Abstract
San Francisco Bay has been contaminated historically by mercury from mine tailings as well as contemporary industrial sources. Native Spartina foliosa and non-native S. alterniflora-hybrid cordgrasses are dominant florae within the SF Bay estuary environment. Understanding mercury uptake and transformations in these plants will help to characterize the significance of their roles in mercury biogeochemical cycling in the estuarine environment. Methylated mercury can be biomagnified up the food web, resulting in levels in sport fish up to one million times greater than in surrounding waters and resulting in advisories to limit fish intake. Understanding the uptake and methylation of mercury in the plant rhizosphere can yield insight into ways to manage mercury contamination.
The transmission x-ray microscope on beamline 6-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) was used to obtain absorption contrast images and 3D tomography of Spartina foliosa roots that were exposed to 1 ppm Hg (as HgCl2) hydroponically for one week. Absorption contrast images of micron-sized roots from S. foliosa revealed dark particles, and dark channels within the root, due to Hg absorption. 3D tomography showed that the particles are on the root surface, and slices from the tomographic reconstruction revealed that the particles are hollow, consistent with microorganisms with a thin layer of Hg on the surface. Hg L3 XANES of ground-up plant roots and Hg L3 micro-XANES from microprobe analysis of micron-sized roots (60–120 microns in size) revealed three main types of speciation in both Spartina species: Hg-S ligation in a form similar to Hg(II) cysteine, Hg-S bonding as in cinnabar and metacinnabar, and methylmercury-carboxyl bonding in a form similar to methylmercury acetate. These results are interpreted within the context of obtaining a “snapshot” of mercury methylation in progress.
Introduction
Many challenges face the health of the San Francisco Bay ecosystem today; among them are high levels of mercury and invasion by exotic species. Mercury is a highly toxic heavy metal and neurotoxin that may bioaccumulate and biomagnify. In florae, mercury can greatly harm or even kill a plant by interrupting photosynthetic processes (1). Wetlands have commonly been regarded as sinks for mercury and other pollutants. Recently, however, wetlands are emerging as significant sources of mercury export into estuarine food webs (2,3).
Little is known about the cycling of mercury and other heavy metals by plants, or how changes in plant community structure might affect mercury bioavailability. Mercury enters the food chain when it is taken up by organisms at the base of the food web, including microbes and plants. Plants, and microbes in their rhizosphere, mobilize nutrients in the sediment environment, shifting chemical speciation of compounds (4,5). Sulfate-reducing bacteria (SRB) (6,7) and iron reducing bacteria (FeRB) (8,9) have been shown to have significant impact on Hg speciation, particularly methylation, in the rhizosphere.
Plant communities inherently shape sediment composition and structure through their functions. Accordingly, the types of plants in a wetland and their abilities to take up metals affect mercury bioavailability (10). It follows then, that invasive plant species might alter the natural biogeochemical cycling of mercury in the Bay. Notably, through unsuccessful competition, native plants in the SF Bay are quickly being replaced by exotic species. Spartina alterniflora-hybrids, including both S. alterniflora and the S. alterniflora × S. foliosa cross, are quickly out-competing and out-breeding the native S. foliosa, California cordgrass (11). S. alterniflora has shown the ability to sequester metals in its above- and below-ground tissues to varying degrees (12), but the definitive relationship between invasive S. alterniflora, or smooth cordgrass, and changes in metal cycling is still unknown.
Despite the close relationship between S. alterniflora and S. foliosa, little research has been performed on the uptake, storage, and translocation of heavy metals by S. foliosa or the hybrid. The purpose of this experiment is to compare mercury uptake of native S. foliosa and non-native S. alterniflora-hybrid, and to study the speciation of Hg in the rhizosphere. Understanding mercury uptake in these plants and speciation in the chemically active rhizosphere will help to characterize their roles in mercury biogeochemical cycling in the estuarine environment.
Mercury speciation can be determined using a variety of techniques, including X-ray Absorption Spectroscopy (XAS). XAS has been used to determine mercury speciation in minerals (13,14,15), and in plants (16,17). Advantages of XAS over the other techniques include: (1) it is element-specific, thus mercury can be analyzed despite other, more dominant species; (2) no chemical or physical alteration is required before sample analysis; and (3) sample quantities can be small (less than 1 g). X-ray absorption near edge structure (XANES) can be very useful to determine speciation by comparison of spectra with appropriate model compounds. XANES has been used to determine the chemical form of mercury in fish (18), the binding of methylmercury to thiol groups in soil (19), and to study mercury methylation in water hyacinth (17).
Imaging techniques such as scanning transmission x-ray microscopy (STXM) and microprobe methods can also contribute information on the spatial conformation, distribution, and speciation of metals in environmental samples (20,21,22), and other materials with 0.5 to 2 µm resolution. Full-field transmission x-ray microscopy (TXM) provides higher resolution than STXM, 40 nm in this work, and 3D tomography reveals the inner structure and 3D orientation. Microprobe can detect 106–107 atoms/µm2 and TXM can image small concentrated areas within a sample even if overall concentration is low. Bulk XANES requires 1–20 ppm Hg depending on matrix. TXM and microprobe, combined with bulk and micro Hg L3 XANES, were used in this study to yield insight into the chemical transformations of Hg occurring within the rhizosphere of S. foliosa.
Experimental Methods
Hg Uptake
The plants studied in this experiment were collected between January 18–28, 2006 from two sites in San Francisco Bay, California, USA. Specimens were selected based on geographic location and positioning within the marsh tidal plain (23). S. foliosa collection was restricted to the North Bay to avoid areas affected by S. alterniflora-hybrid invasion and ensure distinct species. Conversely, S. alterniflora-hybrids were collected from an area dominated by hybrid invasion. The non-native Spartina alterniflora-hybrids were collected from a Central Bay site (CB) in Oakland’s Martin Luther King Jr. Shoreline, where San Leandro Creek enters Arrowhead Marsh. The native Spartina foliosa were gathered from a San Pablo Bay site (SPB) in Pinole Bayfront Park, adjacent to a sewage treatment plant. Distinguishing physical characteristics were also utilized to identify the cordgrasses (24). Plants were collected during low tide with gloved hands and metal hand shovels, and clones were separated at the rhizome.
Plant specimens were rinsed with distilled water and placed into aerated 1000 mL beakers filled with 500 mL of unfiltered composite bay water (1:1 mix of water from both collection sites) and 0.25× Hoagland’s solution (25). All plants acclimated for 1 week, and water level, temperature highs and lows, and relative humidity levels were monitored throughout the experiment. Three days after they were collected, “dry-weight” plants were pulled, patted dry with KimWipes™, weighed to the nearest 0.0001-gram (g) and placed into a 60 degree Celsius oven for 3 days for triplicate determination of dry weight ratios for the roots and shoots of each species.
On day 7, plants were divided into control and test groups using the RAND() random number generator from Microsoft Excel. Control and test plants were placed in fresh composite/Hoagland’s as described above. All “test” plant beakers were spiked with 1 part per million (ppm) Hg(II), as HgCl2. On day 14, all plants were pulled and rinsed thoroughly with distilled water. Plants were patted dry with KimWipes™ and divided into approximate root and shoot sections. All sections were placed into small Ziploc™ baggies and placed in the freezer.
Total Hg determination
For total Hg determination, all plants were lyophilized for 48 hours, ground with mortar and pestle, and digested using modified EPA protocol 245.6. Digested plants were analyzed by Cold Vapor Atomic Absorption (CVAA), with SnCl2 reduction. Gold amalgamation was used for samples with the lowest concentrations. Concentrations in shoots and roots were compared graphically and with general linear model ANOVAs, using MINITAB. Calibration curves, digestion blanks, replicates, spikes and NIST standards were all within EPA limits.
X-ray Absorption Near Edge Structure (XANES)
Bulk XANES Hg L3 edge analysis was utilized to compare the mercury speciation in plant roots of both Spartina species. Only true root parts were removed from the below ground tissues for analysis, consisting of both bulky root parts extending from the stalk and fine matted root sections, occurring near the base of the stalk. All samples were ground to a paste under liquid nitrogen with acid-washed mortar and pestle. Samples were placed directly into 1mm aluminum spacers with Kapton windows, and stored under liquid nitrogen until analysis.
Hg L3 spectra were collected at beam line 10-2 and beam line 9-3 at SSRL, using an Oxford Instruments cryostat to maintain temperature at 10 K. Hg Lα fluorescence was monitored at 9899 eV with a multi-element Ge detector, and calibration was accomplished by simultaneous collection of HgCl2. Data analysis was performed using SIXPack software (26). Following calibration, addition, background removal and normalization, least squares fitting was performed on unsmoothed first derivatives of 15 powder and aqueous models according to methods described by Rajan et al. (17). In this work, however, the spectra for Hg-cysteine and Hg-dicysteine were averaged before fitting because they were not found to be distinct principal components.
Transmission X-ray Microscopy (TXM)
Fine hair-like roots (60–120 microns in size) from washed S. foliosa plants were mounted in windows anchored by Kapton tape for TXM analysis. Images were obtained with the Xradia NanoXCT full-field x-ray microscope at beam line 6-2 at SSRL (27), at 8 keV in phase and absorption contrast, and at 9 keV in absorption contrast. Tomography was obtained using images taken from −75 to +75 degrees, every ½ degree, and reconstructed using Xradia software, and also via an in-house iterative method.
Microprobe and micro-XANES
Fine portions of S. foliosa roots (the same as used for TXM) were further analyzed by microprobe at SSRL BL 2–3 to spatially map the presence, distribution, and size of mercury particles, and to collect micro-XANES. Spatial maps for Hg were collected and processed as a difference of fluorescence above (12300 eV) and below (12250eV) the Hg edge, and analyzed using SMAK software (Sam’s Microprobe Analysis Kit) (26). Hg L3 micro-XANES spectra were collected in regions of highest Hg fluorescence counts. Micro-XANES were determined at several spots with the highest Hg counts in each region. The x-ray micro-beam was moved within each spot for separate scans to avoid photoreduction of Hg. These scans were averaged to determine speciation at each spot. Data analysis was performed in SIXPack, consistent with bulk XANES analysis. Least-squares fitting to spectra of the powder and aqueous model compounds was performed with the same procedures used for bulk XANES.
Results
Hg Uptake
Site conditions were monitored during all collections, and had similar temperature (CB = 12.0–12.5 degrees Celsius, SPB= 12.5–12.7 degrees Celsius), salinity (CB = 1.4–2.2 ppt, SPB = 1.0–1.8 ppt), and conductivity (CB = 2049–4005µS, SPB= 2080–3377µS). Regional Monitoring Program data from 2006 indicated similar methylmercury (MeHg) concentrations at the sites, both for water (CB total MeHg = 0.68 ng/L, SPB total MeHg = .077 ng/L) and sediment (CB MeHg = 0.283 µg/Kg, SPB MeHg = 0.317 µg/Kg) (28). However, total mercury concentrations were two-fold higher in water at the SPB site (CB dissolved Hg = .0008 µg/L, SPB dissolved Hg = .0015 µg/L; CB total Hg = .0031 µg/L, SPB total Hg = 0.0056 µg/L) and three-fold higher in sediment (CB = 0.098 mg/Kg, SPB = 0.332 mg/Kg) (28).
Hg-treated S. foliosa (“test”) roots accumulated higher total mercury values (mean 26.6±3.7 ppm) than Hg-treated S. alterniflora-hybrids (mean 13.0±5.7 ppm). Control (untreated) root concentrations were much closer, with S. foliosa averaging 0.09±0.01 ppm and the non-native averaging 0.10±0.02ppm. Root concentrations of total mercury were significantly different by species (df=1, F= 11.87, P<0.01) and treatment (df= 1, F=100.28, P≪0.001). Moreover, the interaction between species and treatment was also significant, meaning the species responded differently to control and test treatments (df=1, F=11.89, P<0.01).
Notably, shoot concentrations of total mercury were not significantly different by species (df=1, F=0.52, P=0.492), treatment (df=1, F=1.62, P=0.242), or interaction. S. foliosa test shoots averaged 0.21±0.15 ppm, while S. alterniflora-hybrid test shoots averaged 0.65±1.06ppm. Control shoot results were almost identical; S. foliosa control shoots averaged 0.037±0.025ppm, whereas S. alterniflora-hybrid averaged 0.037±0.027ppm.
XANES
Least-squares fitting of Hg L3 XANES showed that mercury speciation was similar in S. foliosa and S. alterniflora (Table 1). Three distinct types of bonding environments were present, including (1) double bonded, inorganic Hg-S coordination, similar to that found in cinnabar and metacinnabar (range 0% – 52.5%); (2) single bonded, organic Hg-S coordination, similar to Hg cysteine and Hg dicysteine (range 40.2%–72.1%); and (3) Hg-O carboxyl coordination, in a form similar to methyl-Hg acetate and Hg diacetate (range 1.9%–17.0%). Generally, Hg-cysteine type bonds are associated with covalent bonds to proteins and phytochelatins. Notably, all methylated species evident in the bulk samples were associated with Hg-O carboxyl coordination, in a form similar to Hg binding to organic acids.
Table 1.
Hg L3 XANES least squares fitting results of S. foliosa roots to model compounds (percent).
| Sample | cinnabar (HgS) |
meta- cinnabar (HgS) |
mercury cysteine and dicysteine |
methyl mercury acetate |
mercury diacetate |
|---|---|---|---|---|---|
| S. alterniflora 1 | —a | 18.8±1.7 | 58.9±2.63 | 11.0±1.8 | — |
| S. alterniflora 2 | 2.6±3.2 | 18.1±1.5 | 67.4±3.5 | 10.9±1.5 | — |
| S. foliosa 1 | — | 14.3±2.0 | 72.1±3.5 | 11.1±2.0 | 1.9±1.6 |
| S. foliosa 2 | 13.2±5.4 | 15.9±2.5 | 61.0±6.2 | 9.2±2.6 | — |
| S. foliosa 3 | — | 16.8±1.3 | 63.4±1.9 | 17.8±1.3 | — |
| S. foliosa 4 | 52.5±4.0 | 2.6±1.6 | 40.2±2.4 | — | 2.4±1.6 |
Less than 1% of total composition.
TXM
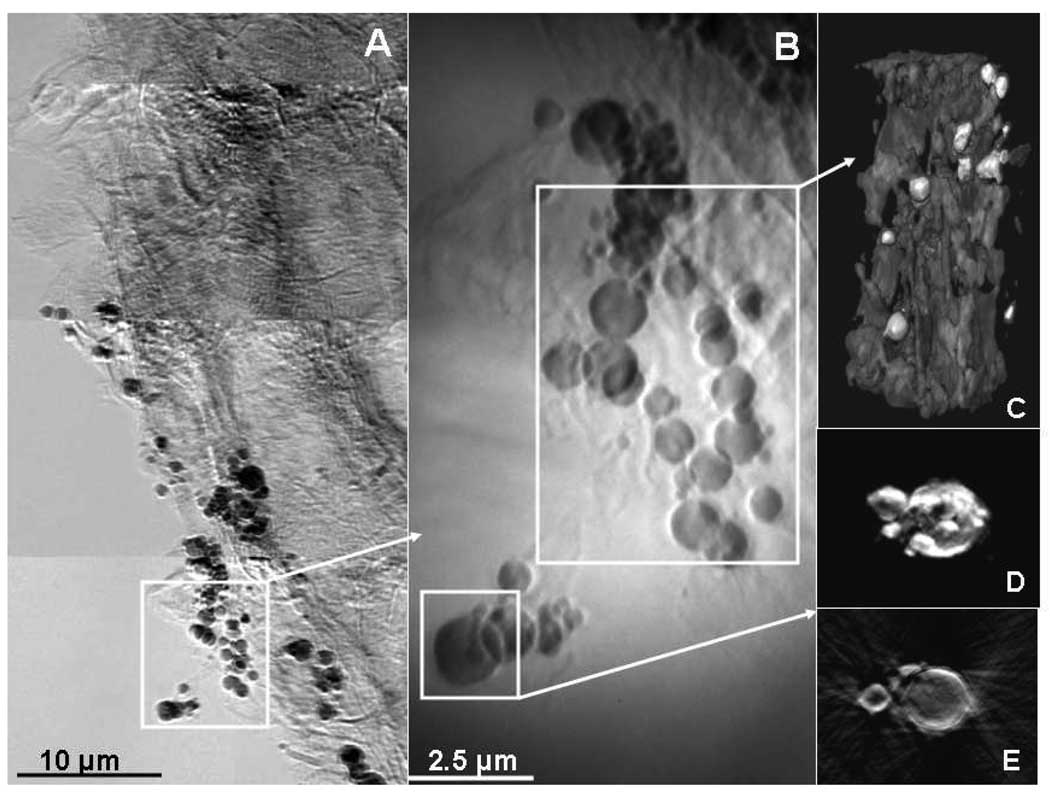
A mosaic TXM image of S. foliosa roots taken in absorption contrast at 9 keV revealed clusters of dark particles and channels of high absorbance within the plant tissue (Figure 1A) due to Hg (discussed below). A closer view of a cluster of the dark particles is shown in Figure 1B. 3D tomography of the cluster in Zernike phase contrast at 8 keV (Fig 1C) revealed that the particles were on the outside of the root, surrounded by less dense tissue, probably biofilm. Tomography of the lower particle (Fig 1D) and a slice from tomographic reconstruction (Fig 1E) revealed that the particle was hollow, with Hg clustered mainly at the surface. All these images indicate that the dark particles within the images are due to Hg at the surface of microbes such as sulfate-reducing bacteria, on the outside of the root, surrounded by biofilm. The dense channels seen in Fig. 1A and Fig. 3A are due to Hg that was taken up by the plant roots.
Figure 1.
TXM mosaic image of S. foliosa roots taken at 9 keV in absorption contrast shows dark particles and dark channels due to absorption by Hg (A). Blowup (B) shows greater detail. 2D stills from tomography of particles from (B) show particles with greatest absorption (lightest), possibly surrounded by biofilms (C). 2D tomographic still (D) and slice (E) of large particle indicate that highest Hg concentrations (lightest intensity) are on the outside of the fairly hollow particles.
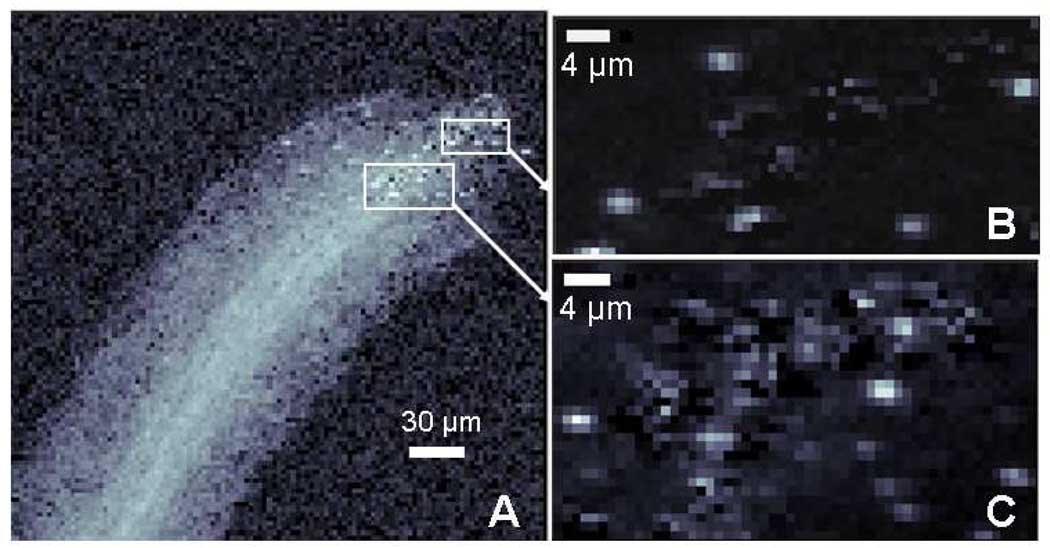
Figure 3.
Microprobe map of Hg fluorescence on logarithmic scale shows size and distribution of Hg within micron-sized S. foliosa roots (A). Micro-XANES points were selected from the highest concentration areas (“hot spots”, indicated white). Fluorescent counts (range 0–603) in insets (B and C) were determined as a difference above the Hg edge (12300 eV), minus below (12250 eV).
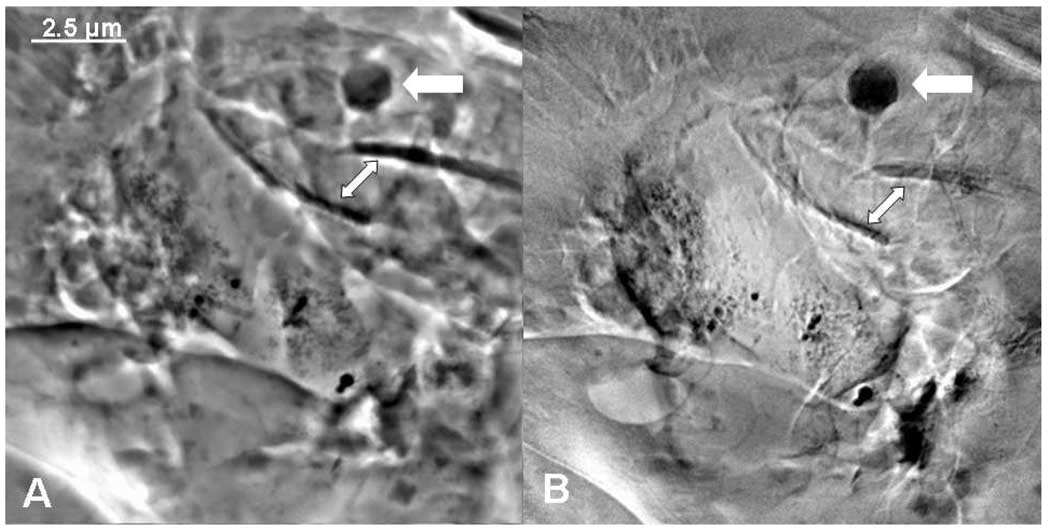
Single averaged images taken at 8 keV in Zernike phase contrast (Figure 2A) and in absorption contrast (Figure 2B) showed smaller nanoparticles in or on the root tissue as well as hexagonal crystals (see the thick white arrows in Fig. 2A and 2B). Phase contrast produced enhanced edges within the biological tissue in Figure 2A, whereas dark areas in the absorption contrast image were due to absorption by heavier elements; Hg in this case. Root hairs can be seen, darkened due to Hg absorption or adsorption (see thin white arrows in Fig. 2A and 2B).
Figure 2.
TXM images taken at 8 keV in phase contrast (A) and absorption contrast (B) of micron-sized S. foliosa roots show smaller Hg nanoparticles in or on the root tissue and hexagonal crystals, probably of metacinnabar (thick white arrows). Root hairs are indicated with thinner white arrows.
Microprobe spatial maps
Microprobe data were collected on the same S. foliosa roots used for TXM to collect elemental maps, to compare the distribution of Hg within the sample, and to verify the presence of Hg in the features portrayed in TXM images (Figure 1 and Figure 2). A typical fluorescent trace from the microprobe data (not shown) indicated the presence mainly of Ca, Mn, Fe, Zn and Hg. Fe was more evenly distributed in the plant root than the dark particles that appeared in the TXM, thus Fe was an unlikely candidate as the element comprising the dark particles. Ca and Zn have poor absorption at 8–9 keV (K-edges of 4038.5 and 9659 eV, respectively), and thus would not show up well in the TXM images. Mn (K-edge of 6539 eV) showed microprobe spots much larger than 1 micron, and has poorer absorption than Hg at 8 and 9 keV. Thus based on particle size, distribution, and absorption at 8 and 9 keV, Hg was determined to be the absorbing element in the dark spots of the TXM images.
Spatial maps consistently showed the highest mercury (“hot spots”), or densely concentrated areas, in the tips of the fine root structures (Figure 3B), but Hg was also found concentrated in channels along the center of the root (Fig. 3A). Distribution and particle size of the Hg hot spots varied. For example, some mercury particles appeared in aggregates up to 2 or 3 microns in size while others appeared smaller in size and more diffusely distributed.
Micro-XANES
Hg L3 micro-XANES spectra collected from the areas of highest Hg concentrations were fit to Hg model compounds using the same least squares method as for the bulk samples. These areas represented more localized environments in the rhizosphere than the bulk root tissue samples. Localized chemical species (Table 2) were proportioned similarly to the bulk results, except for cinnabar which was absent. Hg-S binding as metacinnabar comprised 0%–43.5% of the chemical species, Hg-S binding in a form similar to Hg(II)cysteine represented 20.5%–96.6%, and Hg-O coordination (a sum of methyl mercury acetate and mercury diacetate) represented 0%–36.0%.
Table 2.
Hg L3 micro-XANES least squares fitting results of micron-sized S. foliosa roots to model compounds (percent).
| Sample | cinnabar (HgS) |
meta- cinnabar (HgS) |
mercury cysteine and dicysteine |
methyl mercury acetate |
mercury diacetate |
|---|---|---|---|---|---|
|
S. foliosa root 1 region 1 |
—a | 18.6±2.5 | 57.9±3.9 | 9.6±3.9 | 13.9±1.6 |
|
S. foliosa root 1 region 2 |
— | 29.1±2.0 | 47.0±2.9 | 7.9±3.0 | 16.0±1.2 |
|
S. foliosa root 2 region 1 |
— | — | 96.6±4.1 | 3.4±4.1 | — |
|
S. foliosa root 2 region 2 |
— | 22.8±2.6 | 64.5±2.5 | — | 12.±1.5 |
|
S. foliosa root 3 region 1 |
— | 43.5±1.3 | 20.5±2.0 | 36.0±1.9 | — |
Less than 1% of total composition.
Discussion
Hg Uptake by S. foliosa and S. alterniflora
After growing in the presence of mercury for one week, Spartina roots showed significant accumulation of mercury, and S. foliosa roots took up significantly greater concentrations of mercury than S. alterniflora. Control Hg levels showed no significant differences between species (and therefore site). Accumulation results suggest that S. foliosa is either more efficient at removing mercury from its surroundings, or it has more established microbial populations in the rhizosphere. Whereas our Hg shoot concentrations were low, longer-term studies found that S. alterniflora transported more mercury to the shoots (4,10,12). It is possible that translocation of Hg to shoots would have increased given a longer treatment time. However, control and treated plants of both Spartina species showed signs of distress. While most inner leaves and/or stems were still green and vibrant, outer leaves were yellowing and withering, thus the treatment was ended after 1 week.
Hg speciation and transformation by mircroorganisms
The collection of mercury chemical species found within Spartina root hot spots may provide information about the chemical and biochemical transformations of Hg in the rhizosphere. Least-squares fitting of Hg L3 XANES of bulk roots of S. foliosa and S. alterniflora revealed that Hg speciation was similar in both cordgrasses. The micro and bulk XANES fitting both indicated that most Hg chelation is in a form similar to Hg-S binding in Hg-cysteine and dicysteine. This speciation is consistent with binding to thiol groups of proteins, possibly on the cell walls of microbes (6) or within root channels. Hg was clearly seen on the surface of the microbes in the TXM images (Fig 1D and 1E), thus our results are consistent with observations by Mason et al. (29) that much of HgCl2 made available to microorganisms (diatoms) ends up associated with the cell walls. There is some thought that binding to thiol groups may increase the availability of Hg for methylation (6).
In this study, all methylated mercury was bound to carboxyl groups, possibly to organic acids. In studies of Cd binding to Bacillus subtilis, Cd was bound to carboxyl groups in the cell walls (30). However, Mason et al. (29) found that in diatoms, most of the methylmercury they took up was found in the cytoplasm. The present work does not indicate the exact methylmercury location. TXM images show that most of the Hg associated with microorganisms is exterior to the plant, within microbial cell walls. However, methylmercury could be in the cytoplasm of SRB, or within plant roots. Because methylmercury is found in the same hot spots as cysteine-bound mercury (discussed below), the two species are in close proximity.
Hg as cinnabar, seen in some of the bulk samples, was entirely absent from the micro-XANES. This inconsistency may suggest that the cinnabar had different origins than the species found in the micro-XANES. For example cinnabar found in the bulk samples could have been due to residual sediment from the sampling site, which was caught in root tissue and ground with the sample. The bulk root samples contained thicker root portions as well as micron-sized roots, whereas TXM and microprobe (and micro-XANES) were performed only on micron-sized roots, where sediment was less likely to adhere. However, Hg as metacinnabar was seen in clustered hot spots of microprobe Hg maps via micro-XANES. Clusters of mercury nanoparticles were found next to a metacinnabar crystal in TXM images (Figure 1A top, and Figure 2). In the microprobe maps the Hg clusters appeared more diffuse, because the particles are less than the ~0.5 µm resolution.
Overall, the proximity of various Hg species found in these roots further indicates a chemical and/or biochemical relationship. It is noteworthy that three species of mercury, namely Hg bound to cysteine, methymercury and metacinnabar, were all found in the same regions in the micro-XANES, indicating that they occur in close proximity. As discussed above, Hg-cysteine may be bound to cell wall proteins and thereby more available for methylation. Thus the proximity of methylmercury and cysteine-bound Hg is not surprising. Because metacinnabar was also generally found along with methylmercury, it may be produced from precipitation with reduced sulfur concurrent with methylation.
HgS precipitation and potential biostabilization of Hg
The precipitation of Hg as metacinnabar, if it is occurring naturally in the plant, may provide a route for biostabilization of Hg in the rhizosphere of Spartina. The nanoparticles seen in or on the plant tissue may be due to precipitation of HgS, presumably when sulfate is reduced by SRB to sulfide. It is possible that the HgS precipitated during experimental Hg treatment. This avenue can be explored in future studies of environmental Spartina samples. Precipitation of HgS has also been seen in conditions mimicking the presence of SRB and FeRB (15). The precipitated HgS would most likely be in the form of metacinnabar, the main product of Hg precipitation with sulfides. In SRB, such as Desulfobacterium, mercury methylation was seen only with concurrent sulfate reduction (31), although it has also been seen along with fermentative metabolism (32).
Precipitation of metallic nanoparticles has also been seen in other plants and biofilms. Precipitation of Au has been observed within alfalfa plant roots and shoots (33); and within root cells of Sesbania drummondii (34), which can subsequently be transported to the shoots. Precipitation of ZnS by reduced sulfur has been seen in biolfims populated mainly by SRB (35). Thus, it is reasonable to expect precipitation of HgS in the rhizosphere of S. foliosa, in the presence of microbes surrounded by biofilms.
The TXM images, especially from the tomography in Fig. 1C, indicate the presence of biofilms surrounding microbes. Biofilms, which are networks of microbes interacting with extracellular polymeric substances (36), can affect Hg(II)- methylation by providing a local anoxic environment for SRB (37), even in bulk aerobic conditions. Extracellular biomineralization, as seen in ZnS nanocrystal formation in biofilms (38), has been shown to limit the mobility of nanoparticles formed, opening the possibility of biostabilization. HgS particle formation by SRB in the rhizosphere of Salix viminalis species at a mine site in Poland (39) has also been indicated as having potential for phytostabilization of Hg.
In summary, the use of X-ray microscopy combined with Hg L3 XANES has permitted us to obtain a “snapshot” of mercury methylation and metacinnabar precipitation in S. foliosa and S. alterniflora. Although the native S. foliosa has the capability for greater Hg uptake, perhaps due to its longer adaptation to the Hg-contaminated area, total Hg concentrations are the same in both species in the field, indicating that there is no significant difference in the amount of methylmercury that would be produced by each species. Although concentrations in the field average 0.1 ppm for both Spartina species, these are dominant florae within SF Bay and other locations. If an average of 10% of this Hg is methylated, Spartina must be carefully considered for its role in mercury methylation. Overall, the preferential uptake of mercury through the roots and significant HgS precipitation may show a potential for phytostabilization of mercury by Spartina, in spite of the methylation that occurs. Further studies of binding constants of the methylated mercury would lend further insight into this possibility.
Supplementary Material
Acknowledgments
We thank Susan Opp for her help in statistical treatment of the Hg uptake data; and Sam Webb, Sean Brennan, Jennifer Cassano, and Sarah Hayes for their help with data collection. A. Kahn and B. Mooney were supported by awards from the CSU East Bay Associated Students, and CSU East Bay provided support to J. C. Andrews. SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The transmission x-ray microscope is supported by NIH/NIBIB grant number 5R01EB004321. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
References
- 1.Patra M, Sharma A. Mercury toxicity in plants. Botan. Rev. 2000;66:379–422. [Google Scholar]
- 2.Manolopoulos H, Hurley JP, Babiarz CL, Back RC, Rolfhus KR. Riverine mixing zones as regions of enhanced methylmercury bioaccumulation in Lake Superior. J. Physique IV. 2003;107:805–808. [Google Scholar]
- 3.Wiener JG, Krabbgenhoft DP, Heinz GH, Scheuhammer AM. Ecotoxicology of mercury. In: Hoffman DJ, Rattner BA, Burton GA Jr, Cairns J Jr, editors. Handbook of Ecotoxicology. Boca Raton, FL: CRC Press; 2003. pp. 409–463. [Google Scholar]
- 4.Weis JS, Weis P. Metal uptake, transport and release by wetland plants: implications for phytoremediation and restoration. Environ. Int. 2004;30:685–700. doi: 10.1016/j.envint.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Marvin-DiPasquale MC, Agee JL, Bouse RM, Jaffe BE. Microbial cycling of mercury in contaminated pelagic and wetland sediments of San Pablo Bay, California. Environ. Geol. 2003;43:260–267. [Google Scholar]
- 6.Benoit JM, Gilmour CC, Heyes A, Mason RP, Miller CL. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems. In: Chai Y, Braids OC, editors. Biogeochemistry of Environmentally Important Trace Elements. Washington, DC: American Chemical Society; 2003. pp. 262–297. [Google Scholar]
- 7.Lin C, Jay JA. Mercury methylation by planktonic and biofilm cultures of Desulfovibrio desulfuricans. Environ. Sci. Technol. 2007;41:6691–6697. doi: 10.1021/es062304c. [DOI] [PubMed] [Google Scholar]
- 8.Fleming EJ, Mack EE, Green PG, Nelson DC. Mercury Methylation from Unexpected Sources: Molybdate-Inhibited Freshwater Sediments and an Iron-Reducing Bacterium. Appl. Environ. Microbiol. 2006;72:457–464. doi: 10.1128/AEM.72.1.457-464.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerin EJ, Gilmour CC, Roden E, Suzuki MT, Coates JD, Mason RP. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 2006;72:7929–7931. doi: 10.1128/AEM.01602-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weis P, Windham L, Burke DJ, Weis JS. Release into the environment of metals by two vascular salt marsh plants. Mar. Environ. Res. 2002;54:325–329. doi: 10.1016/s0141-1136(02)00131-9. [DOI] [PubMed] [Google Scholar]
- 11.Ayres DR, Smith DL, Zaremba K, Klohr S, Strong DR. Spread of exotic cordgrasses and hybrids (Spartina sp.) in the tidal marshes of San Francisco Bay, California, USA. Biol. Invas. 2004;6:221–231. [Google Scholar]
- 12.Windham L, Weis JS, Weis P. Uptake and distribution of metals in two dominant salt marsh macrophytes, Spartina alterniflora (cordgrass) and Phragmites australis (common reed) Estuar. Coast. Shelf Sci. 2003;56:63–72. doi: 10.1016/s0025-326x(00)00224-1. [DOI] [PubMed] [Google Scholar]
- 13.Kim CS, Bloom NS, Rytuba JJ, Brown GE., Jr Mercury speciation by x-ray absorption fine structure spectroscopy and sequential chemical extractions: A comparison of speciation methods. Environ. Sci. Technol. 2003;37:5102–5108. doi: 10.1021/es0341485. [DOI] [PubMed] [Google Scholar]
- 14.Slowey AJ, Rytuba JJ, Brown GE., Jr Speciation of mercury and mode of transport from placer gold mine tailings. Environ. Sci. Technol. 2005;39:1547–1554. doi: 10.1021/es049113z. [DOI] [PubMed] [Google Scholar]
- 15.Slowey AJ, Brown GE., Jr Transformations of mercury, iron, and sulfur, during the reductive dissolution of iron oxyhydroxide by sulfide. Geochim. Cosmochim. Acta. 2007;71:877–894. [Google Scholar]
- 16.Riddle SG, Tran HH, DeWitt JG, Andrews JC. Field, laboratory and x-ray absorption spectroscopic studies of mercury accumulation by water hyacinths. Environ. Sci. Technol. 2002;36:1965–1970. doi: 10.1021/es010603q. [DOI] [PubMed] [Google Scholar]
- 17.Rajan M, Darrow J, Hua M, Barnett B, Mendoza M, Greenfield BK, Andrews JC. Hg L3 XANES study of mercury methylation in shredded Eichhornia crassipes. Environ. Sci. Technol. 2008;42:5568–5573. doi: 10.1021/es800284v. [DOI] [PubMed] [Google Scholar]
- 18.Harris HH, Pickering IJ, George GN. The chemical form of mercury in fish. Science. 2003;301:764–766. doi: 10.1126/science.1085941. [DOI] [PubMed] [Google Scholar]
- 19.Skyllberg U, Qian J, Frech A. Combined XANES and EXAFS study on the bonding of methyl mercury to thiol groups in soil and aquatic organic matter. Physica Scripta. 2005;T115:894–896. [Google Scholar]
- 20.Dynes JJ, Tyliszczak T, Araki T, Lawrence JR, Swerhone GDW, Leppard GG, Hitchcock AP. Speciation and quantitative mapping of metal species in microbial biofilms, using scanning transmission x-ray xicroscopy. Environ. Sci. Technol. 2006;40:1556–1565. doi: 10.1021/es0513638. [DOI] [PubMed] [Google Scholar]
- 21.Lobinski R, Moulin C, Ortega R. Imaging and speciation of trace elements in biological environment. Biochimie. 2006;88:1591–1604. doi: 10.1016/j.biochi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Harris HH, Vogt S, Eastgate H, Legnini DG, Hornberger B, Cai Z, Lai B, Lay PA. Migration of mercury from dental amalgam through human teeth. J. Synchr. Radiat. 2008;15:123–128. doi: 10.1107/S0909049507061468. [DOI] [PubMed] [Google Scholar]
- 23.Collins JN. Invasion of San Francisco Bay by smooth cordgrass, Spartina alterniflora: A forecast of geomorphic effects on the intertidal zone. Oakland, CA: San Francisco Estuary Institute; Report to US EPA Region 9: San Francisco, CA. 2002
- 24.Baker AJM. Accumulators and excluders – strategies in the response of plants to heavy-metals. J. Plant Nutr. 1981;3:643–654. [Google Scholar]
- 25.San Francisco Estuary Invasive Spartina Project. [Jan 10, 2006];Invasive Field Identification Guide. Available at http://www.spartina.org/species.htm.
- 26.Webb Sam. Software and documentation. available at: http://www-ssrl.slac.stanford.edu/~swebb.
- 27.Andrews JC, Brennan S, Patty C, Luening K, Pianetta P, Almeida E, van der Meulen MCH, Feser M, Gelb J, Rudati J, Tkachuk A, Yun WB. A high resolution, hard x-ray bio-imaging facility at SSRL. Synchrotron Rad. News. 2008;21:17–26. doi: 10.1080/08940880802123043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.San Francisco Estuary Institute (SFEI) SFEI contribution no. 542. Oakland, CA: San Francisco Estuary Institute; 2007. The 2006 RMP annual monitoring results. [Google Scholar]
- 29.Mason RP, Reinfelder JR, Morel FMM. Uptake, toxicity, and trophic transfer of mercury in a coastal diatom. Water Air Soil Pollut. 1995;80:915–921. [Google Scholar]
- 30.Kelly SD, Boyanov MI, Bunker BA, Fein JB, Fowle DA, Yee N, Kemner KM. XAFS determination of the bacterial cell wall functional groups responsible for complexation of Cd and U as a function of pH. J. Synchrotron Rad. 2001;8:946–948. doi: 10.1107/s0909049500021014. [DOI] [PubMed] [Google Scholar]
- 31.King JK, Kostka JE, Frischer ME, Saunders FM. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl. Environ. Microbiol. 2000;66:2430–2437. doi: 10.1128/aem.66.6.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benoit JM, Gilmour CC, Mason RP. Aspects of bioavailablility of mercury for methylation in pure cultures of Desulfobulbus propionicus. Appl. Environ. Microbiol. 2001;67:51–58. doi: 10.1128/AEM.67.1.51-58.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardea-Torresdey JL, Parsons JG, Gomez E, Peralta-Videa J, Troiani HE, Santiago P, Yacaman MJ. Formation and growth of Au nanoparticles inside live alfalfa plants. Nano Lett. 2002;2:397–401. [Google Scholar]
- 34.Sharma NC, Sahi SV, Nath S, Parsons JG, Gardea-Torresdey JL, Pal T. Synthesis of plant-mediated gold nanoparticles and catalytic role of biomatrix-embedded nanomaterials. Environ. Sci. Technol. 2007;41:5137–5142. doi: 10.1021/es062929a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labrenz M, Banfield JF. Sulfate-reducing bacteria-dominated biofilms that precipitate ZnS in a subsurface circumneutral-pH mine drainage system. Microb. Ecol. 2004;47:205–217. doi: 10.1007/s00248-003-1025-8. [DOI] [PubMed] [Google Scholar]
- 36.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 37.Okabe S, Itoh T, Satoh H, Watanabe Y. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 1999;65:5107–5116. doi: 10.1128/aem.65.11.5107-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreau JW, Weber PK, Martin MC, Gilbert B, Hutcheon ID, Banfield JF. Extracellular proteins limit the disperal of biogenic nanoparticles. Science. 2007;316:1600–1602. doi: 10.1126/science.1141064. [DOI] [PubMed] [Google Scholar]
- 39.Sas-Nowosielska A, Galimska-Stypa R, Kucharski R, Zielonka U, Malkowski E, Gray L. Remediation aspect of microbial changes of plant rhizosphere in mercury contaminated soil. Environ. Monit. Assess. 2008;137:101–109. doi: 10.1007/s10661-007-9732-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.