Abstract
Head and neck squamous cell carcinomas (HNSCC) are commonly resistant to conventional chemotherapy drugs and exhibit overexpression of signal transducer and activator of transcription 3 (STAT3). STAT3 promotes both the proliferation and survival of HNSCC cells. Recent studies have shown that the proteasome inhibitor bortezomib shows cytotoxic activity against HNSCC in vitro and in vivo. We report that treatment of HNSCC cells with bortezomib led to up-regulation of total STAT3 protein and the phosphorylated/activated form of STAT3, as well as an increase in cellular STAT3 activity. This suggested that the ability of bortezomib to kill HNSCC cells may be blunted due to induction of STAT3, and inhibition of STAT3 may be a useful means for improving bortezomib efficacy. Indeed, forced expression of dominant-active STAT3 inhibited bortezomib-induced cell death, whereas expression of dominant-negative STAT3 served to enhance killing by this compound. In addition, specific inhibition of STAT3 with the use of a STAT3 decoy oligonucleotide resulted in enhancement of bortezomib-induced apoptosis signaling and loss of clonogenic survival. Cotreatment of HNSCC cells with bortezomib and guggulsterone, a naturally occurring compound known to inhibit STAT3 activation, led to synergistic activation of cell death and loss of clonogenic survival. In summary, these studies show that bortezomib induces the expression of active STAT3, a key growth-promoting protein in HNSCC cells. Furthermore, our findings suggest that the therapeutic activity of bortezomib against HNSCC may be markedly improved by cotreatment with molecular targeting agents against STATE3.
Introduction
Head and neck squamous cell carcinomas (HNSCC) are malignancies of the upper aerodigestive tract, including the mouth, larynx, and pharynx. HNSCC represents the sixth most common malignancy in the United States and accounts for ~500,000 new cancer cases per year worldwide (1). Surgery, radiation, and chemotherapy are the standard treatment options for HNSCC, but relapse is seen in ~50% of patients and the prognosis for patients with recurrent disease is poor (2, 3). Treatments incorporating radiation or conventional chemotherapy drugs, such as cisplatin, are associated with considerable adverse toxicities. Recently, the addition of cetuximab, an inhibitor of the epidermal growth factor receptor (EGFR), to radiation therapy was found to enhance HNSCC patient survival compared with radiation therapy alone (4). However, despite rapid approval of cetuximab for use in HNSCC by the Food and Drug Administration (FDA), improvement in patient survival with the use of this agent has been only modestly incremental. Thus, alternative strategies are needed for the treatment of HNSCC.
Efforts to devise effective targeted treatment strategies in HNSCC will rely on knowledge of the molecular defects that characterize this disease. Patient-derived primary HNSCC cells and HNSCC cell lines have been shown to overexpress a number of key signaling proteins that contribute to the enhanced growth and survival properties of these cells. HNSCC cells commonly exhibit overexpression and/or hyperactivation of EGFR (5), signal transducer and activator of transcription 3 (STAT3; refs. 6-8), Bcl-XL (9), and nuclear factor κB (10). Overexpression of EGFR has been shown to correlate with poor patient prognosis in HNSCC, whereas overexpression of the antiapoptotic protein Bcl-XL correlates with chemotherapy resistance in HNSCC patients (5, 9). STAT3 is a key downstream target of EGFR. After activation, several tyrosine residues in the cytoplasmic domain of EGFR become phosphorylated, including Tyrl068 and Tyrl086, which serve as recruitment sites for the SH2 domain of STAT3 (11). This leads to phosphorylation of STAT3 on Tyr705 by Janus-activated kinases. The phospho-STAT3 (Tyr705) undergoes dimerization, translocates to the nucleus, and induces the transcription of target genes, including the genes encoding Bcl-XL, which promotes cell survival, and cyclin D1, which promotes proliferation (12). In HNSCC, hyperactivation of STAT3 has been shown to occur through both EGFR-dependent and EGFR-independent mechanisms (8, 12, 13). Treatment with STAT3 antisense oligonucleotides or expression of a dominant-negative (DN) STAT3 mutant results in the inhibition of HNSCC cell proliferation and survival in vitro and suppression of HNSCC xenograft tumors in vivo, supporting a key role for STAT3 in this disease (6, 14, 15).
In view of the importance of STAT3 in HNSCC as well as other cancers, considerable effort has been devoted to identifying and developing molecular targeting agents directed against STAT3 and the STAT3 signaling pathway. Phosphotyrosyl peptides and related peptidomimetic compounds that bind to the STAT3 SH2 domain inhibit the growth of Src-transformed fibroblasts in vitro (16, 17). A duplex decoy oligonucleotide (STAT3 decoy) based on the STAT3 response element in the c-fos promoter potently inhibits the growth of HNSCC cells in vitro and in vivo and enhances cisplatin antitumor effects (18, 19). The STAT3 decoy is 15 bp in length, is readily taken up by cells in culture, and has shown in vivo activity against lung and breast cancer tumors in addition to HNSCC (18, 20, 21). A number of naturally occurring compounds have also been shown to inhibit constitutive and/or inducible STAT3 activation, including guggulsterone, derived from the plant Commiphora mukul and used in traditional Indian Ayurvedic medicine. Treatment with guggulsterone reduces the expression levels of phosphorylated STAT3 in multiple myeloma cells and total STAT3 in colon cancer cells while inducing cell death in both cell types (22, 23). Collectively, studies that use STAT3 inhibitors have suggested that targeting of STAT3 may provide therapeutic benefit in a variety of cancers including HNSCC.
In addition to EGFR and STAT3 targeting, recent studies have suggested potential promise in targeting the proteasome in HNSCC. The proteasome inhibitor bortezomib potently inhibits the growth of HNSCC cells in vitro and in vivo, and these effects are attributed, in part, to the modulation of expression of key signaling molecules that influence the proliferation and survival of HNSCC cells (24-27). Bortezomib is a specific inhibitor of the 26S proteasomal subunit and is approved by the FDA for use in multiple myeloma (28). A clinical trial in HNSCC patients who use bortezomib in combination with radiation has revealed moderate clinical activity (26, 29). On a cellular level, treatment of HNSCC cells with bortezomib results in generation of reactive oxygen species (25), inhibition of cell migration (30), and induction of apoptosis (24, 25, 27). These effects coincide with inhibition of nuclear factor-κB expression (24, 26) and induction of the proapoptotic proteins Bik, Bim, and Noxa (27, 31). Bortezomib treatment also results in potent up-regulation of the antiapoptotic protein Mcl-1L (27), raising the possibility that induction of antiapoptotic proteins may limit the efficacy of this anticancer agent.
In the present study, we investigated the effect of bortezomib on HNSCC cell expression of the prosurvival protein STAT3. We observed marked up-regulation of total STAT3 and phosphorylated/activated STAT3 in response to bortezomib treatment, suggesting that the activity of bortezomib may be attenuated due to up-regulation of this growth-promoting protein. In support of this, forced expression of a dominant-active (DA) STAT3 mutant led to inhibition of bortezomib-induced cell death, whereas expression of DN STAT3 or treatment with STAT3 decoy promoted bortezomib-induced death and loss of clonogenic survival. We further observed that combined treatment of HNSCC cells with bortezomib and guggulsterone yielded synergistic killing effects. We suggest that targeted inhibition of STAT3 may be a useful strategy for improving the therapeutic efficacy of bortezomib in HNSCC.
Materials and Methods
Cell Culture and Reagents
UMSCC-22A and UMSCC-22B cells (hereafter, UM-22A and UM-22B, respectively) were provided by Dr. Thomas Carey (University of Michigan, Ann Arbor, MI). UM-22A and UM-22B originated from the same patient and were derived from primary tumor and a cervical lymph node metastasis, respectively (32). Cells (1483) were kindly provided by Dr. Gary Clayman (M. D. Anderson Cancer Center, Houston, TX) and were derived from a primary tumor of the pharynx (32). Cells were maintained at 37°C and 5% CO2 in DMEM (Mediatech) containing 10% heat-inactivated fetal bovine serum (FBS) and 1× penicillin/streptomycin (Invitrogen).
Chemicals
Bortezomib was purchased from the University of Pittsburgh Cancer Institute Pharmacy, and a stock solution (20 mmol/L) was prepared in DMSO and stored at −80°C. The Z and E stereoisomers of guggulsterone were obtained from Steraloids, Inc., and 20 mmol/L stock solutions were prepared in DMSO and stored at −80°C. For guggulsterone treatments, equimolar mixtures of the Z and E stereoisomers were added to cells to achieve the final total concentration of guggulsterone.
Luciferase Reporter Assays
The cellular activity of STAT3 after treatment with bortezomib was assessed with the use of luciferase reporter assays. UM-22B cells were stably transfected with a luciferase reporter construct, pLucTKSIE (33), containing tandem copies of the STAT3-responsive hSIE element immediately upstream from a luciferase reporter gene. Stably transfected cells were selected and maintained in 0.6 mg/mL G418. For the luciferase assays, 2.5 × 106 UM-22B/pLucTKSIE cells were seeded into 10-cm plates, grown overnight, and then treated with bortezomib for varying lengths of time. Cells were harvested by cell scraping, and assays were done with the use of Dual-Luciferase Reporter Assay System kits (Promega Corp.) according to instructions provided by the manufacturer. Luciferase activities were measured with the use of an AutoLumat LB 953 luminometer (EG&G Berthold).
Cell Viability Assays
Cellular sensitivities to various treatments were determined by 3-4,5-dimethythiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) and trypan blue exclusion assays. MTS assays were done on triplicate wells with the use of CellTiter 96 AQueous One Solution Cell Proliferation Assay kits (Promega). Measurements were done at 490 nm on a VersaMax microplate reader (Molecular Devices). For trypan blue exclusion assays, cells were plated in triplicate wells and, after treatment, a minimum of 300 cells were counted from each well. The plotted data represent the mean of three independent experiments and error bars represent the SE.
Treatment with STAT3 Decoy and Mutant Control Decoy
Sense and antisense oligonucleotides containing the STAT3 decoy and the mutant control decoy were synthesized by the University of Pittsburgh DNA synthesis facility as previously described (18, 19). The sequence of the STAT3 decoy was 5′-CATTTCCCGTAAATC-3′ and 3′-GTAAAGGGCATTTAG-3′ and the sequence of the mutant control decoy was 5′-CATTTCCCTTAAATC-3′ and 3′-GTAAAGGGAATTTAG-5′. Equimolar concentrations of sense and antisense strands were mixed and annealed to generate 1 μmol/L stocks that were stored at −20°C as described previously (19). For transfection into cells, UM-22B cells were first seeded at 4 × 104 per well in 24-well trays. After overnight growth, cells were transfected with STAT3 decoy (6.3 nmol/L) or mutant control decoy (6.3 nmol/L) with the use of Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 4 h, the transfection medium was removed and replaced with fresh DMEM containing 10% heat-inactivated FBS and antibiotics.
Expression of DA or DN STAT3 in HNSCC Cells
UM-22B cells stably transfected with the pLucTKSIE reporter construct were seeded at 2.5 × 105 per well in six-well plates, grown overnight, and then transfected with empty vector (pRcCMV/Neo) or constructs encoding DA STAT3 (STAT3C; ref. 34) or DN STAT3 (STAT3F; ref. 35). For experiments measuring expression of the pLucTKSIE reporter, all cells were also cotransfected with phRL-TK (Promega), which constitutively expresses Renilla luciferase, and cells were normalized for expression of Renilla luciferase. Transfections were done with the use of Lipofectamine 2000 (Invitrogen). After 6 h, the transfection medium was replaced by medium containing 10% FBS and antibiotics, and the transfected cells were left to grow for an additional 48 h. Cells were then either left untreated or treated with 10 nmol/L bortezomib for 24 h, followed by luciferase, MTS, or trypan blue exclusion assays or immunoblotting for poly(ADP)ribose polymerase (PARP).
Colony-Forming Assays
UM-22B cells were seeded at 2.5 × 105 per plate in six-well plates and allowed to grow for 24 h. For experiments combining STAT3 decoy (or mutant control decoy) and bortezomib, cells were transfected for 4 h with 6.3 nmol/L STAT3 decoy or mutant control decoy, followed by replacement of the transfection medium with fresh medium and treatment for an additional 24 h in the absence or presence of bortezomib. For experiments combining bortezomib and guggulsterone, cells were treated simultaneously with both compounds for 48 h. After treatment, cells were rinsed with PBS, collected by trypsinization, subjected to centrifugation, washed again in PBS, replated in six-well plates, and allowed to grow for 10 to 14 d. Colonies were stained for 30 min with the use of a solution of 0.5% crystal violet in 6% glutaraldehyde and washed repeatedly with water, and colonies composed of >50 cells were counted.
Immunoblotting
Whole-cell lysates were prepared by washing adherent cells once in cold PBS, followed by detachment by cell scraping and centrifugation for 5 minat 4°C. The cell pellets were washed once in cold PBS and then resuspended in lysis buffer [50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 0.1% SDS, 1% NP40, 1.5 mmol/L phenylmethylsulfonyl fluoride, 3 μg/mL leupeptin, 20 μg/mL aprotinin]. After incubation on ice for 1 h, the lysates were clarified by centrifugation for 10 min at 13,000 rpm and 4°C, and the protein concentrations were determined with the use of Bio-Rad Protein Assay dye concentrate (Bio-Rad Laboratories). Proteins (40 μg/lane) were electrophoresed on 10% SDS-PAGE gels and transferred onto nitrocellulose membranes for 6 h at 40 V and 4°C. Membranes were blocked for 1 h at room temperature in 5% nonfat dry milk in TBST [10 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 0.1% Tween 20], followed by washing with TBST. The washed filters were incubated with primary antibodies (in 5% nonfat dry milk in TBST) overnight at 4°C. After washing with TBST, incubation with secondary antibody (in 1% nonfat dry milk in TBST) was allowed to proceed for 1 h at room temperature, followed by additional rinsing with TBST. Blots were developed with the use of Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer) and with exposure to BioMax MR film (Kodak). The following antibodies were used for immunoblotting: anti–total STAT3 (clone #9132), anti–phospho-STAT3 (Tyr705; clone #9131), and anti-PARP (Cell Signaling Technology, Inc.); anti–caspase-3 (Stressgen Bioreagents Corp.); and anti–β-actin (Sigma). Horseradish peroxidase–conjugated goat anti-rabbit (Sigma) and goat anti-mouse (Promega) were used as secondary antibodies.
Assessment of Synergy
Cells were seeded at 5,000 per well in triplicate wells in 96-well plates and allowed to grow for 24 h. The cells were then treated for 48 h at 37°C with bortezomib alone, guggulsterone alone, or varying doses of a constant ratio of bortezomib plus gugglesterone. After treatment, MTS assays were done, and data were plotted as the percent of cell metabolic activity relative to untreated cells. Combination indexes were calculated with the use of CalcuSyn v2 software (Biosoft) according to the method of Chou and Talalay (36).
Statistical Analysis
Results were analyzed for statistical significance with the use of SigmaStat software (Jandel Scientific Software). One-way ANOVA followed by Student-Newman-Keuls tests was used for comparison between groups. P < 0.05 was considered statisticaly significant.
Results
Bortezomib Induces STAT3 in HNSCC Cells
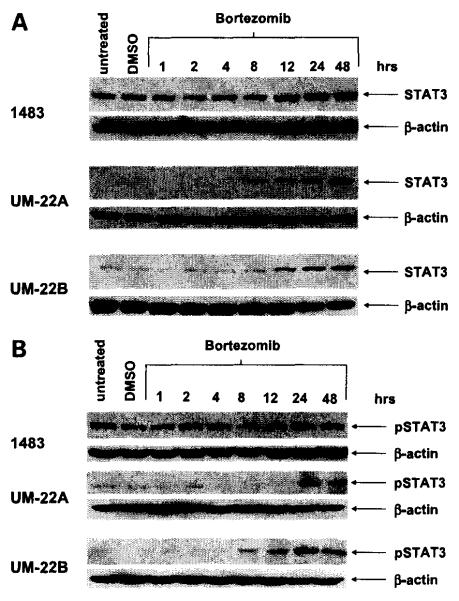
To determine the effect of proteasome inhibition on STAT3 expression in HNSCC cells, we treated three HNSCC cell lines with bortezomib, followed by immunoblotting for total STAT3 protein (Fig. 1A). Cells (1483, UM-22A, and UM-22B) were treated with bortezomib concentrations corresponding to the IC50 values for each cell line (80 nmol/L for 1483, and 20 nmol/L for UM-22A and UM-22B; ref. 27). In all three cell lines, total STAT3 protein was up-regulated within 8 to 12 hours of bortezomib treatment, relative to that seen in untreated cells or control cells treated with 0.1% DMSO (vehicle). The up-regulation of total STAT3 protein was accompanied by increased expression of phosphorylated STAT3, as detected by immunoblotting with anti–phospho-STAT3 (Tyr705; Fig. 1B). Phosphorylation of STAT3 on Tyr705 is known to promote dimerization of STAT3 and represents a critical step in the activation of the protein (12). Thus, bortezomib treatment resulted in increased expression of the active/phosphorylated form of STAT3. Because 1483 cells expressed higher basal levels of total and phospho-STAT3 and exhibited greater resistance to bortezomib (IC50, 80 nmol/L) compared with UM-22A and UM-22B cells (IC50s, 20 nmol/L), we also examined bortezomib sensitivity in a larger panel (n = 9) of HNSCC cell lines expressing varying basal levels of total and phospho-STAT3 (Supplementary Fig. S1). These experiments failed to reveal a correlation between resistance to bortezomib and basal levels of total or phospho-STAT3.
Figure 1.
Bortezomib induces STAT3 in HNSCC cells. A, cells (1483, UM-22A, and UM-22B) were left untreated or were treated for varying lengths of time with bortezomib 11483: 80 nmol/L; UM-22A and UM-22B: 20 nmol/L). As a control, cells were treated with 0.1 % DMSO. After treatment, whole-cell lysates were subjected to immunoblotting for total STAT3 protein, and probing for β-actin was used to show equal loading. B, cells were treated with bortezomib for the indicated lengths of time, followed by immunoblotting with anti–phospho-STAT3 (Tyr705). Experiments were done thrice with similar results each time.
The induction of phospho-STAT3 by bortezomib led us to predict that the total cellular activity of STAT3 transcription factor might be induced. Cellular STAT3 activity was measured with the use of a luciferase-based STAT3 reporter construct. For these studies, UM-22B cells were stably transfected with pLucTKSIE (33), containing tandem copies of the STAT3-responsive hSIE element cloned directly upstream from a luciferase reporter gene. The stably transfected UM-22B/pLucTKSIE cells were then treated with 20 nmol/L bortezomib for varying lengths of time, followed by luciferase assays (Fig. 2). In agreement with results from immunoblotting experiments (Fig. 1), bortezomib markedly induced cellular STAT3 activity. Peak STAT3 activity was seen at 36 hours and corresponded to an 11.4-fold increase relative to untreated UM-22B/pLucTKSIE control cells.
Figure 2.

Bortezomib induces cellular STAT3 activity. UM-22B cells were stably transfected with pLucTKSIE, which contains two copies of the STAT3-responsive hSIE element upstream from a luciferase reporter gene. The UM-22B/pLucTKSIE cells were treated with bortezomib (20 nmol/L) for the indicated lengths of time, followed by luciferase assays. The data are plotted as fold induction of luciferase activity relative to untreated cells. Columns, mean of three independent experiments; bars, SE.
DA STAT3 Inhibits Bortezomib-lnduced Apoptosis whereas DN STAT3 Enhances Bortezomib Killing of HNSCC Cells
Because STAT3 is known to promote the proliferation and survival of HNSCC cells, we reasoned that the induction of STAT3 by bortezomib may act to blunt the ability of this agent to kill HNSCC cells. To investigate this possibility we examined the effect of forced expression of DA STAT3 (34) and DN STAT3 (35) on sensitivity to bortezomib. Prior studies have shown that forced expression of DA STAT3 promotes HNSCC proliferation in vitro and rapid tumor growth in vivo (13), whereas expression of DN STAT3 inhibits the proliferation and survival of HNSCC cells in vitro and in vivo (14, 15). In Fig. 3A, UM-22B/pLucTKSIE cells were transiently transfected with empty vector (RcCMV/Neo), DA STAT3 construct (STAT3C; ref. 34), or DN STAT3 construct (STAT3F; ref. 35). Simultaneously, the cells were cotransfected with a construct that constitutively expresses Renilla luciferase. After normalization for Renilla luciferase activity, the transfected cells were treated for 24 hours in the absence or presence of bortezomib. Immunoblotting of the transiently transfected cells for total STAT3 protein showed roughly equivalent levels of the exogenously expressed DA and DN STAT3 proteins (not shown). As expected, luciferase reporter assays showed bortezomib induction of STAT3 activity in the vector-transfected cells. Additionally, expression of DA STAT3 led to enhanced expression of the luciferase reporter in the absence of bortezomib and even further induction in the presence of bortezomib. On the other hand, DN STAT3 inhibited expression of the luciferase reporter after treatment with bortezomib. These results confirmed the ability of DA STAT3 to enhance cellular STAT3 activity and the ability of DN STAT3 to inhibit the cellular STAT3 induced by bortezomib. In separate experiments, UM-22B and 1483 cells expressing DA STAT3 or DN STAT3 were examined for cellular sensitivity to bortezomib with the use of either MTS assays (Fig. 3B) or trypan blue exclusion assays (Fig. 3C). With the use of either assay, expression of DA STAT3 was found to confer modest resistance to bortezomib-induced cell death relative to vector-transfected control cells. By contrast, DN substantially enhanced sensitivity to bortezomib-induced cell death.
Figure 3.
DA STAT3 inhibits bortezomib-induced cell death whereas DN STAT3 enhances bortezomib action. A, UM-22B cells stably expressing the pLucTKSIE STAT3 reporter construct were transiently transfected with RcCMV/Neo (empty vector) or constructs encoding DA STAT3 or DN STAT3. Simultaneously, the cells were cotransfected with a construct (phRL-TK) directing constitutive expression of Renilla luciferase. After normalization for Renilla luciferase activity, the transfected cells were then left untreated or were treated for 24 h with 10 nmol/L bortezomib. Luciferase assays were done to verify enhancement of cellular STAT3 activity by DA STAT3 and inhibition by DN STAT3. The fold induction of luciferase activity relative to untreated empty vector cells is shown. Columns, mean of three independent experiments; bars, SE. B, UM-22B or 1483 cells transfected with empty vector or constructs encoding DA STAT3 or DN STAT3 were left untreated or were treated for 24 h with 10 nmol/L bortezomib. After treatment, MTS assays were done on triplicate wells, and cellular metabolic activities were plotted relative to untreated empty vector cells. Bars, SD of triplicate wells. Similar resultswere obtained in two independent experiments. C, UM-22B or 1483 cells were transfected and treated as in B, followed by determination of cell viabilities with the use of trypan blue exclusion assays. For each data point, triplicate wells were plated, and a minimum of 300 cells per well were examined. Columns, mean of three independent experiments; bars, SE. D, UM-22B cells transfected with the indicated constructs were left untreated or were treated for 24 h with 10 nmol/L bortezomib. Immunoblotting with anti-PARP was done to detect cleavage of full-length PARP (116 kDa) to an 89-kDa fragment. Probing with anti-β-actin was used to verify equal protein loading. The experiment was done thrice with similar results.
We next examined the effects of DA STAT3 and DN STAT3 on bortezomib-induced apoptosis signaling. UM-22B cells expressing the DA or DN proteins were treated in the absence or presence of bortezomib for 24 hours, followed by immunoblotting to detect cleavage of PARP protein (Fig. 3D). Treatment with bortezomib led to PARP cleavage in vector-transfected control cells, an effect that was substantially reduced in cells expressing DA STAT3. Cells expressing DN STAT3, however, exhibited increased PARP cleavage relative to vector-transfected controls, consistent with the ability of DN STAT3 to confer sensitivity to bortezomib.
STAT3 Decoy Enhances Bortezomib-Induced Apoptosis and Loss of Clonogenic Survival
The findings that DA STAT3 promoted resistance and DN STAT3 promoted sensitivity to bortezomib (Fig. 3B–D) supported the contention that the up-regulation of cellular STAT3 that occurs during treatment with bortezomib (as depicted in Fig. 1) likely serves to blunt the efficacy of this compound against HNSCC cells. Moreover, these findings raised the possibility that inhibition of STAT3 with the use of molecular targeting agents may be a useful means for improving the potency of bortezomib. To investigate this possibility, we evaluated the effect of cotreatment with bortezomib and STAT3 decoy. The STAT3 decoy is a 15-bp duplex oligonucleotide corresponding to the STAT3 response element in the c-fos promoter (18). The STAT3 decoy has been shown to directly bind to activated STAT3, and treatment with STAT3 decoy has been shown to inhibit cellular STAT3 activity, expression of STAT3 target genes, and proliferation and survival of HNSCC cells in vitro and in vivo (18, 19). UM-22B cells were subjected to transfection with STAT3 decoy, followed by treatment in the absence or presence of bortezomib (Fig. 4). As controls, UM-22B cells were transfected with a mutant control decoy (18) containing a single, inactivating point mutation, followed by treatment in the absence or presence of bortezomib. Additionally, mock-transfected cells were treated with either 0.1% DMSO or bortezomib alone. After treatment, all cells were analyzed in clonogenic survival assays. Treatment with STAT3 decoy alone or bortezomib alone resulted in readily detectable loss of clonogenic survival, whereas treatment with mutant control decoy exhibited no apparent effect. Importantly, cotreatment with STAT3 decoy and bortezomib resulted in enhanced loss of clonogenic survival compared with treatment with either agent alone (P < 0.05). By contrast, cotreatment with mutant control decoy and bortezomib failed to increase the loss of clonogenic survival seen with bortezomib alone. Similar results were obtained when cell survival was assessed by MTS assays (not shown).
Figure 4.
STAT3 decoy enhances bortezomib-induced loss of clonogenic survival. UM-22B cells were mock transfected or were transfected for 4 h with 6.3 nmol/L of mutant control decoy or 6.3 nmol/L of STAT3 decoy. Mock-transfected cells were subsequently incubated for 24 h with 0.1% DMSO or 10 nmol/L bortezomib. Cells transfected with mutant control decoy or STAT3 decoy were incubated for 24 h in the absence or presence of 10 nmol/L bortezomib. After treatment, cells were washed twice in PBS, detached from the plate, diluted, and replated in medium containing 10% FBS in six-well plates. After 2 wk, cells were stained with crystal violet. Top, a representative experiment; bottom, the colonies consisting of >50 cells, in each well, were counted. Data represent the percent of colonies relative to mock-transfected, DMSO-treated controls. Columns, mean of four independent experiments; bars, SE.
To determine whether cotreatment with STAT3 decoy and bortezomib would result in enhanced activation of apoptosis signaling, immunoblotting was done to detect cleaved PARP protein and the cleaved/activated form of caspase-3 (Fig. 5). Cotreatment with STAT3 decoy and bortezomib resulted in enhanced PARP cleavage and cleavage/activation of caspase-3, relative to that seen after treatment with STAT3 decoy or bortezomib alone. Again, cotreatment with mutant control decoy and bortezomib did not enhance the activity of bortezomib alone.
Figure 5.

STAT3 decoy enhances bortezomib-induced apoptosis signaling. UM-22B cells were subjected to mock transfection or transfection with mutant control decoy or STAT3 decoy as described in Fig. 4. Mock-transfected cells were then incubated for 24 h with 0.1% DMSO or 10 nmol/L bortezomib, whereas cells transfected with mutant control decoy or STAT3 decoy were incubated for 24 h in the absence or presence of 10 nmol/L bortezomib. The treated cells were subjected to immunoblotting with antibodies directed against PARP or the active form of caspase-3. Probing with anti-β-actin was used to verify equal loading. Similar results were obtained in three independent experiments.
Guggulsterone Synergizes with Bortezomib to Promote Apoptosis and Loss of Clonogenic Survival in HNSCC Cells
To further investigate whether inhibition of STAT3 can be used to enhance the killing activity of bortezomib against HNSCC cells, we cotreated cells with bortezomib and the plant-derived compound guggulsterone (Fig. 6). Recent studies have shown that guggulsterone inhibits constitutive STAT3 activation in HNSCC cells and constitutive and interleukin-6 (IL-6)–induced STAT3 activation in multiple myeloma cells (23). Therefore, UM-22A and UM-22B cells were treated for 48 hours with varying doses of bortezomib alone, guggulsterone alone, or a constant ratio of bortzomib plus guggulsterone (Fig. 6A and B), followed by MTS assays. To assess potential synergism between these agents, combination indexes were calculated according to the method of Chou and Talalay (36). As depicted, the combination of bortezomib plus guggulsterone resulted in combination indexes <1.0 in both cell lines, indicative of synergism.
Figure 6.
Guggulsterone synergizes with bortezomib against HNSCC cells. UM-22A (A) and UM-22B (B) cells were seeded at 5,000 per well in 96-well plates, grown overnight, and then treated for 48 h with the indicated concentrations of bortezomib alone, guggulsterone alone, or bortezomib plus guggulsterone. MTS assays were done, and the data were plotted as the percent of metabolic activity relative to untreated control cells. Points, mean of triplicate wells; bars, SD. Combination indexes were determined as described in Materials and Methods. Similar results were obtained in four independent experiments. C, UM-22B cells were left untreated or were treated for 48 h with 0.1 % DMSO, 3.5 nmol/L bortezomib alone, 10 μmol/L guggulsterone alone, or bortezomib (3.5 nmol/L) plus guggulsterone (10 μmol/L). After treatment, clonogenic assays were done and quantified as described for Fig. 4. Columns, mean of three independent experiments; bars, SE. D, UM-22B cells were left untreated or were treated for 24 h with 0.1 % DMSO, 3.5 nmol/L bortezomib alone, 10 μmol/L guggulsterone alone, or bortezomib (3.5 nmol/L) plus guggulsterone (10 μmol/L). Immunoblotting was done to identify cleaved PARP and the active/processed form of caspase-3. The experiment was done thrice with similar results.
To confirm the observed synergism between bortezomib and guggulsterone, clonogenic survival assays were done. UM-22B cells were left untreated or were treated for 48 hours with 0.1% DMSO, bortezomib alone, guggulsterone alone, or bortezomib plus guggulsterone, followed by determination of survival in colony-forming assays (Fig. 6C). In view of the potent synergism that had been detected in MTS assays (Fig. 6A and B), clonogenic survival studies used a lower concentration of bortezomib (3.5 nmol/L) than was used in earlier experiments. At this concentration, treatment with bortezomib alone resulted in only a 5% loss in clonogenic survival relative to untreated cells. Treatment with guggulsterone (10 μmol/L) alone led to an ~31% loss in clonogenic survival. Strikingly, however, the combination of bortezomib plus guggulsterone yielded a nearly 94% loss of clonogenic survival, markedly greater than that observed with either agent alone (P < 0.05).
The effect of the bortezomib/guggulsterone combination on the induction of apoptosis signaling was again evaluated by immunoblotting for PARP and caspase-3 cleavage (Fig. 6D). Treatment for 24 hours with low concentrations of bortezomib alone or guggulsterone alone failed to result in detectable PARP cleavage or appearance of the cleaved/activated form of caspase-3. By contrast, cleavage of PARP and caspase-3 was clearly detected in cells treated with the bortezomib/guggulsterone combination. Treatment with guggulsterone also was found to abrogate both basal and bortezomib-induced expression of antiapoptotic Bcl-XL, the product of a known STAT3 target gene (Supplementary Fig. S2). Taken together, these results indicate that combined treatment with bortezomib and guggulsterone can be used to achieve synergistic killing of HNSCC cells.
Discussion
HNSCC is a prevalent form of cancer associated with multiple sites in the upper aerodigestive tract. Over the past several decades, there has been little improvement in the 5-year survival rates for HNSCC patients (2). Relapse is common, and recurrent HNSCC tumors are typically resistant to conventional chemotherapy drugs (3). The use of cetuximab provides only modest clinical benefit in this disease (4). These facts underscore the necessity of identifying and developing new strategies for treating HNSCC. One such strategy may involve the use of proteasome inhibitors such as bortezomib. Treatment with bortezomib provides significant therapeutic benefit in multiple myeloma, in which this agent has been approved for use by the FDA (28). Although less success has been achieved against solid tumor malignancies, recent preclinical and clinical evidence shows bortezomib activity against HNSCC. Bortezomib is potently proapoptotic against HNSCC cells in vitro and inhibits the growth of human HNSCC xenograft tumors grown in mice (24-27). Clinical evaluation of bortezomib in combination with radiation in recurrent HNSCC resulted in sustained partial responses in 5 of 18 patients and temporary responses or disease stabilization in additional patients (26, 29). Based on these findings, it is possible that optimization of bortezomib efficacy may yield substantial improvement in the therapy of this disease.
We have previously shown that bortezomib treatment of HNSCC cells results in marked up-regulation of the proapoptotic Bcl-2 family members Bik and Bim (27). Inhibition of Bik and Bim up-regulationwith the use of small interfering RNA partially attenuated bortezomib-induced apoptosis. Similarly, Fribley et al. (31) have reported up-regulation of proapoptotic Noxa after bortezomib treatment and a role for Noxa in mediating bortezomib-induced HNSCC cell death. Bortezomib also inhibits the activity of constitutively active nuclear factor κB, a prosurvival protein in HNSCC (10, 24). Collectively, the ability of bortezomib to induce the expression of proapoptotic proteins and suppress the expression and/or activity of prosurvival proteins likely accounts, in large part, for the apoptosis-inducing activity of this compound in HNSCC. However, we have also reported dramatic up-regulation of Mcl-1L in bortezomib-treated HNSCC cells (27). Mcl-1L is an antiapoptotic member of the Bcl-2 protein family and a known proteasomal substrate (37). Thus, bortezomib also may induce the expression of prosurvival proteins that act to inhibit the killing activity of this compound. The identification of these bortezomib-induced prosurvival proteins will provide attractive targets for strategies aimed at improving bortezomib efficacy.
In this study, we provide evidence that bortezomib induces the expression of STAT3 protein in HNSCC cells. Presumably, this occurs through inhibition of proteasomal degradation. Ubiquitination and proteasome-dependent degradation of STAT3 have been reported in the IL-6–dependent T-cell line KT-3 (38). Bortezomib-induced up-regulation of total STAT3 protein in HNSCC cells coincided with up-regulation of phosphorylated/activated STAT3 and an increase in cellular STAT3 activity. These results contrast with those seen in multiple myeloma, in which treatment with bortezomib leads to inhibition of STAT3 activation/phosphorylation through disruption of IL-6 signaling (39, 40). Thus, the effect of proteasome inhibition on STAT3 expression and activation may depend on the cell type or malignancy being studied.
Because previous studies have revealed important roles for STAT3 in the proliferation and survival of HNSCC cells in vitro and in vivo (6, 14, 15, 41-43), we hypothesized that bortezomib-induced up-regulation of STAT3 may confound the killing activity of this anticancer agent in HNSCC cells. The ability of a DA STAT3 mutant to suppress bortezomib-induced death supported this hypothesis. Further support was obtained when it was determined that inhibition of cellular STAT3 activity through expression of DN STAT3 mutant resulted in enhancement of bortezomib-induced apoptosis signaling and cell death. Moreover, these findings suggested that targeted inhibition of STAT3 might be an effective means for improving bortezomib killing of HNSCC cells.
Molecular targeting of STAT3 was achieved with the use of the STAT3 decoy. In addition to inhibiting cellular STAT3 activity and the growth of HNSCC cells in vitro and in vivo (18, 19), the STAT3 decoy has shown antitumor properties against preclinical models of breast and lung cancer (20, 21). Treatment of HNSCC cells with STAT3 decoy, but not mutant control decoy, enhanced bortezomib-induced killing, confirming results obtained with the DN STAT3 mutant.
Further confirmation that targeted inhibition of STAT3 can be used to enhance bortezomib efficacy was provided by cotreating HNSCC cells with guggulsterone plus bortezomib. Guggulsterone is a plant-derived compound and is one of a number of naturally occurring compounds that have been shown to inhibit constitutive and/or inducible STAT3 activation, and include cucurbitacin I (44), curcumin (45), resveratrol (46), capsaicin (47), ursolic acid (48), and galiellalactone (49). Cotreatment of HNSCC cells with guggulsterone plus bortezomib resulted in synergistic induction of apoptosis signaling and loss of clonogenic survival. However, as with other naturally occurring compounds that inhibit STAT3 activation, guggulsterone is not entirely specific for STAT3 or the STAT3 pathway. Indeed, guggulsterone has been shown to antagonize the farnesoid X receptor (50) and also activates signaling through c-jun NH2-terminal kinase (51, 52). Nonetheless, the potent synergy that was observed with the guggulsterone/bortezomib combination supports the development of a therapeutic strategy involving STAT3 inhibition as a means of optimizing bortezomib activity. In the case of the guggulsterone/bortezomib combination, further optimization may come by identifying which stereoisomer of guggulsterone is important for promoting HNSCC cell death. Guggulsterone is typically isolated as a mixture of Z and E stereoisomers, and our studies used equimolar mixtures of the two stereoisomers. However, Ahn et al. (23) have indicated that inhibition of STAT3 activation can be attributed to the Z, but not the E, stereoisomer.
Future efforts aimed at co-targeting the proteasome and STAT3 also may incorporate small-molecule inhibitors of STAT3 that are identified through high-throughput screening. In this regard, Siddiquee et al. (53) have used virtual screening to identify the small organic molecule NSC 74859 (S3I-201). NSC 74859 inhibits STAT3 DNA-binding activity with an IC50 of 86 μmol/L and inhibits the in vitro and in vivo growth of breast cancer cells harboring constitutively active STAT3. Further refinement of NSC 74859 or isolation of new STAT3 small-molecule inhibitors will hopefully result in highly specific and potent STAT3 inhibitors that can be given as systemic agents.
In conclusion, we have shown that bortezomib induces the expression of activated STAT3 in HNSCC cells, and this diminishes the killing activity of bortezomib against these cells. Inhibition of cellular STAT3 activity enhanced the ability of bortezomib to kill HNSCC cells, with potent synergy observed with the guggulsterone/bortezomib combination. These studies suggest a novel strategy for improving therapeutic efficacy in HNSCC patients.
Supplementary Material
Acknowledgments
Grant support: NIH grants P50 CA097190 and R01 CA101840.
Footnotes
Note: Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 3.Gibson MK, Forastiere AA. Reassessment of the role of induction chemotherapy for head and neck cancer. Lancet Oncol. 2006;7:565–74. doi: 10.1016/S1470-2045(06)70757-4. [DOI] [PubMed] [Google Scholar]
- 4.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 5.Grandis J Rubin, Melhem MF, Gooding WE, et al. Levels of TGF-α and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Nati Cancer Inst. 1998;90:824–32. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 6.Grandis JR, Drenning SD, Zeng Q, et al. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Nati Acad Sci U S A. 2000;97:4227–32. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda M, Suzui M, Yasumatu R, et al. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–5. [PubMed] [Google Scholar]
- 8.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp 130 cytokine system. Cancer Res. 2003;63:2948–56. [PubMed] [Google Scholar]
- 9.Trask DK, Wolf GT, Bradford CR, et al. Expression of Bcl-2 family proteins in advanced laryngeal squamous cell carcinoma: correlation with response to chemotherapy and organ preservation. Laryngoscope. 2002;112:638–44. doi: 10.1097/00005537-200204000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Ondrey FG, Dong G, Sunwoo J, et al. Constitutive activation of transcription factors NF-κB, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and proangiogenic cytokines. Mol Carcinog. 1999;26:119–29. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 11.Shao H, Cheng HY, Cook RG, Tweardy DJ. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63:3923–30. [PubMed] [Google Scholar]
- 12.Song JI, Grandis JR. STAT signaling in head and neck cancer. Oncogene. 2000;19:2489–95. doi: 10.1038/sj.onc.1203483. [DOI] [PubMed] [Google Scholar]
- 13.Kijima T, Niwa H, Steinman RA, et al. STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 2002;13:355–62. [PubMed] [Google Scholar]
- 14.Grandis JR, Drenning SD, Chakraborty A, et al. Requirement of Stat3 but not Statl activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102:1385–92. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandis J Rubin, Zeng Q, Drenning SD. Epidermal growth factor receptor-mediated stat3 signaling blocks apoptosis in head and neck cancer. Laryngoscope. 2000;110:868–74. doi: 10.1097/00005537-200005000-00016. [DOI] [PubMed] [Google Scholar]
- 16.Turkson J, Ryan D, Kim JS, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 17.Turkson J, Kim JS, Zhang S, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–9. [PubMed] [Google Scholar]
- 18.Leong PL, Andrews GA, Johnson DE, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Nati Acad Sci U S A. 2003;100:4138–43. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications forcancer therapy. Oncogene. 2005;24:970–9. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhang J, Wang L, Wei H, Tian Z. Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human lung cancer in xenograft mice. BMC Cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Yao Z, Liu S, Tang H, Yan X. An oligonucleotide decoy for Stat3 activates the immune response of macrophages to breast cancer. Immunobiology. 2006;211:199–209. doi: 10.1016/j.imbio.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Kim ES, Hong SY, Lee HK, et al. Guggulsterone inhibits angiogenesis by blocking STAT3 and VEGF expression in colon cancer cells. Oncol Rep. 2008;20:1321–7. [PubMed] [Google Scholar]
- 23.Ahn KS, Sethi G, Sung B, Goel A, Ralhan R, Aggarwal BB. Guggulsterone, a farnesoid X receptor antagonist, inhibits constitutive and inducible STAT3 activation through induction of a protein tyrosine phosphatase SHP-1. Cancer Res. 2008;68:4406–15. doi: 10.1158/0008-5472.CAN-07-6696. [DOI] [PubMed] [Google Scholar]
- 24.Sunwoo JB, Chen Z, Dong G, et al. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-κB, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin Cancer Res. 2001;7:1419–28. [PubMed] [Google Scholar]
- 25.Fribley A, Zeng Q, Wang CY. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24:9695–704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Waes C, Chang AA, Lebowitz PF, et al. Inhibition of nuclear factor-κB and target genes during combined therapy with proteasome inhibitor bortezomib and reirradiation in patients with recurrent head-and-neck squamous cell carcinoma. Int J Rad Oncol Biol Phys. 2005;63:1400–12. doi: 10.1016/j.ijrobp.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Li R, Grandis JR, Johnson DE. Bortezomib induces apoptosis via Bim and Bik up-regulation and synergizes with cisplatin in the killing of head and neck squamous cell carcinoma cells. Mol Cancer Ther. 2008;7:1647–55. doi: 10.1158/1535-7163.MCT-07-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 29.Allen C, Saigal K, Nottingham L, Arun P, Chen Z, Van Waes C. Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-κB subunits in head and neck cancer. Clin Cancer Res. 2008;14:4175–85. doi: 10.1158/1078-0432.CCR-07-4470. [DOI] [PubMed] [Google Scholar]
- 30.Lorch JH, Thomas TO, Schmoll HJ. Bortezomib inhibits cell-cell adhesion and cell migration and enhances epidermal growth factor receptor inhibitor-induced cell death in squamous cell cancer. Cancer Res. 2007;67:727–34. doi: 10.1158/0008-5472.CAN-06-2162. [DOI] [PubMed] [Google Scholar]
- 31.Fribley AM, Evenchik B, Zeng Q, et al. Proteasome inhibitor PS-341 induces apoptosis in cisplatin-resistant squamous cell carcinoma cells by induction of Noxa. J Biol Chem. 2006;281:31440–7. doi: 10.1074/jbc.M604356200. [DOI] [PubMed] [Google Scholar]
- 32.Lin CJ, Grandis JR, Carey TE, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–88. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 33.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 35.Tokumaru S, Sayama K, Yamasaki K, et al. SOCS3/CIS3 negative regulation of STAT3 in HGF-induced keratinocyte migration. Biochem Biophys Res Commun. 2005;327:100–5. doi: 10.1016/j.bbrc.2004.11.145. [DOI] [PubMed] [Google Scholar]
- 36.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 37.Opferman JT. Unraveling MCL-1 degradation. Cell Death Differ. 2006;13:1260–2. doi: 10.1038/sj.cdd.4401978. [DOI] [PubMed] [Google Scholar]
- 38.Daino H, Matsumura I, Takada K, et al. Induction of apoptosis by extracellular ubiquitin in human hematopoietic cells: possible involvement of STAT3 degradation by proteasome pathway in interleukin 6-dependent hematopoietic cells. Blood. 2000;95:2577–85. [PubMed] [Google Scholar]
- 39.Hideshima T, Chauhan D, Hayashi T, et al. Proteasome inhibitor PS-341 abrogates IL-6 triggered signaling cascades via caspase-dependent downregulation of gp130 in multiple myeloma. Oncogene. 2003;22:8386–93. doi: 10.1038/sj.onc.1207170. [DOI] [PubMed] [Google Scholar]
- 40.Voorhees PM, Chen Q, Kuhn DJ, et al. Inhibition of interleukin-6 signaling with CNTO 328 enhances the activity of bortezomib in preclinical models of multiple myeloma. Clin Cancer Res. 2007;13:6469–78. doi: 10.1158/1078-0432.CCR-07-1293. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Ricker JL, Malhotra PS, et al. Differential bortezomib sensitivity in head and neck cancer lines corresponds to proteasome, nuclear factor-κB and activator protein-1 related mechanisms. Mol Cancer Ther. 2008;7:1949–60. doi: 10.1158/1535-7163.MCT-07-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TL, Yeh J, Friedman J, et al. A signal network involving coactivated NF-κB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. Int J Cancer. 2008;122:1987–98. doi: 10.1002/ijc.23324. [DOI] [PubMed] [Google Scholar]
- 43.Lee TL, Yeh J, Van Waes C, Chen Z. Epigenetic modification of SOCS-1 differentially regulates STAT3 activation in response to interleukin-6 receptor and epidermal growth factor receptor signaling through JAK and/or MEK in head and neck squamous cell carcinomas. Mol Cancer Ther. 2006;5:8–19. doi: 10.1158/1535-7163.MCT-05-0069. [DOI] [PubMed] [Google Scholar]
- 44.Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–9. [PubMed] [Google Scholar]
- 45.Bharti AC, Donato N, Aggarwal BB. Curcumin (diferuloylmethanel inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol. 2003;171:3863–71. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 46.Kotha A, Sekharam M, Cilenti L, et al. Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein. Mol Cancer Ther. 2006;5:621–9. doi: 10.1158/1535-7163.MCT-05-0268. [DOI] [PubMed] [Google Scholar]
- 47.Bhutani M, Pathak AK, Nair AS, et al. Capsaicin is a novel blocker of constitutive and interleukin-6-inducible STAT3 activation. Clin Cancer Res. 2007;13:3024–32. doi: 10.1158/1078-0432.CCR-06-2575. [DOI] [PubMed] [Google Scholar]
- 48.Pathak AK, Bhutani M, Nair AS, et al. Ursolic acid inhibits STAT3 activation pathway leading to suppression of proliferation and chemosensitization of human multiple myeloma cells. Mol Cancer Res. 2007;5:943–55. doi: 10.1158/1541-7786.MCR-06-0348. [DOI] [PubMed] [Google Scholar]
- 49.Hellsten R, Johansson M, Dahlman A, Dizeyi N, Sterner O, Bjartell A. Galiellalactone is a novel therapeutic candidate against hormone-refractory prostate cancer expressing activated Stat3. Prostate. 2008;68:269–80. doi: 10.1002/pros.20699. [DOI] [PubMed] [Google Scholar]
- 50.Urizar NL, Liverman AB, Dodds DT, et al. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science. 2002;296:1703–6. doi: 10.1126/science.1072891. [DOI] [PubMed] [Google Scholar]
- 51.Singh SV, Choi S, Zeng Y, Hahm ER, Xiao D. Guggulsterone-induced apoptosis in human prostate cancer cells is caused by reactive oxygen intermediate dependent activation of c-Jun NH2-terminal kinase. Cancer Res. 2007;67:7439–49. doi: 10.1158/0008-5472.CAN-07-0120. [DOI] [PubMed] [Google Scholar]
- 52.Shishodia S, Sethi G, Ahn KS, Aggarwal BB. Guggulsterone inhibits tumor cell proliferation, induces S-phase arrest, and promotes apoptosis through activation of c-Jun N-terminal kinase, suppression of Akt pathway, and downregulation of antiapoptotic gene products. Biochem Pharmacol. 2007;74:118–30. doi: 10.1016/j.bcp.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siddiquee K, Zhang S, Guida WC, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Nati Acad Sci U S A. 2007;104:7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.