Abstract
Objective
A recent admixture mapping analysis identified interleukin 6 (IL6) and IL6 receptor (IL6R) as candidate genes for inflammatory diseases. In the airways during allergic inflammation, IL6 signaling controls the production of proinflammatory and anti-inflammatory factors. In addition, albuterol, a commonly prescribed asthma therapy, has been shown to influence IL6 gene expression. Therefore, we reasoned that interactions between the IL6 and IL6R genes might be associated with bronchodilator drug responsiveness to albuterol in asthmatic patients.
Methods
Four functional IL6 single nucleotide polymorphisms (SNPs) and a nonsynonymous IL6R SNP were genotyped in 700 Mexican and Puerto Rican asthma families and in 443 African-American asthma cases and controls. Both family-based association tests and linear regression models were used to assess the association between individual SNPs and haplotypes with bronchodilator response. Gene–gene interactions were tested by using multiple linear regression analyses.
Results
No single SNP was consistently associated with drug response in all the three populations. However, on the gene level, we found a consistent IL6 and IL6R pharmacogenetic interaction in the three populations. This pharmacogenetic gene–gene interaction was contextual and dependent upon ancestry (racial background). This interaction resulted in higher drug response to albuterol in Latinos, but lower drug response in African-Americans. Herein, we show that there is an effect modification by ancestry on bronchodilator responsiveness to albuterol.
Conclusion
Genetic variants in the IL6 and IL6R genes act synergistically to modify the bronchodilator drug responsiveness in asthma and this pharmacogenetic interaction is modified by the genetic ancestry.
Keywords: asthma genetics, African-Americans, ancestry, effect modification, gene–gene interaction, IL6, IL6 receptor, latinos
Introduction
The interleukin-6 gene (IL6) pathway has been implicated in the pathogenesis of asthma, because it controls the production of proinflammatory and anti-inflammatory factors in the airways during allergic inflammation [1]. Compared with healthy controls, individuals with asthma have increased circulating levels of both IL6 and soluble IL6 receptor (sIL6R), which are further increased during an asthma attack or antigen inhalation [2,3].
A recent admixture mapping analysis of an African-American cohort showed an association between European ancestry on chromosome 1 and sIL6R levels [4]. Fine mapping of this region determined that a single coding variant of the IL6R gene, which resulted in a nonsynonymous amino acid substitution (rs8192284), was strongly associated with sIL6R and IL6 serum levels in Blacks and Whites. Interestingly, the frequency of this variant was highest in Europeans (35%) and lowest in West Africans (4%). In addition, genetic variants in the IL6 gene have been shown to affect IL6 gene expression and IL6 serum levels [5–7]. Furthermore, albuterol, a commonly used asthma medication, has been shown to increase IL6 gene expression in vivo and in vitro [8–10].
On the basis of the racial-specific genetic differences and the biological evidence, we hypothesized that genetic variants in the IL6 gene pathway interact to modify the bronchodilator drug responsiveness between Latino and African-American individuals with asthma. To test this hypothesis, we analyzed individuals from the Genetics of Asthma in Latino Americans (GALA) Study and the Study of African-Americans, Asthma, Genes, and Environments (SAGE) [11,12].
Methods
Study participants
Recruitment and patient characteristics have been described earlier elsewhere [11,12]. Briefly, 700 Latinos consisting of 301 Mexicans and 399 Puerto Ricans with asthma (probands) and their biological parents were enrolled to the GALA study. African-American asthmatic patients (nλ=λ267) and healthy controls (nλ=λ176) were enrolled to the SAGE study. In both GALA and SAGE studies, ethnicity was self-reported and patients were enrolled only if both biological parents and all four biological grandparents were of the same ethnic background. Patients were 8–40 years of age, had a current physician diagnosis of asthma, and had experienced asthma symptoms (wheezing, cough, or shortness of breath) over the last 2 years. Local institutional review boards approved all studies, and all participants provided written, age-appropriate informed consent.
Pulmonary function tests
Spirometry was performed according to the ATS standards [13]. Pulmonary function test results were expressed as a percentage of the predicted normal value using age-adjusted prediction equations from Hankinson et al. [14]. Baseline lung function was determined by the forced expiratory volume in 1λs expressed as Pre-FEV1. Albuterol was administered by a standard metered dose inhaler through a spacer device: 180λµg (two puffs) for patients aged below 16 years, and 360λµg (four puffs) for patients aged above or equal to 16 years. A quantitative measure of bronchodilator drug responsiveness (ΔFEV1) was calculated as the percentage change in baseline lung function (Pre-FEV1) after albuterol administration using the formula: ([post-FEV1 (in liters)– pre-FEV1 (in liters)]×100)/pre-FEV1 (in liters).
Individual ancestry estimation
To determine the individual genetic ancestry estimates, we genotyped 104 ancestry informative markers (AIMs) in all Mexicans, Puerto Ricans, and African-American asthmatic patients. The AIMs were selected to be distributed across the genome and encompassed large allele frequency differences between Native American, West African, and European ancestral populations. Genotyping data of these AIMs were imputed into the program STRUCTURE 2.1 to determine the individual genetic ancestry estimates in each patient. To correct for population stratification, individual genetic ancestry estimates were included in the regression models [15].
Controlling for population admixture
Population substructure and admixture can confound genetic association studies in unrelated individuals, thus resulting in spurious associations [16]. Given that our selected sample sets were drawn from highly admixed populations, we addressed this possible confounding by using a family-based design (Transmission Disequilibrium Test), which is not susceptible to confounding because of population structure and/or recent admixture [16]. The Transmission Disequilibrium Test includes both parents and an affected child, where an allele is tested for association with a phenotype based on over or under transmission [16]. We also measured genetic ancestry and controlled for genetic confounding between unrelated individuals.
Selection of single nucleotide polymorphisms
The SNPs were selected based on their potential to alter gene function. Of the selected SNPs, two SNPs in the IL6 gene promoter [−174C/G (rs1800795) and −572C/G (rs1800796)] have been extensively studied and have been shown to affect the IL6 gene expression and serum levels [5–7]. The two IL6 coding variants, Pro32Ser (rs2069830) and Asp162Glu (rs13306435), have been recently described to affect IL6 binding to gp130 [17,18]. The genetic variant IL6R Asp358Ala (rs8192284), which was identified through admixture mapping, corresponds to the proteolytic cleavage site of the IL6R and affects serum levels of sIL6R and IL6 [4,19].
Before carrying out large-scale genotyping, we sought to determine the allele frequencies of these SNPs in a subset of 72 unrelated asthmatic patients, 24 of each ethnic group (Mexican, Puerto Rican, and African-American). The inclusion of 48 chromosomes for each group provides greater than 80% power to detect any polymorphic variants with a minor allele frequency (MAF) of greater than 5% with a significance level (α) of less than 0.05. The genetic variants with a MAF greater than 5% were further selected to be genotyped in the following populations: IL6R rs8192284 and IL6 rs1800796 were genotyped in all the three populations; IL6 rs1800795 in both Puerto Ricans and Mexicans; IL6 rs2069830 in both Puerto Ricans and African-Americans; and IL6 rs13306435 in Mexicans only.
Genotyping
The SNPs were genotyped using the AcycloPrime-FP (PerkinElmer, Waltham, Massachusetts, USA) method [20]. The PCR cocktail included: 2.4–4.0λng of genomic DNA, 0.1–0.2λµmol/l of primers, 2.5λmmol/l of MgCl2, 50λµmol/l of dNTPs, 6λµl volume with Platinum Taq PCR buffer, and 0.1–0.2λU Platinum Taq (Invitrogen, Carlsbad, California, USA). PCR cycling conditions were as follows: one cycle 95°C for 2λmin, 35 cycles of 92°C for 10λs, 58°C for 20λs, 68°C for 30λs, and final extension at 68°C for 10λmin. We used AcycloPrime-FP kits for enzymatic cleanup and single-base extension genotyping reactions. Plates were read on an EnVision fluorescence polarization plate reader (PerkinElmer).
Statistical analyses
In the GALA trios, Mendelian inconsistencies were identified using PedCheck [21]. Families with Mendelian inconsistencies were excluded from further analyses. The Hardy–Weinberg equilibrium (HWE) was calculated by means of the χ2 goodness-of-fit tests for each ethnic group. Pair-wise linkage disequilibrium (LD) was estimated by using the program LD Plotter, which uses an iterative expectation–maximization algorithm to calculate r2 values [22].
Single SNP associations
Family-based association tests [23] and family-based tests for associating haplotypes (Haplotype-Based Association Testing, HBAT) [24] were used to assess the association between individual SNPs and haplotypes with the bronchodilator drug responsiveness in Latino asthmatic patients. Haplotypes were determined using HBAT [24]. Multiple linear regression models were used to assess the association between individual SNPs with the bronchodilator drug response in African-Americans. In all the three populations, we tested for an association with bronchodilator responsiveness (defined by the quantitative phenotype of ΔFEV1) and with either high or low bronchodilator responsiveness (defined by the qualitative phenotype of ΔFEV1 greater or less than 12%, respectively) [25]. A dominant SNP model was used for the analyses to provide greater statistical power by generating tests with few degrees of freedom and allowing for tests of interaction between SNPs of low allele frequency. Given the number of comparisons made, a permutation test implemented in the program PLINK was used to assess the reliability of the results [26]. This permutation tests calculates empirical P values based on the number of times the permuted test is greater than the observed test.
Gene–gene interactions
The effects of gene–gene interaction on drug response (ΔFEV1) were determined by using multiple linear regression models. Specifically, SNPs of the IL6 gene were paired with the IL6R SNP to model the effect of gene–gene interaction on the bronchodilator drug responsiveness. The following variables were tested for their inclusion in a backward stepwise regression approach based on their potential for confounding: age, sex, ancestry, ethnicity, asthma duration, pre-FEV1 and use of short-acting β2-agonists, long-acting β2-agonists, inhaled corticosteroids, oral corticosteroids, leukotrienes, theophylline, and cromolyn. An F-test determined which of these aforementioned variables significantly affected the gene–gene-interaction model. The variables included in the final model were age, ancestry, ethnicity, asthma duration, and pre-FEV1. All regression analyses were performed using the statistical software package STATA/SE 9.0 (StataCorp, College Station, Texas, USA). Analyses were adjusted for baseline level of lung function, that is, Pre-FEV1.
Pharmacogenetic effect modification by genetic ancestry
Prior genetic analyses showed ancestry-specific genetic differences at the IL6R locus, which were associated with circulating serum levels of both the IL6 and sIL6R [4]. Circulating levels of these cytokines significantly differed between African-Americans and Whites. Therefore, we reasoned that the pharmacogenetic effect resulting from the gene–gene interaction might be modified by genetic ancestry. A three-way gene–gene–ancestry interaction term was generated to model the effect of gene–gene–ancestry pharmacogenetic interaction on ΔFEV1. For this analysis, we analyzed SNPs, IL6 rs1800796 and IL6R rs8192284, which were genotyped in all the three populations. To illustrate the effect of ancestry on the pharmacogenetic interaction, mean bronchodilator drug response (ΔFEV1) resulting from the interaction between the C allele of IL6 rs1800796 and the C allele of IL6R rs8192284 was generated by obtaining mean ΔFEV1 for every quartile of the percentage of European and Native American ancestry in each individual.
Results
Characteristics of the patients
Demographic and clinical characteristics of the 700 probands with asthma (301 Mexicans and 399 Puerto Ricans) and 267 African-Americans with asthma are shown in Table 1. The median age of the Mexican, Puerto Rican, and African-American asthmatic patients was 16.5, 14, and 19 years, respectively. Both Mexican and Puerto Rican asthmatic populations had approximately 50% male and female patients, whereas African-American asthmatic populations had 39.3% male patients. The median baseline lung function (Pre-FEV1) was 88.9, 83.7, and 91.5% in Mexican, Puerto Rican, and African-American asthmatic patients, respectively. As described earlier, the bronchodilator responsiveness measured by the ΔFEV1 was significantly greater for Mexican asthmatic patients than for Puerto Rican or African-American asthmatic patients (P<0.0001) [11].
Table 1.
Demographic and clinical characteristics of the Mexican and the Puerto Rican individuals with asthma from the GALA study and the African-American individuals with asthma from the SAGE study
| Mexicans (nλ=λ301) | Puerto Ricans (nλ=λ399) |
African-Americans (nλ=λ267) |
|
|---|---|---|---|
| λCharacteristics: | |||
| νAge (years) | 16.5 (11λ:λ22) | 14.0 (9λ:λ19) | 19.3 (13λ:λ26) |
| νSex (% male) | 53.8 | 55.9 | 39.3 |
| νBMI (kg/m2) | 24.5 (20λ:λ29) | 22.3 (18λ: λ27) | 26.9 (22λ:λ32) |
| νPlasma IgE (IU/ml) | 470.4 (50λ:λ891) | 487.9 (58λ:λ918) | 292.4 (0λ:λ590) |
| Baseline Spirometry: | |||
| νPre-FEV1 (% predicted) | 88.9 (76λ:λ101) | 83.7 (72:95) | 91.5 (79λ:λ103) |
| νPre-FEV1 (% predicted) <80% | 30.7 | 39.9 | 30.7 |
| Bronchodilator responsiveness: | |||
| νΔFEV1 (relative % change) | 10.1 (2λ:λ19) | 6.2 (0λ:λ15) | 9.8 (3λ:λ17) |
Values are expressed as median (25thλ:λ75th percentile) and were missing for some individualss.
BMI, body mass index, Pre-FEV1, baseline forced expiratory volume in 1λs expressed as percentage of predicted, ΔFEV1, relative percent change in Pre-FEV1 after albuterol administration; GALA, Genetics of Asthma in Latino Americans; IL6, interleukin 6; SAGE, Study of African-Americans, Asthma, Genes and Environments.
Allele frequencies, Hardy–Weinberg equilibrium, and linkage disequilibrium
The MAFs of IL6R and IL6 SNPs for Mexicans, Puerto Ricans, and African-Americans are listed in Table 2. The observed distribution of genotypes within each ethnic group was in the HWE and the frequencies did not differ from those reported in public databases (Table 2 lists the frequencies available in public databases in different ethnic groups for comparison). Similar to the allele frequencies noted in public databases and to Reich et al. [4], we observed, that the C allele of IL6R SNP rs8192284 was overrepresented in Native Americans in comparison with other ethnic groups. Among our study participants, Native American ancestry, on average, was highest in Mexicans (52%), intermediate in Puerto Rican (18%), and lowest in African-Americans (<1%). The allele frequency of the IL6R SNP rs8192284 paralleled Native American ancestry. Mexicans had the highest frequency of the C allele of IL6R rs8192284 (0.54), whereas Puerto Ricans had an intermediate frequency (0.41), and African-Americans had the lowest frequency (0.13). No significant LD was observed between the IL6 SNPs (r2<0.7, Table 3).
Table 2.
Minor allele frequencies and P value for HWE analysis for IL6 and IL6R variants in Mexican and Puerto Rican GALA probands and parents and African-American SAGE cases and controls
| Mexicans | Puerto Ricans | African-Americans | HapMapb | SNP500Cancer Databaseb |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Probands | Parents | Probands | Parents | Cases | Controls | Caucasians | Africans | Hispanics | |
| λIL6 | |||||||||
| rs1800795 | |||||||||
| C allele | 0.13 | 0.14 | 0.23 | 0.24 | NAa | NAa | 0.53 | 0 | 0.22 |
| HWE (P value) | 0.28 | 0.46 | 0.34 | 0.84 | |||||
| IL6 | |||||||||
| rs1800796 | |||||||||
| C allele | 0.34 | 0.31 | 0.13 | 0.14 | 0.10 | 0.12 | 0.04 | 0.09 | 0.28 |
| HWE (P value) | 0.34 | 0.70 | 0.33 | 0.52 | 0.74 | 0.82 | |||
| IL6 | |||||||||
| rs13306435 | |||||||||
| A allele | 0.11 | 0.13 | NAa | NAa | NAa | NAa | 0.0 | 0.03 | 0.02 |
| HWE (P value) | 0.68 | 0.06 | |||||||
| IL6 | |||||||||
| rs2069830 | |||||||||
| T allele | NAa | NAa | 0.03 | 0.02 | 0.08 | 0.10 | 0.0 | 0.09 | |
| HWE (P value) | 0.62 | 0.52 | 0.25 | 0.22 | |||||
| IL6R | |||||||||
| rs8192284 | |||||||||
| Ala | 0.54 | 0.55 | 0.41 | 0.41 | 0.13 | 0.12 | 0.35 | 0.04 | 0.48 |
| HWE (P value) | 0.41 | 0.31 | 0.95 | 0.06 | 0.49 | 0.07 | |||
HWE, Hardy–Weinberg equilibrium; GALA, Genetics of Asthma in Latino Americans; IL6, interleukin 6; SAGE, Study of African-Americans, Asthma, Genes and Environments.
NA, not applicable, because of a low minor allele frequency (<5%) in the populations.
Allele frequencies from public database are listed for comparison.
Table 3.
Pairwise linkage disequilibrium (r2) between the interleukin 6 (IL6) variants in Mexicans, Puerto Ricans, and African-Americans
| Mexicans | |||
|---|---|---|---|
| rs1800795 | rs1800796 | rs13306435 | |
| λrs1800795 | — | ||
| rs1800796 | 0.067 | — | |
| rs13306435 | 0.022 | 0.063 | — |
| Puerto Ricans | |||
| rs1800795 | rs1800796 | rs2069830 | |
| rs1800795 | — | ||
| rs1800796 | 0.045 | — | |
| rs2069830 | 0.008 | 0.004 | — |
| African-Americans | |||
| rs1800796 | rs2069830 | ||
| rs1800796 | — | ||
| rs2069830 | 0.011 | — | |
Association analysis of IL6 and IL6R genotypes and haplotypes with the bronchodilator drug response
Among Mexicans, the IL6 SNPs rs1800796, in the promoter region, and rs13306435, in the exon region, were significantly associated with lower drug response, as defined by a ΔFEV1 less than 12% (Table 4). The C allele of rs1800796 conferred an OR of 1.40 for lower drug response among Mexicans (95% confidence intervalλ=λ1.00–1.96, Pλ=λ0.02). In addition, the A allele of rs13306435 conferred an OR of 0.57 for lower drug response among Mexicans (95% confidence intervalλ=λ0.36–0.90, Pλ=λ0.002). These results in Mexicans remained statistically significant in SNP rs1800796 and SNP rs13306435 after 360 and 1014 permutation tests, respectively. In contrast, no significant association with the bronchodilator drug response was found for SNP rs1800796 in Puerto Ricans or African-Americans (Table 4), although the frequency of the C allele of this SNP was lower (0.14 in Puerto Ricans and 0.12 in African-Americans vs. 0.34 in Mexicans, Table 2). There was no significant association between IL6R genotypes with the bronchodilator response in the three populations.
Table 4.
Association analyses of IL6 and IL6R variants with bronchodilator response (ΔFEV1) among Mexican trios, Puerto-Rican trios and African-American cases and controls
| IL6 variants | IL6R variant | ||||
|---|---|---|---|---|---|
| rs1800795 | rs1800796 | rs13306435 | rs2069830 | rs8192284 | |
| λMexicans (301 trios) | |||||
| ΔFEV1 | (−) 0.43 | (−) 0.22 | (+) 0.58 | NA | (−) 0.29 |
| ΔFEV1 >12% | (−) 0.18 | (+) 0.93 | (+) 0.12 | (−) 0.84 | |
| ΔFEV1 <12% | (+) 0.25 | (+) 0.02* | (−) 0.002* | (−) 0.53 | |
| Puerto Ricans (399 trios) |
|||||
| νΔFEV1 | (−) 0.65 | (−) 0.24 | NA | (−) 0.72 | (+) 0.46 |
| νΔFEV1 >12% | (+) 0.56 | (−) 0.28 | NA | (+) 0.15 | |
| νΔFEV1 <12% | (−) 0.33 | (−) 0.96 | (+) 0.16 | (+) 0.47 | |
| African-Americans | |||||
| (267 cases, 176 controls) | |||||
| νΔFEV1 | NA | (−) 0.10 | NA | (−) 0.23 | (+) 0.68 |
| νΔFEV1 >12% | (−) 0.85 | (−) 0.32 | (−) 0.46 | ||
| νΔFEV1 <12% | NA | NA | NA | ||
Analyses were performed using a dominant model. For the trios (Mexicans and Puerto Ricans), family-based association analyses were performed, where (+) and (−) indicate the direction of the association. For the quantitative trait ΔFEV1, (+) means that the genotype is associated with a higher value of the trait and (−) with a lower value. For the qualitative traits, ΔFEV1 greater or less than 12%, (+) means that the genotype is overtransmitted to the probands and (−) undertransmitted.
For the cases and controls (African-Americans), linear regression analyses were performed, where (+) and (−) indicate the direction of the association: (+) represents genotypes associated with higher value of the trait and (−) with lower value.
IL6,interleukin 6; IL6R, interleukin 6 receptor.
P values <0.05. For definition of abbreviations, see Table 1.
A total of four different IL6 haplotypes (frequency of > 1%) were observed in Mexican, Puerto Rican, and African-American asthmatic patients. The haplotype association results were consistent with those of the individual SNPs. Among Mexicans, the haplotypes which carried either the C allele of SNP rs1800796 or the A allele of SNP rs13306435 were associated with lower drug response, as defined by a ΔFEV1 less than 12% (Pλ=λ0.05 and 0.002, respectively).
IL6 and IL6R gene–gene interaction analyses
We tested for the effects of a gene–gene interaction on bronchodilator drug response (ΔFEV1, Table 5). All of the SNPs genotyped were used to analyze the IL6R/IL6 gene–gene interaction (i.e. IL6R SNP rs8192284 modeled with IL6 SNP rs1800795, rs1800796, and rs13306435 among Mexicans; IL6R SNP rs8192284 modeled with IL6 SNP rs1800795, rs1800796, and rs2069830 among Puerto Ricans; and IL6R SNP rs8192284 modeled with IL6 SNP rs1800796 and rs2069830 among African-Americans). The alleles of the SNPS in the gene–gene interaction were found to be significantly associated with modified bronchodilator drug response among Mexicans and African-Americans. Among Mexicans, the C allele of IL6R rs8192284 significantly interacted with the C allele of IL6 rs1800795, the C allele of IL6 SNP rs1800796, and the A allele of IL6 SNP rs13306435 to be associated with increased bronchodilator drug responsiveness (Pλ=λ0.005, 0.009 and 0.005, respectively). Among Puerto Ricans, the interaction of the C allele of IL6R rs8192284 with the C allele of IL6 SNP rs1800796, and the T allele of IL6 SNP rs2069830 were marginally associated with increased bronchodilator drug response (Pλ=λ0.08 and 0.06, respectively). In contrast, the interaction of C allele of IL6R rs8192284 with the T allele of IL6 SNP rs2069830 significantly interacted to be associated with decreased bronchodilator drug response among African-Americans (Pλ=λ0.04).
Table 5.
P values for gene–gene interaction on drug responsiveness (ΔFEV1) among Mexican, Puerto Rican, and African-American individuals with asthma
| Gene–gene interaction | ||||
|---|---|---|---|---|
| IL6 | IL6R | Mexicans | Puerto Ricans | African-Americans |
| λrs1800795 | rs8192284 | (+) 0.005* | (+) 0.22 | NA |
| rs1800796 | rs8192284 | (+) 0.009* | (+) 0.08 | (−) 0.28 |
| rs13306435 | rs8192284 | (+) 0.005* | NA | NA |
| rs2069830 | rs8192284 | NA | (+) 0.06 | (−) 0.04* |
Multiple linear regression analysis was performed with genetic variants categorized in a dominant model. For ΔFEV1, positive (+) direction of association indicates that the gene–gene interaction is associated with an increased drug response and negative (−) direction of association indicates that the gene–gene interaction is associated with a decreased drug response.
IL6, interleukin 6; IL6R, interleukin 6 receptor.
P values <0.05. For definition of abbreviations, see Table 1.
Pharmacogenetic effect modification by genetic ancestry
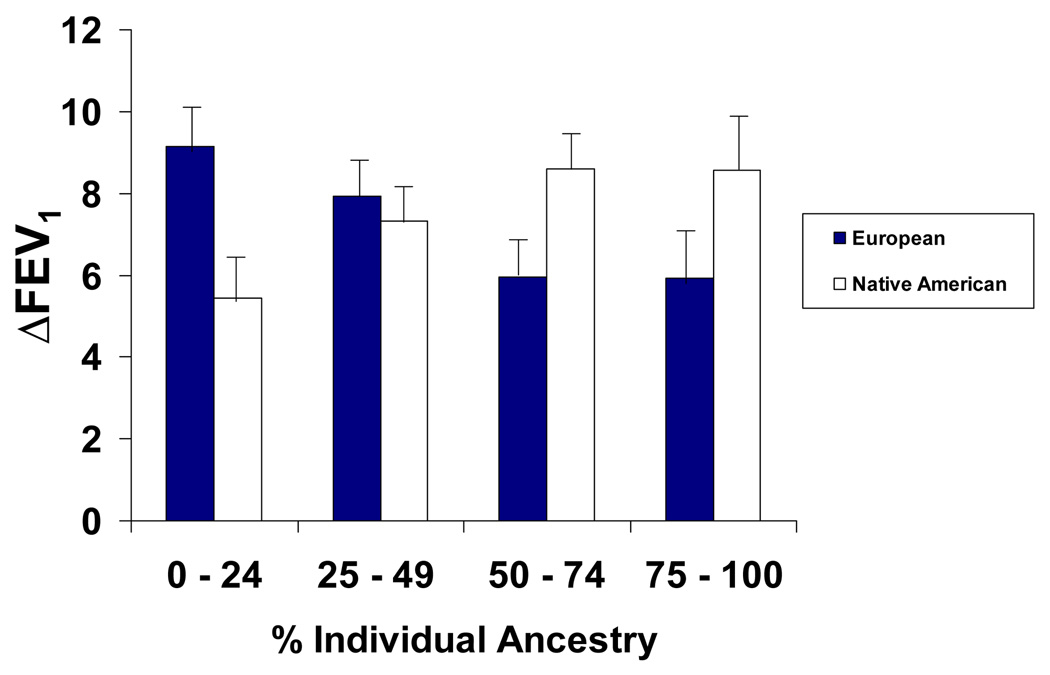
The IL6 and IL6R pharmacogenetic gene–gene interaction resulted in higher drug response to albuterol in Latinos, but lower drug response in African-Americans. We reasoned that these results were, in part, contextual and dependent upon the ancestral background of the population. Therefore, we tested for effect modification by genetic ancestry on drug response. Significant effect modification was observed with Native American ancestry within the Mexican population. Specifically, among Mexican asthmatic patients, the C allele of IL6R rs8192284 and the C allele of IL6 SNP rs1800796 interacted with Native American ancestry to result in a significantly increased bronchodilator drug response (Pλ=λ0.03) and a trend toward interaction with European ancestry that resulted in decreased bronchodilator drug response (Pλ=λ0.09). Although the pharmacogenetic interaction between the C allele of the IL6R SNP rs8192284 and the C allele of the IL6 SNP rs1800796 with ancestry did not reach statistical significance in Puerto Rican or African-American asthmatic patients, a similar trend in the same direction was observed; the C allele of IL6R SNP rs8192284 and the C allele of IL6 SNP rs1800796 pharmacogenetic interaction with Native American ancestry resulted in a higher bronchodilator drug response in Puerto Rican asthmatic patients, whereas this same pharmacogenetic interaction with European ancestry resulted in a lower bronchodilator drug response in both Puerto Rican and African-American asthmatic patients. On the basis of this similar trend of effect modification by ancestry along with similar frequencies in the HWE in both Mexican and Puerto Rican asthmatic patients, we combined these two populations together to show the mean bronchodilator drug response (ΔFEV1) for the C allele of IL6R SNP rs8192284 and the C allele of IL6 SNP rs1800796 pharmacogenetic interaction with ancestry (Fig. 1). To control for possible marginal effects of ancestry on drug response, ancestry was included as an independent variable in the regression model. The mean ΔFEV1 for the C allele of IL6R SNP rs8192284 and the C allele of IL6 SNP rs1800796 pharmacogenetic interaction increased with increasing amounts of Native American ancestry; whereas, the mean ΔFEV1 for the same pharmacogenetic interaction decreased with increasing amounts of European ancestry.
Fig. 1.
The mean ΔFEV1 for the combination of the C allele of IL6 rs1800796 with the C allele of IL6R rs8192284 gene–gene interaction among Mexican and Puerto Rican asthmatic patients with effect modification by ancestry. Data are represented as mean±SEM. FEV1, forced expiratory volume in 1λs.
Discussion
In this study, we identified the involvement of the IL6 and IL6R genes in the bronchodilator drug response in Latino and African-American asthmatic patients. The inclusion of three ethnically diverse populations (Mexicans, Puerto Ricans, and African-Americans) has allowed us to identify an effect modification by ancestry on the IL6 and IL6R genetic interaction of drug response amongst asthmatic patients. Although no single SNP was consistently associated with drug response in the three populations, on the gene level, we have found a consistent gene–gene pharmacogenectic interaction. Among admixed populations, complex genetic associations may be more informative on the gene level rather than on the level of individual SNPs [27]. Furthermore, cross-ethnic validation provides strong evidence for true genetic effects on disease especially after controlling for population structure by family-based design [28]. These findings are further supported by the fact that both IL6 and IL6R have been implicated in the pathogenesis of asthma [29]. Together, as a whole, these results suggest that there is a pharmacogenetic interaction between IL6 and IL6R genes and bronchodilator drug response in asthma.
Results of an admixture mapping analysis for inflammatory markers led us to explore the IL6 pathway as a potential modifier of bronchodilator drug response [4]. In the analysis of inflammatory markers, Reich et al. [4] showed that a missense mutation, IL6R SNP rs8192284, fully accounted for an admixture peak. Furthermore, this genetic variant influenced circulating markers of inflammation. The IL6R SNP rs8192284 corresponds to the proteolytic cleavage site of membrane-bound IL6R that has been shown to affect sIL6 and IL6 serum levels [4,19,30]. In asthma, sIL6R serum and airway levels are increased and, in mice, the blockade of sIL6R suppresses airway inflammation [31,32]. Furthermore, β-2 agonists such as albuterol have been shown to modulate IL6 serum levels in asthma [8–10]. On the basis of these results, we considered IL6R SNP rs8192284 as a plausible biological modifier for bronchodilator drug response. Despite these suggestive studies, we found no significant association between IL6 SNP rs8192284 and the drug response among the three populations, Mexican, Puerto Rican, and African-American asthmatic patients. Similar to earlier studies, the C allele of the IL6R SNP rs8192284 was overrepresented in Native Americans in comparison with other ethnic groups and this allele frequency paralleled the pattern of Native American ancestry in our study participants [4].
To further explore the IL6 pathway, we selected functional IL6 gene variants to investigate the interaction with the IL6R gene. Although IL6 plays a major role in the inflammation process, the exact pathogenetic mechanism of IL6 in asthma remains unclear. Our results revealed that IL6 variants (rs1800796 and rs13306435) and haplotypes significantly modified bronchodilator drug response in Mexican asthmatic patients. These associations remained statistically significant even after permutation testing, which accounts for multiple testing. The proliferation of the mucosal Th2 cells, thought to play a major role in asthma, depends on IL6 trans-signaling through the sIL6R. In contrast, suppression of regulatory T cells and differentiation of CD4+ cells into Th2 within the lung depends on gp130 signaling mediated by the membrane-bound IL6R [31]. Moreover, in comparison with wild-type mice, IL6 deficient mice manifested exaggerated inflammation, whereas mice overexpressing IL6 manifested diminished inflammation in the bronchial airways [32]. These studies suggest that abnormalities in the production and/or effector functions of IL6 can contribute to the generation, severity, and chronicity of asthma. Furthermore, albuterol has been shown to increase IL6 gene expression in vivo and in vitro. On the basis of these findings, we studied IL6 functional variants that affect IL6 expression, production or binding to gp130, as a potential bronchodilator drug modifier in asthma. To the best our knowledge, this study is the first to investigate the IL6 and IL6R genes association with asthma among ethnically diverse populations.
On the gene level, we found a consistent pharmacogenetic gene–gene interaction in the three admixed populations. The pharmacogenetic interaction resulted in differential drug responsiveness to albuterol, which was most likely attributable to IL6 cytokine binding to the IL6 receptor, thus resulting in changes of the inflammatory process. This observed pharmacogenetic interaction, was contextual, however, and dependent upon individual genetic ancestry. Our results showed that polymorphisms of IL6 and IL6R occurring together resulted in higher drug response in Latinos, but lower drug response in African-Americans. To explain this discrepancy in drug response, our analysis suggested that genetic ancestral background might modify the effects of the gene–gene interaction on bronchodilator drug response. In Latino asthmatic patients, when the minor alleles of polymorphisms IL6R rs8192284 and IL6 rs1800796 occurred together, increased European ancestry resulted in decreased drug response, whereas increased Native American ancestry resulted in increased drug response. Although statistical significance for the effect modification by ancestry was achieved only in Mexican asthmatic patients (Pλ=λ0.03), parallel trends were observed for effect modification by ancestry in Puerto Rican asthmatic patients as well. This lack of significance in Puerto Rican asthmatic patients may be explained by lower frequencies of each allele, which have resulted in decreased statistical power. The interaction of genes and ancestry has been observed in both murine models and humans. In murine models, strain background has been shown to modify the effect of loci associated with airway hyperresponsiveness [33]. In humans, genetic ancestral background has been shown to modify the genetic effects of risk on inflammatory diseases [34]. Taken together, our results coupled with earlier results suggest higher-level gene–gene interactions.
In conclusion, this study provides evidence of significant pharmacogenetic gene–gene interaction between IL6 and IL6R that modifies bronchodilator drug response in three ethnically diverse asthmatic populations. Further functional studies will be needed to evaluate the biological interaction between IL6 and IL6R genetic variants to elucidate the casual mechanism behind differential bronchodilator drug responseOn the basis of these findings, future studies should focus on the potential clinical implications of genetic variants on drug response amongst asthmatic patients.
Acknowledgements
All of the authors are aware and agree to the content of the paper and approve its submission. They acknowledge the families and the patients for their participation. They also thank the numerous healthcare providers and community clinics for their support and participation in the GALA and SAGE studies. Finally, they especially thank Jeffrey M. Drazen, MD; Scott Weiss, MD; Ed Silverman, MD, PhD; Homer A. Boushey, MD; Jean G. Ford, MD; and Dean Sheppard for all of their effort toward the creation of the GALA Study.
This study was supported by the National Institutes of Health (RO1 HL078885, Flight Attendants Medical Research Institute, NCMHD Health Disparities Scholar, Extramural Clinical Research Loan Repayment Program for Individuals from Disadvantaged Backgrounds, to E.G.B.); American Thoracic Society Breakthough Opportunites for Lung Disease (BOLD) grant (ATS-05-078) and Tobacco-Related Disease Research Program New Investigator Award (15KT-0008) to S.C.; Ernest S. Bazley Grant to Northwestern University to P.C.A; SFGH General Clinical Research Center M01RR00083-4, U01-HL 65899, UCSF-Children’s Hospital of Oakland Pediatric Clinical Research Center (M01 RR01271), Oakland, California, Sandler Center for Basic Research in Asthma and the Sandler Family Foundation. Fondation pour la Recherche Medicale (FRM), Societe de Pneumologie de Langue Francaise (SPLF), Chancellerie des Universités (Legs Poix), Assistance Publique-Hôpitaux de Paris (APHP).
Footnotes
Conflicts of interest: none of the authors have any commercial or other associations that might pose a conflict of interest.
References
- 1.Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukoc Biol. 2006;80:227–236. doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama A, Kohno N, Fujino S, Hamada H, Inoue Y, Fujioka S, et al. Circulating interleukin-6 levels in patients with bronchial asthma. Am J Respir Crit Care Med. 1995;151:1354–1358. doi: 10.1164/ajrccm.151.5.7735584. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama A, Kohno N, Sakai K, Kondo K, Hirasawa Y, Hiwada K. Circulating levels of soluble interleukin-6 receptor in patients with bronchial asthma. Am J Respir Crit Care Med. 1997;156:1688–1691. doi: 10.1164/ajrccm.156.5.9610070. [DOI] [PubMed] [Google Scholar]
- 4.Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, et al. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet. 2007;80:716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brull DJ, Montgomery HE, Sanders J, Dhamrait S, Luong L, Rumley A, et al. Interleukin-6 gene •174g>c and •572g>c promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler Thromb Vasc Biol. 2001;21:1458–1463. doi: 10.1161/hq0901.094280. [DOI] [PubMed] [Google Scholar]
- 6.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 8.Ferrada MA, Gordon EL, Jen KY, He HZ, Lu X, Barone LM, et al. (R)-albuterol decreases immune responses: role of activated T cells. Respir Res. 2008;9:3. doi: 10.1186/1465-9921-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strandberg K, Palmberg L, Larsson K. Effect of formoterol and salmeterol on IL-6 and IL-8 release in airway epithelial cells. Respir Med. 2007;101:1132–1139. doi: 10.1016/j.rmed.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Tan KS, Nackley AG, Satterfield K, Maixner W, Diatchenko L, Flood PM. Beta2 adrenergic receptor activation stimulates pro-inflammatory cytokine production in macrophages via PKA- and NF-kappaB-independent mechanisms. Cell Signal. 2007;19:251–260. doi: 10.1016/j.cellsig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169:386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 12.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet. 2006;119:547–557. doi: 10.1007/s00439-006-0169-2. [DOI] [PubMed] [Google Scholar]
- 13.American Thoracic Society. Standardization of Spirometry, 1994 Update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 14.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 15.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118:652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 16.Ziv E, Burchard EG. Human population structure and genetic association studies. Pharmacogenomics. 2003;4:431–441. doi: 10.1517/phgs.4.4.431.22758. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Berthier-Schaad Y, Fallin MD, Fink NE, Tracy RP, Klag MJ, et al. IL-6 haplotypes, inflammation, and risk for cardiovascular disease in a multiethnic dialysis cohort. J Am Soc Nephrol. 2006;17:863–870. doi: 10.1681/ASN.2005050465. [DOI] [PubMed] [Google Scholar]
- 18.Noponen-Hietala N, Virtanen I, Karttunen R, Schwenke S, Jakkula E, Li H, et al. Genetic variations in IL6 associate with intervertebral disc disease characterized by sciatica. Pain. 2005;114:186–194. doi: 10.1016/j.pain.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 19.Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun. 2004;5:513–516. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell JR, Weeks DE. PedCheck a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 23.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of. Genet Epidemiol. 2000;19 Suppl 1:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 24.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 25.Global Strategy for Asthma Management and Prevention. Global Initiative for Asthma (GINA) 2007 Available from: http://www.ginasthma.org. [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgenson E, Witte JS. A gene-centric approach to genome-wide association studies. Nat Rev Genet. 2006;7:885–891. doi: 10.1038/nrg1962. [DOI] [PubMed] [Google Scholar]
- 28.Mountain JL, Risch N. Assessing genetic contributions to phenotypic differences among ‘racial’ and ‘ethnic’ groups. Nat Genet. 2004;36:S48–S53. doi: 10.1038/ng1456. [DOI] [PubMed] [Google Scholar]
- 29.Barnes PJ, Chung KF, Page CP. Inflammatory mediators of asthma: an update. Pharmacol Rev. 1998;50:515–596. [PubMed] [Google Scholar]
- 30.Mullberg J, Oberthur W, Lottspeich F, Mehl E, Dittrich E, Graeve L, et al. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol. 1994;152:4958–4968. [PubMed] [Google Scholar]
- 31.Doganci A, Eigenbrod T, Krug N, De Sanctis GT, Hausding M, Erpenbeck VJ, et al. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. J Clin Invest. 2005;115:313–325. doi: 10.1172/JCI22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Homer RJ, Chen Q, Elias JA. Endogenous and exogenous IL-6 inhibit aeroallergen-induced Th2 inflammation. J Immunol. 2000;165:4051–4061. doi: 10.4049/jimmunol.165.7.4051. [DOI] [PubMed] [Google Scholar]
- 33.Ackerman KG, Huang H, Grasemann H, Puma C, Singer JB, Hill AE, et al. Interacting genetic loci cause airway hyperresponsiveness. Physiol Genomics. 2005;21:105–111. doi: 10.1152/physiolgenomics.00267.2004. [DOI] [PubMed] [Google Scholar]
- 34.Helgadottir A, Manolescu A, Helgason A, Thorleifsson G, Thorsteinsdottir U, Gudbjartsson DF, et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2006;38:68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]