Abstract
Using high-resolution oligonucleotide CGH arrays, we evaluated chromosomal copy number changes in a series of 16 breast cancers, selected on the basis of highly similar pathological and molecular features characteristic of the “basal-like” phenotype. Each of these cancers showed numerous gains and losses, reflecting multiple chromosomal rearrangements during the development of these high-grade cancers. Chromosomal losses were particularly prevalent on chromosomal arms 5q, 8p, 9q, 12q, 17p, 19p, and Xq, and gains were commonly seen on chromosomal arms 1q, 8q, and 17q. Particularly remarkable were regions of high-level amplification (> 8-fold copy number change) on 4q12, 8q23.3, 19p12, and 19q13.2. These regions included candidate oncogenes cKIT, JUND, and AKT2., and immunohistochemistry confirmed that these particular genes were highly expressed in the cancers harboring the specific amplifications. However, each of these amplifications was observed only in individual cases, and no particular chromosomal alteration appeared to generally characterize this group of cancers. Thus, genomic changes among breast cancers with basal-like features appear to be very heterogeneous. Distinct high-level amplifications may provide new targets for treating some of these cancers, but copy number changes do not reveal a distinctive genomic fingerprint for this proposed class of breast cancers.
Introduction
Breast cancer is a heterogeneous disease, and the diversity of phenotypes defies comprehensive classification by histology alone. Because of widespread expectations that advances in cancer taxonomy will lead to improved clinical management as well as an improved understanding of disease pathogenesis, molecular methods are increasingly being used to further characterize address this complexity and to supplement morphology for breast cancer classification.
One of the breast cancer phenotypes proposed by both traditional histopathological studies [1, 2] and gene expression profiling [3–5] is that of the “basal-like” phenotype, which appears to be characterized by a relatively unfavorable prognosis and lack of a specific therapeutic target. There is no universally accepted definition for this phenotype, and in general, gene expression array classification correlates well with classification according to molecular features that can be measured by immunohistochemistry [6]. These features include absence of expression for estrogen and progesterone receptors, low expression (with no amplification) of the HER2 oncogene, expression of cytoskeletal proteins typical for basal cells (e.g., high molecular weight cytokeratins, vimentin), and frequently high expression of the epidermal growth factor receptor (EGFR) [7, 8]. In addition, many breast cancers with this molecular profile have a characteristic morphologic features that commonly include poorly differentiated cells (including cells with squamous differentiation or spindled shape), growth in nests with pushing borders, comedo necrosis, and high mitotic index [7, 9–12].
Patterns of chromosomal changes, if distinctive, could also help improve the classification of basal-like (and other) breast cancers, as well as provide a foundation for phenotype-specific molecular studies. Several previous studies that have included basal-like breast cancers in genomic analysis have not revealed a consistent pattern of chromosomal changes for this phenotype [13–18], but this could reflect variable methodologies that were used as well as inconsistent definitions of the basal-like phenotype. To further address this issue, we used high-resolution array comparative genomic hybridization to examine chromosomal changes in a subset of breast cancers, selected on the basis of aggressive histopathological features and basal-like expression characteristics. For purposes of this study, we selected only invasive breast cancer cases with distinctive basaloid morphology, immunohistochemical staining patterns consistent with the reported basal-like expression profile, and high Ki67 staining to determine whether such distinctive chromosomal changes could be identified for basal-like cancers, or perhaps an even more narrowly defined subset of aggressive cancers with basal-like properties.
Materials and Methods
Breast cancer samples
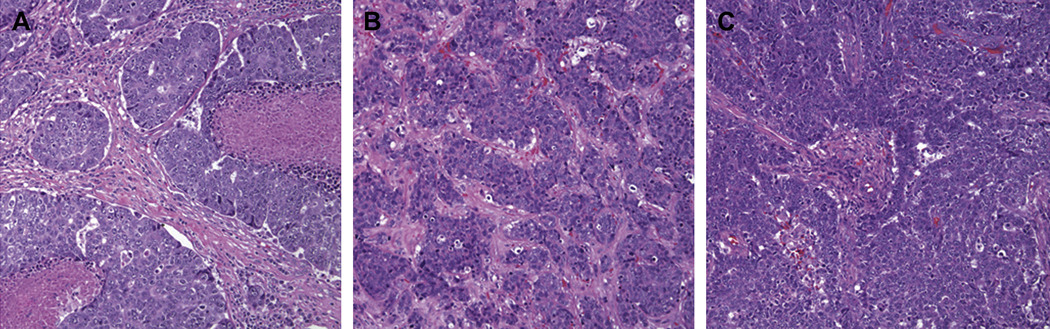
Snap-frozen breast cancer samples were obtained from tissues submitted for routine pathological examination. All samples used for research represented excess of what needs for routine diagnosis, and collection and use of these tissues was conducted with approval of the Institutional Review Board for Human Subjects. Selection of cases was made on the basis of negative immunohistochemical staining for ER, PR, and HER2 proteins, and expression of one or more markers considered to be characteristic of basal-like breast cancers (CK5/6 or EGFR). In addition, we further restricted our set of cases by selecting only those with high (>50%) percentage of cells reacting to Ki67 and morphologic features commonly described for basal-like cancers, including pushing borders and large, highly atypical cells. Typical microscopic features of cases used in the study are shown in figure 1.
Figure 1. Typical histology of basal-like breast cancers.
All cancers used in this study were selected on the basis of molecular features (negative for estrogen receptor, progesterone receptor, and HER2 expression, and positive for CK5/6 or EGFR expression) and also on the morphology represented in these three panels, with highly atypical cells, pushing borders, and minimal ductal differentiation. Note the areas of necrosis in the case shown on the left. The three cases shown in this figure are the same as those shown in figure 5 below.
Preparation of DNA for analysis
Isolated genomic DNA was assessed for concentration and quality using a spectrophotometer (NanoDrop Technologies, Wilmington, DE) to assure absence of contaminating protein (260/A280 ratio is greater than or equal to 1.8) and absence of carbohydrate, lipid and residual phenol (260/A230 ratio greater than or equal to 2.0). Integrity of genomic DNA was confirmed by low voltage 0.6% agarose gel electrophoresis with mean band size of about 50Kb. For array analysis, 10 ng of genomic DNA was amplified with phi29 DNA polymerase (Repli-G Amplification Kit, Qiagen) at 30C for 16 hours following the manufacturer- recommended protocol. Amplified DNA was then digested with 50 units of Alu I and Rsa I (Promega) for 2 hours at 37°C and samples were purified by using QIAQuick PCR clean-up Kit (Qiagen). Labeling reactions were performed with 7 ug aliquots of this DNA for 3 hours at 37C using a BioPrime Array CGH Genomic Labeling Module (Invitrogen). All samples were labeled with Cy5-dUTP (Perkin Elmer) and a pooled female normal human genomic DNA (Promega) was labeled with Cy3-dUTP. Labeled samples were purified, concentrated on a Centricon YM-30 column, and then mixed with 10x blocking agent and 2x hybridization buffer (Agilent Technologies).
Array hybridization, data acquisition, and analysis
Hybridization mixtures were first denatured at 95°C for 3 min and then immediately transferred to 37°C for 30 min. To remove any precipitates, the mixtures were centrifuged at 14,000g for 5 min. DNA from individual tumor samples and normal human female reference samples were co-hybridized on human CGH arrays (G4410B) containing about 43,000 unique probes with a median probe spatial resolution of 43Kb. These mixtures were then hybridized to microarrays for 40 hours at 65°C in a rotating oven (Robbins Scientific, Mountain View, CA) at 10 rpm. Hybridized microarrays were washed and dried according to manufacturer’s protocols, and imaged with Agilent G2565BA microarray scanner using default settings. Data were extracted using Feature Extraction Software v8.1 (Agilent Technologies).
Noise was then estimated for each sample array by calculating the spread of the log ratio differences between consecutive probes (dLRsd) along all chromosomes, and dividing by the square-root of 2, to counteract the effect of noise averaging. Aberrant regions (gains or losses) were then identified using the Aberration Detection Method 2 (ADM-2), with a threshold of 6. This algorithm uses an iterative procedure to find intervals of consistent high or low log ratios within an ordered set of probes and considers the log-ratio error information (quality-weighted interval score).
Immunohistochemical analysis of candidate genes within amplicons
Briefly, four-micrometer tissue sections were cut from tissue blocks and mounted on poly-L-lysine-coated slides. Sections were deparaffinized in xylene and hydrated in graded alcohols. Heat-induced epitope retrieval was done by microwave or for 15 minutes in ethylene diamine tetraacetic acid (EDTA) buffer (pH 8.0) or sodium citrate buffer (pH 6.0). Endogenous peroxidase activity was inactivated by incubation in 4% H2O2 for 5 minutes. After rinsing sections in phosphate-buffered saline, tissue sections were then incubated with primary antibodies for 60 minutes at room temperature and developed using the DAKO EnVision Plus-HRP detection system (DAKO, Glostrup, Denmark) according to the manufacturer's instructions. Primary antibodies included c-Kit (DAKO, 1:50 dilution), JUND (Santa Cruz, 1:50 dilution) and AKT2
Results
Numerous chromosomal copy number changes in high-grade basal-like breast cancers
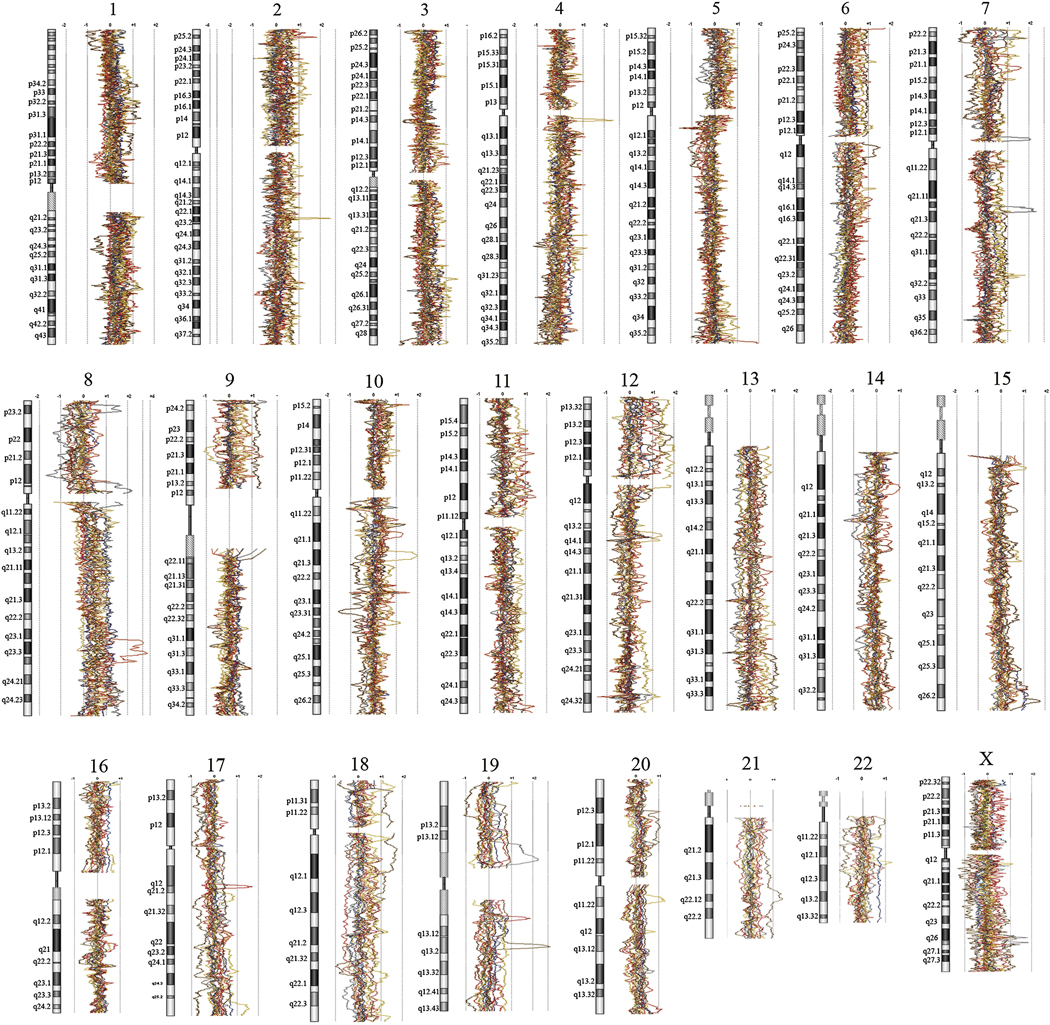
As typical for high-grade malignant neoplasms, these basal-like breast cancers showed numerous chromosomal copy number alterations. In these 16 cases, we noted a mean of 75 (± 27) chromosomal regions showing gain detectable over baseline, and a mean of 50 (± 27) regions of chromosomal loss per case. Remarkably, the use of high-density CGH arrays allows definition of copy number changes at high resolution, and our analysis revealed that the chromosomal copy number changes were often complex, involving discontinuous chromosomal segments. Specific regions of gains and losses for specific chromosomal loci are diagrammed for each case in figure 2, and data for all specific probes has been deposited with the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo//query/acc.cqi?acc=GSE12659). Frequencies of gains and losses for chromosomal segments are also summarized in a histogram (figure 3).
Figure 2. Chromosomal copy number changes for each chromosome in basal-like breast cancers with aggressive features.
For each sample, genomic quantity (compared to DNA from diploid human cells) is plotted as a log2 moving average along each chromosomal axis. Gains are represented by deviations of plotted lines to right, and losses are represented by deviations of plotted lines to the left.
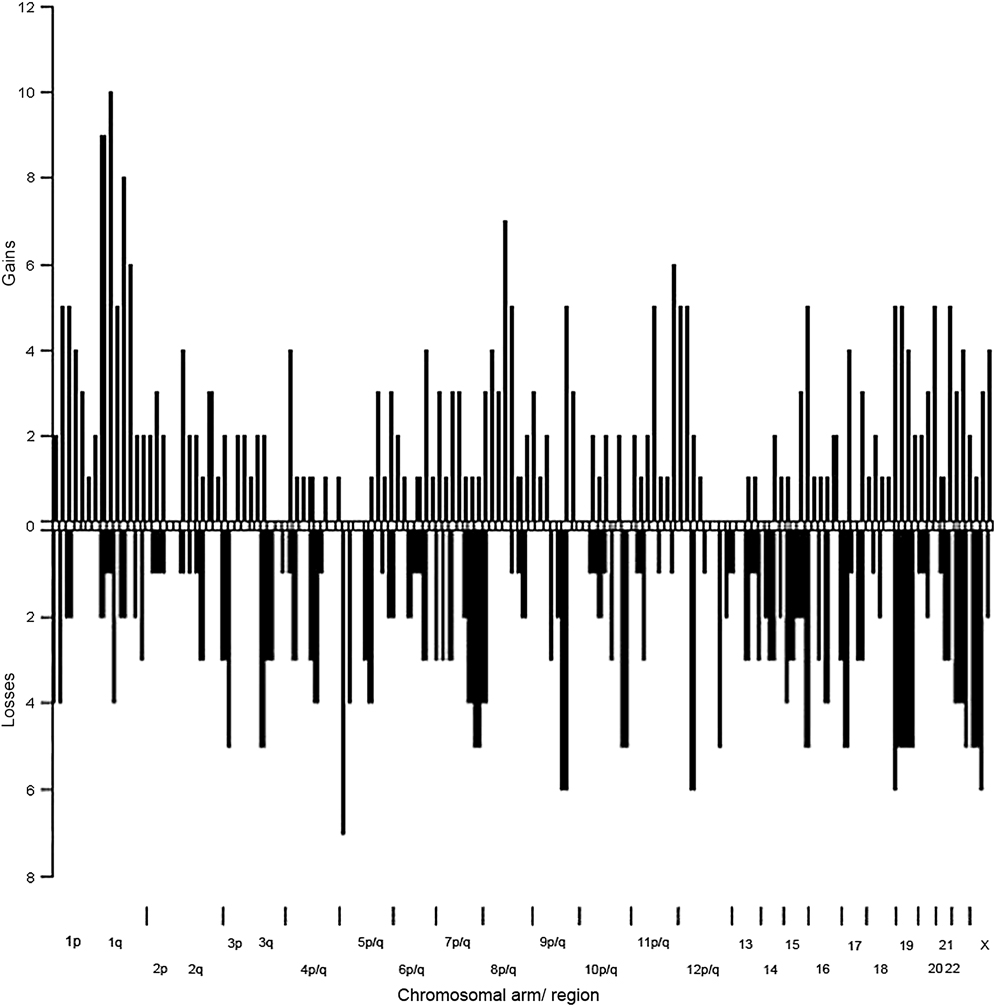
Figure 3. Summary of chromosomal copy number changes in basal-like breast cancers with aggressive features.
Chromosomal gains and losses were determined for each case as described in methods and plotted as histogram for designated chromosomal regions.
Chromosomal changes are highly variable in high-grade basal-like breast cancers
Overall, we observed remarkably little consistency for chromosomal changes among these breast cancers, selected to have highly similar phenotypes. The chromosomal arm most commonly affected by chromosomal losses in these cases is 5q, corroborating findings of previous studies that reported loss involving this chromosomal arm 5q as common in basal-like breast cancers [13, 14, 18]. In our samples, losses of chromosomal arm 5q were observed in 7 of the 16 cases, but these losses, extending from 5q11.2 to 5q14.2, did not involve any single non-overlapping region in more than 6 of the 16 cases. Losses affecting other chromosomal arms were also found to be relatively common among these cancers. For example, losses in 6 of the 16 cases were observed for chromosomal arms 8p (overlapping in 5 cases at approximately 8p21.3), 9q (overlapping in all 6 cases at approximately 9q33.3), 12q (overlapping in 5 cases at 12q12–q14.1 and in 5 cases and 5q24.21–q24.32), 17p (overlapping in all cases at approximately 17p13.2), 19p (overlapping in all cases at 19p13.3-term), and the X chromosome (overlapping in 4 cases at Xq13.2). Thus, while several loci are commonly affected by chromosomal loss in these cancers, no loss of a particular locus stands out as distinctive for these cancers.
Low level gains (≥log2 1.0) were most commonly observed affecting chromosomal arm 1q, with 12 of 16 cases demonstrating regions of copy number gain on this chromosomal arm. As noted in general, the gains on this chromosomal arm also appear to be complex and discontinuous, with a narrow region of overlap affecting 1q23.2 in 10 of the cases. Gains were also observed in 10 of 16 cases on chromosomal arm 8q. Again, gains on this chromosomal arm are complex and discontinuous, with regions from approximately 17q22.2–17q23.1 and 17q25-term both showing gain in 7 of the cases. Thus, no single chromosomal copy gain can be identified as a “fingerprint” of these poorly differentiated ER-/PR-/HER2- breast cancers.
Multiple loci of high-level gene amplification in high-grade basal-like breast cancers
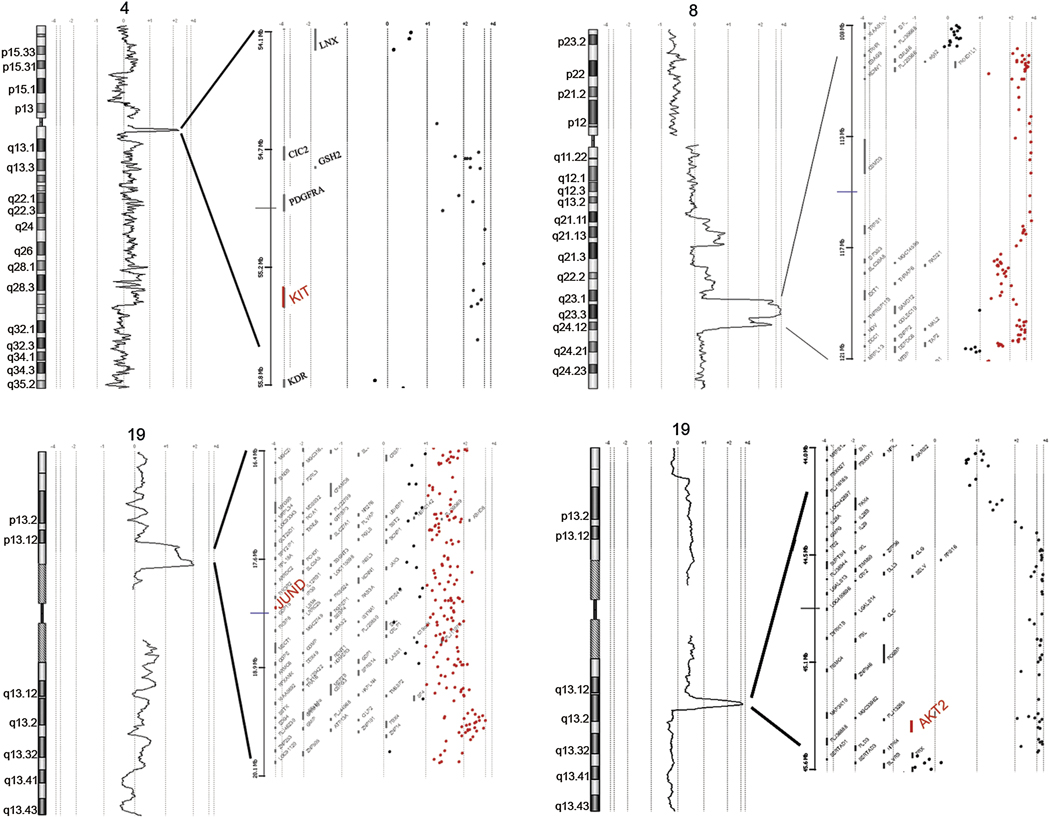
In addition to these regions of gain or loss seen at high frequency in these cancers, several distinct regions of amplification were seen in individual cases. Particularly remarkable are four independent high-level amplifications (greater than 8-fold copy number gain), occurring in four different cancers. One amplification involves a segment of about 1 Mb on chromosomal arm 4q, another a segment of about 3.7 Mb on chromosomal arm 19p, the third a segment of about 1.2 Mb on chromosomal arm 19q, and the fourth a segment of about 12 Mb on chromosomal arm 8q. Expanded images of these amplifications are shown in figure 4, with representative landmark genes designated for each amplicon (data for all genes available at http://www.ncbi.nlm.nih.gov/projects/geo//query/acc.cqi?acc=GSE12659). These amplicons include several candidate oncogenes, including cKIT on 4q, AKT2on 19q, and JUN-D on 19p. The amplicon on chromosomal arm 8q is somewhat larger than the other amplicons and notably does not appear to include the MYC oncogene at 8p24.2.
Figure 4. High level amplifications in individual breast cancer cases.
As above, relative genomic quantity is plotted as a log2 moving average along each chromosomal axis. Regions with high-level amplifications are expanded, showing values for individual probes (dots) and relative positions of genes mapped to the amplified regions. Selected genes within amplified regions are designated, and candidate oncogenes are highlighted in large font.
Immunohistochemical measurements of candidate oncogenes within amplicons
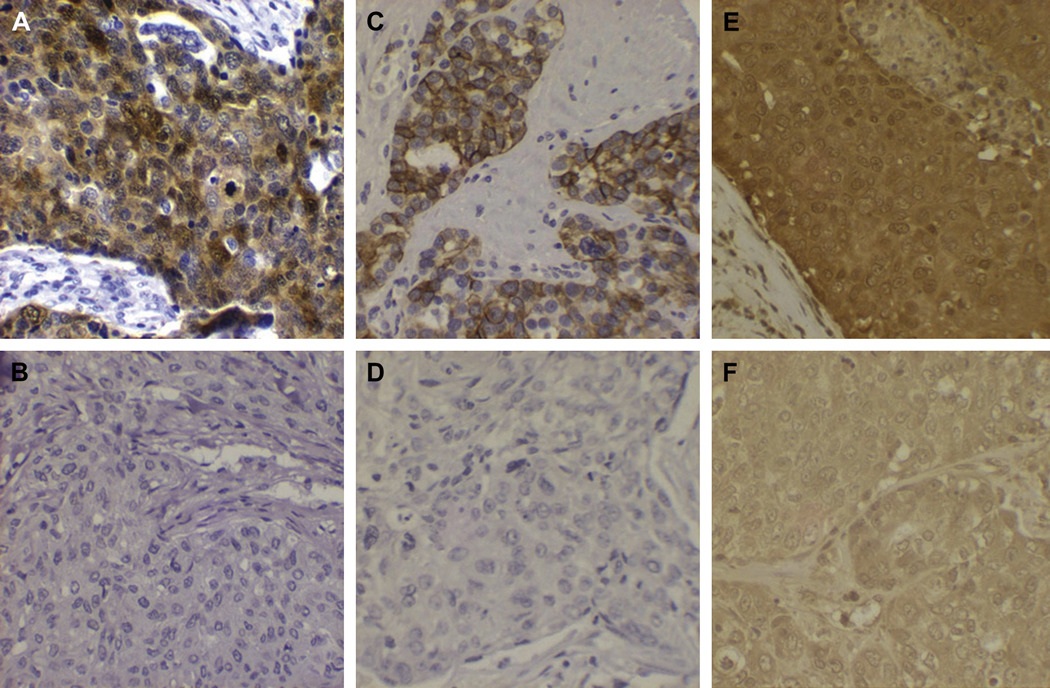
As noted above, three of the amplicons observed in these samples include previously recognized candidate oncogenes. The inclusion of recognized oncogenes in the amplicons does not specifically implicate those genes in the pathogenesis of the respective cancers, however, and thus we further evaluated the potential roles of those genes by measuring the levels of expression in the relevant tumors by immunohistochemistry. As shown in figure 4, the cancers with amplifications of cKIT, AKT2, and JUND all showed markedly increased levels of those respective proteins, compared to normal tissues and other breast cancers that do not have the specific amplifications. Based on inclusion of these genes within narrow regions of amplifications, high levels of protein expression, and known functions that are consistent with oncogenic function, these three genes appear to be strong candidate oncogenes for their respective tumors.
Discussion
This study was designed to provide high-resolution measurements of chromosomal copy numbers in cases of high-grade breast cancer, all selected on the basis of similar histologic appearance and immunohistochemical expression patterns characteristic of the “basal-like” phenotype. Our results show these aggressive breast cancers with basal-like features to have numerous and complex chromosomal alterations, with frequently discontinuous gains and losses. In addition, several high-level amplifications were noted, including chromosomal regions that include AKT-2, JUND, and cKit.
The involvement of these specific genes in the pathogenesis of these breast cancers is further suggested by findings that the corresponding proteins (Akt-2, Jun-D, and c-Kit) are highly expressed in the respective tumors. Amplifications of Akt-2 have been reported previously in 3 of 106 (2.8%) breast carcinomas in a distribution that appears to be independent of HER2 amplifications [19]. The c-kit protein is expressed in normal breast [20] and several studies have described high levels of c-kit expression at both the mRNA and protein level in breast cancers that have basal-like expression patterns [21]. Although this gene has been previously reported to be amplified in some testicular cancers and gliomas [22–24], to our knowledge, this is the first report of this gene being amplified in breast cancer. JUND is a member of the JUN family, and a functional component of the AP1 transcription factor complex [25, 26]. Although JUND has not been previously implicated as a potential oncogene in breast cancer, JUN (the first member of the family that was identified) is amplified in some aggressive sarcomas [27], and high expression of this gene has been shown to have functional significance in experimental models of breast cancer [28, 29]. Based on our observation of high expression in the specific breast cancer that harbors a JUND amplification, and the functional significance of the JUN gene family in cancer, it is reasonable to hypothesize that JUND represents a critical gene in this particular amplicon.
Analysis of additional samples will be needed to determine the frequencies of whether amplifications of these genes in breast cancers, and whether these amplifications are more common in cancers with basal-like features than in other types of breast cancer. Furthermore, additional studies will be needed to determine whether these amplifications are restricted - or even more prevalent - in high grade or ER-negative breast cancers. Notably, an association between amplification (of any gene) and high grade has been previously noted [30], with sites of localized high-level DNA amplification including known oncogenes, such as 7p12 (EGFR), 8q24 (MYC), 11q13 (CCND1), 12q14 (MDM2), 17q12 (ERBB2), 20q12 (AIB1), and 20q13 (ZNF217).
Overall, our study is consistent with previously reported data on chromosomal changes in basal-like breast cancers from the perspective of being unable to define a distinctive fingerprint for these cancers. Several studies used conventional metaphase comparative genomic hybridization (CGH) to investigate losses and gains of chromosomal regions in basal-like breast tumors, including Korsching et al., who clustered primary breast tumors based on phenotypic and CGH profiles and found that cancers expressing myoepithelial/ basal markers have a higher number of genetic alterations relative to other breast cancers [31]. In this study, no unique alterations in this basal-like subclass were found to differentiate them from other breast cancers. Subsequently, Jones et al [32] analyzed breast cancers by CGH and found a lower mean number of changes (2.1, range 0–4) in cancers described as myoepithelial, compared to the numbers seen in unselected ductal carcinomas (mean of 8.6, range 3.6–13.8). The same group subsequently compared CGH profiles of 43 cases of CK-14 positive breast cancers to grade- and age-matched CK-14 negative cancers and found a mean of 6.5 changes for the CK-14 positive cancers, compared to a mean of 10.3 changes for the CK-negative cases.
More recently, several studies have used array-based methods for CGH analysis of breast cancer copy numbers. For example, one study used oligonucleotide arrays designed for SNP analysis (Affymetrix platform) to measure LOH in breast cancers that had been previously classified according to gene expression patterns and found LOH more frequently on chromosomal arms 5q, 4p, and 18q in the basal like cancers than in tumors from other subclasses [18]. In another array-based CGH project [14], higher total numbers of gains and losses were found in cancers classified (by gene expression profiles) as basal-like, including losses at 3q12, 4p15–p32, 4q31–q35, 5q11–q31, and 14q22–q23, and gains at 1q12–q41, 6p12–p25, 7q22–q36, 10p12–p15, 17q25, and 21q22.
Using the same CGH array platform used in our project, Adelaide et al [13] found fewer gains (with a log2 ratio >1) in basal-like breast cancers than found in our study. Furthermore, these gains targeted somewhat different chromosomal regions (8p11, 8q11-tre, 10q11, 11q13–14, and 17q12–13) than the regions of highest gain found in our analysis. Interestingly, this study also observed amplifications of small regions that include several known or candidate oncogenes, namely EGFR, PIK3CA, FGFR2, BCL2L2, IGF1R, and CCNE1. However, these samples did not display amplification of the KIT, JUND, or AKT2 genes, as we find in our analysis. However, these data are consistent with those of our present study in demonstrating indicate that basal-like breast cancers are highly heterogeneous, with a multitude of chromosomal loci that are affected by losses and gains. Moreover, a variety of different genes appear to be amplified among these cancers, with no individual gene representing a dominant characteristic of these cancers. Thus, multiple targeted therapeutic agents may need to be developed to effectively treat these cancers.
In summary, considerable data now suggests that basal-like breast cancers are highly heterogeneous from both molecular and phenotypic perspectives. Our study of genomic changes focused on a set of cancers that appeared to have relatively similar histologic features and a high rate of cell replication, in addition to similar expression of basal cell cytokeratins or EGFR. Yet, these cancers displayed a heterogeneous pattern of chromosomal losses and gains, including different regions of high-level amplification. Interestingly, surveys of mutations in breast cancers at the nucleotide sequence level also found considerable heterogeneity in the genes mutated in various tumors, with very few genes mutated at high frequency across many tumors [33, 34]. This complexity poses significant challenges to developing a full understanding of these cancers at the molecular level.
Figure 5. High expression of candidate oncogenes in breast cancers with amplifications.
Protein expression of candidate oncogene products was measured using immunohistochemistry, as described in Materials and Methods. Panels A, C, and E represent high staining for these proteins in cases with amplification of the AKT2, cKIT, and JUND genes, respectively. Panels B (AKT2), D (cKIT), and E (JUND) represent breast cancer cases stained with the same antibodies that do not have the specific genes amplified.
Acknowledgements
The authors acknowledge support by grants U54CA091409 and P50CA088843 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murad TM. A proposed histochemical and electron microscopic classification of human breast cancer according to cell of origin. Cancer. 1971;27:288–299. doi: 10.1002/1097-0142(197102)27:2<288::aid-cncr2820270207>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 2.Dairkee SH, Ljung BM, Smith H, Hackett A. Immunolocalization of a human basal epithelium specific keratin in benign and malignant breast disease. Breast Cancer Res Treat. 1987;10:11–20. doi: 10.1007/BF01806130. [DOI] [PubMed] [Google Scholar]
- 3.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fadare O, Tavassoli FA. The phenotypic spectrum of basal-like breast cancers: a critical appraisal. Adv Anat Pathol. 2007;14:358–373. doi: 10.1097/PAP.0b013e31814b26fe. [DOI] [PubMed] [Google Scholar]
- 7.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 8.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 9.Fulford LG, Easton DF, Reis-Filho JS, Sofronis A, Gillett CE, Lakhani SR, Hanby A. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology. 2006;49:22–34. doi: 10.1111/j.1365-2559.2006.02453.x. [DOI] [PubMed] [Google Scholar]
- 10.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Bartelink H, Van de Vijver MJ. Gene expression profiling and histopathological characterization of triple negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol. 2006;208:495–506. doi: 10.1002/path.1916. [DOI] [PubMed] [Google Scholar]
- 12.Rakha EA, El-Sayed ME, Green AR, Paish EC, Lee AH, Ellis IO. Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology. 2007;50:434–438. doi: 10.1111/j.1365-2559.2007.02638.x. [DOI] [PubMed] [Google Scholar]
- 13.Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, Schoenmakers E, Viens P, Jacquemier J, Birnbaum D, Bertucci F, Chaffanet M. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 14.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 15.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Jones C, Ford E, Gillett C, Ryder K, Merrett S, Reis-Filho JS, Fulford LG, Hanby A, Lakhani SR. Molecular cytogenetic identification of subgroups of grade III invasive ductal breast carcinomas with different clinical outcomes. Clin Cancer Res. 2004;10:5988–5997. doi: 10.1158/1078-0432.CCR-03-0731. [DOI] [PubMed] [Google Scholar]
- 17.Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, Zaks TZ, Weber BL. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–R1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang ZC, Lin M, Wei LJ, Li C, Miron A, Lodeiro G, Harris L, Ramaswamy S, Tanenbaum DM, Meyerson M, Iglehart JD, Richardson A. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 2004;64:64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- 19.Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, Wan M, Dubeau L, Scambia G, Masciullo V, Ferrandina G, Benedetti Panici P, Mancuso S, Neri G, Testa JR. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 20.Simon R, Panussis S, Maurer R, Spichtin H, Glatz K, Tapia C, Mirlacher M, Rufle A, Torhorst J, Sauter G. KIT (CD117)-positive breast cancers are infrequent and lack KIT gene mutations. Clin Cancer Res. 2004;10:178–183. doi: 10.1158/1078-0432.ccr-0597-3. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 22.Puputti M, Tynninen O, Sihto H, Blom T, Maenpaa H, Isola J, Paetau A, Joensuu H, Nupponen NN. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol Cancer Res. 2006;4:927–934. doi: 10.1158/1541-7786.MCR-06-0085. [DOI] [PubMed] [Google Scholar]
- 23.Joensuu H, Puputti M, Sihto H, Tynninen O, Nupponen NN. Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J Pathol. 2005;207:224–231. doi: 10.1002/path.1823. [DOI] [PubMed] [Google Scholar]
- 24.McIntyre A, Summersgill B, Grygalewicz B, Gillis AJ, Stoop J, van Gurp RJ, Dennis N, Fisher C, Huddart R, Cooper C, Clark J, Oosterhuis JW, Looijenga LH, Shipley J. Amplification and overexpression of the KIT gene is associated with progression in the seminoma subtype of testicular germ cell tumors of adolescents and adults. Cancer Res. 2005;65:8085–8089. doi: 10.1158/0008-5472.CAN-05-0471. [DOI] [PubMed] [Google Scholar]
- 25.Berger I, Shaul Y. Structure and function of human jun-D. Oncogene. 1991;6:561–566. [PubMed] [Google Scholar]
- 26.Ryder K, Lanahan A, Perez-Albuerne E, Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci U S A. 1989;86:1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariani O, Brennetot C, Coindre JM, Gruel N, Ganem C, Delattre O, Stern MH, Aurias A. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11:361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Smith LM, Wise SC, Hendricks DT, Sabichi AL, Bos T, Reddy P, Brown PH, Birrer MJ. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene. 1999;18:6063–6070. doi: 10.1038/sj.onc.1202989. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Pu X, Shi M, Chen L, Song Y, Qian L, Yuan G, Zhang H, Yu M, Hu M, Shen B, Guo N. Critical role of c-Jun overexpression in liver metastasis of human breast cancer xenograft model. BMC Cancer. 2007;7:145. doi: 10.1186/1471-2407-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, Spichtin H, Maurer R, Mirlacher M, Kochli O, Zuber M, Dieterich H, Mross F, Wilber K, Simon R, Sauter G. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- 31.Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola J, van Diest PJ, Brandt B, Boecker W, Buerger H. Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest. 2002;82:1525–1533. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- 32.Jones C, Foschini MP, Chaggar R, Lu YJ, Wells D, Shipley JM, Eusebi V, Lakhani SR. Comparative genomic hybridization analysis of myoepithelial carcinoma of the breast. Lab Invest. 2000;80:831–836. doi: 10.1038/labinvest.3780087. [DOI] [PubMed] [Google Scholar]
- 33.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 34.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Willson S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]