. A Strategy for the Determination of Enzyme Kinetics Using Electrospray Ionization with an Ion Trap Mass Spectrometer. Anal Chem. 2001;73:5078–5082. doi: 10.1021/ac0105890. [DOI] [PubMed] [Google Scholar]
- 36.Pi N, Armstrong JI, Bertozzi CR, Leary JA. Kinetic Analysis of Nodst Sulfotransferase Using an Electrospray Ionization Mass Spectrometry Assay. Biochemistry. 2002;41:13283–13288. doi: 10.1021/bi020457g. [DOI] [PubMed] [Google Scholar]
- 37.Gao H, Leary JA. Multiplex Inhibitor Screening and Kinetic Constant Determinations for Yeast Hexokinase Using Mass Spectrometry Based Assays. J Am Soc Mass Spectrom. 2003;14:173–181. doi: 10.1016/S1044-0305(02)00867-X. [DOI] [PubMed] [Google Scholar]
- 38.Pi N, Hoang MB, Gao H, Mougous JD, Bertozzi CR, Leary JA. Kinetic Measurements and Mechanism Determination of Stf0 Sulfotransferase Using Mass Spectrometry. Anal Biochem. 2005;341:94–104. doi: 10.1016/j.ab.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Gao H, Leary JA. Kinetic Measurements of Phosphoglucomutase by Direct Analysis of Glucose-1-Phosphate and Glucose-6-Phosphate Using Ion/Molecule Reactions and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal Biochem. 2004;329:269–275. doi: 10.1016/j.ab.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Singer SS. Enzymatic Sulfation of Steroids. Vi. A Simple, Rapid Method for Routine Enzymatic Preparation of 3'-Phosphoadenosine-5'-Phosphosulfate. Anal Biochem. 1979;96:34–38. doi: 10.1016/0003-2697(79)90550-5. [DOI] [PubMed] [Google Scholar]
- 41.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 42.Gibson BW, Cohen P. Liquid Secondary Ion Mass Spectrometry of Phosphorylated and Sulfated Peptides and Proteins. Methods Enzymol. 1990;193:480–501. doi: 10.1016/0076-6879(90)93434-m. [DOI] [PubMed] [Google Scholar]
- 43.Rappsilber J, Steen H, Mann M. Labile Sulfogroup Allows Differentiation of Sulfotyrosine and Phosphotyrosine in Peptides. J Mass Spectrom. 2001;36:832–833. doi: 10.1002/jms.195. [DOI] [PubMed] [Google Scholar]
- 44.Vargas F, Frerot O, Tuong MD, Schwartz JC. Characterization of a Tyrosine Sulfotransferase in Rat Brain Using Cholecystokinin Derivatives as Acceptors. Biochemistry. 1985;24:5938–5943. doi: 10.1021/bi00342a037. [DOI] [PubMed] [Google Scholar]
- 45.Rens-Domiano S, Roth JA. Characterization of Tyrosylprotein Sulfotransferase from Rat Liver and Other Tissues. J Biol Chem. 1989;264:899–905. [PubMed] [Google Scholar]
- 46.Lin WH, Roth JA. Characterization of a Tyrosylprotein Sulfotransferase in Human Liver. Biochem Pharmacol. 1990;40:629–635. doi: 10.1016/0006-2952(90)90566-4. [DOI] [PubMed] [Google Scholar]
- 47.Kasinathan C, Sundaram P, Slomiany BL, Slomiany A. Identification of Tyrosylprotein Sulfotransferase in Rat Gastric Mucosa. Enzyme. 1992;46:179–187. doi: 10.1159/000468786. [DOI] [PubMed] [Google Scholar]
- 48.Sundaram P, Slomiany A, Slomiany BL, Kasinathan C. Tyrosylprotein Sulfotransferase in Rat Submandibular Salivary Glands. Int J Biochem. 1992;24:663–667. doi: 10.1016/0020-711x(92)90344-z. [DOI] [PubMed] [Google Scholar]
- 49.Sane DC, Baker MS. Human Platelets Possess Tyrosylprotein Sulfotransferase (Tpst) Activity. Thromb Haemost. 1993;69:272–275. [PubMed] [Google Scholar]
- 50.Suiko M, Fernando PH, Sakakibara Y, Kudo H, Nakamura T, Liu MC. Characterization of Bovine Heart Sulfotransferase Catalyzing the Sulfation of Tyrosine-Containing Peptides. J Nutr Sci Vitaminol (Tokyo) 1997;43:485–490. doi: 10.3177/jnsv.43.485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.