Summary
Transcriptional regulatory networks direct the development of specialized cell types. The transcription factors Stat4 and T-bet are required for the development of T helper 1 cells, although the hierarchy of activity by these factors has not been clearly defined. In this report we show that these factors are not in a linear pathway and that each factor plays a unique role in programming chromatin architecture for Th1 gene expression, with subsets of genes depending on Stat4, T-bet, or both for expression in Th1 cells. T-bet is not able to transactivate expression of Stat4-dependent genes in the absence of endogenous Stat4 expression. Thus, T-bet requires Stat4 to achieve complete Th1 fate determination.
INTRODUCTION
The proper development and function of T helper cells is a central requirement for the generation of appropriate immune responses to pathogens and foreign molecules. The differentiation of T helper cells to effector subsets is directed by transcription factors that are capable of programming the expression of genes that are required for specialized functions of a subset of cells (Murphy and Reiner, 2002; Ansel et al., 2003). Th1 differentiation is promoted by stimulation with IL-12 and the subsequent activation of Stat4 (Hsieh et al., 1993; Kaplan et al., 1996; Thierfelder et al., 1996). The T-box transcription factor T-bet has been termed a master regulator of Th1 development (Szabo et al., 2000; Szabo et al., 2002), and expression is induced during Th1 differentiation by IFNγ stimulated Stat1 activation (Lighvani et al., 2001; Afkarian et al., 2002). The susceptibility of Stat4- and T-bet-deficient mice to intracellular pathogens, and the resistance of these mice to the development of inflammatory disease support a model wherein Stat4 and T-bet are required for the normal development and/or function of Th1 cells (Szabo et al., 2002; Kaplan, 2005; Sullivan et al., 2005).
The development of specialized cells requires networks of transcription factors that work together to mediate changes in gene expression to determine cell fate (Laiosa et al., 2006; Rothenberg, 2007). While Stat4 and T-bet are required for development of Th1 cells, the coordination of Th1 gene programming by these factors has not been well studied. In the absence of Stat4 or T-bet, there is decreased histone acetylation and increased DNA methylation of Th1 genes including Ifng and Il18r1 (Avni et al., 2002; Fields et al., 2002; Chang and Aune, 2005; Yu et al., 2007) and ectopic T-bet expression can induce histone modification and chromatin remodeling, even in the absence of Stat4 (Mullen et al., 2001; Shnyreva et al., 2004; Tong et al., 2005). It is not clear, however, if Stat4 and T-bet operate in linear or parallel pathways to the Th1 phenotype. In a linear pathway model represented by IL-12-Stat4-IFNγ-Stat1-T-bet-IFNγ, Stat4 provides a transient increase in IFNγ that is then able to induce T-bet expression, which in turn potentiates Ifng expression (Usui et al., 2003). In a linear pathway, it is also possible that Stat4 has transient effects on chromatin that allows access to other factors that mediate sustained gene programming. Indeed, Stat4 mediates transient histone acetylation of the Il2ra, IL12RB2 and Il18r1 genes (O'Sullivan et al., 2004; Letimier et al., 2007; Yu et al., 2007). A separate, though not mutually exclusive pathway, suggests that TCR and IFNγ signaling promote T-bet expression and induce Il12rb2 expression to facilitate IL-12 and Stat4 function (Mullen et al., 2001). However, several reports suggest that these models do not completely define the relative roles of Stat4 and T-bet in Th1 differentiation. First, it is not clear that T-bet is required for Il12rb2 expression (Usui et al., 2006). Moreover, while overexpression of T-bet in Stat4-deficient T cells can induce IFNγ expression and histone acetylation, it does not recapitulate wild-type IFNγ expression or Ifng acetylation levels by itself (Mullen et al., 2001; Fields et al., 2002). Despite the proposal that Stat4 mainly plays a role in Th1 expansion or survival downstream of T-bet (Mullen et al., 2001; Murphy and Reiner, 2002; Ansel et al., 2003), Stat4 is activated in T-bet-deficient cells and transduction of Stat4 into differentiating T-bet-deficient T cells results in increased IFN production, suggesting that Stat4 has some effects even in the absence of T-bet (Usui et al., 2006; Zhang and Boothby, 2006). Thus, the functional relationship between Stat4 and T-bet in developing Th1 cells may be more complex than is currently appreciated.
In this report, we examine the relative roles of Stat4 and T-bet in Th1 gene programming. In examining many genes associated with the Th1 program, we find subsets that require both Stat4 and T-bet, or only one of the factors. Stat4-dependent gene expression could not be rescued by supplemental IFNγ, and T-bet was capable of binding some common target genes in the absence of Stat4. Chromatin modifications to Th1 genes were altered in the absence of either factor, though specific modifications were affected more by one factor than the other. Moreover, ectopic T-bet expression was able to rescue Th1 gene expression and histone acetylation in T-bet-deficient T cells but not in Stat4-T-betdouble- deficient T cells, supporting a model wherein both Stat4 and T-bet are required for complete activation of the Th1 phenotype.
RESULTS
Stat4 and T-bet regulation of Th1 gene expression
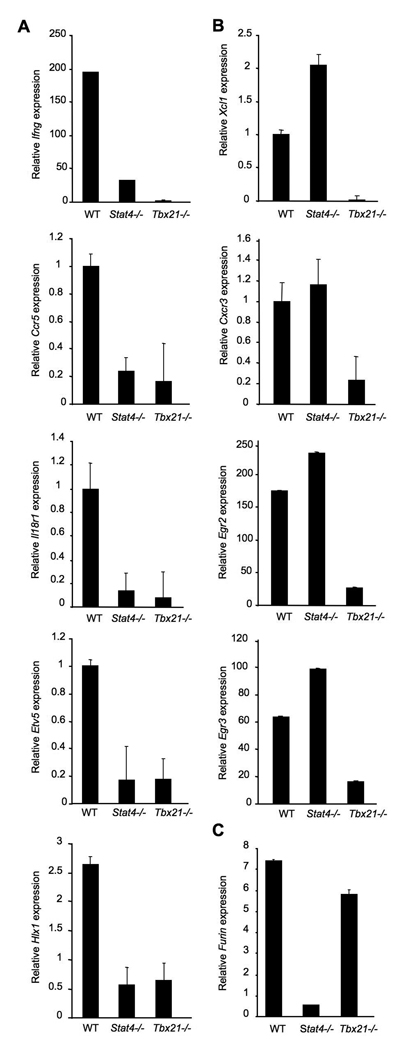
To define the relative roles of Stat4 and T-bet in the differentiation of Th1 cells we systematically analyzed the mRNA levels of genes previously described as having Th1 restricted expression (Table S1). CD4+ T cells from C57BL/6, Stat4−/− or Tbx21−/− mice were differentiated under Th1 culture conditions for five days and gene expression was analyzed using quantitative RT-PCR either in resting Th1 cells or in Th1 cells activated for six hours with anti-CD3, the latter when expression levels were low or undetectable in resting cells. We found that the expression of Ifng, Ccr5, Il18r1, Hlx1 and Etv5 were largely dependent upon the presence of both Stat4 and T-bet (Fig. 1A). In contrast, Xcl1, Cxcr3, Egr2 and Egr3 were decreased in Tbx21−/− cultures but had normal expression in the absence of Stat4 (Fig. 1B). Furin, which has previously been shown to be Stat4- dependent (Pesu et al., 2006), is independent of T-bet (Fig. 1C). Similar results are observed in cultures of naïve CD4+ T cells (Fig. S1A). These results demonstrate that Stat4 and T-bet regulate both overlapping and distinct subsets of Th1 genes.
Figure 1. Contribution of Stat4 and T-bet to expression of genes in Th1 cells.
Wild type, Stat4-deficient (Stat4−/−) and T-bet-deficient (Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL−12 + anti-IL-4) for five days. RNA was isolated from cells either before (Ccr5, Il18r1, Etv5, Cxcr3) or six hours after (Ifng, Hlx1, Xcl1, Egr2, Egr3, Furin) re-stimulation of cells with anti-CD3. Quantitative PCR using TaqMan primers specific for each gene was performed and results were normalized to expression of beta2-microglobulin. Results are the average ± SD of replicate samples and are representative of four experiments with similar results.
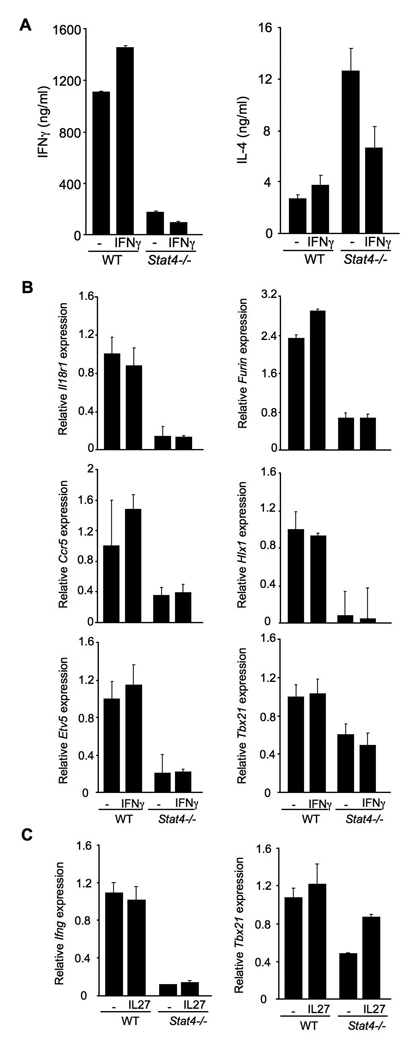
Whether T-bet expression is actually decreased in Stat4-deficient Th1 cultures has been somewhat controversial (Mullen et al., 2001; White et al., 2001; Afkarian et al., 2002; Hoey et al., 2003), and it is possible that the decreased endogenous IFNγ in Stat4-deficient Th1 cultures contributes to decreased Th1 development in the absence of Stat4 (Usui et al., 2003). To directly test whether decreased IFNγ levels are responsible for the phenotype of Stat4−/− cultures we incubated wild type or Stat4−/− T cells under Th1 conditions in the presence or absence of supplemental IFNγ. After five days of culture cells were washed and re-stimulated before levels of IFNγ production were assessed by ELISA and gene expression assessed by quantitative PCR. The addition of IFNγ to Stat4- deficient cultures did not alter the production of IFNγ from re-stimulated Stat4-deficient Th1 cultures (Fig. 2A). Adding IFNγ activated Stat1 and decreased the amount of IL-4 produced from Stat4-deficient cultures (Fig. 2A and data not shown), agreeing with previous reports on increased IL-4 production in Stat4−/− Th1 cultures, the ability of IFNγ to repress IL-4 in Th1 cultures (Kaplan et al., 1996; Zhang et al., 2001), and confirms that IFNγ added to these cultures was present at biologically active levels. Adding exogenous IFNγ to Stat4-deficient T cell cultures did not rescue gene expression of Il18r1, Ccr5, Etv5, Furin, or Hlx1 (Fig. 2B). We did observe a modest decrease in T-bet expression in the absence of Stat4, and expression levels were not recovered by the addition of IFNγ.
Figure 2. IFNγ or IL-27 do not rescue gene expression in Stat4-deficient Th1 cells.
(A) Wild type and Stat4-deficient (Stat4−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) in the presence or absence of 100 ng/ml recombinant IFNγ for five days. Cells were re-stimulated with anti-CD3 for 18 hours and supernatants were analyzed for levels of IFNγ and IL-4 using ELISA.
(B) Cells cultured as in (A) were analyzed for gene expression using qPCR as described in Figure 1. Results in (A) and (B) the average ± SD of replicate samples and are representative of four experiments with similar results.
(C) Wild type and Stat4-deficient (Stat4−/−) CD4+ T cells were cultured under Th1 conditions (IL−12 + anti-IL-4) in the presence or absence of 100 ng/ml recombinant IL-27 for five days. Expression of genes was determined after activation with anti-CD3 for four hours. Results are representative of two experiments with similar results.
Since T-bet expression was modestly decreased in these cultures and not rescued by IFNγ addition to the culture, we wanted to further test whether the addition of IL-27, another cytokine implicated in Th1 development that induces T-bet expression in a Stat1- dependent manner (Takeda et al., 2003), to Stat4-deficient cultures could recover any of the phenotype. Despite IL-27-induced Tbx21, IL-27 did not increase Ifng expression in Stat4-deficient Th1 cultures (Fig. 2C). Thus, neither a lack of endogenous IFNγ production, nor the modest decrease in T-bet expression, is the sole defect in Th1 generation by Stat4−/− T cells.
Stat4 and T-bet remodel Hlx1 and other Th1 gene loci
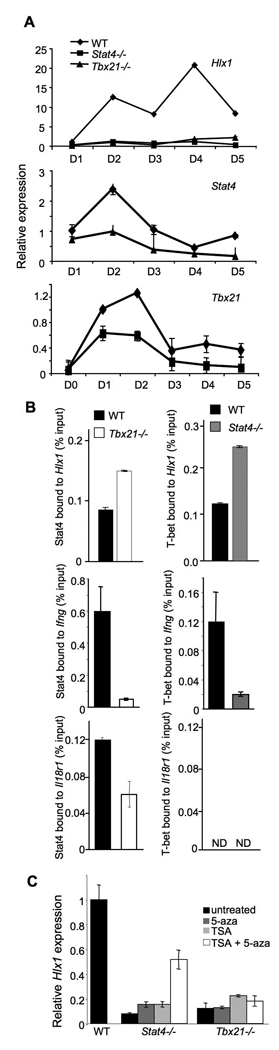
These experiments suggest that Stat4 and T-bet function independently, in parallel pathways promoting Th1 development. To further explore the relative roles of these factors in programming Th1 gene expression, we chose one gene, Hlx1, for detailed study. Hlx cooperates with T-bet in IFNγ production, even in the absence of Stat4, and likely plays an important role in Stat4 and T-bet-dependent programming of Ifng expression (Mullen et al., 2002; Martins et al., 2005). Expression of Hlx1 is decreased in Stat4- and T-bet-deficient cultures throughout the period of Th1 differentiation (Fig. 3A). The peaks of Hlx1 expression at days 2 and 4 likely represent direct induction by Stat4 correlating to IL-12 being added to cultures on the first and third days of culture. Through the same time period, we also noted the lower expression of Stat4 in T-bet-deficient cultures (Underhill et al., 2005; Usui et al., 2006) and observed that like T-bet expression in the absence of Stat4, decreased expression is most dramatic early during differentiation (Fig. 3A).
Figure 3. Stat4 and T-bet bind to the Hlx1 locus.
(A) Wild type, Stat4-deficient (Stat4−/−) and T-bet-deficient (Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and RNA was isolated from cells during each day of culture. Expression of Hlx1, Stat4 and Tbx21 were assessed in each of the samples using qPCR. Results are representative of two experiments.
(B) Wild type, Stat4-deficient (Stat4−/−) and T-bet-deficient (Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and chromatin was isolated for ChIP assay. ChIP was performed for Stat4 bound to the promoter of Hlx1, Ifng or Il18r1 in wild type and T-bet-deficient cells (left) or for T-bet bound to the same regions in wild type or Stat4-deficient cells (right). QPCR was performed using TaqMan primers specific for each promoter. Transcription factor bound to the locus is expressed as the percent of the input used for the ChIP assay. Results are the average ± SD of replicate samples and are representative of three experiments for Hlx1 and two experiments for binding to other promoters with similar results. ND, not detected.
(C) Wild type, Stat4-deficient (Stat4−/−) and T-bet-deficient (Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days in the presence or absence of 20 nM trichostatin A (TSA) and/or 10 µM 5-aza-deoxycytidine (5-aza). RNA was isolated for analysis of Hlx1 gene expression as described in Figure 1. Results are representative of two experiments.
We next defined the ability of Stat4 and T-bet to bind to the Hlx1 promoter, as well as the characterized promoters of Ifng and Il18r1 in the absence of the reciprocal factor (Chang and Aune, 2007; Schoenborn et al., 2007; Yu et al., 2007). There are a number of conserved non-coding sequences in the promoter and intron 3 of Hlx1 that we used for primer design (Fig. S2). Using ChIP and qPCR to detect binding to the Hlx1 promoter and intron 3 in Th1 cultures at day 3, Stat4 binding was detected at both regions, and binding was increased in the absence of T-bet (Fig. 3B and data not shown). Similarly, T-bet binding was detected at both regions and binding was enhanced in the absence of Stat4 (Fig. 3B and data not shown). However, binding patterns of Stat4 and T-bet were distinct at other promoters. There was significantly decreased binding of Stat4 and T-bet to Ifng, respectively, in Tbx21−/− and Stat4−/− Th1 cultures, compared to levels in control cultures (Fig. 3B). Stat4 binding to Il18r1 was partially affected by T-bet-deficiency and we did not observe binding of T-bet to Il18r1, even in wild type cells (Fig. 3B). To further illustrate specificity for Stat4 and T-bet binding, we also tested the association of other STAT and T-box factors to Hlx1. While we observed association of Stat1 at less than 50% of the level observed for Stat4, we did not observe association of Stat6 or Tbx5 at the Hlx1 promoter (data not shown). These results suggest there are gene-specific effects of Stat4- or T-bet-deficiency on the binding of other factors to target loci and that the ability of Stat4 to bind a gene and promote histone modifications is not required to allow accessibility for T-bet at all loci.
Since both factors bind to Hlx1 in the absence of the reciprocal factor, and both factors can mediate epigenetic modifications, we next tested whether altering the levels of epigenetic modifications to DNA and histones would recover expression or support a role for either factor in directly promoting transcription. Stat4−/− and Tbx21−/− T cells were cultured in the absence, presence or combination of histone deacetylase and DNA methylation inhibitors in parallel with wild type Th1 cultures. While each inhibitor had modest effects on Hlx1 mRNA levels in Stat4−/− cells, the combination of both inhibitors increased mRNA to about half of that observed in wild type cells (Fig. 3C). The inhibitors had less of an effect in the Tbx21−/− cells, suggesting that T-bet may be required for regulating transcription as well as chromatin remodeling.
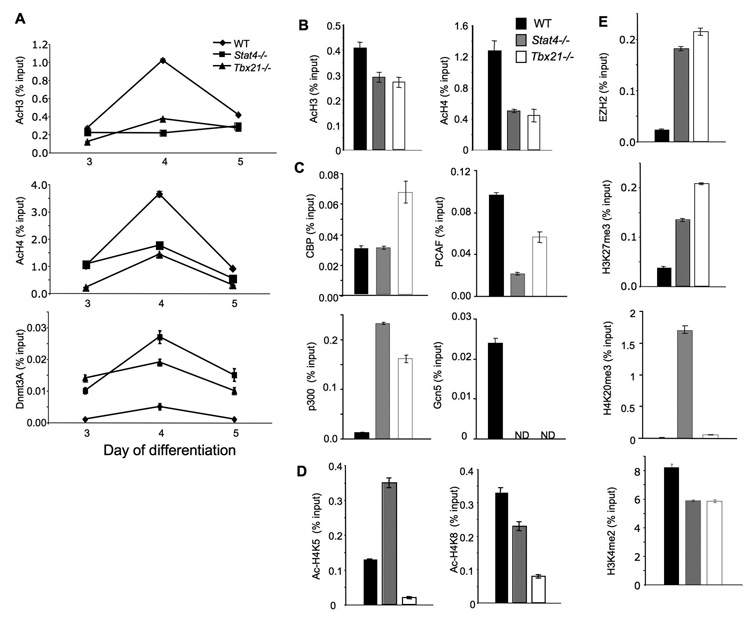
If T-bet and Stat4 were in a linear pathway, we would expect that chromatin modifications mediated by either factor would be similar. Conversely, distinct changes mediated by each factor would support a model wherein they act independently. To define the changes in chromatin that are mediated by Stat4 and T-bet, we examined the levels of histone modifications and histone modifying enzymes at the Hlx1 locus in wild type, Stat4- and T-bet-deficient cells. Total aceytlation of H3 and H4 was decreased at the Hlx1 promoter in the absence of either Stat4 or T-bet on days 3–5 of differentiation (Fig. 4A and B). To determine if the decrease in histone acetylation was due to a decrease in the association of specific histone acetyl-transferases (HATs), we performed ChIP assays for HATs at the Hlx1 promoter. In wild type cells, the association of HATs at the Hlx1 promoter did not vary greatly over the days 3–5 of differentiation (data not shown). Levels of Hlx1-associated CBP were not decreased in the absence of Stat4 and were increased in the absence of T-bet compared to wild type cells (Fig. 4C). Levels of Hlx1- associated p300 were increased in both Stat4- and T-bet-deficient cultures, compared to wild type cells. In contrast, levels of PCAF and Gcn5 were respectively decreased and undetectable in Stat4- and T-bet-deficient Th1 cultures (Fig. 4C). The differential effects of Stat4- and T-bet-deficiency were also detected in the changes of acetylation at specific histone lysine residues. While overall H4 acetylation was decreased in Stat4-deficient cells, acetylation of H4K5 was, compared to wild type cells, decreased in T-bet-deficient cells and increased in Stat4-deficient cells (Fig. 4D). Acetylation of H4K8 was lower in T-bet-deficient cultures than in Stat4-deficient or wild type cultures. Thus, while the absence of Stat4 and T-bet results in decreased histone acetylation, each factor has distinct effects on the acetylation of specific histone residues.
Figure 4. Stat4- and T-bet-dependent chromatin remodeling at the Hlx1 locus.
(A-E) Wild type, Stat4-deficient (Stat4−/−) and T-bet-deficient (Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and chromatin was isolated for ChIP assay. ChIP was performed for acetylated-H3, -H4 and DNMT3a on days 3–5 of culture (A) or day 5 only (B), the histone acetyltransferases CBP, p300, PCAF and Gcn5 on day 5 of culture (C), acetylated H4K5 and K8 on day 5 of culture (D), or EZH2, H3K27me3, H4K20me3 and H3K4me2 on day 5 of culture (E) using qPCR primers for the Hlx1 promoter. Results are the average ± SD of replicate samples and are representative of 3–5 experiments for each modification or enzyme with similar patterns. ND, not detected.
We have recently shown that one of the effects of Stat4 activity is to reduce the association of DNMT3a with target loci (Yu et al., 2007). We also observe that DNMT3a has increased association with Hlx1 in T-bet-deficient cells on days 3–5 of differentiation, though effects were greater in Stat4-deficient cells (Fig. 4A). The polycomb group protein EZH2 is involved in gene repression through methylaton of H3K27, also had increased association with Hlx1 in the absence of Stat4, and to a greater level in Tbx21−/− cells, correlating with increased levels of H3K27me3 (Fig. 4E). The H4K20me3 modification is also associated with gene repression and was only increased in the absence of Stat4 (Fig. 4E). Decreases in H3K4me2 were similar in Stat4- and T-bet-deficient cells and similar results were observed in cultures initiated from naïve CD4 T cells (Fig. 4E and Fig. S1B). These data demonstrate that Stat4 and T-bet mediate distinct but overlapping changes in chromatin in programming a gene for expression in Th1 cells.
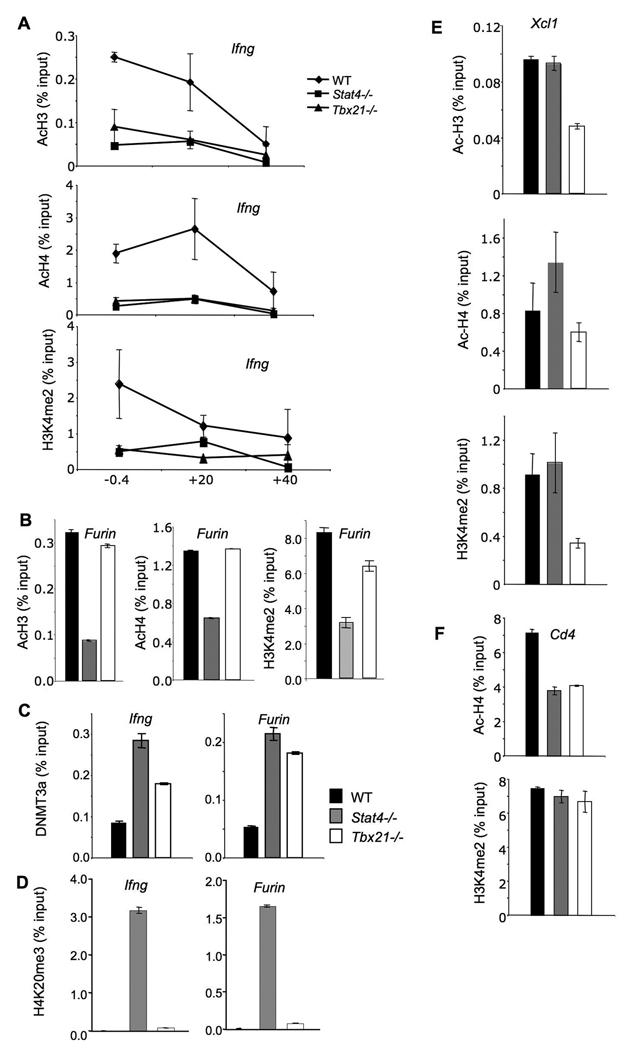
As only a subset of the Th1 genes have been analyzed for changes in chromatin structure in the absence of Stat4 or T-bet, we wanted to see if the transcription factor-dependent changes we observed at the Hlx1 locus were also seen at other loci. We examined the Ifng gene, which is dependent on both T-bet and Stat4, the Furin gene, which was more dependent on Stat4 than T-bet, and the Xcl1 gene, which was T-bet-dependent but Stat4-independent. In agreement with previous reports, acetylated H3 and H4 were decreased in the absence of either Stat4 or T-bet at several sites across the Ifng locus (Fig. 5A). Moreover, H3K4me2 levels were highest at the Ifng promoter but were decreased in T-bet- and Stat4-deficient cells (Fig. 5A). While Stat4-deficiency resulted in decreased levels of Ac-H3, Ac-H4, and H3K4me2 at the Furin promoter, these modifications showed only minor changes in the absence of T-bet (Fig. 5B). The increased DNMT3a associated with the Ifng gene in Stat4- or T-bet-deficient cells was similar to increases observed at the Hlx1 locus, with slightly greater effect of Stat4-deficiency than T-bet deficiency (Fig. 5C). Despite the relative T-bet-independence of Furin expression, T-bet deficiency also increased the level of DNMT3a present at the Furin locus, suggesting that some effects of T-bet-deficiency could be the result of broader changes in factor recruitment (Fig. 5C). The increase in H4K20me3 observed at the Hlx1 locus in Stat4- deficient cells was also seen at the Ifng and Furin loci (Fig. 5D). In contrast to the Stat4-restricted effects on Furin, we observed T-bet-dependent effects on Xcl1 (Fig. 5E). Histone acetylation and H3K4 di-methylation were decreased in T-bet-deficient cells but not in cells lacking Stat4 expression. Thus, Stat4 and T-bet not only have distinguishable effects on common target genes, but further have specific effects on genes that require only one factor for expression in Th1 cells.
Figure 5. Stat4- and T-bet-dependent chromatin remodeling at target loci.
(A-F) Wild type, Stat4-deficient (Stat4−/−) and T-bet-deficient (Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and chromatin was isolated for ChIP assay. ChIP assay was performed for acetylated-H3, -H4 and H3K4me2 at the Ifng promoter (−0.4 kb), and at sites +20 kb and +40 kb from the transcriptional start site (A), and at the Furin promoter (B). ChIP assay was performed for DNMT3a (C) and H4K20me3 (D) at the Ifng and Furin promoters. ChIP assay was performed for acetylated-H3, -H4 and H3K4me2 at the Xcl1 promoter (E) and for acetylated-H4 and H3K4me2 at intron 1 of Cd4 (F). Results are the average ± SD of replicate samples and are representative of 2–4 experiments for each modification or enzyme with similar patterns.
To determine if there are global changes in chromatin modifications and enzyme association, we examined Cd4 as a common gene that should be independent of Stat4 and T-bet. We did observe a decrease in the level of Cd4 histone acetylation, though it is important to note that the overall level of acetylation at this locus is 5-fold higher than for the Th1 genes examined (Fig. 5F). However, H3K4 methylation levels were unchanged and DNMT3a association was undetectable at this locus (Fig. 5F and data not shown). Moreover, acetylation of the Il4 locus, a gene that is similarly repressed in these cells, was unchanged among wild type, Stat4−/− and Tbx21−/− Th1 cells (data not shown). Thus, there are not global changes in chromatin modifications, though some effects can be observed at genes that do not seem to be direct targets for Stat4 or T-bet.
The ability of T-bet to activate the Th1 genetic program requires Stat4
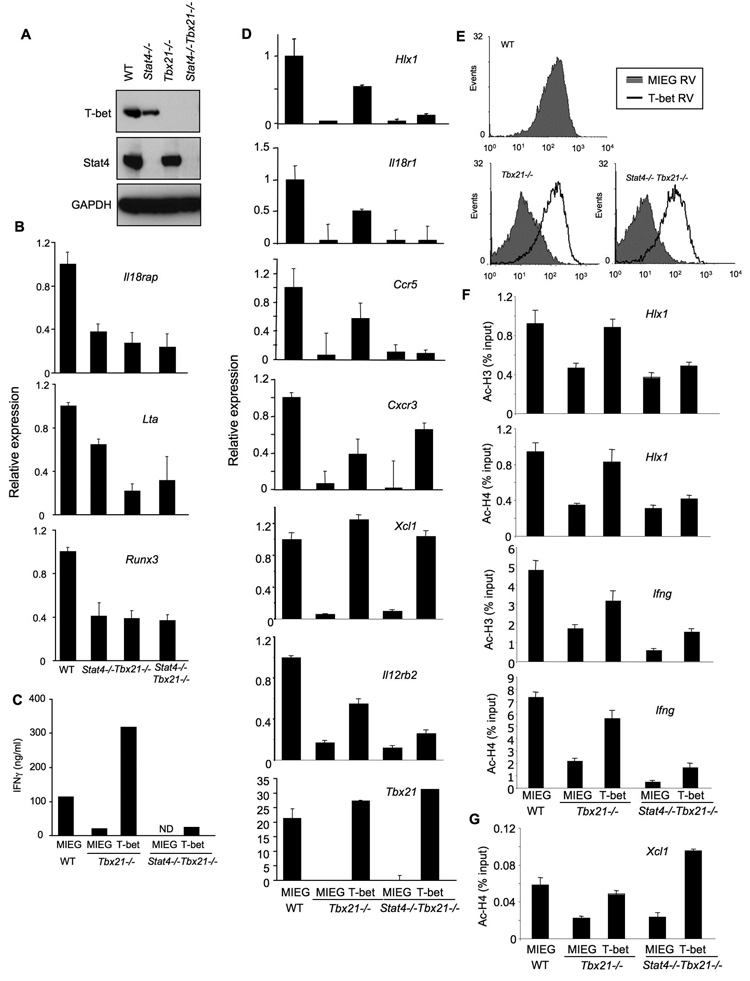
The experiments described thus far indicate that Stat4 and T-bet have separable functions in programming Th1 gene expression. To directly test the ability of T-bet to function in the absence of Stat4, we generated Stat4-T-bet-double deficient mice. These mice developed normally and had normal thymic and splenic cellularity. Normal T cell development in the thymus and T cell numbers in the periphery were observed in Stat4-T-bet-double-deficeint mice (Fig.S3). The decreased numbers of NK and NKT cells in the absence of T-bet was also observed in Stat4-T-bet-double deficient mice, but was not affected by additional deficiency in Stat4. Deficiency in protein expression was confirmed by western blot of protein extracts from wild type, Stat4-, T-bet- and Stat4-T-bet- deficient Th1 cultures (Fig. 6A). We then examined the expression of Th1 genes in these cultures to determine if there were any redundant functions of Stat4 and T-bet in the expression of genes that were only partially affected by the absence of Stat4 and/or T-bet (Lta, IL18rap, Runx3). However, there was not a cumulative effect of deficiency in both Stat4 and T-bet in the expression of any of the Th1 genes examined (Fig. 6B).
Figure 6. Stat4 requirement in T-bet function.
(A) Wild type, Stat4-deficient (Stat4−/−), T-bet-deficient (Tbx21−/−) and Stat4-T-bet-double deficient (Stat4-Tbx21−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and total cell extracts were immunoblotted for T-bet, Stat4 and GAPDH as a control.
(B) Cells cultured as in (A) were assessed for the expression of Th1 genes before (Il18rap, Runx3) or after (Lta) re-stimulation with anti-CD3.
(C-G) Wild type, T-bet-deficient (Tbx21−/−) and Stat4-T-bet-double deficient (Stat4−/− Tbx21−/−) CD4+ T cells were cultured under Th1 conditions. On day 2 of the culture period, cells were transduced with a bicistronic retrovirus expressing EGFP only (MIEG) or T-bet and EGFP (T-bet). At the end of the culture, cells were sorted for EGFP expression and stimulated for 18 hours with anti-CD3. Supernatants were analyzed for IFNγ levels using ELISA (C). RNA was isolated from each population to determine the expression levels of the indicated genes using qPCR (D). Surface expression of CXCR3 was determined using flow cytometry (E). ChIP assay was performed for acetylated-H3 or -H4 at the Hlx1 and Ifng promoters (F) or acetylated-H4 at the Xcl1 promoter (G). Results are the average ± SD of replicate samples and are representative of 2–3 experiments with similar results.
If Stat4 and T-bet perform truly independent functions in the programming of Th1 genes in that each are required for expression, we would expect that T-bet would not be able to activate gene expression in the absence of Stat4. To test this, we transduced T-bet-deficient or Stat4-T-bet-deficient T cells with control or T-bet expressing retrovirus and compared gene expression to levels seen in control retrovirus transduced wild type cells. Retroviral T-bet expression was fully capable of inducing IFNγ production from T-bet-deficient cells but had only minor effects in transduced Stat4-T-bet-deficient cells (Fig. 6C) despite expression of T-bet that was similar to wild type Th1 cells (Fig. 6D). Similarly, while ectopic T-bet expression could rescue expression of Hlx1, Il18r1 and Ccr5 in T-bet-deficient cultures, it had little if any effect in Stat4-T-bet-deficient cultures (Fig. 6D). Ectopic T-bet expression minimized the decrease in Il12rb2 expression observed in Tbx21−/− cells, though had less of an effect in double-deficient cells, correlating with partial Stat4-dependence of this gene (Fig.6D)(Lawless et al., 2000). In contrast, retroviral T-bet expression was able to induce expression of Xcl1 and Cxcr3, Stat4-independent genes, in both T-bet-deficient and Stat4-T-bet-deficient cultures (Fig. 6D and E).
To demonstrate that recovery in gene expression correlates with recovery of histone acetylation mediated by ectopic expression of T-bet, we performed ChIP analysis of Ac-H3 and Ac-H4 in wild type, T-bet-deficient or Stat4-T-bet double deficient cells transduced with control or T-bet-expressing retrovirus. Histone acetylation was decreased in Stat4-T-bet double deficient cells, compared to T-bet-deficient cells at the Hlx1 and Ifng promoters (Fig. 6F). Ectopic expression of T-bet increased Ac-H3 and Ac-H4 levels at the Hlx1 and Ifng promoter in Tbx21−/− cells to levels similar to wild type, though T-bet expression had only minor effects on histone acetylation in double-deficient cells (Fig. 6F). In contrast, T-bet expression was capable of increasing histone acetylation at the Xcl1 locus in both single- and double-deficient Th1 cells, similar to the ability of T-bet to promote Xcl1 expression in Tbx21−/− and Stat4−/−Tbx21−/− Th1 cells (Fig. 6G). These results demonstrate that there is an intrinsic difference in the ability of T-bet to function at Stat4-dependent and –independent loci, and that T-bet requires Stat4 activity to promote chromatin modification and gene expression of the complete Th1 phenotype.
DISCUSSION
Transcription factors are critical in regulating the development of effector T cell subsets. Stat4 and T-bet have been extensively characterized for their role in Th1 development, but how they functionally interact in the programming of the Th1 genetic signature has not been documented. Many factors have been termed “master regulators” of developmental pathways, and while these factors are clearly important, it is becoming apparent that they are only part of more complex transcriptional networks. In this report we have determined that Stat4 and T-bet are not in a linear pathway. Moreover, a decrease of Stat4 expression in T-bet-deficient cells or T-bet in Stat4-deficient cells (Fig. 3A and 6A) does not alone account for defects in gene expression. First, the identification of genes that depend solely on Stat4 or T-bet suggests that each factor has biological function in the absence of the other factor. Second, the addition of IL-27 does not increase Ifng expression in Stat4−/− Th1 cultures, despite induction of Tbx21. Third, binding of Stat4 or T-bet to Hlx1 is not compromised in the reciprocal gene-deficient cells. We further demonstrate that while T-bet is able to induce chromatin modifications and mRNA of T-bet-dependent, Stat4-independent genes, T-bet is unable to activate Stat4-dependent genes in the absence of Stat4. This demonstrates that both transcription factors are needed for the development of the complete Th1 phenotype.
These data raise the question of the temporal requirements for Stat4 and T-bet to function as chromatin remodeling factors or as factors that interact with the transcriptional machinery in the appropriate chromatin environment. Following Stat4 binding to a gene, it mediates histone hyperacetylation and alters other histone modifications and chromatin associated enzymes (O'Sullivan et al., 2004; Yu et al., 2007; Yu et al., 2008)(this report). Through these functions, Stat4 also results in increased transcription of target loci (O'Sullivan et al., 2004; Yu et al., 2007). During the differentiation period, the addition of IL-12 on the third day of culture results in an increase in mRNA and acetylated histone levels of Il18r1, Hlx1 and likely other genes as well (Yu et al., 2007)(Fig. 3 and 4). However, as Stat4 is only transiently activated, it is unlikely that Stat4 needs to remain bound to target loci to maintain gene expression. The role of T-bet is less clear with reports showing T-bet-dependence and –independence of epigenetic modification of the Ifng locus (Avni et al., 2002; Mullen et al., 2002; Usui et al., 2006). Our studies demonstrate the ability of T-bet to induce histone acetylation in the context of Stat4 (Fig. 6). A recent report using an inducible form of T-bet suggested that stable but not transient T-bet activity was required to maintain gene expression (Matsuda et al., 2007). Thus, T-bet may induce remodeling, but is also a direct activator of transcription. Indeed, we observed that in the presence of inhibitors that block repressive chromatin and DNA modifications, Hlx1 gene expression is increased in Stat4−/− but not Tbx21−/− Th1 cultures (Fig. 3C), suggesting that transcription depends upon the presence of T-bet.
The degree to which Stat4 and T-bet regulate each others expression has been examined in a number of reports. One particularly contentious point is whether T-bet expression is decreased in the absence of Stat4 (Mullen et al., 2001; White et al., 2001; Afkarian et al., 2002; Hoey et al., 2003), and there are several explanations for discrepancies among these reports. First, while IFNγ and Stat1 efficiently induce Tbx21 expression, recent reports do support a lesser role for Stat4 in activating Tbx21 (Usui et al., 2006; Yang et al., 2007). Second, the time during differentiation and the activation state of the cells have an impact on the level of difference in expression levels (Fig. 3). Furthermore, differences in culture systems, such as purified T cells versus the use of TCR transgenics where APCs are present, may affect results. APCs might provide cytokines, including IL-27 (Fig. 2), or other co-stimulatory signals that affect Tbx21 expression levels independent of the IL-12/Stat4 signal. Similarly, culture conditions and the cytokine environment might affect Stat4 expression in the absence of T-bet (Usui et al., 2006). Importantly, even in conditions where decreases in Stat4 or T-bet expression are observed, changes are not dramatic. Moreover, as we have shown, the modest decreases in the expression of either factor do not negatively affect the ability of each factor to bind at least one target gene, Hlx1, in the absence of the reciprocal factor, or induce the subsets of Th1 genes and chromatin modifications that are differentially dependent on either factor.
Changes in chromatin that mediate gene programming are necessarily complex. While routine examination of acetylated histones H3 and H4 define overall acetylation of the protein, which largely correlates with transcription at the locus, these analyses lack the resolution of examining specific chromatin modifications and the recruitment of chromatin modifying complexes to specific loci. While we observed that overall histone acetylation was decreased in the absence of Stat4 or T-bet, and that retroviral expression of T-bet induced histone acetylation when endogenous Stat4 was present, specific H4 residues actually had increased acetylation in the absence of Stat4 (Fig. 4B and D). Moreover, the increase in Hlx1-associated p300 in Stat4- and T-bet-deficient cells, and the increase in Hlx1-associated CBP in T-bet-deficient cells highlight that the recruitment of these enzymes is not dependent on either factor at this locus, and that associated CBP or p300 levels do not always correlate with total acetylation or gene expression (Fig. 4B and C). In contrast, both Stat4 and T-bet contributed to the recruitment of PCAF and Gcn5, components of large histone remodeling complexes including STAGA, TFTC and PCAF (Lee and Workman, 2007; Nagy and Tora, 2007) and levels of these factors correlated with the overall acetylation of H3, H4 and specifically H4K8 (Fig. 4B-D). This is similar to the ability of Gcn5/PCAF but not CBP/p300 to acetylate H4K8 in the context of the IFNβ gene (Agalioti et al., 2002). Thus, the recruitment of specific HAT complexes is required for Hlx1 gene expression.
In addition to regulating histone acetylation, Stat4 and T-bet also regulate the recruitment of other enzymes that generate chromatin modifications associated with either gene activation or gene repression. Stat4, but not T-bet, mediates the recruitment of Brg1-containing SWI/SNF complex to the Ifng locus (Zhang and Boothby, 2006). Moreover, T-bet recruits H3K4 methylases, which include Set7/9, to the Ifng and Cxcr3 loci (Shnyreva et al., 2004; Lewis et al., 2007). We observe a similar requirement for T-bet in mediating this modification at Hlx1, Ifng and Xcl1 genes, and also show that Stat4 promotes H3K4 methylation at target loci (Fig. 4 and 5). T-bet limits the association of repressive complex proteins such as mSin3a, while T-bet and Stat4 prevents the recruitment of DNA methyltransferases and DNA methylation of target loci, though DNMT3a association is more affected by Stat4-deficiency (Mullen et al., 2002; Tong etal., 2005; Yu et al., 2007)(Fig. 4 and 5). In the absence of either Stat4 or T-bet there are increases in EZH2 associated with the locus and increased H3K27me3 while increases in H4K20me3 levels were specifically found in the absence of Stat4 (Fig. 4 and 5). Changes to target loci are the result of transcription factors changing the equilibrium of positively-and negatively-acting actors associated with local chromatin. Moreover, chromatin alterations that affect gene transcription can occur at a distance as evidenced by recent extensive analyses of the Ifng locus (Chang and Aune, 2007; Schoenborn et al., 2007), and may depend on transient changes to a target locus as well. A further understanding of the hierarchy of chromatin modifier association to target loci, in the presence or absence of Stat4 and T-bet, should provide insight into how genes are programmed during T cell differentiation.
These data further suggest that Th1 gene expression and function could be heterogeneous depending on the cytokine environment that developing cells are exposed to. In a milieu with high IFNγ, and therefore high T-bet, but low IL-12, T cells should still be programmed with expression of the Stat4-independent genes. Cells derived in this environment would have low expression of IFNγ, but normal expression of Egr2/3, which promote FasL expression (Rengarajan et al., 2000). Moreover, they would express CXCR3 and XCL1 allowing them to be recruited, and to recruit to, sites of inflammation. It is not yet clear if there is a gradient or a threshold for gene programming by these factors. If a threshold exists, we would expect distinct cell states, cells with sufficient T-bet activation to program Th1 genes but not sufficient Stat4 activation, cells with sufficient Stat4 activation but with reduced T-bet and cells with sufficient activation of both factors. However, a model where there could be gradient of effects of either factor would predict even greater heterogeneity in the Th1 response allowing increased programmatic flexibility in responding to specific pathogens.
T-bet has been termed a “master regulator” of the Th1 phenotype. In this report, we demonstrate that T-bet does not act alone; that Stat4 is also required to modify chromatin and establish the full Th1 phenotype. The need for Stat4 in this process may be direct, by binding to genes and altering the chromatin environment, and also indirect by, for example inducing the expression of Hlx and Runx3, which have been shown to functionally cooperate with T-bet in promoting Ifng expression (Mullen et al., 2002; Djuretic et al., 2007). In addition to their complementary roles in Th1 differentiation, Stat4 and T-bet regulate the development of other Th subsets including Th17 (Mathur et al., 2006; Mathur et al., 2007; Furuta et al., 2008), as well as playing important roles in innate immune cells. It will be important to define the precise mechanisms of transcriptional regulation that involve Stat4 and T-bet in these other cell types to determine if they are similar, or if context dependent functions result in appropriate transcriptional regulation.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 Stat4−/− and Tbx21−/− (Taconic Farms, Germantown NY) mice have been previously described (Kaplan et al., 1996; Szabo et al., 2002). Wild type C57BL/6 mice were purchased from Harlan Bioproducts (Indianapolis, IN). Stat4-Tbx21−/− mice generated by intercrossing single-deficient mice. Mice were maintained under specific pathogen free conditions. All experiments were performed with the approval of the Indiana University IACUC.
In vitro T cell differentiation and analysis of gene expression
CD4 cells were isolated from spleen and lymph nodes of mice using magnetic beads (Miltenyi Biotec). For Th differentiation, CD4 cells (1 × 106 cells/ml) were cultured with plate bound anti-CD3 (4 µg/ml), 0.5 µg/ml soluble anti-CD28, under Th1 (2 ng/ml IL-12 and 10 µg/ml anti-IL-4) or Th2 (10 ng/ml IL-4 and 10 µg/ml anti-IFNγ) skewing conditions and expanded after three days. In some experiments 100 ng/ml IFNγ or 100 ng/ml IL-27 were added as described. After 5 days of culture, cells were harvested for gene expression or re-stimulated with anti-CD3 for ELISA. Quantitative RT-PCR was performed as described (Mathur et al., 2006). Message levels were analyzed using TaqMan PCR reagents specific for each of the indicated genes (Applied Biosystems, Foster City, CA). Cycle numbers of duplicate samples were normalized to expression of β2-microglobulin. Expression of some genes was examined after activation with anti-CD3 for six hours (Ifng, Xcl1, Egr2, Egr3, Furin, Lta) when mRNA levels in resting cells was very low or undetectable. Immunoblot and ELISA were performed using standard methods (Mathur et al., 2006; Yu et al., 2007).
Chromatin immunoprecipitation
ChIP assay was performed as previously described (Yu et al., 2007) with minor modification. In brief, cross-linking of protein-chromatin complexes was achieved by adding formaldehyde into cell cultures to a final concentration of 1%. Cells were washed in PBS, resuspended in cell lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA and 1% SDS) and incubated 10 minutes on ice. An ultrasonic processor (Vibra-Cell) was used to shear genomic DNA (150–300 bp fragments), with 10 10-second 70W bursts. Cell extracts were diluted in ChIP buffer, pre-cleared with salmon sperm DNA, BSA and protein A agarose bead slurry (50%) at 4°C for 1 hour. The supernatant was incubated in the presence or absence of 5 µg antibody (anti-Stat4, anti-T-bet, anti-Dnmt3a, anti-HAT, anti-p300, anti-CBP and anti-PCAF (Santa Cruz Biotechnology, Santa Cruz, CA), anti-acetylated H3, anti-acetylated H4 and anti-H4K20me3 (Millipore, Billerica, MA), anti-H4K5, anti-H4K8 and anti-Ezh2 (Abcam, Cambridge, MA)) at 4°C overnight. The immunocomplex was precipitated with protein A agarose beads at 4°C for 2 hour followed by centrifugation. The supernatant from the control precipitation was used as input material. The beads were washed consecutively with low salt wash buffer, high salt wash buffer, LiCl wash buffer, and twice in TE buffer. Bound DNA was eluted from the beads twice with elution buffer (0.1M NaHCO3, 1% SDS) by rotating at room temperature for 15 minutes. The supernatant was collected, supplemented with 2 mM EDTA, 20 mM Tris-Cl, 10 mg/ml Proteinase K and incubated at 37°C. DNA crosslinks were reversed by incubating precipitates at 65°C for 16 hours. DNA was purified by phenol/chloroform extraction and ethanol precipitation, and was resuspended in H2O. Real time quantification of ChIP assay was done as previously described using TaqMan primer sequences previously reported (Yu et al., 2007) or primers for SYBR Green as listed in Table S2. To quantify chromatin immunoprecipitates, a standard curve was generated from serial dilutions of a known amount of sonicated Th1 cells DNA. To calculate ChIP results as a percentage of input, the amount of the immunoprecipitated DNA from the isotype control antibody was subtracted from the amount of the immunoprecipitated DNA from the specific antibody ChIP followed by normalizing against the amount of the input DNA using quantitative PCR. Data are shown as percent input from a representative of 2–4 experiments.
Retroviral transduction
Purified CD4+ T cells were culture under Th1 conditions and on day 2, cells were transduced with a bicistronic retrovirus expressing EGFP only (MIEG) or T-bet and EGFP (T-bet) in the presence of 20 units/ml of IL-2 as previously described (Chang etal., 2005; Mathur et al., 2006). After transduction, cells were rested at 37°C for 2 hrs and cultured under Th1 conditions for another 3 days prior to flow cytometry or cell sorting for ELISA and real time PCR application. Flow cytometric analysis was performed using standard methods with a PE-labeled anti-CXCR3 (R&D Systems, Minneapolis, MN)(Mathur et al., 2007). For analysis of histone acetylation, cells were fixed directly after sorting and ChIP analysis was performed as described above with the addition of normalizing results to control analysis of Cd4.
Supplementary Material
Acknowledgements
This work was supported by U.S. Public Health Service Awards AI45515 and AI57459 (to M.H.K.) from the National Institutes of Health. We thank GS Kansas, CH Chang, RN Laribee and A Finnegan for careful review of this manuscript, R. Kapur for help with the retroviral transduction system, and GS Kansas for supplying the T-bet retrovirus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci U S A. 2005;102:17095–17100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Aune TM. Dynamic changes in histone-methylation 'marks' across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. Cutting Edge: Changes in Histone Acetylation at the IL-4 and IFN-gamma Loci Accompany Th1/Th2 Differentiation. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- Furuta S, Kagami S, Tamachi T, Ikeda K, Fujiwara M, Suto A, Hirose K, Watanabe N, Saito Y, Iwamoto I, Nakajima H. Overlapping and distinct roles of STAT4 and T-bet in the regulation of T cell differentiation and allergic airway inflammation. J Immunol. 2008;180:6656–6662. doi: 10.4049/jimmunol.180.10.6656. [DOI] [PubMed] [Google Scholar]
- Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH. Distinct requirements for the naturally occurring splice forms Stat4α and Stat4β in IL-12 responses. EMBO J. 2003;22:4237–4248. doi: 10.1093/emboj/cdg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C-S, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of Th1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- Kaplan MH. STAT4: A critical regulator of inflammation in vivo. Immunologic Research. 2005;32:231–241. doi: 10.1385/IR:31:3:231. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun Y-L, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- Lawless VA, Zhang S, Ozes ON, Bruns HA, Oldham I, Hoey T, Grusby MJ, Kaplan MH. Stat4 regulates multiple components of IFN-γ-inducing signaling pathways. J Immunol. 2000;165:6803–6808. doi: 10.4049/jimmunol.165.12.6803. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. Embo J. 2007;26:1292–1302. doi: 10.1038/sj.emboj.7601586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet's ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O'Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins GA, Hutchins AS, Reiner SL. Transcriptional activators of helper T cell fate are required for establishment but not maintenance of signature cytokine expression. J Immunol. 2005;175:5981–5985. doi: 10.4049/jimmunol.175.9.5981. [DOI] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Kapur R, Belladonna ML, Kansas GS, Kaplan MH. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood. 2006;108:1595–1601. doi: 10.1182/blood-2006-04-015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O'Malley JT, Kapur R, Levy DE, Kansas GS, Kaplan MH. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, George TC, Hagman J, Gapin L. Temporal dissection of T-bet functions. J Immunol. 2007;178:3457–3465. doi: 10.4049/jimmunol.178.6.3457. [DOI] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, Reiner SL. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat Immunol. 2002;3:652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- O'Sullivan A, Chang HC, Yu Q, Kaplan MH. STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J Biol Chem. 2004;279:7339–7345. doi: 10.1074/jbc.M309979200. [DOI] [PubMed] [Google Scholar]
- Pesu M, Muul L, Kanno Y, O'Shea JJ. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood. 2006;108:983–985. doi: 10.1182/blood-2005-09-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD, Glimcher LH. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity. 2000;12:293–300. doi: 10.1016/s1074-7613(00)80182-x. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BM, Jobe O, Lazarevic V, Vasquez K, Bronson R, Glimcher LH, Kramnik I. Increased susceptibility of mice lacking T-bet to infection with Mycobacterium tuberculosis correlates with increased IL-10 and decreased IFN-gamma production. J Immunol. 2005;175:4593–4602. doi: 10.4049/jimmunol.175.7.4593. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Takeda A, Hamano S, Yamanaka A, Hanada T, Ishibashi T, Mak TW, Yoshimura A, Yoshida H. Cutting edge: Role of IL-27/WSX-1 signaling for induction of T-bet through activation of Stat1 during initial Th1 commitment. J Immunol. 2003;170:4886–4890. doi: 10.4049/jimmunol.170.10.4886. [DOI] [PubMed] [Google Scholar]
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DAA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12 mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- Tong Y, Aune T, Boothby M. T-bet antagonizes mSin3a recruitment and transactivates a fully methylated IFN-gamma promoter via a conserved T-box half-site. Proc Natl Acad Sci U S A. 2005;102:2034–2039. doi: 10.1073/pnas.0409510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill GH, Zisoulis DG, Kolli KP, Ellies LG, Marth JD, Kansas GS. A crucial role for T-bet in selectin ligand expression in T helper 1 (Th1) cells. Blood. 2005;106:3867–3873. doi: 10.1182/blood-2005-03-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O'Shea JJ, Strober W. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Underhill GH, Kaplan MH, Kansas GS. Cutting edge: differential requirements for Stat4 in expression of glycosyltransferases responsible for selectin ligand formation in Th1 cells. J Immunol. 2001;167:628–631. doi: 10.4049/jimmunol.167.2.628. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110:2494–2500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chang HC, Ahyi AN, Kaplan MH. Transcription factor-dependent chromatin remodeling of Il18r1 during Th1 and Th2 differentiation. J Immunol. 2008;181 doi: 10.4049/jimmunol.181.5.3346. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Thieu VT, Kaplan MH. Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Ralpha gene during Th1 differentiation. Embo J. 2007;26:2052–2060. doi: 10.1038/sj.emboj.7601653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006;203:1493–1505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Apilado R, Coleman J, Ben-Sasson S, Tsang S, Hu-Li J, Paul WE, Huang H. Interferon gamma stabilizes the T helper cell type 1 phenotype. J Exp Med. 2001;194:165–172. doi: 10.1084/jem.194.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.