Abstract
The recognition of the importance of angiogenesis in tumor progression has led to the development of antiangiogenesis as a new strategy for cancer treatment and prevention. By modulating tumor microenvironment and inducing angiogenesis, the proinflammatory cytokine interleukine (IL)-1 β has been reported to promote tumor development. However, the factors mediating IL-1β-induced angiogenesis in non-small cell lung cancer (NSCLC) and the regulation of these angiogenic factors by IL-1β are less clear. Here, we report that IL-1β upregulated an array of proangiogenic CXC chemokine genes in NSCLC cell line A549 and in normal human tracheobronchial epithelium (NHTBE) cells, as determined by microarray analysis. Further analysis revealed that IL-1β induced much higher protein levels of CXC chemokines in NSCLC cells than in NHTBE cells. Conditioned medium from IL-1β treated A549 cells markedly increased endothelial cell migration, which was suppressed by neutralizing antibodies against CXCL5 and CXCR2. We also found that IL-1β-induced CXC chemokine gene overexpression in NSCLC cells was abrogated with the knockdown of CREB or NF-κB. Moreover, the expression of the CXC chemokine genes as well as CREB and NF-κB activities were greatly increased in tumorigenic NSCLC cell line compared with normal, premalignant immortalized or non-tumorigenic cell lines. A disruptor of the interaction between CREB-binding protein (CBP) and transcription factors such as CREB and NF-κB, 2-naphthol-AS-E-phosphate (KG-501), inhibited IL-1β-induced CXC chemokine gene expression and angiogenic activity in NSCLC. We propose that targeting CREB or NF-κB using small molecule inhibitors, such as KG-501, holds promise as a preventive and/or therapeutic approach for NSCLC.
Keywords: Angiogenesis, chemoprevention, CREB, NF-κB, CXC Chemokine, lung cancer
Introduction
Lung cancer is the leading cause of cancer deaths both in the United States and worldwide (1). The growth and development of lung cancer as well as other solid tumors are critically dependent on a functional vascular supply. Numerous evidence have shown that angiogenesis, the formation of new blood vessels from the pre-existing vasculature, is one of the critical steps in the entire process of cancer development from tumor growth to distant metastasis (2–5). Also, a study showed that a solid tumor would remain dormant at a volume of only 2 to 3mm3 in the absence of neovascularization and would be unable to metastasize (6). For solid tumors to develop and to metastasize, they need to secrete a number of proangiogenic factors to induce the formation of new blood vessels to overcome the physical limitations on the diffusion of nutrients and oxygen within the tumor—a process known as angiogenic switch (3). Recognizing the importance of angiogenesis in the growth of tumors has led to the development of antiangiogenesis as a new strategy for cancer therapy and prevention, a novel concept termed “angioprevention” (7–9). Actually, a series of molecules proposed as chemopreventive agents have been shown to have potent antiangiogenic properties when tested in in vitro and in vivo angiogenesis models (7).
Angiogenesis can be regulated by various growth factors and cytokines, including vascular endothelial growth factor (VEGF), basic fibroblast growth factor, transforming growth factors α and β, platelet-derived endothelial cell growth factors, chemokines and interleukine (IL)-1β (10–14). Recent studies have shown the importance of the tumor microenvironment in facilitating angiogenesis and promoting tumor invasion and metastasis (15–19). Once a tumor is vascularized, the tumor-associated antigens can be recognized by the immune system and the tumor is infiltrated by leukocytes. Although leukocyte infiltration in tumors is often considered to be associated with better prognosis and overall survival, studies have also shown that inflammatory cells can promote tumor cell proliferation, angiogenesis, metastasis and hence, tumor development (15, 16). Leukocyte infiltration can influence angiogenesis in tumors, because some subsets of leukocytes, especially the tumor-associated macrophages, can secrete both angiostatic and angiogenic factors (17, 18). IL-1 is a proinflammatory cytokine produced mainly by monocytes and macrophages. There are two IL-1 agonistic proteins, IL-1α and IL-1β. IL-1α is a precursor or membrane-associated molecule and is primarily a regulator of intracellular events and a mediator of local reactions. On the other hand, IL-1β acts as a systemic, hormone-like mediator and is only active in a secreted mature form. However, once these two proteins bind to their receptors, they have similar biological activities (20). Both IL-1α and IL-1β can promote tumor angiogenesis, but the role of IL-1β is more evident (14). IL-1 has been shown to contribute to the production of proangiogenic factors VEGF, hepatocyte growth factor, tumor necrosis factor and CXC chemokines (14, 21). Members of a subfamily of CXC chemokines sharing a characteristic glutamatelecine-arginine (ELR) motif near the N-terminus of the molecule are chemoattractants for neutrophils and are important for wound repair. The ELR-positive chemokines, including CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7 and CXCL8, are pro-angiogenic, whereas members of another subfamily lacking the ELR motif—ELR-negative chemokines, such as CXCL4, CXCL9, CXCL10, and CXCL11—are in general interferon-inducible and are potential inhibitors of angiogenesis. Generally, CXCR2 is the receptor for angiogenic CXC chemokine-mediated angiogenesis, and CXCR3 is the receptor for angiostatic interferon-inducible CXC chemokine inhibition of angiogenesis (13). CXC chemokine ligands and receptors have been demonstrated to play important roles in mediating NSCLC-associated angiogenesis and organ-specific metastases (13). Recently, it has been reported that CXCL5 and CXCL8 protein level were elevated in tumor specimens freshly isolated from patients with NSCLC and that these two ELR-positive CXC chemokines are important mediators of angiogenesis during NSCLC tumorigenesis (22, 23). Compared with CXCL8, CXCL5 was reported to have a higher degree of correlation with NSCLC-derived angiogenesis (23). In a model system of human NSCLC tumorigenesis in severe combined immunodeficiency mice, CXCL5 expression was found to be directly correlated with tumor growth, tumor-derived angiogenesis, and metastatic potential. Depletion of CXCL5 in this model system resulted in attenuation of both tumor growth and spontaneous metastasis due to the inhibition of angiogenesis (23).
Being a product of tumor infiltrated macrophages, IL-1β is known to increase angiogenesis. However, in NSCLC, what angiogenic factors are induced by IL-1β and how they are regulated by IL-1β are still not clear. To elucidate these critical issues, we performed a microarray analysis to determine the effect of IL-1β on global gene expression in the NSCLC adenocarcinoma cell line A549 and in normal human tracheobronchial epithelium (NHTBE) cells. We found that IL-1β dramatically induced the expression of an array of proangiogenic CXC chemokine genes and significantly augmented the angiogenic activity of NSCLC. In addition, we found that transcription factors CREB and NF-κB both play critical roles in the regulation of IL-1β-induced CXC chemokine gene expression and angiogenic activity.
Materials and Methods
Cell cultures, chemicals and conditioned media (CM)
NHTBE cells were purchased from Cambrex (Walkersville, MD) and cultured in six-well plates as described previously (24–28). Human umbilical vein endothelial Cells (HUVECs) were purchased from Cambrex and maintained in EGM complete endothelial growth medium. HUVECs were used at passage 4. The human NSCLC cell lines A549, H1734, H226, and H2170, were obtained from the American Type Culture Collection (Manassas, VA) and grown in RPMI 1640 medium containing 10% fetal bovine serum, 100 units/ml penicillin, and 100µg/ml streptomycin. The BEAS-2B, 1799, 1198, and 1170-I human bronchial epithelial (HBE) cell lines were obtained from Dr. R. Lotan (The University of Texas M. D. Anderson Cancer Center, Houston, TX) and Dr. A. Klein-Szanto (Fox Chase Cancer Center, Philadelphia, PA) and grown in Keratinocyte Serum-Free Medium (Life Technologies, Inc., Gaithersburg, MD) containing epidermal growth factor and bovine pituitary extract (29). All of the cells were cultured at 37°C in a humidified atmosphere of 95% air and 5% CO2. Conditioned media (CM) were generated as follows: NSCLC cells (3 × 105/ml) were cultured in RPMI 1640 medium containing 0.5% fetal bovine serum with or without IL-1β and/or 2-naphthol-AS-E-phosphate (KG-501) at different concentrations for 24 h. Cell-free supernatants were collected and stored at −70°C until use.
Reagents and antibodies
IL-1β, Quantikine® CXCL5 and CXCL8 Enzyme-Linked ImmunoSorbent Assay (ELISA) kits and neutralizing antibodies against CXCL5, CXCL8, CXCR2, and VEGF were purchased from R&D Systems (Minneapolis, MN). KG-501 was purchased from Sigma-Aldrich (St. Louis, MO) and dissolved in dimethyl sulfoxide. Antibodies against NF-κB p65, CREB and phospho-CREB (Ser133) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Upstate (Billerica, MA), respectively. A monoclonal antibody against β-actin was from Sigma-Aldrich. Transwell chambers with polyethylene terephthalate membranes containing 8-µm pores were obtained from BD Biosciences (Bedford, MA).
Microarray analysis
After confluence, NHTBE and A549 cells were treated with control medium or the same medium containing 2.5 ng/ml of IL-1β for 8 h prior to total RNA extraction. Total RNA was isolated using the RNeasy mini Kit (Qiagen, Valencia, CA). The integrity of mRNA and the relative rRNA contamination were analyzed using the RNA 6000 Nano LabChip (Agilent Technologies, Santa Clara, CA) and the Agilent 2100 bioanalyzer (Agilent Technologies). The RNA from control group was amplified and labeled with cyanine 3 and the RNA from IL-1β treated cells with cyanine 5. Equal amount of the differently labeled RNAs were then mixed and hybridized with 44K whole human genome oligonucleotide microarrays (Agilent Technologies). After hybridization, the arrays were scanned and the resulting images were analyzed using the Agilent feature extraction software program (GE2, version 5.91; Agilent Technologies).
Quantitative RT-PCR (qPCR)
Validation of the differentially expressed genes in NHTBE and NSCLC cells was performed using an iCycler real-time PCR detection system (Bio-Rad, Hercules, CA) with gene-specific primers. Primers for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the reference gene, were 5'-TGCACCACCAACTGCTTAGC (forward) and 5'-GGCATGGACTGTGGTCATGAG (reverse), for CXCL1 were 5'-AGTGACAAATCCAACTGACC (forward) and 5'-GATGCTCAAACACATTAGGC (reverse), for CXCL2 were 5'-CCCAAGTTAGTTCAATCCTG (forward) and 5'-TTCCTCAGCCTCTATCACAG (reverse), for CXCL3 were 5'-CTTGTCTCAACCCCGCATCC (forward) and 5'-TCTGGTAAGGGCAGGGACCA (reverse), for CXCL5 were 5'-TCCAATCTCCGCTCCTCCAC (forward) and 5'-AGCAGCAGCAGCACCAACAG (reverse), for CXCL6 were 5'-GTTTGTCTGGACCCGGAAGC (forward) and 5'-TCCGCTGAAGACTGGGCAAT (reverse), for CXCL8 were 5'-GCATAAAGACATACTCCAAACC (forward) and 5'-ACTTCTCCACAACCCTCTG (reverse), and for CREB were 5'-AAGCTGAAAACCAACAAATGACAGTT (forward) and 5'-TGAACTGTCTGCCCATTGG (reverse). Single-stranded cDNAs were synthesized in 50 µl of reverse transcription (RT) mix containing 1 µg of total RNA using the GeneAmp RNA PCR Core Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. PCR analysis was performed using 25 µl of volumes with SYBR Green PCR Core Reagents (Applied Biosystems). Primers (200 nM) and RT mix (2 µL) were used in each PCR. Each sample was assayed in triplicate per PCR run, and the experiment was repeated three times. The cycling conditions were an initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, and elongation at 60°C for 60 s. The real-time PCR data were analyzed using the comparative Ct method.
Transfection of small-interfering RNA against NF-κB p65 and transduction of lentiviral short hairpin RNA against CREB
SMARTpool-sequenced small-interfering RNA (siRNA) targeting human NF-κB p65 (GenBank accession no. NM_021975) and nonspecific control pool siRNA were purchased from Dharmacon RNA Technologies (Lafayette, CO) and diluted to 20 µM. NSCLC cells at 50% confluence were transfected with siRNA for NF-κB p65 or control siRNA at a final concentration of 50 nM and 100 nM using the LipofectAMINE 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Seventy two hours after transfection, the cells were treated with IL-1β for 8 h. Total protein and RNA were collected from each sample.
For CREB-targeting viral short hairpin RNA (shRNA) delivery, the lentiviral plasmid pLKO.1 with a shRNA clone against CREB (Clone ID: TRCN0000011085) was purchased from Open Biosystems (Huntsville, AL). The pLKO.1 plasmid with a scrambled shRNA sequence and virus packaging plasmids (psPAX2 and pseudo-typing plasmid pMD2.G) were obtained from Addgene (Cambridge, MA). HEK 293T cells were obtained from the American Type Culture Collection. HEK 293T cells at 50% confluence were co-transfected with pLKO.1 and virus packaging plasmids using FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN). After transfection for 16 h, the medium containing the transfection agent was replaced with fresh growth medium. Viral particles were then collected from the medium every 24 h for twice. The virus titer in the pooled suspension was determined by counting the puromycin-resistant colonies in the virus-transduced culture. For knockdown of CREB, cells at 50% confluence were incubated with viral suspension at a multiplicity of infection of about 50 for 16 h. Four days after transduction, the cells were treated with IL-1β for 24 h, and total protein and RNA were collected from the cells.
Migration assay
HUVECs (5 × 104) were suspended in serum-free RPMI 1640 medium and seeded in the transwell chambers coated with gelatin. CM from NSCLC cells were applied in the outer chambers. HUVECs were incubated at 37°C in 5% CO2 for 16 h. Following the incubation, cells were fixed in 90% ethanol and stained with 0.1% crystal violet. Nonmigrated cells on the upper surface of the chamber filters were removed by swabbing, and the cells that had migrated through the filter were photographed under a microscope and quantified using the ImageJ software program (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij).
Western blot analysis
Western blot analysis of target proteins was performed as described previously (30). Equal amounts of protein (30 µg) were resolved using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The mouse monoclonal antibody against human NF-κB p65 was diluted in 5% nonfat milk at a ratio of 1:200 and rabbit polyclonal antibodies against human CREB and pCREB were diluted at a ratio of 1:1000 and incubated with the membranes overnight at 4°C. Proteins reactive with the primary antibody were visualized with a horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibody and enhanced chemiluminescence reagents (Amersham Bioscience, Piscataway, NJ).
Measurement of CXC chemokine protein secretion
The levels of secreted CXC chemokines in the conditioned media were measured with immuno-dot blotting and ELISA. Immuno-dot blotting was performed as described before (24, 28). Briefly, the conditioned media were applied to a nitrocellulose membrane using the Manifold I Dot-Blot System (Scheleicher & Schuell, Keene, NH). The membrane was then probed with anti-human CXCL5 and CXCL8 antibodies and the target proteins were detected using chemiluminescence and quantified with densitometry. ELISA was performed with Quantikine® CXCL5 and CXCL8 ELISA kits (R&D Systems) according to manufacturer’s instruction. Both measurements were normalized against the cell number.
Chromatin immunoprecipitation (ChIP) analysis
ChIP assay was performed using EZ ChIP kits (Upstate) according to manufacture’s instruction. Briefly, NHTBE, BEASE-2B, 1799, 1198, and 1170-1 cells were grown in plate with normal media and chromatins were cross-linked by reaction with 1% formaldehyde for 10 min. The cross-linked chromatins were fragmented by sonication and subsequently immunoprecipitated with anti-CREB (Upstate) or anti-NF-κB (p65, Santa Cruz). The DNA in the precipitate was purified and used as the template for PCR. The primers for CXCL5 promoter with NF-κB binding site were 5'-TAGAGGTGCACGCAGCTCCT (forward) and 5'-GAGCACTGTGGCTTCCTCGT (reverse); for CXCL5 promoter with CREB binding site were 5'-CTGGACACACGTATACTTGC (forward) and 5'-GGCAGGTCATTCTAGGTTTC (reverse); for CXCL8 promoter with both CREB and NF-κB binding sites were 5'-AAAACTTTCGTCATACTCCG (forward) and 5'-AAAGTTTGTGCCTTATGGAG (reverse). PCR products were then separated in 1.2% agarose gel and stained with GelRed (Biotium, Hayward, CA).
Statistical analysis
Each experiment presented in the figures was repeated three or more times. The data are presented as the mean ± standard error (SE). Comparisons between groups were evaluated using analysis of variance and a two-tailed Student’s t-test. P<0.05 were considered statistically significant.
Results
Microarray analysis revealed an array of CXC chemokine genes upregulated by IL-1β in NHTBE and A549 cells
To identify IL-1β-responsive genes involved in angiogenesis and tumorigenesis in NSCLC, we used total RNA isolated from NHTBE and A549 cells treated with or without 2.5 ng/ml of IL-1β for 8 hours to generate cRNA for microarray hybridization and analysis using 44K whole human genome oligonucleotide microarrays. Analysis of the resulting microarray images using the Agilent feature extraction software program showed that expression of six CXC chemokine genes, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL8, was commonly upregulated in response to IL-1β in both NHTBE and A549 cells (p<0.001). As expected, we also saw that the expression of another angiogenic gene, VEGF, was upregulated by IL-1β in both NHTBE and A549 cells (Table 1). To confirm that these microarray data reflect the IL-1β-induced gene expression, we examined the expression of CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL8 genes in NHTBE and A549 cells using qPCR. Indeed, expression of all of these CXC chemokine genes was strongly induced after stimulation by IL-1β, and the qPCR results agreed with the microarray analysis results (Table 1).
Table 1.
Analysis of IL-1β induction of CXC chemokine gene expression in NHTBE and A549 cells using microarray and real-time PCR analysis.
| Gene Name | Systematic Name | NHTBE (fold change) |
A549 (fold change) |
|||
|---|---|---|---|---|---|---|
| Gene Description | microarray | PCR | microarray | PCR | ||
| CXCL1 | NM_001511 | Homo sapiens chemokine (C-X-C motif) ligand 1 | 18.8 | 11.9 | 32.5 | 26.9 |
| CXCL2 | NM_002089 | Homo sapiens chemokine (C-X-C motif) ligand 2 | 18.8 | 12.9 | 23.3 | 14.9 |
| CXCL3 | NM_002090 | Homo sapiens chemokine (C-X-C motif) ligand 3 | 18.2 | 29.9 | 19.2 | 11.7 |
| CXCL5 | NM_002994 | Homo sapiens chemokine (C-X-C motif) ligand 5 | 173.3 | 95.9 | 12.0 | 8.2 |
| CXCL6 | NM_002993 | Homo sapiens chemokine (C-X-C motif) ligand 6 | 31.3 | 80.6 | 13.9 | 28.8 |
| CXCL8 | NM_000584 | Homo sapiens chemokine (C-X-C motif) ligand 8 | 11.9 | 17.6 | 40.8 | 32.4 |
| VEGF | NM_001025366 | Homo sapiens vascular endothelial growth factor | 2.49 | 1.93 | 3.2 | 2.14 |
IL-1β differentially regulates CXCL5 and CXCL8 protein secretion in NHTBE and NSCLC cells
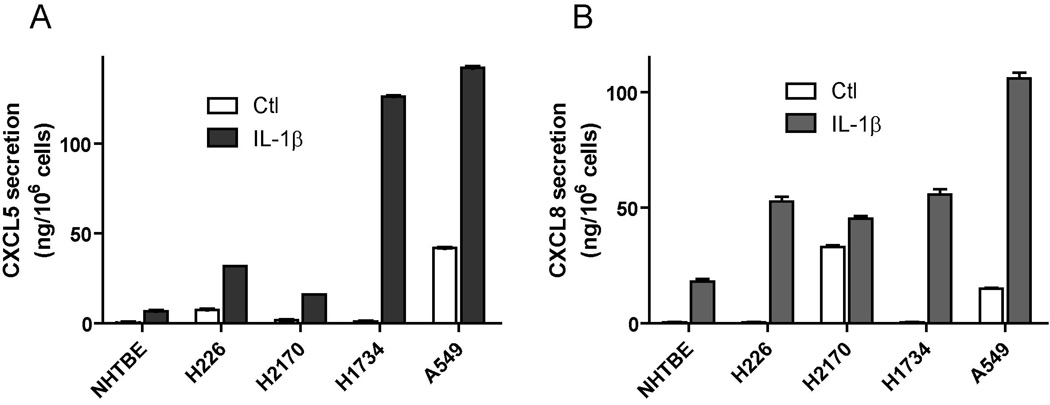
According to the microarray and qPCR data, IL-1β stimulated the gene expression of angiogenic CXC chemokines in both NHTBE and A549 cells. We next verified whether such induction of gene expression was indeed translated into the protein secretion in these cells. For this purpose, we focused on CXCL5 and CXCL8, as they reportedly have dominant angiogenic effects in lung cells. Using the immuno-dot blot assay, we detected the secretion of CXCL5 and CXCL8 in the CM from all four NSCLC cell lines (H226, H2170, A549 and H1734) treated with or without IL-1β. However, by the same method, the secretion of CXCL-8 in NHTBE cells was only detected in IL-1β treated cells, but not in the untreated cells, and the CXCL5 protein was not detectable even with IL-1β treatment (data not shown). We verified such differential expression of angiogenic CXC chemokines between tumor and normal cells with ELISA (Fig. 1A, B). Again, we observed a significantly greater expression of CXCL5 and CXCL8 in tumor cells than in normal cells, especially, with the induction of IL-1β (Fig. 1B). This result indicated that although IL-1β increases CXCL5 mRNA level in both normal epithelial cells and lung cancer cells, only the lung cancer cells produce significant level of CXCL5 chemokine protein. As A549 cells produced substantial amount of both CXCL5 and CXCL8, we used A549 cells as a model system to further explore the effect of IL-1β on angiogenesis.
Figure 1. IL-1β differentially regulates CXCL5 and CXCL8 protein secretion in NHTBE and NSCLC cells.
NHTBE, H226, H2170, H1734 and A549 cells were treated with 2.5 ng/ml of IL-1β for 24 hours. CM was collected from above cell lines. The concentrations of (A) CXCL5 and (B) CXCL8 in CM were measured using ELISA. Results were normalized according to the cell number. Data were expressed as the mean ± SE.
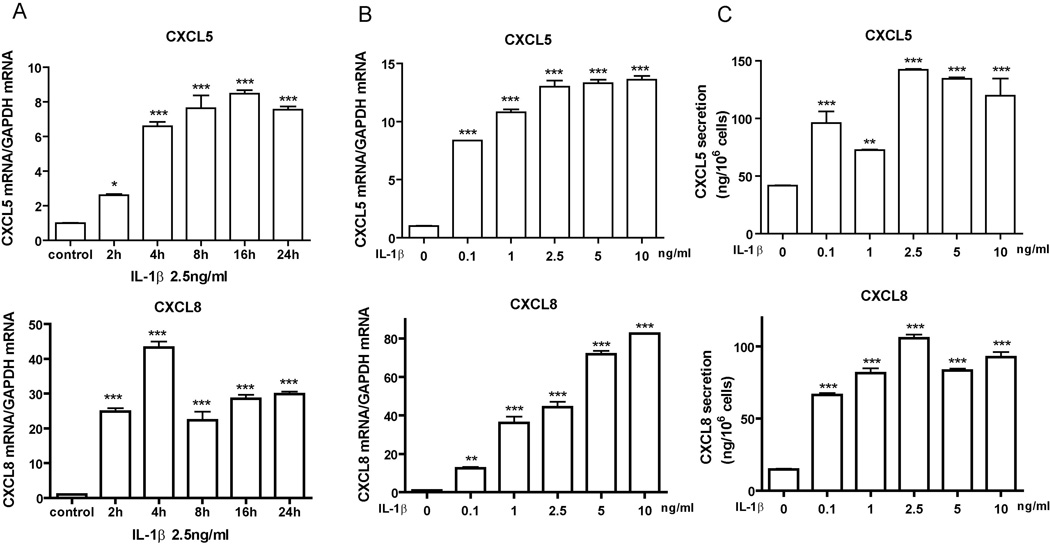
To further assess the response of CXCL5 and CXCL8 gene expression to IL-1β treatment, we measured the time and dose effect of IL-1β on A549 cells. The level of CXCL5 and CXCL8 transcription peaked at 4 h after IL-1β stimulation (Fig. 2A), indicating that the cells responded to IL-1β by rapidly inducing the expression of CXC chemokine genes. We found that CXCL5 mRNA expression was induced significantly even with IL-1β at 0.1 ng/mL. CXCL8 mRNA expression reached a plateau at 2.5 to 5.0 ng/ml of IL-1β treatment (Fig. 2B). We confirmed this dose-dependent response by measuring the CXCL protein secretion with ELISA (Fig. 2C).
Figure 2. IL-1β stimulates CXCL5 and CXCL8 gene expression and protein secretion in A549 cells in a time- and dose-dependent manner.
A, A549 cells were incubated with control medium or with medium containing 2.5 ng/ml of IL-1β for the indicated times. Total RNAs were collected from the cells and expression of the indicated genes was measured using qPCR. GAPDH was used as an internal control for normalizing the RNA loading. B, A549 cells were incubated with IL-1β at the indicated concentrations for 8 h. Total RNAs were collected from the cells for qPCR. C, Culture media were collected from A549 cells with treatment as in (B) and the concentrations of CXCL5 and CXCL8 were measured with ELISA Data were expressed as the mean ± SE. *, p<0.05; **, p<0.01; ***, p<0.001 compared with untreated control.
IL-1β significantly augments the angiogenic activity of NSCLC by inducing the expression of angiogenic CXC chemokine genes
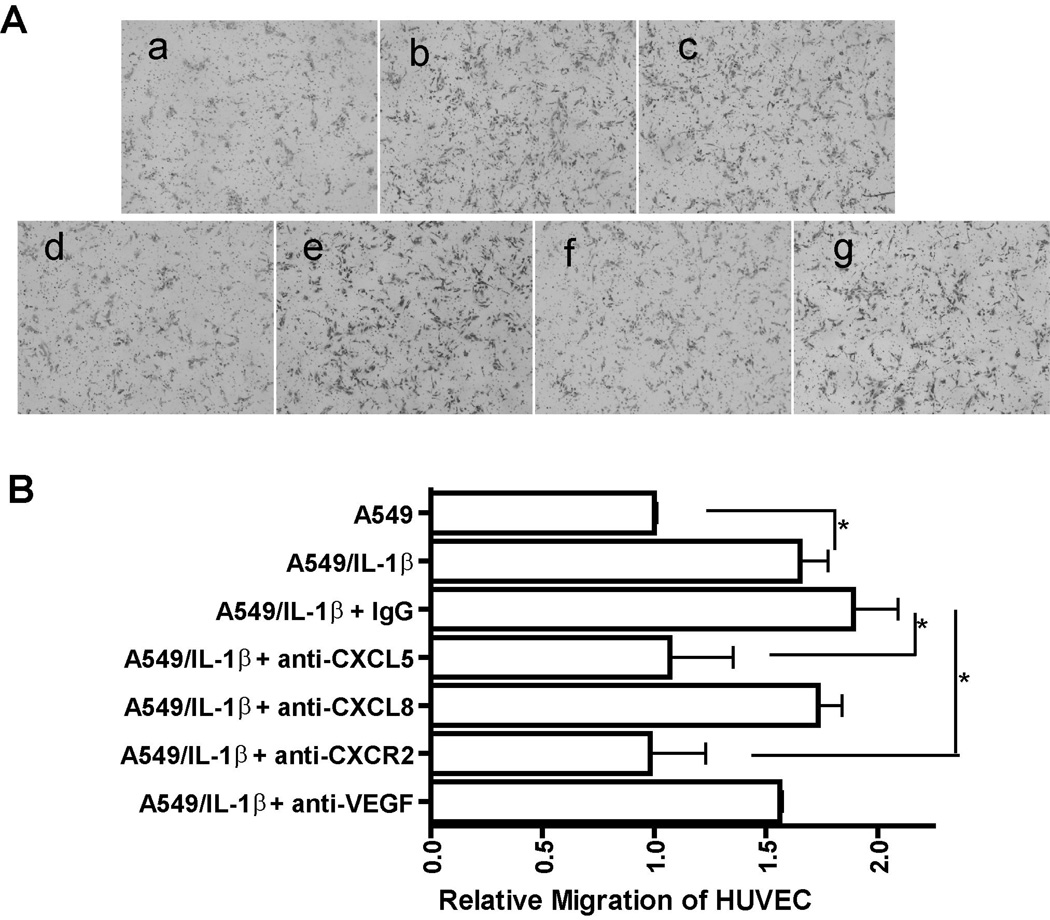
As IL-1β up-regulates the expression of important angiogenic CXC chemokine genes in NSCLC, we inferred that IL-1β may enhance the net angiogenic activity of NSCLC. To test this possibility, we performed an endothelial cell (HUVEC) migration assay with CM from A549 NSCLC cell lines treated with or without IL-1β. As shown in Fig. 3A (a and b), compared with the CM from untreated A549 cells, the CM from IL-1β treated cells (A549/IL-1β) markedly increased the angiogenic activity as assessed by migration of HUVECs. Similar results were observed with CM prepared from IL-1β treated H1734 cells (data not shown). These data clearly indicated that IL-1β significantly augments the net angiogenic activity of NSCLC as measured by the chemotactic activity of endothelial cell. Next, we hypothesized that the increased migration of HUVECs in response to CM from A549/IL-1β cells was attributable to the observed increased expression of angiogenic CXC chemokine genes in NSCLC cells induced by IL-1β. To test this, we performed a HUVECs migration assay with CM from A549/IL-1β cells in the presence of neutralizing antibodies against CXCL5, CXCL8, CXCR2, and VEGF. We found that migration of HUVECs was reduced significantly in the presence of neutralizing antibodies against CXCL5 and CXCR2 (Fig. 3A [d and f]). However, we did not observe the similar effect on antibodies against CXCL8 and VEGF (Fig. 3A [e and g]). These results suggested that IL-1β induced-angiogenic activity of NSCLC is mainly attributable to the induced expression of CXCR2-dependent CXC chemokines, most likely CXCL5.
Figure 3. IL-1β significantly augments the angiogenic activity of NSCLC by inducing the expression of angiogenic CXC chemokine genes.
A, CM was collected from A549 cells treated with either control medium (a) or medium containing 2.5 ng/ml of IL-1β (b) for 24 h and CM from IL-1β-treated A549 cells was preincubated with 10 µg/mL of non-immune IgG (c) or an anti-CXCL5 monoclonal antibody (d) or an anti-CXCL8 monoclonal antibody (e) or an anti-CXCR2 monoclonal antibody (f) or an anti-VEGF monoclonal antibody (g) at 37°C for 1 h. A migration assay was performed by stimulating HUVECs with the indicated CM as described in Materials and Methods. Migrated dells were photographed after 16 h of incubation at 37°C. B, Image analysis of HUVECs migration was quantified using the ImageJ software program. Data were expressed as the mean ± SE. *, p<0.05.
NF-κB and CREB mediate IL-1β-induced CXC chemokine gene expression
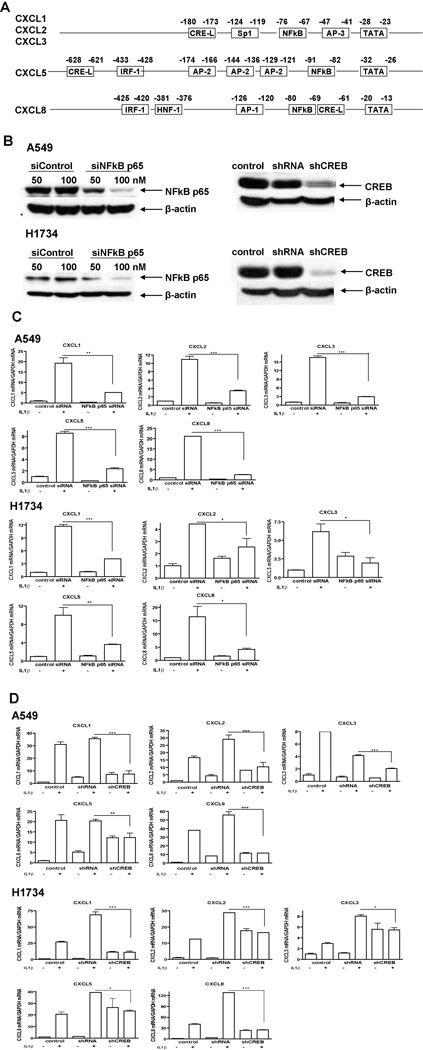
To investigate the mechanism underlying the IL-1β-induced CXC chemokine gene expression, we focused on the transcription factors mediating this effect. Sequence analysis of the promoters of these CXC chemokine genes indicated the potential binding sites for NF-κB, AP-1, AP-2, AP-3, Sp1, interferon regulatory factor-1 (IRF-1), hepatic nuclear factor 1 (HNF-1), and CREB (Fig. 4A) (31–34). Because IL-1β activates NF-κB and CREB, and the NF-κB site and CRE-like sites are located in the promoters of CXCL1, CXCL2, CXCL3, CXCL5, and CXCL8 genes, we sought to determine whether CREB and NF-κB mediate IL-1β-induced CXC chemokine gene expression in NSCLC cells. For this purpose, we abrogated NF-κB and CREB gene expression in A549 and H1734 cells by either transfecting the cells with a siRNA for NF-κB p65 or transducing the cells with a lentivirus containing the shRNA for CREB (shCREB). We confirmed that the NF-κB and CREB protein levels were knocked down by more than 80% after siRNA transfection or viral transduction in these two cell lines (Fig. 4B). We then treated the cells with IL-1β and measured the CXC chemokine gene expression levels in the cells. Real-time PCR results showed that IL-1β-induced CXC chemokine gene expression decreased significantly after knockdown of NF-κB (Fig. 4C). At the same time, shCREB-transduced cells had no or a much less significant response to IL-1β in the induction of CXC chemokine gene expression when compared with the non-transduced control or scrambled shRNA-transduced cells (Fig. 4D). These findings suggested that both CREB and NF-κB mediate IL-1β-induced CXC chemokine gene expression in NSCLC cells.
Figure 4. NF-κB and CREB mediate IL-1β-induced CXC chemokine gene expression.
A, Transcription factor binding domains located in the gene promoters of CXCL1, CXCL2, CXCL3, CXCL5, and CXCL8. B, A549 cells were transfected with siRNA for NF-κB p65 or transduced with shCREB lentivirus; none-transduced cells and cells transfected with control siRNA/transduced with scrambled shRNA were used as controls. Proteins were extracted from the cells 72 h after transfection or 96 h after transduction. NF-κB and CREB protein expression levels were detected using western blot analysis. β-actin protein was probed as a loading control. C, A549 and H1734 cells were transfected with siRNA for NF-κB p65, and cells transfected with control siRNA were used as control. Seventy-two hours later, the cells were treated either with control medium or medium containing 2.5 ng/ml of IL-1β for 8h. D, A549 and H1734 cells were transduced with shCREB, and non-transduced cells and cells transduced with scrambled shRNA were used as controls. Ninety-six hours later, the cells were treated with or without 2.5 ng/ml of IL-1β for 24h. Total RNAs were isolated from the cells in (C) and (D). Real-time PCR was performed with specific primers for the indicated genes. Data were expressed as the mean ± SE. *, p<0.05; **, p<0.01; ***, p<0.001.
The expression level of CXC chemokine genes is associated with the development of lung cancer
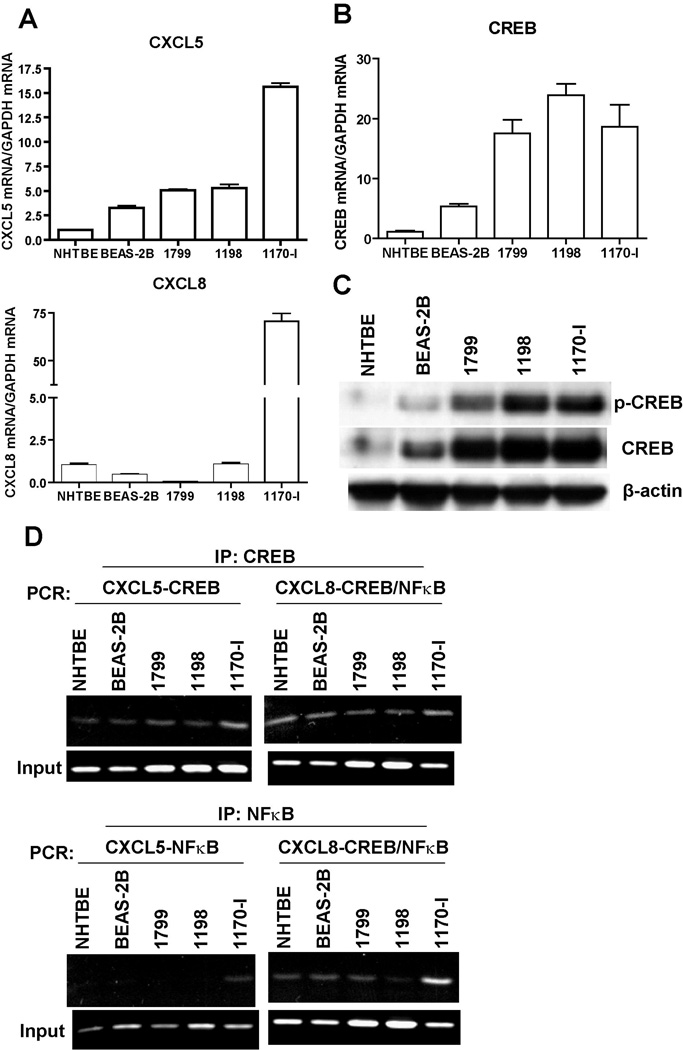
To investigate if the angiogenic CXC chemokine genes are involved in the development of lung cancer, we measured the expression level of CXCL5 and CXCL8 in an in vitro lung carcinogenesis model (IVLCM) which includes normal (NHTBE), immortalized (BEAS-2B and 1799), transformed (1198) and tumorigenic (1170-I) HBE cells. The qPCR results (Fig. 5A) showed that these two CXC chemokine genes were differentially expressed in the cell lines of IVLCM, with progressive increase from NHTBE to BEAS-2B and 1799, and then further to 1198 and 1170-I. As we demonstrated that CREB and NF-κB regulate CXC chemokine gene expression, we examined whether the activities of these transcription factors correspond to the expression levels of CXCL5 and CXCL8 in these IVLCM cells. Western blot analysis and qPCR data showed that both the activity and the expression level of of CREB were gradually increased with the progression of tumor development in the IVLCM (Fig. 5B and C). The similar pattern was also observed with NF-κB level (data not shown). Such trend of change was consistent with the tendency of their expression levels of CXCL5 and CXCL8. We further performed ChIP assay to determine whether CREB and NF-κB regulate the gene expression of these two chemokines in the IVLCM cells. The results of ChIP assay (Fig. 5D) demonstrated that CREB bound to the promoter regions of both CXCL5 and CXCL8 in all these cells; and that while NF-κB also bound to the CXCL8 promoter in all these cells, its binding to the CXCL5 promoter was only detected in the tumorigenic (1170-I) cells. These results indicated that the expression of the angiogenic CXC chemokine genes in theses cells is well correlated with the progression of lung cancer from normal to invasive phenotype and which might further associated with their inherent activities of CREB and NF-κB.
Figure 5. The expression of CXC chemokine genes and CREB is associated with the development of lung cancer.
A and B, Total RNAs from NHTBE and sub-confluent cultures of BEAS-2B, 1799, 1198 and 1170-I were extracted and the expression of designated genes were quantitated with real-time PCR using specific primers. Each specific gene expression was normalized with the expression level of GAPDH. Data were expressed as the mean ± SE. C, Whole cell lysates (20 µg/lane) were isolated from NHTBE and IVLCM cell lines, and the levels of phospho-CREB (p-CREB) and total CREB were measured by immunoblot analysis. D, CREB and NF-kB binding on CXCL 5 and CXCL 8 promoter. ChIP assay was performed with chromatins prepared from NHTBE, BEASE-2B, 1799, 1198, and 1170-1 cells. The binding of CREB or NFkB to the CXCL promoter was detected by the visualization of PCR product. The single bands detected in input samples indicate the specificity of the PCR primers.
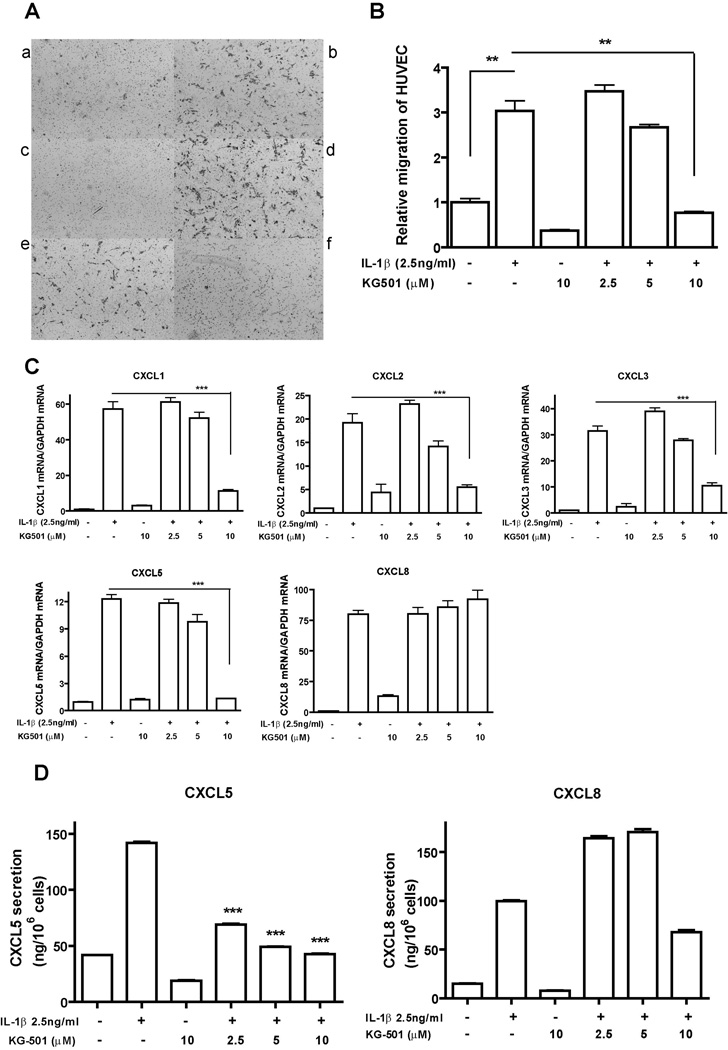
KG-501 inhibits the endothelial cell migration induced by CM from IL-1β-treated NSCLC cells via the suppression of CXC chemokine gene expression in NSCLC cells
In looking for inhibitors that can block IL-1β-induced endothelial cell migration, we focused on the agents targeting the transcription factors NF-κB and CREB. It has been reported that KG-501 is a small molecule which binds to the transcription coactivator CBP and blocks the interaction of CBP with the active form of CREB, phospho-CREB (35). KG-501 can also inhibit NF-κB transcription activity because NF-κB also utilize CBP as a co-factor to regulate gene expression (36). Based on our results that CREB and NF-κB could mediate IL-1β-induced CXC chemokine gene expression in NSCLC cells, we hypothesized that KG-501 could suppress the expression of these CXC chemokine genes and inhibit the endothelial cell migration induced by CM from NSCLC/IL-1β cells. As shown in Figure 6A and B, the migration of HUVECs induced by CM from A549 cells treated with IL-1β plus 10 µM of KG-501 was significantly lower than that induced by CM from A549 cells treated with IL-β1 alone (p<0.05). Next, we evaluated the effect of KG-501 on the transcriptional and proteomic level of CXC chemokines induced by IL-1β. At 10 µM, KG-501 suppressed the expression of all of the IL-1β-induced CXC chemokine genes except CXCL8 (Fig. 6C). For the protein level, we measured the expression of CXCL5 and CXCL8, in A549 cells. KG-501 significantly suppressed IL-1β-induced CXCL5 protein secretion. However, its effect on IL-1β-induced CXCL8 protein secretion was not consistent with its concentration, with a stimulatory effect at low concentrations, but a slightly inhibitory effect at high concentration. (Fig. 6D). Similar effects of KG-501 were also observed in H1734 cell line (data not shown).
Figure 6. KG-501 suppresses NSCLC/IL-1β CM-induced migration of HUVECs by regulating IL-1β-induced CXC chemokines gene expression in NSCLC cells.
A, HUVECs migration was induced with CM from A549 cells treated with control medium (a) or medium containing 2.5 ng/ml of IL-1β (b) or 10 µM of KG-501 (c) or 2.5 ng/mL of IL-1β plus 2.5 µM of KG-501 (d) or 2.5 ng/mL of IL-1β plus 5 µM of KG-501 (e) or 2.5 ng/mL of IL-1β plus 10 µM of KG-501 (f) in serum-free medium for 24h. B, Quantification of image analysis for the HUVECs migration assay. C, Total RNAs were isolated from cells with the treatment described in (A), and real-time PCR was performed to measure the expression level of the indicated genes. D, The concentration of CXCL5 and CXCL8 protein in the CM was measured with ELISA. Results were normalized according to the cell number. Data were expressed as the mean ± SE. **, p<0.01; ***, p<0.001.
Discussion
In the present study, we demonstrated that the proinflammatory cytokine IL-1β upregulates expression of an array of proangiogenic CXC chemokine genes in the NSCLC cell lines and that both of the transcription factors, CREB and NF-κB can mediate this upregulation. IL-1β augments the angiogenic activity of NSCLC as manifested by the ability of CM from IL-1β treated cells to induce endothelial cell migration. Our finding that the transcription factor CREB and NF-κB mediates IL-1β-induced CXC chomokine gene expression extends our knowledge about the mechanism of the angiogenic factors gene regulation and provides new potential target for angioprevention. Finally, the findings that the small molecule KG-501 significantly suppressed IL-1β-induced CXC chemokine gene expression and in turn reduced the CM-induced endothelial cell migration indicate that KG-501 may have therapeutic and preventive potential for NSCLC.
Our data showing that IL-1β upregulates the expression of angiogenic CXC chemokine genes and augments the angiogenic activity of NSCLC are consistent with the previous reports on the role of CXCR2 and its ligands in promoting tumor-associated angiogenesis and early development of NSCLC (22, 23, 37–40). In an in vivo study using murine Lewis lung cancer heterotopic and orthotopic tumor model system with CXCR2−/− versus CXCR2+/+ mice, researchers demonstrated that the tumors in CXCR2−/− mice exhibited reduced growth, increased necrosis, inhibited tumor-associated angiogenesis, and reduced metastatic potential (37). Similar to our finding that a neutralizing antibody against CXCR2 blocked CM-induced endothelial cell migration, the report showed that a specific neutralizing antibody against CXCR2 inhibited tumor growth, increased necrosis, and reduced tumors vessel density in CXCR2+/+ mice (37). Furthermore, studies showed that CXCL5 and CXCL8 play dominant role in promoting angiogenesis in patients with NSCLC (22, 23, 40). While CXCL8 was the first angiogenic ELR-positive CXC chemokine discovered in NSCLC, CXCL5 reportedly has a higher degree of correlation with NSCLC-derived angiogenesis (23). In our study, using the neutralizing antibodies, we observed that CXCL5 neutralization inhibited the migration of endothelial cells to the same degree as did CXCR2 neutralization. We failed to see this inhibitory effect using the CXCL8-neutralizing antibody, indicating that CXCL8 produced by A549 cells in response to IL-1β may not be sufficient to induce endothelial cell migration. Our experiment using KG-501 further supported such observation, as this small molecule blocked endothelial cell migration without affecting the CXCL8 level in CM. In addition, another angiogenic factor, VEGF, may play only a minor role in inducing endothelia cell migration in NSCLC, as neutralization of which could not inhibit the migration of HUVECs. Because high levels of CXCL5 and CXCL8 protein expression were detected in IL-1β-treated NSCLC cells, but only a very low level of CXCL5 protein was induced in NHTBE cells, we speculated that IL-1β may induce an angiogenic response only in NSCLC tumor cells but not in surrounding normal cells.
It is well documented that IL-1β upregulates the expression of the proangiogenic CXC chemokine genes and that NF-κB is the common transcription factor that mediates this effect (31, 41, 42). All of the angiogenic CXC chemokine gene promoters contain a putative cis-element that is recognized by the NF-κB family of transcriptional factors (31–33, 43). Consistent with these findings, our results showed that after knockdown of NF-κB p65 by siRNA transfection, both basal and IL-1β-induced expression of CXC chemokine genes decreased dramatically in A549 and H1734 cells. IL-1β can also regulate gene expression through the transcription factor CREB. Previously, we reported that IL-1β activates the mitogen-activated protein kinase (ERK1/2)/mitogen- and stress-activated protein kinase/CREB pathway and regulates MUC5AC gene expression in human airway epithelial cells (30). Sequence analysis of the CXC chemokine gene promoters identified a CRE or CRE-like domain in the gene promoters of CXCL1, CXCL2, CXCL3, CXCL5, and CXCL8 (31, 32, 34). In the present study, we showed that NSCLC cells with CREB knockdown were much less responsive to IL-1β than cells without such knockdown in terms of the induction of angiogenic CXC chemokine gene expression. We further confirmed the binding of CREB to the CXCL5 and CXCL8 promoters with ChIP assay. These data suggested that CREB can also regulate the expression of CXC chemokine genes. It have been reported that cyclooxygenase (COX)-2 is critical for IL-1β-induced angiogenesis both in vitro and in vivo through the production of prostanoids, such as PGE2 and thromboxane A2 (44). In addition, Pold et al. (40) reported that COX-2 contributes to the progression of NSCLC tumorigenesis by enhancing the expression of CXCL5 and CXCL8. Since COX-2 expression is also regulated by CREB in many cell types, including lung cell lines (45–48), CREB may regulate CXC chemokine gene expression through multiple mechanisms which need to be further delineated. Also, the interaction between CREB and NF-κB in regulating the expression of these chemokine genes should be further investigated, as these two transcription factors may compete with each other for the binding to CBP or work synergistically to affect the outcome of gene regulation (49). Additionally, the response elements for CREB and NF-κB on the promoter of CXCL8 gene are consecutively located, which further complicated the interaction of these two transcription factors and might have also contribute to the perplexing pattern of CXCL8 expression in response to KG-501 treatment.
In the IVLCM cell lines, we detected that the expression of the angiogenic CXC chemokine genes CXCL5 and CXCL8 increased progressively from normal (NHTBE), immortalized (BEAS-2B and 1799) and transformed (1198) to tumorigenic (1170-I) HBE cells. This is not unexpected because the angiogenic CXC chemokines CXCL5 and CXCL8, have been reported to be elevated in NSCLC tissues and their expression is related to tumor progression (23, 50, 51). Recently, accumulating evidences show that CXC chemokines induce tumorigenesis through stimulating cell proliferation, mediating cell survival, promoting angiogenesis, and facilitating tumor cell migration and invasion (13, 52). Also, we detected that the expression and activation of CREB increased correspondingly with the tumorigenicity in these IVLCM cell lines. The consistency of the expression and activation of these factors with the expression of angiogenic CXC chemokine genes in IVLCM confirmed the regulation of CXC chemokine genes by CREB and NF-κB, and lent support to their involvement in lung carcinogenesis. CREB has been known for its role in cell proliferation and survival (53–55). We have recently reported that the basal activity and expression level of CREB are commonly higher in a number of NSCLC cell lines versus normal cells, and the inhibition of CREB transcription activity induces apoptosis in these NSCLC cells by suppressing the expression of CREB-regulated genes that are involved in cell proliferation (56). Moreover, we have accumulated data from archived tumor tissue specimens showing that the CREB and p-CREB level are commonly higher in the lung tumor tissues versus the adjacent normal tissues (57). By demonstrating the role of CREB in tumor angiogenesis, the current study further suggests that CREB can be an effectual target for therapy as well as prevention of NSCLC.
KG-501 is a small molecule that binds to the KIX domain of CBP (35). It disrupts the interaction of CBP with CREB and inhibits the CREB-dependent activation of cellular genes. KG-501 can also disrupt the interaction of other factors with CBP, such as NF-κB (35). Our finding that KG-501 reduced the NSCLC/IL-1β conditioned medium-induced migration of HUVECs by down-regulating the expression of the ELR-positive CXC chemokine genes in NSCLC cells indicates that KG-501 can be used as therapeutic and/or preventive agent for inhibiting tumor-associated angiogenesis in NSCLC. Although the CXCL8 promoter contains both a NF-κB-binding site and a CRE-like motif, and our knockdown experiments shows that the depletion of either factors reduced IL-1β-induced CXCL8 expression (Fig. 4C and D), KG-501 was not able to effectively inhibit CXCL8 gene expression and protein secretion. Since KG-501 disrupts CREB- or NF-κB-CBP interaction, the regulation of CXCL8 gene by these factors may be mediated via a CBP-independent mechanism. A recent study demonstrated that the transcriptional activity of CREB on CXCL8 promoter requires a different coactivator, termed transducer of regulated CREB (TORC1) (34), suggesting a different regulatory mechanism beyond the binding of CREB to CRE. Nevertheless, CXCL8 did not seem to play a critical role in the induction of HUVECs migration as evidenced by the results of CXCL8 neutralization, therefore KG-501 can still effectively inhibit the NSCLC/IL-1β CM-induced HUVECs migration.
Our findings also implicated a positive association between chronic obstructive pulmonary disease (COPD) and lung cancer. COPD is a product of chronic inflammation which leads to tissue damage and physiological adaptations (58, 59). It has been known for years that local inflammation in the lungs plays an important role in airway remodeling and parenchymal destruction which are effects typified by COPD (59). It is now well recognized that in addition to lung inflammation, patients with COPD frequently demonstrate persistent low-grade systemic inflammation, with the characteristic release of pro-inflammatory mediators, such as IL-1β and tumor necrosis factor-α, into the circulation (60). Considerable evidences have associated chronic inflammation with cancer development. Our finding that CREB and NF-κB regulation of proangiogenic CXC chemokines in response to proinflammatory cytokine IL-1β may provide novel mechanistic linkage between COPD and the development of lung cancer.
In summary, IL-1β increases the angiogenic activity of NSCLC by upregulating the expression of an array of proangiogenic CXC chemokine genes which subsequently induce endothelial cell migration. The transcription factors CREB and NF-κB both can mediate this effect, suggesting that these two transcription factors are involved in tumor-associated angiogenesis, and therefore could be potential targets for the angioprevention in NSCLC. We also conclude that the small molecule KG-501 neutralizes the effect of NSCLC/IL-1β-conditioned medium on endothelial cell migration by inhibiting CREB and NF-κB transcriptional activity which resulted in the down-regulation of CXC chemokine genes expression in NSCLC cells, suggesting that KG-501 may be used as a therapeutic and angiopreventive agent for NSCLC.
Acknowledgments
We acknowledge Don Norwood for critical editing of this manuscript.
Grant support: Department of Defense VITAL grant W81XW-04-1-0142 (to J.S. Koo); National Heart, Lung, and Blood Institute Grant R01-HL-077556 (to J.S. Koo); Head and Neck Specialized Program of Research Excellence grant P50 CA097007 (PI: S.M. Lippman) HN SPORE Developmental Research Program Grant (to J.S. Koo); and National Cancer Institute Cancer Center Support Grant CA-16672 (to The University of Texas M. D. Anderson Cancer Center).
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 4.Liotta LA, Stracke ML. Tumor invasion and metastases: biochemical mechanisms. Cancer Treat Res. 1988;40:223–238. doi: 10.1007/978-1-4613-1733-3_10. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren L, O'Reilly MS, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 6.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 7.Tosetti F, Ferrari N, De Flora S, Albini A. Angioprevention': angiogenesis is a common and key target for cancer chemopreventive agents. Faseb J. 2002;16:2–14. doi: 10.1096/fj.01-0300rev. [DOI] [PubMed] [Google Scholar]
- 8.Bisacchi D, Benelli R, Vanzetto C, Ferrari N, Tosetti F, Albini A. Anti-angiogenesis and angioprevention: mechanisms, problems and perspectives. Cancer Detect Prev. 2003;27:229–238. doi: 10.1016/s0361-090x(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 9.Albini A, Tosetti F, Benelli R, Noonan DM. Tumor inflammatory angiogenesis and its chemoprevention. Cancer Res. 2005;65:10637–10641. doi: 10.1158/0008-5472.CAN-05-3473. [DOI] [PubMed] [Google Scholar]
- 10.Yoneda J, Kuniyasu H, Crispens MA, Price JE, Bucana CD, Fidler IJ. Expression of angiogenesis-related genes and progression of human ovarian carcinomas in nude mice. J Natl Cancer Inst. 1998;90:447–454. doi: 10.1093/jnci/90.6.447. [DOI] [PubMed] [Google Scholar]
- 11.Herbst RS, Fidler IJ. Angiogenesis and lung cancer: potential for therapy. Clin Cancer Res. 2000;6:4604–4606. [PubMed] [Google Scholar]
- 12.Bremnes RM, Camps C, Sirera R. Angiogenesis in non-small cell lung cancer: the prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood. Lung Cancer. 2006;51:143–158. doi: 10.1016/j.lungcan.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Strieter RM, Belperio JA, Burdick MD, Sharma S, Dubinett SM, Keane MP. CXC chemokines: angiogenesis, immunoangiostasis, and metastases in lung cancer. Ann N Y Acad Sci. 2004;1028:351–360. doi: 10.1196/annals.1322.041. [DOI] [PubMed] [Google Scholar]
- 14.Voronov E, Shouval DS, Krelin Y, et al. IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci U S A. 2003;100:2645–2650. doi: 10.1073/pnas.0437939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin EY, Pollard JW. Role of infiltrated leucocytes in tumour growth and spread. Br J Cancer. 2004;90:2053–2058. doi: 10.1038/sj.bjc.6601705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brigati C, Noonan DM, Albini A, Benelli R. Tumors and inflammatory infiltrates: friends or foes? Clin Exp Metastasis. 2002;19:247–258. doi: 10.1023/a:1015587423262. [DOI] [PubMed] [Google Scholar]
- 17.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 18.Dirkx AE, Oude Egbrink MG, Wagstaff J, Griffioen AW. Monocyte/macrophage infiltration in tumors: modulators of angiogenesis. J Leukoc Biol. 2006;80:1183–1196. doi: 10.1189/jlb.0905495. [DOI] [PubMed] [Google Scholar]
- 19.Nakao S, Kuwano T, Tsutsumi-Miyahara C, et al. Infiltration of COX-2-expressing macrophages is a prerequisite for IL-1 beta-induced neovascularization and tumor growth. J Clin Invest. 2005;115:2979–2991. doi: 10.1172/JCI23298. Epub 005 Oct 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 21.Saijo Y, Tanaka M, Miki M, et al. Proinflammatory cytokine IL-1 beta promotes tumor growth of Lewis lung carcinoma by induction of angiogenic factors: in vivo analysis of tumor-stromal interaction. J Immunol. 2002;169:469–475. doi: 10.4049/jimmunol.169.1.469. [DOI] [PubMed] [Google Scholar]
- 22.Arenberg DA, Kunkel SL, Polverini PJ, Glass M, Burdick MD, Strieter RM. Inhibition of interleukin-8 reduces tumorigenesis of human non-small cell lung cancer in SCID mice. J Clin Invest. 1996;97:2792–2802. doi: 10.1172/JCI118734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arenberg DA, Keane MP, DiGiovine B, et al. Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. J Clin Invest. 1998;102:465–472. doi: 10.1172/JCI3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 25.Kim SW, Cheon K, Kim CH, et al. Proteomics-based identification of proteins secreted in apical surface fluid of squamous metaplastic human tracheobronchial epithelial cells cultured by three-dimensional organotypic air-liquid interface method. Cancer Res. 2007;67:6565–6573. doi: 10.1158/0008-5472.CAN-06-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodziejski PJ, Musial A, Koo JS, Eissa NT. Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc Natl Acad Sci U S A. 2002;99:12315–12320. doi: 10.1073/pnas.192345199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo JS, Jetten AM, Belloni P, Yoon JH, Kim YD, Nettesheim P. Role of retinoid receptors in the regulation of mucin gene expression by retinoic acid in human tracheobronchial epithelial cells. Biochem J. 1999;338(Pt 2):351–357. [PMC free article] [PubMed] [Google Scholar]
- 28.Koo JS, Yoon JH, Gray T, Norford D, Jetten AM, Nettesheim P. Restoration of the mucous phenotype by retinoic acid in retinoid-deficient human bronchial cell cultures: changes in mucin gene expression. Am J Respir Cell Mol Biol. 1999;20:43–52. doi: 10.1165/ajrcmb.20.1.3310. [DOI] [PubMed] [Google Scholar]
- 29.Lacroix L, Feng G, Lotan R. Identification of genes expressed differentially in an in vitro human lung carcinogenesis model. Cancer Biol Ther. 2006;5:665–673. doi: 10.4161/cbt.5.6.2870. [DOI] [PubMed] [Google Scholar]
- 30.Song KS, Lee WJ, Chung KC, et al. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J Biol Chem. 2003;278:23243–23250. doi: 10.1074/jbc.M300096200. [DOI] [PubMed] [Google Scholar]
- 31.Anisowicz A, Messineo M, Lee SW, Sager R. An NF-kappa B-like transcription factor mediates IL-1/TNF-alpha induction of gro in human fibroblasts. J Immunol. 1991;147:520–527. [PubMed] [Google Scholar]
- 32.Corbett MS, Schmitt I, Riess O, Walz A. Characterization of the gene for human neutrophil-activating peptide 78 (ENA-78) Biochem Biophys Res Commun. 1994;205:612–617. doi: 10.1006/bbrc.1994.2709. [DOI] [PubMed] [Google Scholar]
- 33.Mukaida N, Shiroo M, Matsushima K. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J Immunol. 1989;143:1366–1371. [PubMed] [Google Scholar]
- 34.Iourgenko V, Zhang W, Mickanin C, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best JL, Amezcua CA, Mayr B, et al. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci U S A. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci U S A. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keane MP, Belperio JA, Xue YY, Burdick MD, Strieter RM. Depletion of CXCR2 inhibits tumor growth and angiogenesis in a murine model of lung cancer. J Immunol. 2004;172:2853–2860. doi: 10.4049/jimmunol.172.5.2853. [DOI] [PubMed] [Google Scholar]
- 38.Wislez M, Fujimoto N, Izzo JG, et al. High expression of ligands for chemokine receptor CXCR2 in alveolar epithelial neoplasia induced by oncogenic kras. Cancer Res. 2006;66:4198–4207. doi: 10.1158/0008-5472.CAN-05-3842. [DOI] [PubMed] [Google Scholar]
- 39.Addison CL, Daniel TO, Burdick MD, et al. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol. 2000;165:5269–5277. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
- 40.Pold M, Zhu LX, Sharma S, et al. Cyclooxygenase-2-dependent expression of angiogenic CXC chemokines ENA-78/CXC Ligand (CXCL) 5 and interleukin-8/CXCL8 in human non-small cell lung cancer. Cancer Res. 2004;64:1853–1860. doi: 10.1158/0008-5472.can-03-3262. [DOI] [PubMed] [Google Scholar]
- 41.Keates AC, Keates S, Kwon JH, et al. ZBP-89, Sp1, and nuclear factor-kappa B regulate epithelial neutrophil-activating peptide-78 gene expression in Caco-2 human colonic epithelial cells. J Biol Chem. 2001;276:43713–43722. doi: 10.1074/jbc.M107838200. [DOI] [PubMed] [Google Scholar]
- 42.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B-and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J Biol Chem. 1990;265:21128–21133. [PubMed] [Google Scholar]
- 43.Rovai LE, Herschman HR, Smith JB. Cloning and characterization of the human granulocyte chemotactic protein-2 gene. J Immunol. 1997;158:5257–5266. [PubMed] [Google Scholar]
- 44.Kuwano T, Nakao S, Yamamoto H, et al. Cyclooxygenase 2 is a key enzyme for inflammatory cytokine-induced angiogenesis. Faseb J. 2004;18:300–310. doi: 10.1096/fj.03-0473com. [DOI] [PubMed] [Google Scholar]
- 45.Wardlaw SA, Zhang N, Belinsky SA. Transcriptional regulation of basal cyclooxygenase-2 expression in murine lung tumor-derived cell lines by CCAAT/enhancer-binding protein and activating transcription factor/cAMP response element-binding protein. Mol Pharmacol. 2002;62:326–333. doi: 10.1124/mol.62.2.326. [DOI] [PubMed] [Google Scholar]
- 46.Tang Q, Chen W, Gonzales MS, Finch J, Inoue H, Bowden GT. Role of cyclic AMP responsive element in the UVB induction of cyclooxygenase-2 transcription in human keratinocytes. Oncogene. 2001;20:5164–5172. doi: 10.1038/sj.onc.1204667. [DOI] [PubMed] [Google Scholar]
- 47.Han S, Sidell N, Roser-Page S, Roman J. Fibronectin stimulates human lung carcinoma cell growth by inducing cyclooxygenase-2 (COX-2) expression. Int J Cancer. 2004;111:322–331. doi: 10.1002/ijc.20281. [DOI] [PubMed] [Google Scholar]
- 48.Chen JJ, Huang WC, Chen CC. Transcriptional regulation of cyclooxygenase-2 in response to proteasome inhibitors involves reactive oxygen species-mediated signaling pathway and recruitment of CCAAT/enhancer-binding protein delta and CREB-binding protein. Mol Biol Cell. 2005;16:5579–5591. doi: 10.1091/mbc.E05-08-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shenkar R, Yum HK, Arcaroli J, Kupfner J, Abraham E. Interactions between CBP, NF-kappaB, and CREB in the lungs after hemorrhage and endotoxemia. Am J Physiol Lung Cell Mol Physiol. 2001;281:L418–L426. doi: 10.1152/ajplung.2001.281.2.L418. [DOI] [PubMed] [Google Scholar]
- 50.Yuan A, Yang PC, Yu CJ, et al. Interleukin-8 messenger ribonucleic acid expression correlates with tumor progression, tumor angiogenesis, patient survival, and timing of relapse in non-small-cell lung cancer. Am J Respir Crit Care Med. 2000;162:1957–1963. doi: 10.1164/ajrccm.162.5.2002108. [DOI] [PubMed] [Google Scholar]
- 51.White ES, Flaherty KR, Carskadon S, et al. Macrophage migration inhibitory factor and CXC chemokine expression in non-small cell lung cancer: role in angiogenesis and prognosis. Clin Cancer Res. 2003;9:853–860. [PubMed] [Google Scholar]
- 52.Yeudall WA, Miyazaki H. Chemokines and squamous cancer of the head and neck: targets for therapeutic intervention? Expert Rev Anticancer Ther. 2007;7:351–360. doi: 10.1586/14737140.7.3.351. [DOI] [PubMed] [Google Scholar]
- 53.Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 2005;15:457–459. doi: 10.1016/j.tcb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Shankar DB, Sakamoto KM. The role of cyclic-AMP binding protein (CREB) in leukemia cell proliferation and acute leukemias. Leuk Lymphoma. 2004;45:265–270. doi: 10.1080/1042819031000151095. [DOI] [PubMed] [Google Scholar]
- 55.Abramovitch R, Tavor E, Jacob-Hirsch J, et al. A pivotal role of cyclic AMP-responsive element binding protein in tumor progression. Cancer Res. 2004;64:1338–1346. doi: 10.1158/0008-5472.can-03-2089. [DOI] [PubMed] [Google Scholar]
- 56.Aggarwal S, Kim SW, Ryu SH, Chung WC, Koo JS. Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer Res. 2008;68:981–988. doi: 10.1158/0008-5472.CAN-06-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo H-S, Liu D, Bekele B, et al. CREB Overexpression: A Feature Associated with Negative Prognosis in Never-Smokers with NSCLC. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-07-5376. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szilasi M, Dolinay T, Nemes Z, Strausz J. Pathology of chronic obstructive pulmonary disease. Pathol Oncol Res. 2006;12:52–60. doi: 10.1007/BF02893433. [DOI] [PubMed] [Google Scholar]
- 59.Jeffery PK. Remodeling and inflammation of bronchi in asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2004;1:176–183. doi: 10.1513/pats.200402-009MS. [DOI] [PubMed] [Google Scholar]
- 60.Hegab AE, Sakamoto T, Saitoh W, et al. Polymorphisms of TNFalpha, IL1beta, and IL1RN genes in chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 2005;329:1246–1252. doi: 10.1016/j.bbrc.2005.02.099. [DOI] [PubMed] [Google Scholar]