Summary
E6 is an oncoprotein produced by human papillomavirus (HPV). The E6 protein from high-risk HPV type 16 (HPV-16) contains two zinc-binding domains with two C-x-x-C motifs each. E6 exerts its transforming functions through formation of a complex with E6AP, which binds p53 and stimulates its degradation. There have been few biophysical and structural studies due to difficulty in preparation of soluble protein and here we describe preparation of soluble E6 constructs including the two separated zinc-binding domains of E6. These proteins are used to examine the extent to which the two domains cooperate to mediate E6 function, how zinc influences the behavior of E6 protein, and which domains mediate aggregation. We demonstrate that these soluble proteins are active using p53 degradation, E6AP binding, and hDlg PDZ binding assays and that they are folded and stable using NMR, circular dichroism, and fluorescence spectroscopies. We show that the separated N-terminal and C-terminal domains interact but non-productively for E6 function. The two domains bind zinc differently with higher affinity associated with the C-terminal domain. Analyses using surface plasmon resonance and circular dichroism and fluorescence spectroscopies show that aggregation is mediated more through the N-terminal domain than the C-terminal domain. These studies allow a model in which the C-terminal zinc-binding domain of E6 recruits a target protein such as hDlg and the N-terminal domain is mostly responsible for recruiting a ubiquitin ligase to mediate target protein degradation.
Keywords: HPV, E6, protein-protein interaction, zinc-binding, biophysical methods
Introduction
Human papillomaviruses (HPV) are small DNA tumor viruses that infect epithelial cells and are strongly associated with the development of cervical cancer.1 The E6 oncoprotein from high-risk HPV type 16 (HPV-16) is a 151 amino acid residue protein that contains two zinc-binding domains with two C-x-x-C motifs each. E6 protein has activities that are mediated by recognition of cellular proteins which have been well-summarized in recent reviews.2-4 The complex of E6 with the cellular 100 kDa E6-associated protein, E6AP, binds p53 and stimulates its ubiquitination and degradation through the proteasome, thereby allowing E6 to exert its transforming functions.5-7 Proteins other than p53 are also targeted.8
In contrast to the many reports on the biological functions of E6, there have been few biophysical and structural studies due to difficulty in preparation of soluble protein. Several approaches have been tried. The full-length protein, either by itself or His6-tagged, can be produced in inclusion bodies9 and some percentage refolded,10 and E611 has been reported to also be expressed in a soluble form.11-13 However in each case, the solubility has been insufficient for biophysical characterization.11-13 The minimal segment still functional for p53 degradation comprises residues 1-142 (E6Δ143-151).12 E6Δ143-151 has been purified by cleaving a GST fusion, although the maximum concentration obtained was only low micromolar despite some optimization of buffer conditions and therefore insufficient for extensive biophysical characterization.12 Its N-terminal half forms a domain (E6N) and its C-terminal half forms a second domain (E6C). The two zinc ions of E6 are coordinated by conserved cysteine residues 30, 33, 63, and 66 in the N-terminal half, and residues 103, 106, 136, and 139 in the C-terminal half. Mutating the six non-conserved cysteines to serine improves solubility but the concentration of purified protein remains low.11 Similar approaches have been used on the E6 protein from other papillomavirus types with similar results.14
The zinc-binding domains of E6 are more soluble when separated and both have been characterized.12,13 Cleavage of the GST-fusion of the N-terminal Zn2+-binding domain (E6N) results in a protein that is soluble in the hundreds of micromolar range, but sedimentation studies indicate a monomer-dimer equilibrium at only 53 μM concentration.12 Studies with the C-terminal Zn2+-binding domain (E6C) with its four non-conserved cysteines mutated to serine have been more successful. A highly concentrated E6C sample was obtained for multidimensional NMR experiments13 and its structure solved.15 The structure of the N-terminal domain of E6 could be modeled using the shared sequence similarity to the C-terminal domain of E6. The E6N and E6C domains have been shown to directly interact by NMR methods.15 A model for full-length E6 was then built by burying hydrophobic contact points between the two domains and combining 3-stranded β-sheets from each domain to create a 6-stranded sheet. The details of the inter-domain arrangement may be different as the effects of some mutations and the location of some conserved residues in the structure cannot be readily explained.2
These studies on the separated halves of E6 have contributed greatly to our understanding of their structures, but the structure of full-length E6 and the mechanism by which it is poorly soluble has not been ascertained. In addition, the extent to which the two domains cooperate to mediate E6 function is unknown. Although E6 protein contains two zinc ions, EGTA-treated E6 protein contains only one zinc ion, yet remains active for p53 degradation.16 The identity of the domain from which zinc can be sequestered by EGTA is unknown. How zinc influences the behavior of E6 protein and its domains has also not been extensively characterized. The prerequisite to such studies is to obtain enough purified E6 protein and both of its separated domains.
The fusion of poorly soluble proteins to other highly soluble domains has been used to increase protein solubility as well as providing a convenient method for purification. Many fusion tags have been evaluated for their competency to increase the solubility of target proteins as the optimal fusion partner varies depending on the physicochemical properties of the target protein. 17-19 Although GST, MBP, and TRX have been used as solubilizing agents, they are large (>100 amino acids) and hinder direct NMR studies and biophysical characterization of the corresponding fusion proteins.20 Cleavage of the fusion proteins often re-introduces problems with solubility and stability. For example, although GST-E6 and MBP-E6 can be over-expressed in E. coli in a soluble form and then purified using the appropriate single-step affinity procedure, removal of the fusion partner leads to rapid precipitation of the E6 protein.21,22 Among the better solubilization partners, GB1 has considerable advantages as it is sufficiently small (56 amino acids) that the resultant fusion protein can be studied using NMR methods. The domain has been fused to E6 proteins16,23 and other proteins such as GCN4.24
In this paper, we employed GB1 fusions of HPV-16 E6 and its two zinc-binding domains as well as a construct with an altered linker between the domains. We demonstrate the extent to which these soluble proteins are active using p53 degradation, E6AP binding, and hDlg PDZ binding assays. The folding of the proteins were examined by UV, CD, fluorescence and NMR spectroscopies and their stability by thermal and urea denaturation and limited proteolysis. Finally, we examined the role of zinc in E6 folding, stability, and aggregation. In doing so, we identified different behaviors of the zinc-binding domains with respect to stability and function.
Results
Preparation of soluble HPV-16 E6 protein and related domains
In this study to solubilize E6, we used a GB1 fusion tag (GBF)23 with mutations at three positions (D22N, D36R and E42K) that has an estimated pI of 8.0 (instead of 4.5) designed to prevent intra-molecular electrostatic interactions while fused to basic proteins such as E6 (G. Wagner, personal communication). Residues 1 to 142 is the minimal region competent for p53 degradation12. Although the six non-conserved cysteines (residues 16, 51, 80, 97, 111, and 140) are not necessary for E6-mediated p53 degradation activity,15,25 we have found that mutations of two of these cysteines (residues 16 and 51) disrupts E6BP binding,26 and therefore in our construct they were left unchanged and other four non-conserved cysteines were converted to serine. The discrepancy in binding may result from differential binding of E6AP versus E6BP by E6. In this research, the E6 constructs of GBF-fusion were: GBF-E6 comprising residues 2-142 of HPV-16 E6 with 4 Cys to Ser mutations at 80, 97, 111, and 140, GBF-E6C comprising residues 79 to 142 with the same mutations, and GBF-E6N comprising residues 2 to 74 of E6 with Cys16 and Cys51 unchanged (Figure 1).
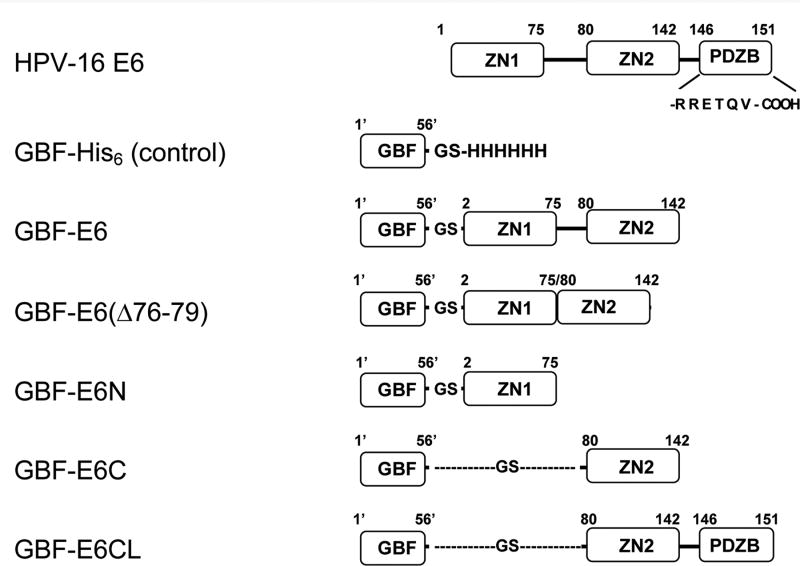
Figure 1.
The E6 constructs used in this study. The E6 protein has two zinc-binding domains (ZN1 and ZN2) and a PDZ-binding motif (PDZB). The residues of the PDZ-binding motif, RRETQV, are indicated. The residues of GBF are numbered as 1′ to 56′. GBF-E6 comprises F2Q3D4…S140R141S142 of wild-type HPV-16 E6 with 4 Cys to Ser mutations at 80, 97, 111 and 140); GBF-E6(Δ76-79) comprises F2Q3D4…S140R141S142 with the same 4 Cys to Ser mutations, but without Y76R77H78Y79; GBF-E6N comprises residues F2Q3D4…I73S74E75 of wild-type HPV-16 E6; GBF-E6C comprises residues S80Y81S82…S140R141S142 of wild-type HPV-16 E6 with the 4 Cys to Ser mutations; GBF-E6CL comprises residues S80Y81S82…T149Q150V151 with the 4 Cys to Ser mutations. GBF-E6 and GBF-E6C constructs with a Trp to Phe mutation in the GBF region (W43′F) were also prepared.
All GBF-tagged E6 constructs showed high levels of expression in E. coli BL21(DE3) upon induction by IPTG at 30 °C. Cells were harvested and lysed, and sufficient protein was found in the supernatant for subsequent purification. DTT was added to prevent the oxidation of cysteine. We found both a deficiency and an excess of free zinc ions in the buffer reduced the solubility of E6, consistent with observations made previously.27 We obtained the most soluble protein by binding the GBF-E6 in low salt to a cationic exchange column, then adding saturating amounts of Zn2+ (100 μM ZnSO4), followed by washing without Zn2+. Although the resultant GBF-E6 could be concentrated to ∼1 mg/ml, samples precipitated within several hours to several days at 4°C. A calibrated size exclusion gel filtration column was performed to check the aggregation of the soluble samples. In the presence of 200 mM NaCl and 50 mM phosphate (pH 7.50), at least half of the loaded protein eluted as a monomer. When using a low salt buffer (NaCl <100 mM), the sample precipitated in the column.
Concentration of the monomeric form of GBF-E6 protein was monitored by UV and CD spectroscopy. The absorption intensities at 280 nm in UV spectra and at the minima of 208 nm and 222 nm in CD spectra remained inversely proportional to the volume during concentration, whereas samples prepared in low salt or without the fusion tag did not. Light scattering indicative of aggregation was monitored by measuring the absorption above 300 nm, which remained low throughout concentration. Therefore, in high salt, the GBF-fusion appeared to significantly increase the solubility of E6 protein while remaining monomeric.
The concentrated GBF-E6 (about 1 mg/ml) did however precipitate over time, and the buffer system was therefore optimized prior to gel filtration.28 The results showed that a further increase in NaCl from 200 mM to 1 M did not significantly affect the solubility, but lower salt (< 100 mM) precipitated the protein readily. While Tris (pH 8.0) seemed best for the early stages of purification, phosphate (pH 7.50) was better for long-term storage. Both were much better than either MOPS (pH 7.50) or Hepes (pH 7.50). Although addition of 5% glycerol, 0.1% SDS, 0.5 mg/ml dodecyl maltoside (DDM), 0.1% Triton X-100, or 100 mM imidazole did not improve solubility, adding 5 mM DTT, 2 M urea, or 10 mM CHAPS helped to prevent precipitation. On gel filtration, the addition of 5 mM DTT and 10 mM CHAPS to the loaded protein increased the percentage of the monomer (from roughly 10% to 50%) and the ability to concentrate the protein, which reached as high as 5 mg/ml (∼ 200 μM). We evaluated several other domains derived from E6, such as an N-terminal half (GBF-E6N) and two versions of C-terminal halves. The shorter C-terminal half, GBF-E6C, comprises residues 80-142, and the longer one, GBF-E6CL, comprises residues 80-151 that includes the PDZ domain binding motif of E6. To probe whether the properties of E6 were sensitive to the linker between the N and C-terminal domains, we also examined a form of GBF-E6 in which 4 amino acids were deleted, GBF-E6 (Δ76-79).
GBF-E6 (Δ76-79) purified in a way similar to GBF-E6, whereas the shorter domains all showed high solubility in the lysate of harvested cells. The GBF-fused C-terminal domains are highly basic like GBF-E6, and were purified using the same procedures. GBF-E6N has a calculated isoelectric point of 8.2, and therefore did not bind to cation or anion exchange columns at pH 8.0, although the use of such a column allowed efficient removal of contaminating proteins. After gel filtration, >95% pure GBF-E6N could be obtained. The final concentrations of GBF-E6N and GBF-E6C were in the millimolar range (∼20 mg/ml) in 50 mM phosphate buffer (pH 6.50). The buffer conditions for GBF-E6N and GBF-E6C were further optimized for NMR spectroscopy by monitoring increases in the intensities of dispersed cross peaks in their respective 15N, 1H-HSQC spectra. The optimized conditions were 50 mM phosphate (pH 6.50) for GBF-E6C and 50 mM phosphate (pH 7.50) with 500 mM NaCl and 5 mM CHAPS for GBF-E6N.
Assessment of activity
To ensure that the proteins were properly folded prior to assessment using biophysical methods, several different types of activity measurements were used such as the ability to interact with either E6AP29 or a PDZ domain30 or to degrade p53.31
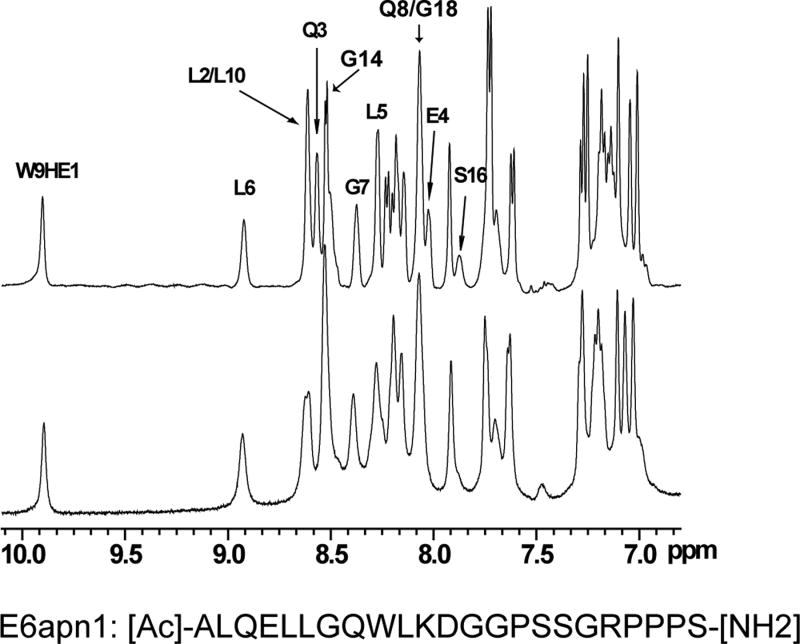
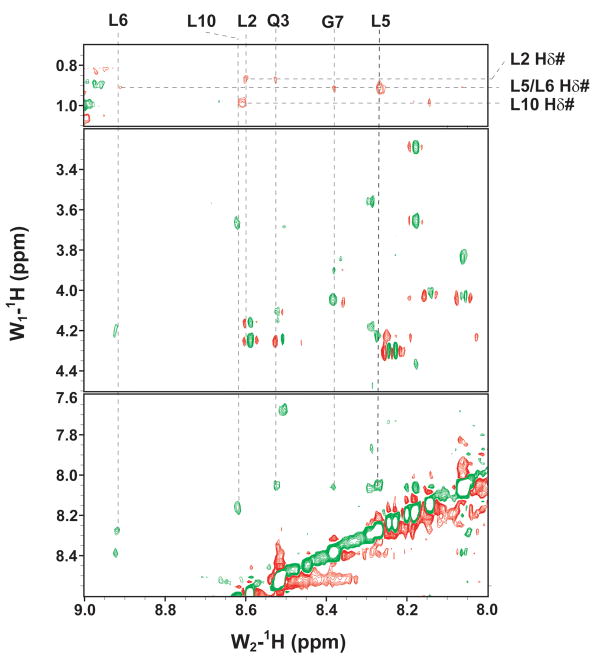
NMR methods were used to validate the binding of E6 and examine the folds of E6N and E6C. The activity of GBF-E6 was indicated by its ability to bind to a peptide, E6apn1, used to mimic E6AP29 (Figure 2). E6apn1 contains the sequence L2Q3E4L5L6G7Q8, which has the E6-binding consensus sequence, LhxφLsh, where the bolded letters indicate conserved leucine residues, h is an amino acid residue capable of accepting hydrogen bonds (D, E, Q, or N), x denotes any amino acid, φ is hydrophobic, and s represents a small amino acid residue (G or A).32 We previously showed that the unbound E6-binding motif is helical.29,32 Several resonances in the 1D spectrum could be readily followed as GBF-E6 protein was added (Figure 2A). In particular, the line widths of resonances L6 NH and G7 NH in the consensus sequence increased from 14 and 17 Hz to 20 and 23 Hz in the presence of GBF-E6 protein. Other resonances were also observed to broaden, together demonstrating that purified GBF-E6 is able to interact with E6apn1 peptide containing the E6-binding motif (Figure 2A). The chemical shifts of E6apn1 did not change significantly in the presence of 10% GBF-E6 protein, suggesting the peptide structure retained its α-helical conformation. A NOESY difference spectrum of E6apn1 with and without E6 protein was calculated (Figure 2B). Overall, the NOE cross-peaks of E6apn1 with or without GBF-E6 were similar, in that there were strong NHi-NHi+1 correlations from residues L2 to K11. Whereas most cross-peaks broadened on the addition of GBF-E6 and thus became weaker, cross-peaks from the sidechain methyls to the backbone amides of three hydrophobic leucines of E6apn1 (L2, L5, and L6) that are crucial for binding E6 increased in the presence of GBF-E6. Intensity increases were observed between the methyls of L2 and the NH of Q3; the methyl of L6 and the NH of G7, and the intra-residue methyl of L5 to its own NH. The increased intensity suggests either that these leucine sidechains became more ordered on binding to E629 or that spin-diffusion effects have become more efficient. Conformational changes may involve mostly side-chain reorientation because in near-UV CD spectroscopy over a wide range of peptide:protein ratios (0:1 to 18:1) we observed no spectral changes suggesting no changes in backbone secondary structure (data not shown).
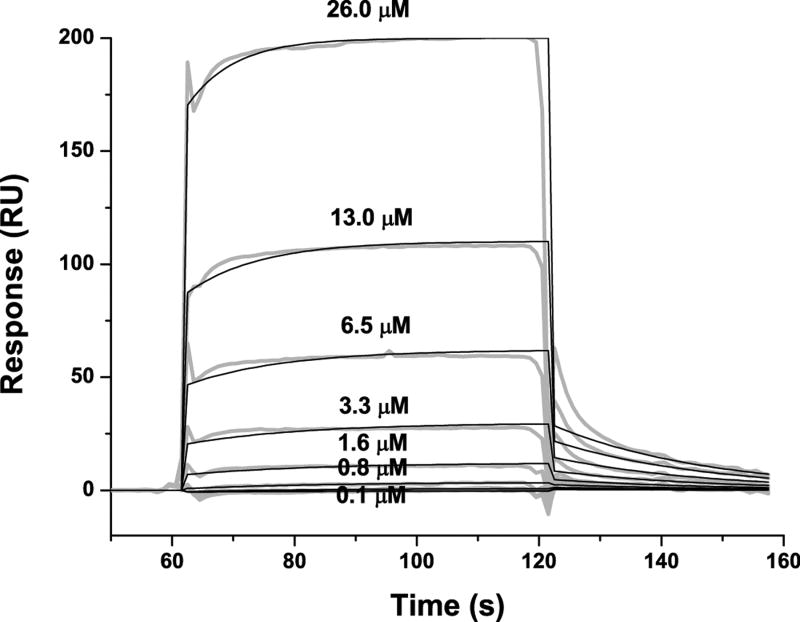
Figure 2.
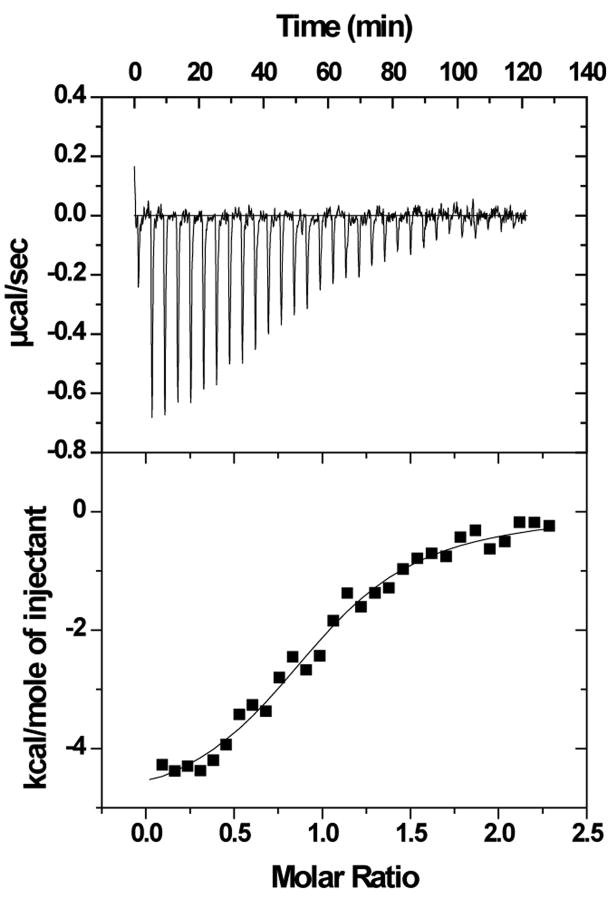
The binding of purified E6 proteins to E6 targets. (A) One-dimensional NMR spectra of E6apn1 peptide (2.0 mM) in the absence (top) and in the presence of (bottom) of GBF-E6 protein (200 μM) in 50 mM phosphate (pH 7.50) with 200 mM NaCl at 5 °C. The indicated assignments are based on our previous report.29 (B) The NOESY difference spectrum of E6apn1 peptide with and without GBF-E6 protein. Increases in peak intensity are shown in red and decreases in peak intensity are in green. (C) SPR analysis of the interaction of GBF-E6 and E6apc2 peptide in 20 mM phosphate (pH 6.50). E6apc2 was immobilized to the CM5 sensor surface and a series of GBF-E6 solutions allowed to flow over the surface. The experimental curves are shown in gray, the fits shown in black. (D) ITC analysis of the interaction of hDlg PDZ2 domain and E6CL peptide in 20 mM phosphate (pH 6.50). A 500 μM solution of hDlg PDZ2 was injected 30 times in 10 μl aliquots into the 1.4 ml sample cell containing GBF-E6CL at a concentration of 50 μM at 20 °C.
Because the NMR binding experiments used only a single peptide and protein construct, we turned to surface plasmon resonance, which uses much less material, to test a variety of constructs and controls to determine the extent of specificity. SPR has been used before for measuring E6-peptide interactions where affinities were in the low micromolar range.33 SPR data using E6apn1 gave complex kinetic profiles, but profiles with a related E6-binding peptide E6apc2 that contains the E6-binding consensus sequence at its C-terminus29 gave a profile consistent with obvious binding (Figure 2C and Table S1). E6N bound E6apc2 with weak affinity and E6C did not bind. The C-terminus of E6 in GBF-E6CL is able to bind to various PDZ domains30 and also showed a strong binding profile with the PDZ2 domain of hDlg protein (Table S1 and Figure S1). The SPR data did not fit well to the classical 1:1 Langmuir model as shown in Figures 2C and S1, and therefore accurate kinetic constants for binding could not be obtained. Also because the association curves for most binding reactions do not reach saturation (Figure S1), we did not fit for equilibrium dissociation constants. The poor fits for some interactions are most likely due to protein aggregation. The interaction of E6CL with the hDlg PDZ2 domain were measured by isothermal titration calorimetry (ITC) and yielded a comparable dissociation constant of 6.6±1.0 μM with the expected stoichiometry of 1.0:1.0 (Figure 2D). This affinity was the same as with a short 6-mer peptide from the C-terminus of HPV-16 E6 protein, which we found to have a dissociation constant of 6.5±0.8 μM,30 suggesting that residues N-terminal to the C-terminal 6 residues of E6 do not contribute to binding, which is consistent with the crystal structure and the results with E6-mediated degradation of hDlg.34,35 Similar binding studies with MAGI-1 PDZ1 show that the 11 most C-terminal residues of E6 mimics the C-terminal zinc-binding domain.36 GBF-E6N and GBF-E6 were not sufficiently stable for study by ITC.
GBF-E6 promoted p53 degradation in vitro while GBF-E6(Δ76-79) had ∼40% activity of wild-type (Figure S2). As expected, the other domains, GBF-E6N, GBF-E6C, and a mixture of GBF-E6N and GBF-E6C, had no activity.
Assessment of protein fold
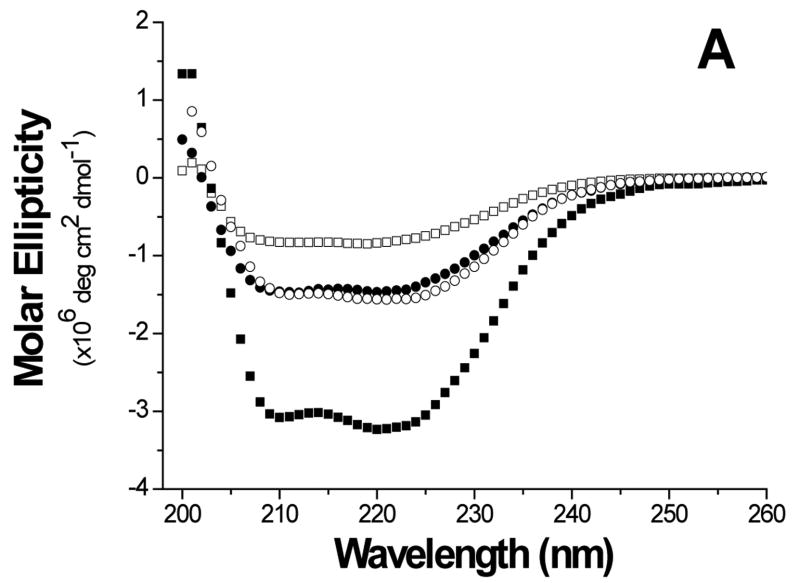
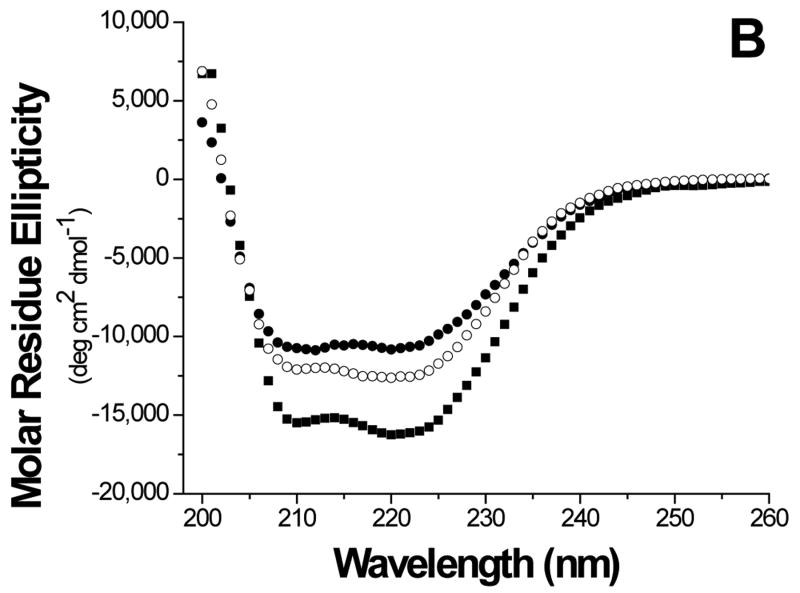
The folds of the E6 constructs were assessed using biophysical methods. The circular dichroism (CD) curve of GBF-E6 displayed two negative minima at 208 and 222 nm. The molar ellipticity contribution of the GBF fragment in GBF-fused E6 constructs could be visualized using the GBF-His6 control (Figure 3A). The significant negative minima in addition to the contribution of GBF portion indicate that the E6 portion of the fusion was folded well. The molar ellipticity of GBF-E6 was more negative than either GBF-E6N or GBF-E6C halves, suggesting that additional secondary structure in E6 is generated through interaction of the two halves (Figure 3B).
Figure 3.
Comparison of secondary structure content of E6 proteins. A. The molar ellipticities of GBF-E6 (solid squares), GBF-E6N (solid circles), and GBF-E6C (open circles). A construct representing the fusion tag, GBF, is also plotted (open squares). B. The molar residue ellipticities of GBF-E6, GBF-E6N and GBF-E6C are shown with the same symbols. The proteins were between 5 and 10 μM in 50 mM sodium phosphate (pH 7.50) with 200 mM NaCl at 25 °C.
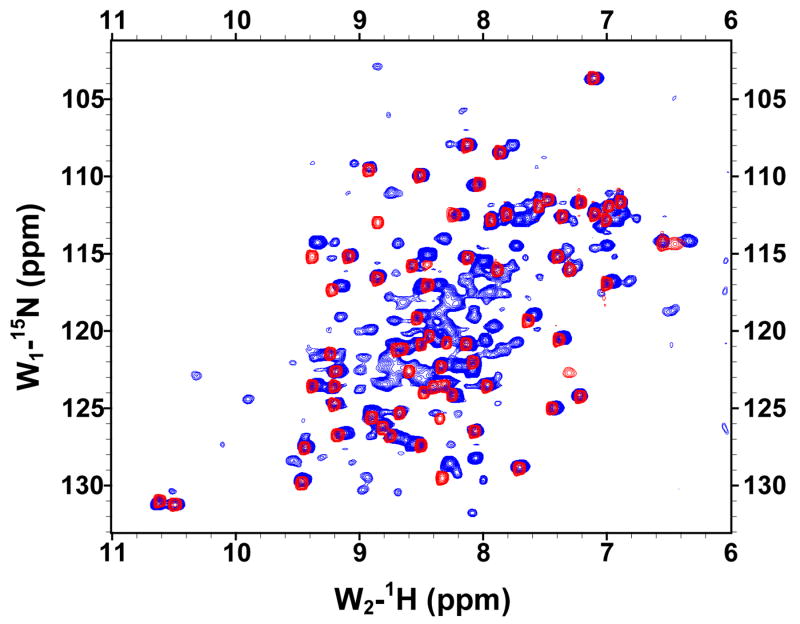
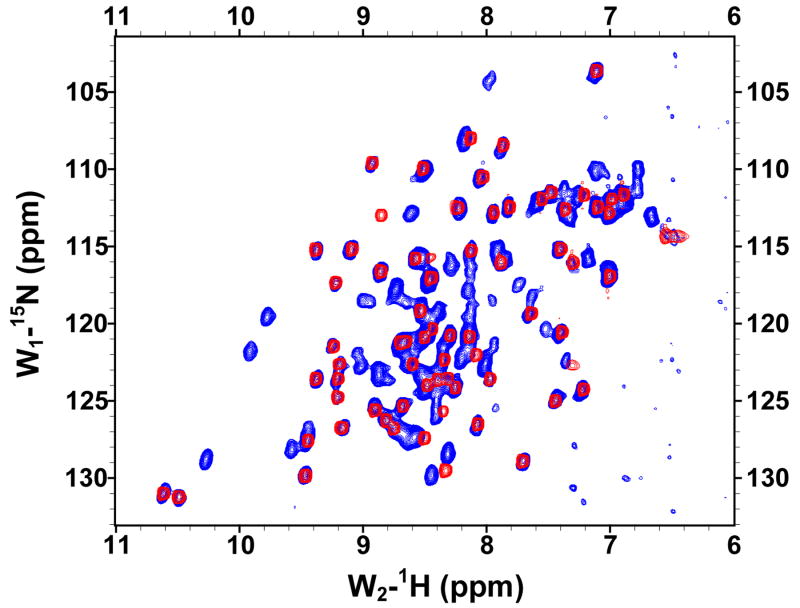
The solubilities of the GBF fusions of the E6 halves were sufficiently high for recording 1H, 15N-HQSC NMR spectra (Figure 4). The peaks deriving from the GBF portion could be readily discerned as they appeared with the same chemical shifts in both fusion protein and the GBF-His6 control. Thus, the folding of GBF domain did not appear to be influenced by the presence of either half of E6. The cross-peaks from the E6 halves showed good dispersion. The peaks from E6N were obviously weaker than those from the GBF tag. Those from E6C were about 30% less intense than those from the GBF tag. The weaker peak intensity indicates flexibility on the chemical shift timescale even when folded or possible aggregation of the E6 halves that does not extend across the flexible linker to the GBF tag.
Figure 4.
The 15N,1H-HSQC spectra of N-terminal and C-terminal halves of HPV-16 E6 protein indicated that the domains are well folded. (A) Overlay of GBF-E6N (blue) and GBF-His6 (red). (B) GBF-E6C (blue) and GBF-His6 (red). The protein concentrations were about 2.0 mM and data was collected at 20 °C. GBF-E6N was in 50 mM phosphate (pH 7.50), 500 mM NaCl, 5 mM CHAPS, and GBF-E6C and GBF-His6 were in 20 mM phosphate (pH 6.50).
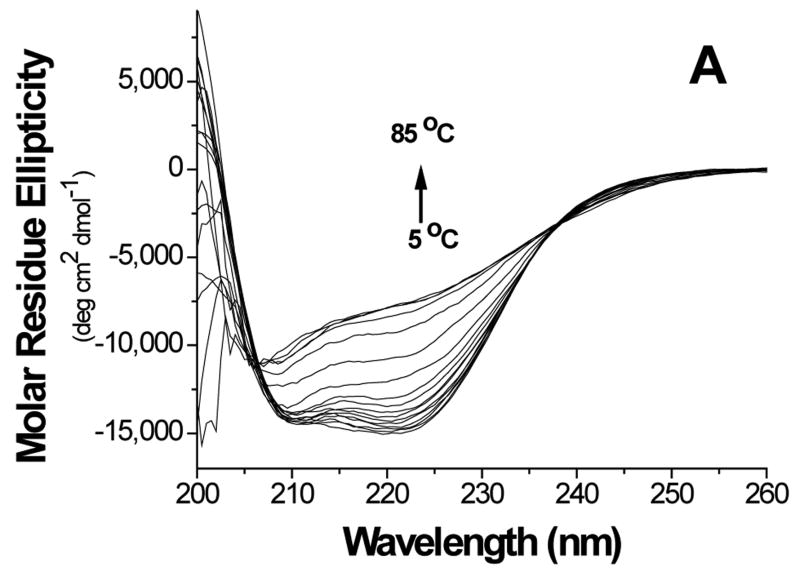
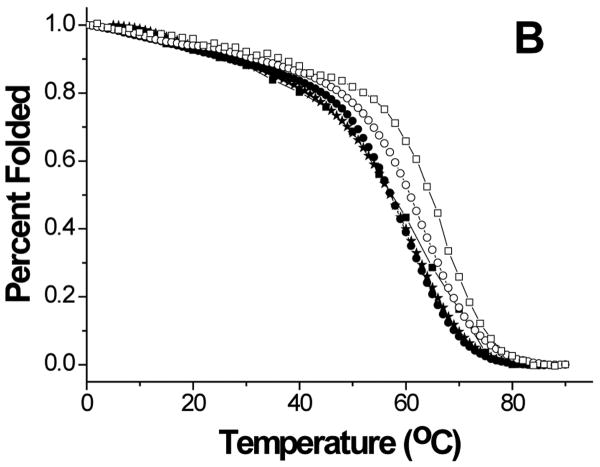
Thermal stability of the proteins was investigated by monitoring the secondary structure using CD (Figure 5). The data showed cooperative melting for GBF-His6 and GBF-fused E6 constructs. GBF-His6 was fit using a two-state equilibrium between folded and unfolded states and a Tm 66.2 °C was obtained. In theory, GBF-E6N and GBF-E6C each contain two independent domains. Through deconvolution of denaturation curves with the Tm value of the GBF portion, Tm values for E6N and E6C were determined to be 52.0 °C and 58.0 °C, respectively. The denaturation of GBF-E6, which should contain three independent domains, could be fit using the three Tm values of its individual domains. We also fit the denaturation of E6 assuming that the E6N and E6C did not denature independently and obtained an apparent Tm 55.4 °C for the E6 portion. Using a similar analysis, GBF-E6(Δ76-79) had an apparent Tm of 52.5 °C (Figure S3). Because the actual Tm determined varied with the temperature range studied, we do not believe that there is a significant difference between the Tm values of E6 and E6(Δ76-79). The existence of cooperative melting indicates that all proteins were well folded.
Figure 5.
Thermal denaturation of GBF-E6 and its mutants. (A) CD spectra of GBF-E6 as the temperature was increased in steps of 5°C. (B) The thermal unfolding of GBF-E6 (solid squares), GBF-E6(Δ76-79) (stars), GBF-E6N (solid circles), GBF-E6C (open circles) and GBF-His6 (open squares). The proteins were 5 to 10 μM in 50 mM phosphate buffer (pH 6.50) containing 200 mM NaCl. The proteins were considered to be 100% folded at 0°C.
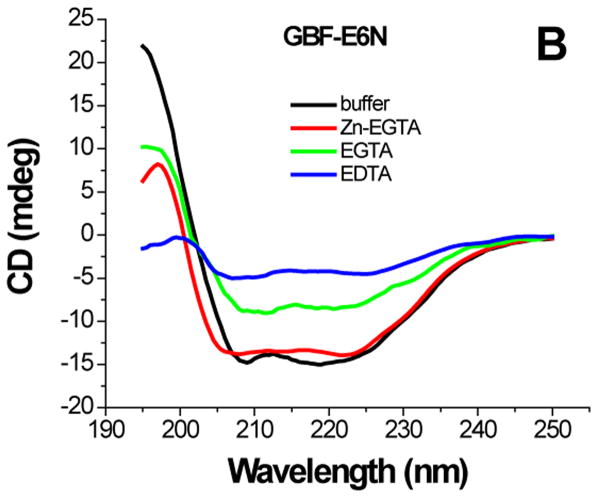
Having established that the proteins were well folded, zinc contents were determined in order to compare with values previously reported for full-length proteins.16 GBF-E6C contained one ion (1.0±0.05) while GBF-E6 and GBF-E6N contained slightly less than the expected 2 ions (1.6±0.08) and 1 (0.75±0.08) ion, respectively, suggesting a lower ability of the N-terminal domain of E6 to bind zinc. In a version of E6, GBF-E6(Δ76-79) in which the linker between the two domains is shortened, the stoichiometric ratio of zinc to protein was around 0.8 (0.8±0.08), instead of the expected 2.0, suggesting an altered fold relative to E6.
Earlier reports showed that EGTA-treated E6 was still functional for promoting the degradation of p53, although it bound only one zinc ion.16 After dialysis against EGTA, GBF-E6 and GBF-E6C proteins still contained 0.6 zinc per molecule (0.57±0.11, 0.52±0.16), but the zinc contents of GBF-E6(Δ76-79) and GBF-E6N were near zero. Therefore EGTA appeared to sequester one of the two zinc ions binding to E6 protein, and only that of the N-terminal domain. These observations suggest that the C-terminal zinc binding domain of E6 binds to zinc more strongly than the N-terminal domain.
To ask whether the fold was perturbed in the construct with the shortened linker, GBF-E6 and GBF-E6(Δ76-79) were assessed by limited proteolysis using chymotrypsin (Figure S5). While 90% of GBF-E6 was digested within 1 hour, 5 hours were needed for 90% digestion of GBF-E6(Δ76-79). For GBF-E6(Δ76-79), only 50% was digested within one hour. Removal of the linker in GBF-E6(Δ76-79) thus appears to lose a preferential exposed digestion site or possibly induce a formation of an aggregate that is stable to digestion. Both GBF-E6 and GBF-E6(Δ76-79) produced similarly sized bands that were stable as they resisted further digestion for at least 8 hours. The similar digestion patterns indicate that the structures of the N- and C-terminal domains were not significantly affected by the deletion of the linker. N-terminal sequencing and mass spectral analysis indicated that the largest stable proteolytic fragment had the same N-terminal residues as GBF domain and that the preferred site of cleavage was at or near the Y76RHY79 linker instead of between the GBF fusion and the N-terminal domain.
The proteolytic sensitivity of the Y76RHY79 linker between the E6 domains, and its proposed location as a loop in the full-length E6 structure13 would suggest that its exact structure would not be important. However, from both the activity measurements of the E6 constructs and the zinc-binding results, the linker appears to play an important role in orienting the two zinc binding domains with respect to each other that is needed for biological activity of E6. Because the GBF-E6(Δ76-79) had lowered biological activity, lowered zinc content, and altered proteolytic stability, it was not characterized further in this study.
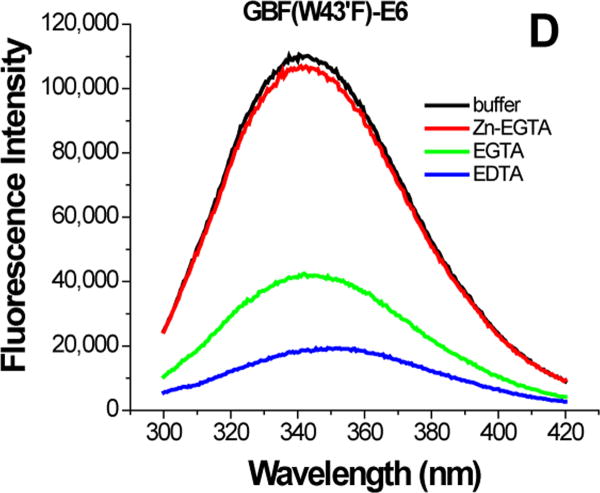
To confirm the initial observation that the C-terminal domain of E6 binds zinc more tightly than the N-terminal domain, we took advantage of the fact that E6 has only one tryptophan that could be monitored using fluorescence spectroscopy. The only tryptophan is in the C-terminal domain (residue 132) and relatively close to the coordinating cysteine residues (residues 103, 106, 136 and 139 and its associated zinc ion), but not the N-terminal domain (residues 30, 33, 63, 66 and its associated zinc ion). A preliminary model suggests the tryptophan is 18 Å to the zinc of the C-terminal domain and 29 Å to the zinc of the N-terminal domain.15 The flexible nature of the linker joining the domains and the distances suggests that removal of the C-terminal zinc ion would affect the environment of the tryptophan residue more than removal of the N-terminal zinc ion. GBF has one tryptophan (W43′), whose intrinsic fluorescence would not be expected to be sensitive to the presence of zinc ion. Where needed, the fluorescence contribution from the GBF tryptophan was eliminated by preparing proteins with a W43′F mutation.
Samples were prepared in a series of buffers starting with a standard buffer containing 50 mM phosphate buffer (pH 6.50) and 200 mM NaCl. A Zn-EGTA buffer system contained 10 mM each of Zn2+ and EGTA that provides approximately 28 μM of free zinc ion as calculated by the WEBMAXC program. Buffered 10 mM EGTA and 10 mM EDTA were also used to compete with E6 for zinc binding. Samples of each GBF-E6, GBF-E6N and GBF-E6C were prepared by diluting the corresponding protein stock into each of the four buffers, and after incubating at room temperature for 24 hours, the UV, CD and fluorescence properties of the samples were examined. As a control, GBF-His6 was checked using the same buffer conditions. The maximum of intrinsic fluorescence of GBF-His6 was about 353 nm, consistent with its partially exposed location on the surface of the protein.37 Its fluorescence spectrum was not significantly affected by EDTA or EGTA, consistent with the lack of interaction with metal ions (data not shown). Its CD spectra were also insensitive to the presence of chelating agents (data not shown).
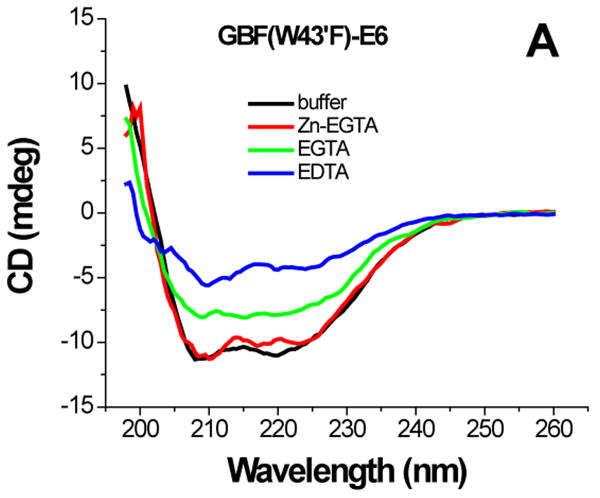
For GBF-E6 protein, the presence of Zn-EGTA (that supplies free zinc) did not significantly change the shape and minima of the CD curve (Figure 6A) or the fluorescence spectrum (Figure 6D). However, EGTA and EDTA significantly decreased the fluorescence intensity, and shifted the maximum emission wavelength from 341 nm to 345 nm and 353 nm respectively. The red shift observed in the fluorescence spectra is indicative of a loss of burial of the Trp 132 in E6 upon chelation with EGTA and more so with EDTA. A lower UV absorption at 280 nm and higher intensity above 300 nm corresponding to the scattering of light of E6 constructs suggests that the apparent decrease of the far-UV CD and fluorescence signals caused by removal of zinc ions is due to some degree of protein aggregation in addition to the loss in structure. Our observations on GBF-fused E6 are similar to the detailed observations with a maltose-binding protein (MBP) fusion of E6.11
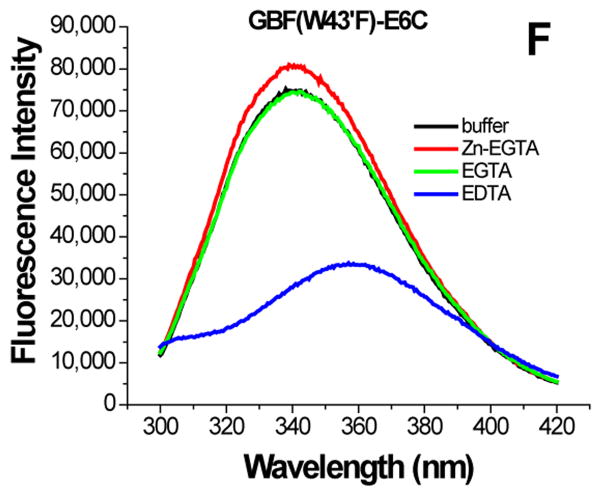
Figure 6.
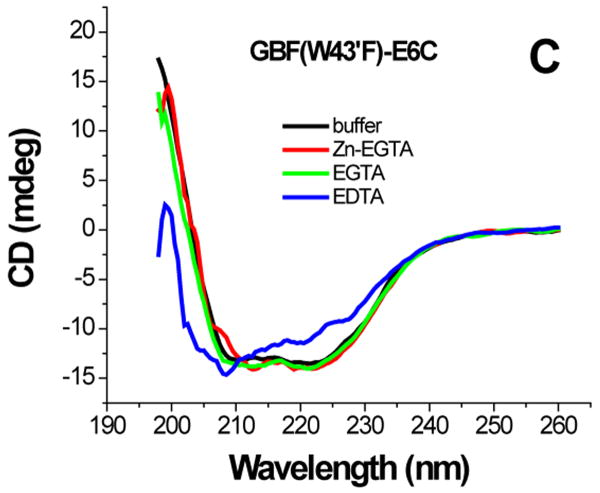
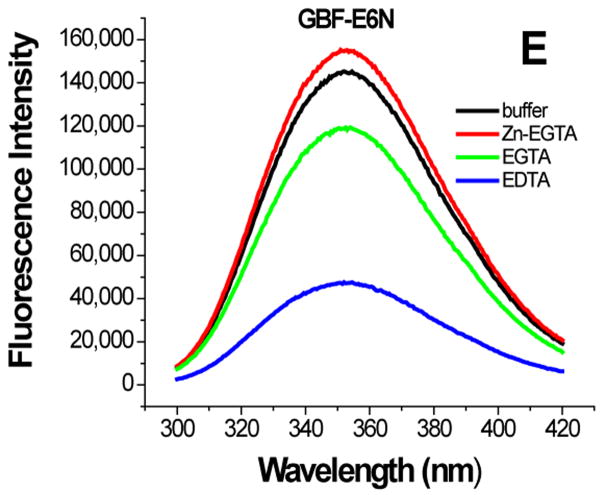
The effect of zinc binding on HPV E6 structure. (A), (B), and (C) show the CD spectra of GBF-E6, GBF-E6N, and GBF-E6C and (D), (E), and (F) show the fluorescence spectra of GBF(W43′F)-E6, GBF-E6N, and GBF(W43′F)-E6C. GBF(W43′F)-E6 was 5 μM and GBF-E6N and GBF(W43′F)-E6C were 10 μM. The buffers used were 50 mM phosphate, pH 6.50 (black), 50 mM phosphate, pH 6.50 with 8 mM Zn-EGTA (red), 50 mM phosphate, pH 6.50 with 8 mM EGTA (green), 50 mM phosphate, pH 6.50 with 8 mM EDTA (blue).
The GBF-E6N protein behaved similarly with respect to additions of Zn-EGTA, EGTA and EDTA. 10 mM Zn-EGTA did not significantly change the shape and minima of the CD curve whereas EGTA decreased the signal and EDTA decreased the signal further (Figure 6B). In the fluorescence experiments, Zn-EGTA slightly increased the intensity, but EGTA and EDTA significantly decreased the intensity, but did not shift the maximum emission wavelength at 353 nm of the single tryptophan within the GBF portion (Figure 6E). Because control experiments using EGTA or EDTA showed no effect on GBF-His6, it appears that the E6N portion of GBF-E6N promotes aggregation by EGTA or EDTA. Lower UV absorption at 280 nm and higher UV intensities above 300 nm consistent with light scattering also confirmed the presence of protein aggregation caused by metal chelation by EGTA or EDTA.
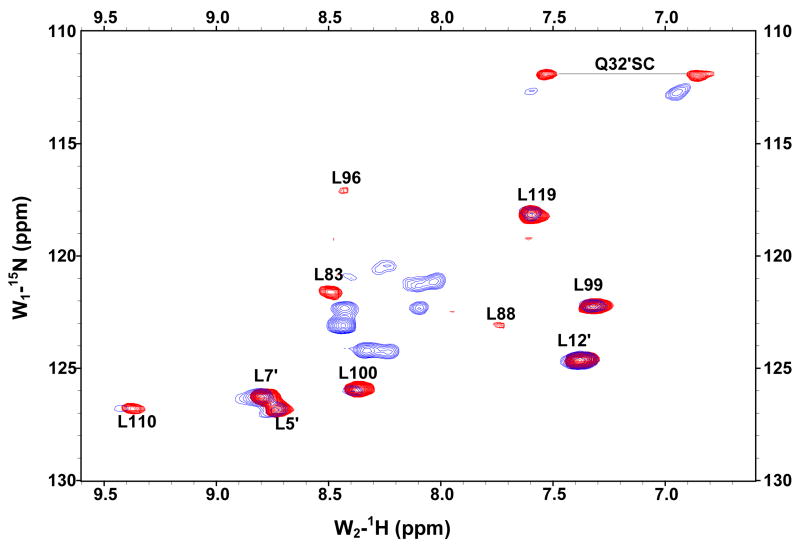
The GBF-E6C protein behaved differently with respect to Zn-EGTA, EGTA and EDTA. Neither Zn-EGTA nor EGTA significantly changed the shape and minima of the CD curves while EDTA changed the shape of the CD curve showing a decreased minimum at 222 nm (Figure 6C). In the presence of EGTA or EDTA, GBF-E6C did not show precipitation or aggregation, which was confirmed by UV measurements. Therefore, the loss of signal in the presence of EDTA was commensurate with a loss in secondary structure. In the presence of Zn-EGTA, the fluorescence of GBF(W43′F)-E6C (with the only tryptophan residue in the E6 portion) increased slightly whereas EGTA had no change. EDTA significantly decreased the fluorescence intensity and shifted the λmax from 341 nm to 359 nm (Figure 6F). These data suggest that EGTA does not remove the zinc from the C-terminal domain of E6, while EDTA does remove the zinc ion and allows the tryptophan to become exposed to solvent, indicating a loss of tertiary structure. We verified the effect of zinc chelation in GBF-E6C by using NMR. To simplify the 15N, 1H-HSQC spectrum we specifically labeled the protein with leucine (Figure 7). Of the total of ten leucines, three are from GBF (L5′, L7′ and L12′) and seven are from the C-terminal domain of E6 (L83, L88, L96, L99, L100, L110 and L119) for which published assignments are available.27 In the presence of 2 mM EDTA, the peaks of L99, L100, and L119 became weaker and the dispersed peaks corresponding to L83, L88, L96, and L110 apparently shifted as they disappeared and, characteristic of a less well-packed protein, new peaks reappeared near the center of the spectrum. The peaks from the three leucines of the GBF domain did not change chemical shift or intensity, suggesting that EDTA did not affect the GBF portion and that the GBF-E6C fusion protein remained soluble.
Figure 7.
Overlaid 15N,1H-HSQC spectra of 15N-Leu-labeled GBF-E6C fusion protein in the absence of (red) and in the presence of 2 mM EDTA (blue) at 25°C. L5, L7 and L12 are from the GBF part, and L83, L88, L96, L99, L100, L110 and L119 are from the E6C part. The assignments of GBF are based on our own data and published results.46 The numberings for E6C are from published assignments of a similar construct.27
Characterization of the aggregation of E6 proteins
The losses of intensity in the CD and fluorescence spectra of GBF-E6 and GBF-E6N in the presence of EGTA or EDTA (Figure 6) were likely due to changes in solubility. To investigate the extent of aggregation of the soluble fraction of the E6 proteins after zinc ion chelation, proteins were checked by gel filtration using loading concentrations of 5 to 10 μM (Figure S5). Four buffer conditions were used as described above. In all conditions, GBF-E6 appeared with a molecular weight of ∼10 kDa that was smaller than that expected for its monomer mass of 24 kDa, consistent with retention by the Sephacryl column. GBF-E6C eluted as a monomer of its predicted monomeric size (∼14 kDa) in all conditions. GBF-E6N (theoretical monomeric mass of ∼15 kDa) eluted at a volume corresponding to that between that of a monomer and a dimer, with some variation depending on the buffer conditions. Therefore, the soluble fractions of GBF-E6, GBF-E6N and GBF-E6C did not change their oligomeric state significantly at low micromolar concentration whether or not zinc ions were present. The slightly different behavior for GBF-E6 and -E6N versus what was observed using spectroscopic methods may arise from the lower protein concentration during elution from the column.
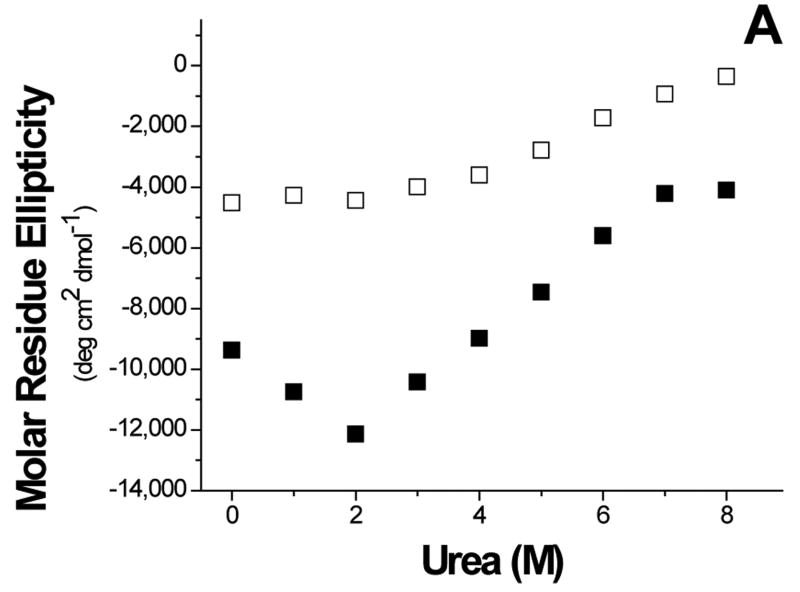
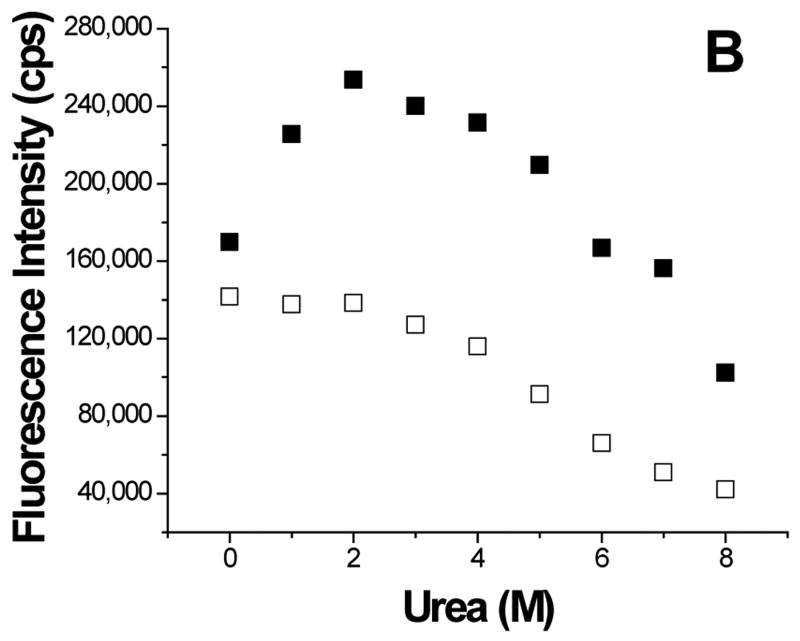
To further understand the aggregation of GBF-E6, it was denatured using urea (Figure 8). The magnitude of the minimum at 222 nm in CD spectra decreased with high concentrations of urea, indicating denaturation. Interestingly, at low amounts of urea, the 222 nm signal became more intense, reaching a maximum at 2 M urea. Because the CD spectra of GBF-His6 did not change using urea concentrations below 2 M, the changes in GBF-E6 were likely due to the properties of the E6 portion rather than that of GBF. Similarly, the fluorescence intensity of GBF-E6, but not GBF, decreased when the concentration of urea was higher than 2 M, but increased when urea was added at lower concentrations (< 2 M). The larger CD signal at low urea concentration and the higher emission intensity in fluorescence spectra may result from disrupting soluble aggregates or, possibly increased order in the secondary and tertiary structure. The disruption of aggregates is consistent with our observation that adding 2 M urea clarifies cloudy NMR protein samples at higher protein concentrations. Alternatively, some contribution to higher fluorescence signals could be caused by changes in quenching as the fluorophores in large non-ordered aggregates may be partially quenched due to a heterogeneous hydrophobic/polar environment. Our results are consistent with observations made with MBP-E6 where 1.5 M guanidine hydrochloride disrupted the aggregation.38
Figure 8.
Urea denaturation. (A) The molar residue ellipticity of GBF-E6 (solid squares) measured on CD spectra at 222 nm and (B), the GBF-E6 fluorescence intensity (solid squares) at 350 nm vs. urea concentration. The GBF-His6 control is shown for comparison (open squares). The protein concentrations were 4.0 μM for CD and about 1.0 μM for fluorescence.
To investigate the extent of aggregation of E6 proteins, surface plasmon resonance methods were employed (Figure S1 and Table S1). GBF-E6 bound itself and the N-terminal domain. Consistent with observations that the most C-terminal residues of E6 change chemical shift on binding the E6N domain,15 we found different behavior with respect to the longer E6CL and the shorter E6C domain, as E6CL bound GBF-E6, but not E6C. Likewise, when GBF-E6N was immobilized, it bound the GBF-E6CL slightly stronger than E6C, as well as itself. This indicates that E6N mainly contributes to the aggregation of E6. E6N also displayed higher affinity than E6C to E6apc2 peptide (derived from E6AP protein), suggesting that it contributes more to E6-E6AP binding than E6C.
Discussion
We achieved high solubility and stability of HPV-16 E6 protein by attaching an N-terminal GB1 protein as a non-cleavable solubility-enhancement tag which allowed characterization using a wide range of biochemical and biophysical properties. The full-length GBF-E6 protein was shown to be competent in binding of a peptide representing E6AP as well as to be active for promoting the degradation of p53. The longer C-terminal subdomain of E6 that had RRETQV as its six C-terminal amino acid residues (residues 146-151), bound with the same ratio and affinity to a PDZ domain (hDlg PDZ2) as a six residue peptide.30 The similarity in binding suggests that HPV-16 E6 uses only its C-terminal domain for binding hDlg consistent with a model of globular C-terminal zinc-binding domain with a unstructured C-terminus that extends away from it as proposed earlier.15
The viral E6 protein comprises two domains (E6N and E6C) that likely require a particular orientation to each other for biological activity. The structure of the C-terminal domain has been determined, and a model for the N-terminal domain has been proposed based on the ca. 15% homology in amino acid sequence.15 Little information is available about the relative orientation of the two domains, but there is sufficient evidence that they do interact. For example, in an 1H, 15N-HSQC experiment of 15N-labeled E6CL, the cross-peak intensities of the C-terminal tail of E6CL (R141, R147, E148 and T149) decrease on addition of non-labeled E6N.15 Using SPR, we also found evidence that E6N binds to E6C. We found that the mixture of E6N and E6C was not able to promote p53 degradation, suggesting that the separated domains do not interact productively to support biological activity. Furthermore, the GBF-E6(Δ76-79) protein construct, missing 4 amino acids in the linker region between the E6N and E6C domains, had decreased p53 degradation activity, presumably due to an altered interaction between the domains. Likewise, the different solubility of E6 from its separated domains suggests that the interactions of separated E6N and E6C is different from the intramolecular interaction of the N- and C-terminal domains within the full-length E6 protein as we found that GBF-E6N and GBF-E6C could be concentrated to high (millimolar) levels but E6, as an E6N-fused E6C, is much less soluble.
Because we observed that the GBF-E6N had more of a tendency to aggregate than the GBF-E6C, the aggregation of E6 appears to be mediated via aggregation of its N-terminal domain, with the caveat that inter-domain interactions in E6 may not reflect the properties of the separated domains. Our observations on GBF-fused proteins are consistent with other E6 constructs, as MBP-fused E6 exists as a soluble agglomerate11,38 and unfused E6N aggregates at concentrations above 20 to 50 μM12 while unfused E6CL is soluble to high concentrations.13 Our model is that aggregation is mediated by hydrophobic interaction centered on E6N that may compete with E6AP binding, which has been shown to be through hydrophobic contacts.32
Comparison of the activities of chimeric E6 proteins (that contain the E6N of HPV-6 fused to E6C of HPV-16, and vice versa) for p53 binding and degradation suggest that the positively-charged C-terminal domain of high-risk E6 is important for p53 binding and the N-terminal domain is important for p53 degradation.39 HPV-16 E6 binds to its PDZ-containing target proteins such as hDlg through its C-terminus.30,40-42 The SPR experiment suggests that HPV-16 E6 binds to E6AP protein through its N-terminal domain that would lead to E6-promoted hDlg and p53 degradation. Recent mutagenesis results indicate that the residues of E6 that are required for binding a target protein may not be the same as those need to induce its degradation, at least in the case of p53.35 Therefore we propose that the main function of the C-terminal zinc-binding domain is to recruit a target protein, such as hDlg or p53, and the N-terminal E6 domain is mostly responsible for recruiting a ubiquitin ligase, such as E6AP, to mediate target protein degradation.
The structures of the two zinc-binding domains of E6 were found to be zinc dependent and the two domains bind to zinc ions with different affinities. EGTA appears to be able to remove the zinc from the N-terminal domain, but that removal of zinc does not significantly impair the interaction with E6AP because E6 protein still possesses p53 degradation activity.16 Commercial reagents such as rabbit reticulocyte lysate used for supplying cellular components in the p53 degradation assay typically contains 4 mM EGTA, so published in vitro p53 degradation assays are likely to use a form of E6 that contains only one zinc ion (in its C-terminal domain). Zinc-ejecting inhibitors of HPV-16 E6 have been described that represent potential drugs against cervical cancer.43 These organic disulfides release about 50% of the zinc from HPV-16 E6 but they have different effects on the inhibition of binding of E6 to E6AP (or the related E6BP protein) and the loss of p53 degradation. Our results indicate that removal of zinc ion using EGTA, which does not affect p53 degradation, is from the N-terminal zinc-binding domain. Therefore we suggest, but have not experimentally verified, that the different activities of the zinc-ejecting inhibitors with respect to E6AP binding and p53 degradation may originate from a different selection of the zinc ion removed from E6 protein.
Experimental procedures
Plasmid Construction
A plasmid expressing the mutant form of the B1 domain from Streptococcal G protein with a C-terminal His6-tag was a gift from Dr. G. Wagner.23 The minimal region of HPV-16 E6 protein in p53 degradation covers residues 1 to 14212 and the six non-conserved cysteines (residues 16, 51, 80, 97, 111, and 140) are not necessary for E6-mediated p53 degradation activity and four-way DNA (Holliday) junction binding.15,25 Using a similar construct, we found that mutations of two of these cysteines (residues 16 and 51) disrupted E6BP binding,26 and therefore in our construct they were left unchanged and other four non-conserved cysteines were converted to serine. The sequences encoding the E6 protein and its fragments were generated by PCR and cloned into pET30a so that they contained an N-terminal GBF fusion linked by glycine-serine (Figure 1). GBF-E6N encoded residues 2 to 74 of E6, GBF-E6C encoded residues 79 to 142, and GBF-E6CL encoded residues from 79 to 151. The C-terminal His6-tagged GBF was used as a control. To focus on the tryptophan in the C-terminus of E6 in intrinsic fluorescence experiments, the other tryptophan (W43′) in the GBF region was mutated to phenylalanine in the appropriate constructs. Sequences were verified by DNA sequencing.
Protein Expression and purification
Overnight cell cultures were inoculated into 1 liter of LB medium containing 50 μg/ml of kanamycin and grown at 37°C for 3 hours until the A600 was 0.5 to 0.8. Protein production was induced with 0.4 mM IPTG for 20 hours at 28 °C. Cells were harvested by centrifugation and stored at -80°C until needed. The cell pellet of GBF-E6 was resuspended in 80 ml of 50 mM Tris-HCl (pH 8.0) with 10 mM DTT, 1 ml of 19 mg/ml PMSF 2-propanol solution and 0.5 ml EDTA-free protease inhibitor cocktail (Calbiochem), lysed by sonication, and centrifuged at 14,000 rpm for 35 min. The supernatant was loaded onto a SP-Sepharose column (1×16 cm) equilibrated with 50 mM Tris-HCl (pH 8.0) with 50 mM NaCl, then washed stepwise using (1), 150 ml 50 mM Tris-HCl (pH 8.0) with 50 mM NaCl; (2), 50 mM Tris-HCl (pH 8.0) with 5 mM DTT; (3), 50 mM Tris-HCl (pH 8.0) with 100 μM ZnSO4; and (4), 50 mM Tris-HCl (pH 8.0) with 100 mM NaCl. GBF-E6 protein was eluted by 50 mM Tris-HCl (pH 8.0) with 200 mM NaCl. The pooled fractions were concentrated to ∼3 ml using a Centriprep-30 centrifugal filter (Amicon), and DTT added to 5 mM and CHAPS to 10 mM. Further purification used a Superdex-75 (1.0 × 70 cm) gel filtration in 50 mM sodium phosphate (pH 7.50) with 200 mM NaCl, and fractions corresponding to monomeric protein were pooled. The purified protein was checked by SDS-PAGE and concentrated as above. Freshly purified proteins were used for all biophysical experiments. The expression and purification of GBF-E6(Δ76-79), GBF-E6C, and GBF-E6CL used the same protocol.
The purification of GBF-E6N protein was similar, except that after lysing the cell pellet, the supernatant was loaded to SP-Sepharose column and the flow-through was collected and concentrated. The pooled fraction was then loaded to a Q-Sepharose column at pH 8.0. The GBF-E6N protein was in the flow-through, which was pooled, concentrated and further purified by gel filtration column pre-equilibrated with 50 mM phosphate buffer (pH 6.50).
GBF-His6 was expressed as described for GBF-E6 and purified by using a Ni2+-HiTrap column in 50 mM Tris-HCl (pH 8.0) and further purified on a gel filtration column in 50 mM phosphate buffer (pH 6.50). The protein sample was desalted using G-50 Sepharose column and lyophilized.
The homogeneity of the protein was > 90% by SDS-PAGE under reducing conditions. The concentration of the proteins were determined by UV absorption at 280 nm using molar extinction coefficients of 28020 M-1cm-1 for GBF-E6, 25460 M-1cm-1 for GBF-E6(Δ76-79), 15930 M-1cm-1 for GBF-E6N, 19060 M-1cm-1 for GBF-E6C and GBF-E6CL, and 9530 M-1cm-1 for GBF-His6, respectively. The molar extinction coefficients of the constructs with a W43′F mutation in GBF were adjusted by subtracting the contribution by one tryptophan (5500 M-1cm-1).
Determination of zinc content
The zinc content was determined by using the 4-(2-pyridylazo)-resorcinol (PAR) assay.44 Zinc ions were released by treating the proteins with hydrogen peroxide (H2O2).43 Either the metal standard solution (100 μM) or the protein solution (for the experimental data) was mixed with guanidine–HCl (4 M, final concentration) and H2O2 (0.6%) in a volume of 500 μl, and incubated at room temperature for about 4 min. 500 μl of a freshly prepared solution of PAR (concentration 200 μM) was then added and kept at room temperature for 10 min. The absorbance of each sample was determined at 500 nm using a HP 8453 UV-Vis spectrometer. The zinc contents of the various monomeric GBF-fused E6 proteins were determined according the absorbance against the standard curve obtained using ZnSO4 solutions.
To verify the effect of zinc-binding of the E6 proteins, samples of GBF-E6, GBF-E6N and GBF-E6C were treated by dialyzing each against 3 different buffers overnight: 50 mM phosphate buffer (pH 6.50) alone, 50 mM phosphate buffer (pH 6.50) containing 10 mM EGTA or 10 mM EDTA. The samples were then dialyzed against 50 mM phosphate buffer (pH 6.50) to remove the additional chelating reagents before zinc determination.
NMR spectroscopy
15N-labeled samples were expressed in a modified M9 medium with 1 gram 15N-labeled Celtone-N (Spectra Stable Isotopes, USA) per liter, and purified as described above. 15N-Leu-labeled GBF-E6C was expressed in LB (with 50 mg 15N-labeled Leucine per liter) as described previously.45 15N, 1H –HSQC spectra of GBF-E6N and GBF-E6C were collected at 25 °C on Bruker Avance-300 or Avance-600 spectrometers. A 23-mer E6-binding peptide, E6apn1 (Sequence: [Ac]-ALQELLGQWLKDGGPSSGRPPPS-[NH2]) that we characterized previously,29 was used to verify binding activity of the purified GBF-E6 protein. The 1D and 2D NOESY spectra of E6apn1 (2.0 mM) and its mixture with GBF-E6 (200 μM) were collected in 50 mM phosphate (pH 7.50) with 200 mM NaCl at 5 °C on a Bruker AMX-500 spectrometer. A NOESY difference spectrum was obtained by subtracting spectrum of the peptide from that in the presence of GBF-E6 protein.
Isothermal titration calorimetry
ITC experiments were performed using samples at 20 °C in 20 mM phosphate buffer (pH 6.50) using a VP-ITC system (MicroCal). Proteins were dialyzed against buffer, centrifuged, and degassed. Typically, a 500 μM peptide solution of hDlg PDZ2 was injected 30 times in 10 μl aliquots into the 1.4 ml sample cell containing GBF-E6CL at a concentration of 50 μM. Data were fit with a nonlinear least-squares routine using a single-site binding model with Origin for ITC v.7.0 (MicroCal), varying the stoichiometry (n), the enthalpy of the reaction (ΔH) and the association constant (Ka).
SPR experiments
The interaction of proteins was measured by SPR analysis on a Biacore 3000 instrument. Ligands to be immobilized were diluted to 30 μg/ml in 10 mM sodium acetate, pH 4.0 and 5.0 and immobilized onto the dextran matrix of a CM5 sensor chip (Biacore) using the amine coupling method. All binding experiments were performed at 25 °C in 50 mM phosphate (pH 6.50) containing 200 mM NaCl. Pulses of 10 mM glycine (pH 3.0) were used to regenerate the surfaces between injections. A 33-mer peptide, E6apc2 that binds E6, and a 29-mer peptide, E6apc1 that does not bind E6, were used as controls which were characterized previously.29 SPR data were analyzed using BIAevaluation 4.1 software (Biacore). The data were fitted by nonlinear curve fitting analysis after correction for non-specific binding using a 1:1 Langmuir binding model.
UV, CD and Fluorescence spectroscopy
UV spectra were collected using a HP 8453 UV-visible spectrometer and far-UV CD spectra were recorded in a 0.1 cm cuvette on a JACSO model 810 spectropolarimeter using two scans at room temperature. Spectra were baseline corrected by buffer subtraction. Fluorescence measurements were performed at 25 °C with a SPEX Fluorolog-3 spectrofluorometer (SPEX Industries, Inc., Edison, NJ) equipped with a 450W Xe lamp, a double-grating excitation monochromator, and a double-grating emission monochromator. The high voltages were fixed at 950 V. The Xenon lamp spectrum was calibrated using the water Raman spectra. The slit widths were set to 1.25 nm for both excitation and emission. The solution (400 μl) was placed in a 0.2 × 1 cm cuvette (0.2 cm for excitation, 1 cm for emission and excited at 293 nm. Spectra were baseline corrected by buffer subtraction. Thermal stabilities were monitored by CD using a temperature range of 0 to 90 °C. Tm values were estimated by fitting the denaturation curve using Boltzmann sigmoid curves. For some data, GBF-His6 was fit first and its denaturation parameters used for deconvolution of the curves for the multi-domain constructs (Figure S3). Urea denaturations at 25 °C were performed by diluting the protein stock to the corresponding urea concentration and collecting CD or fluorescence spectra.
Analytical Gel Filtration
The aggregation of the protein samples were analyzed by using an analytical Sephacryl H100 gel filtration column (1×18 cm) in 50 mM phosphate buffer (pH 6.50) with 200 mM NaCl. The flow rate of 1.2 ml/min was controlled by Varian ProStar HPLC system and the elution of the proteins was detected by absorbance at 280 nm.
Limited proteolysis of GBF-E6 and GBF-E6(Δ76-79)
The purified protein GBF-E6 or GBF-E6(Δ76-79) was diluted to a concentration of 5 μM in a buffer with 50 mM Tris (pH 8.0) and 200 mM NaCl and incubated with chymotrypsin, sampling at various times. At each time point an aliquot of the reaction mixture was added to SDS-PAGE loading buffer at 90°C to stop the digestion. The reaction products were separated by SDS-PAGE on a 20% polyacrylamide gel, silver-stained, and quantified using the ImageQuant 5.2 Program. For sequencing, bands were transferred to PVDF membranes and submitted to the Tufts University Core Facility for N-terminal sequencing by the Edman degradation reaction.
Supplementary Material
1 table summarizing SPR results and 5 figures showing E6-dependent p53 degradation, SPR data analysis, analyses of denaturation profiles, limited proteolysis of GBF-E6 and GBF-E6(Δ76-79), and molecular size analyses of GBF-E6, GBF-E6N and GBF-E6C by gel filtration.
Acknowledgments
This work was supported by NIH grant CA119904. We thank Dr. Eric Sundberg at Boston Biomedical Research Institute for help with collecting and analyzing SPR data.
Abbreviations used
- MBP
maltose-binding protein
- CD
circular dichroism spectroscopy
- CHAPS
3-[(3-Chloromidopropyl)dimethylammonio]-1-propanesulfonate
- DDM
n-Dodecyl β-D-maltoside
- DTT
dithiothreitol
- hDlg/SAP
human homolog of the Drosophila discs large tumor suppressor protein/synapse-associated protein
- HPV
human papillomavirus
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- GST
glutathione-S-transferase
- NOESY
nuclear Overhauser effect spectroscopy
- NP40
ethylphenyl-polyethylene glycol (Nonidet P-40)
- PAR
4-(2-pyridylazo)-resorcinol
- PDZ
Postsynaptic density protein/Disc large/Zonula occludens
- PMSF
phenylmethanesulfonylfluoride
- SPR
surface plasmon resonance
- TCEP
Tris(2-Carboxyethyl) Phosphine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dell G, Gaston K. Human papillomaviruses and their role in cervical cancer. Cell Mol Life Sci. 2001;58:1923–1942. doi: 10.1007/PL00000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Baleja JD. Structure and function of the papillomavirus E6 protein and its interacting proteins. Front Biosci. 2008;13:121–134. doi: 10.2741/2664. [DOI] [PubMed] [Google Scholar]
- 3.Tungteakkhun SS, Duerksen-Hughes PJ. Cellular binding partners of the human papillomavirus E6 protein. Arch Virol. 2008;153:397–408. doi: 10.1007/s00705-007-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–1017. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- 5.Huibregtse JM, Scheffner M, Howley PM. E6-AP directs the HPV E6-dependent inactivation of p53 and is representative of a family of structurally and functionally related proteins. Cold Spring Harb Symp Quant Biol. 1994;59:237–245. doi: 10.1101/sqb.1994.059.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Huibregtse JM, Howley PM. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massimi P, Gammoh N, Thomas M, Banks L. HPV E6 specifically targets different cellular pools of its PDZ domain-containing tumour suppressor substrates for proteasome-mediated degradation. Oncogene. 2004;23:8033–8039. doi: 10.1038/sj.onc.1207977. [DOI] [PubMed] [Google Scholar]
- 9.Imai Y, Tsunokawa Y, Sugimura T, Terada M. Purification and DNA-binding properties of human papillomavirus type 16 E6 protein expressed in Escherichia coli. Biochem Biophys Res Commun. 1989;164:1402–1410. doi: 10.1016/0006-291x(89)91826-3. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Alai MM, Dantur KI, Smal C, Pietrasanta L, Prat-Gay G. High-Risk HPV E6 Oncoproteins Assemble into Large Oligomers that Allow Localization of Endogenous Species in Prototypic HPV-Transformed Cell Lines. Biochemistry. 2007;46:341–349. doi: 10.1021/bi602457q. [DOI] [PubMed] [Google Scholar]
- 11.Nomine Y, Ristriani T, Laurent C, Lefevre JF, Weiss E, Trave G. A strategy for optimizing the monodispersity of fusion proteins: application to purification of recombinant HPV E6 oncoprotein. Protein Eng. 2001;14:297–305. doi: 10.1093/protein/14.4.297. [DOI] [PubMed] [Google Scholar]
- 12.Lipari F, McGibbon GA, Wardrop E, Cordingley MG. Purification and biophysical characterization of a minimal functional domain and of an N-terminal Zn2+-binding fragment from the human papillomavirus type 16 E6 protein. Biochemistry. 2001;40:1196–1204. doi: 10.1021/bi001837+. [DOI] [PubMed] [Google Scholar]
- 13.Nomine Y, Charbonnier S, Ristriani T, Stier G, Masson M, Cavusoglu N, Van Dorsselaer A, Weiss E, Kieffer B, Trave G. Domain substructure of HPV E6 oncoprotein: biophysical characterization of the E6 C-terminal DNA-binding domain. Biochemistry. 2003;42:4909–4917. doi: 10.1021/bi026980c. [DOI] [PubMed] [Google Scholar]
- 14.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, Sibler AP, Desplancq D, Atkinson RA, Weiss E, Orfanoudakis G, Kieffer B, Trave G. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 16.Degenkolbe R, Gilligan P, Gupta S, Bernard HU. Chelating agents stabilize the monomeric state of the zinc binding human papillomavirus 16 E6 oncoprotein. Biochemistry. 2003;42:3868–3873. doi: 10.1021/bi027390h. [DOI] [PubMed] [Google Scholar]
- 17.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Hall JP. Applying fusion protein technology to E. coli. 2007 Biopharm International On line journal: http://biopharminternational.findpharma.com/biopharm/Article/Applying-Fusion-Protein-Technology-to-E-coli/ArticleStandard/Article/detail/423193.
- 19.Hammarstrom M, Hellgren N, van Den BS, Berglund H, Hard T. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 2002;11:313–321. doi: 10.1110/ps.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N Y) 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 21.Kukimoto I, Aihara S, Yoshiike K, Kanda T. Human papillomavirus oncoprotein E6 binds to the C-terminal region of human minichromosome maintenance 7 protein. Biochem Biophys Res Commun. 1998;249:258–262. doi: 10.1006/bbrc.1998.9066. [DOI] [PubMed] [Google Scholar]
- 22.Lechner MS, Laimins LA. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J Virol. 1994;68:4262–4273. doi: 10.1128/jvi.68.7.4262-4273.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR. 2001;20:11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- 24.Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ristriani T, Masson M, Nomine Y, Laurent C, Lefevre JF, Weiss E, Trave G. HPV oncoprotein E6 is a structure-dependent DNA-binding protein that recognizes four-way junctions. J Mol Biol. 2000;296:1189–1203. doi: 10.1006/jmbi.2000.3527. [DOI] [PubMed] [Google Scholar]
- 26.Be X. PhD thesis. Sackler School of Graduate Biomedical Sciences, Tufts University; 2001. Characterization of human papillomaviral E6 protein and structural studies of E6-E6AP interaction and antibody surrogate light chain with NMR. [Google Scholar]
- 27.Nomine Y, Charbonnier S, Miguet L, Potier N, Van Dorsselaer A, Atkinson RA, Trave G, Kieffer B. 1H and 15N resonance assignment, secondary structure and dynamic behaviour of the C-terminal domain of human papillomavirus oncoprotein E6. J Biomol NMR. 2005;31:129–141. doi: 10.1007/s10858-004-7802-y. [DOI] [PubMed] [Google Scholar]
- 28.Bagby S, Tong KI, Liu D, Alattia JR, Ikura M. The button test: a small scale method using microdialysis cells for assessing protein solubility at concentrations suitable for NMR. J Biomol NMR. 1997;10:279–282. doi: 10.1023/a:1018359305544. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Liu Z, Androphy E, Chen J, Baleja JD. Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus. Biochemistry. 2004;43:7421–7431. doi: 10.1021/bi049552a. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Henry GD, Hegde RS, Baleja JD. Solution structure of the hDlg/SAP97 PDZ2 domain and its mechanism of interaction with HPV-18 papillomavirus E6 protein. Biochemistry. 2007;46:10864–10874. doi: 10.1021/bi700879k. [DOI] [PubMed] [Google Scholar]
- 31.Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 32.Be X, Hong Y, Wei J, Androphy EJ, Chen JJ, Baleja JD. Solution structure determination and mutational analysis of the papillomavirus E6 interacting peptide of E6AP. Biochemistry. 2001;40:1293–1299. doi: 10.1021/bi0019592. [DOI] [PubMed] [Google Scholar]
- 33.Zanier K, Charbonnier S, Baltzinger M, Nomine Y, Altschuh D, Trave G. Kinetic analysis of the interactions of human papillomavirus E6 oncoproteins with the ubiquitin ligase E6AP using surface plasmon resonance. J Mol Biol. 2005;349:401–412. doi: 10.1016/j.jmb.2005.03.071. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Dasgupta J, Ma RZ, Banks L, Thomas M, Chen XS. Structures of a HPV-E6 polypeptide bound to MAGUK proteins: mechanisms of targeting tumor suppressors by a high-risk HPV oncoprotein. J Virol. 2007;87:3618–3626. doi: 10.1128/JVI.02044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas M, Dasgupta J, Zhang Y, Chen X, Banks L. Analysis of specificity determinants in the interactions of different HPV E6 proteins with their PDZ domain-containing substrates. Virology. 2008;376:371–378. doi: 10.1016/j.virol.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Charbonnier S, Stier G, Orfanoudakis G, Kieffer B, Atkinson RA, Trave G. Defining the minimal interacting regions of the tight junction protein MAGI-1 and HPV16 E6 oncoprotein for solution structure studies. Protein Expr Purif. 2008;60:64–73. doi: 10.1016/j.pep.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 38.Nomine Y, Ristriani T, Laurent C, Lefevre JF, Weiss E, Trave G. Formation of soluble inclusion bodies by HPV E6 oncoprotein fused to maltose-binding protein. Protein Expr Purif. 2001;23:22–32. doi: 10.1006/prep.2001.1451. [DOI] [PubMed] [Google Scholar]
- 39.Crook T, Tidy JA, Vousden KH. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 40.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SS, Weiss RS, Javier RT. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spanos WC, Geiger J, Anderson ME, Harris GF, Bossler AD, Smith RB, Klingelhutz AJ, Lee JH. Deletion of the PDZ motif of HPV16 E6 preventing immortalization and anchorage-Independent growth in human tonsil epithelial cells. Head Neck. 2008;30:139–147. doi: 10.1002/hed.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beerheide W, Bernard HU, Tan YJ, Ganesan A, Rice WG, Ting AE. Potential drugs against cervical cancer: zinc-ejecting inhibitors of the human papillomavirus type 16 E6 oncoprotein. J Natl Cancer Inst. 1999;91:1211–1220. doi: 10.1093/jnci/91.14.1211. [DOI] [PubMed] [Google Scholar]
- 44.McCall KA, Fierke CA. Colorimetric and fluorimetric assays to quantitate micromolar concentrations of transition metals. Anal Biochem. 2000;284:307–315. doi: 10.1006/abio.2000.4706. [DOI] [PubMed] [Google Scholar]
- 45.Lee KM, Androphy EJ, Baleja JD. A novel method for selective isotope labeling of bacterially expressed proteins. J Biomol NMR. 1995;5:93–96. doi: 10.1007/BF00227474. [DOI] [PubMed] [Google Scholar]
- 46.Byeon IJ, Louis JM, Gronenborn AM. A protein contortionist: core mutations of GB1 that induce dimerization and domain swapping. J Mol Biol. 2003;333:141–152. doi: 10.1016/s0022-2836(03)00928-8. [DOI] [PubMed] [Google Scholar]
- 47.Sherman L, Jackman A, Itzhaki H, Stoppler MC, Koval D, Schlegel R. Inhibition of serum- and calcium-induced differentiation of human keratinocytes by HPV16 E6 oncoprotein: role of p53 inactivation. Virology. 1997;237:296–306. doi: 10.1006/viro.1997.8778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1 table summarizing SPR results and 5 figures showing E6-dependent p53 degradation, SPR data analysis, analyses of denaturation profiles, limited proteolysis of GBF-E6 and GBF-E6(Δ76-79), and molecular size analyses of GBF-E6, GBF-E6N and GBF-E6C by gel filtration.