Abstract
OBJECTIVE
Fasting plasma glucose and risk of type 2 diabetes are higher among Indian Asians than among European and North American Caucasians. Few studies have investigated genetic factors influencing glucose metabolism among Indian Asians.
RESEARCH DESIGN AND METHODS
We carried out genome-wide association studies for fasting glucose in 5,089 nondiabetic Indian Asians genotyped with the Illumina Hap610 BeadChip and 2,385 Indian Asians (698 with type 2 diabetes) genotyped with the Illumina 300 BeadChip. Results were compared with findings in 4,462 European Caucasians.
RESULTS
We identified three single nucleotide polymorphisms (SNPs) associated with glucose among Indian Asians at P < 5 × 10−8, all near melatonin receptor MTNR1B. The most closely associated was rs2166706 (combined P = 2.1 × 10−9), which is in moderate linkage disequilibrium with rs1387153 (r2 = 0.60) and rs10830963 (r2 = 0.45), both previously associated with glucose in European Caucasians. Risk allele frequency and effect sizes for rs2166706 were similar among Indian Asians and European Caucasians: frequency 46.2 versus 45.0%, respectively (P = 0.44); effect 0.05 (95% CI 0.01–0.08) versus 0.05 (0.03–0.07 mmol/l), respectively, higher glucose per allele copy (P = 0.84). SNP rs2166706 was associated with type 2 diabetes in Indian Asians (odds ratio 1.21 [95% CI 1.06–1.38] per copy of risk allele; P = 0.006). SNPs at the GCK, GCKR, and G6PC2 loci were also associated with glucose among Indian Asians. Risk allele frequencies of rs1260326 (GCKR) and rs560887 (G6PC2) were higher among Indian Asians compared with European Caucasians.
CONCLUSIONS
Common genetic variation near MTNR1B influences blood glucose and risk of type 2 diabetes in Indian Asians. Genetic variation at the MTNR1B, GCK, GCKR, and G6PC2 loci may contribute to abnormal glucose metabolism and related metabolic disturbances among Indian Asians.
The World Health Organization estimates that by 2025 there will be 57 million Indian Asians with type 2 diabetes, (1) by which time one-quarter of type 2 diabetic patients globally will be Indian Asian. Migration data among Indian Asians show that the prevalence of type 2 diabetes is ∼20% among Indian Asians in the U.K. and North America compared with ∼15% in urban Indian and ∼6% in rural India (2–5). Although these studies emphasize the importance of environmental triggers such as sedentary lifestyle and excess consumption of energy-dense foods, genetic factors are also likely to be important (6). Understanding the genetic mechanisms influencing glucose metabolism among Indian Asians may provide insight into the increased risk of type 2 diabetes among Indian Asians. Genome-wide association studies have recently identified common genetic variation in and around the genes GCK, GCKR, G6PC2, and MTNR1B as determinants of glucose levels in European populations (7–11). We have carried out a genome-wide association study of fasting plasma glucose levels in a population-based cohort of Indian Asian men and women.
RESEARCH DESIGN AND METHODS
Genome-wide association for fasting plasma glucose was performed among 5,089 nondiabetic Indian Asians (fasting glucose levels <7.0 mmol/l; free from pharmacologic or dietary treatment for diabetes) (12) and genotyped using the Illumina Hap610 BeadChip, and a separate sample of 2,385 Indian Asians was genotyped with the Illumina Hap300 BeadChip. Results for the Hap610 sample were analyzed separately, followed by meta-analysis of the results from Hap610 and Hap300 genotyping arrays. To examine the possible contribution of single nucleotide polymorphisms (SNPs) identified to raised glucose in Indian Asians, allele frequencies and effect sizes were compared with results for 4,462 European Caucasian participants of the Northern Finland Birth Cohort of 1966 (NFBC1966).
All Indian Asian participants were recruited though the London Life Sciences Prospective Population (LOLIPOP) Study. LOLIPOP is an ongoing population-based cohort study of ∼30,000 Indian Asian and European Caucasian men and women aged 35–75 years and recruited from the patient records among 58 general practitioners in West London, U.K. (13). Indian Asians were identified as having all four grandparents born on the Indian subcontinent. Response rates averaged 62%; there were no material differences between responders and nonresponders with respect to age, sex, comorbidity, and available data for risk factors. All participants gave written informed consent, including for genetic studies. The study is approved by the local Research Ethics Committee.
An interviewer-administered questionnaire was used to collect data on medical history, family history, current prescribed medication, and cardiovascular risk factors. Physical assessment included anthropometric measurements (height, weight, waist circumference, and hip circumference) and blood pressure. Blood was collected after an 8-h fast for biochemical analysis, including glucose, insulin, total and HDL cholesterol, and triglycerides, and whole blood was taken for extraction of DNA. Insulin sensitivity and pancreatic β-cell function were estimated using the homeostatic model assessment (HOMA) method (14) where insulin sensitivity (HOMA-S) = 22.5 (fasting plasma insulin × fasting plasma glucose) and β-cell function (HOMA-B) = (20 × fasting plasma insulin)/(fasting plasma glucose − 3.5).
We studied 5,089 nondiabetic Indian Asians genotyped with the Illumina Hap610 BeadChip (Hap610 sample) and 2,385 Indian Asians genotyped with the Hap300 BeadChip (Hap300 sample). The Hap300 sample included 698 Indian Asians with type 2 diabetes, enabling testing of SNPs against type 2 diabetic case subject or control subject status.
All European Caucasians were participants of NFBC1966, a study of factors affecting preterm birth, low birth weight, and subsequent morbidity and mortality. Participants comprise 12,068 mothers and their 12,231 births in the provinces of Oulu and Lapland during 1966. At age 31 years, all individuals still living in the Helsinki area or northern Finland were asked to participate in a detailed biological and medical examination (n = 6,007) including measurement of fasting glucose. For the current study, genotype and fasting glucose measurements were available for 4,462 NFBC1966 participants (2,116 male and 2,346 female). Study methods and characteristics of participants have been described previously (11). The University of Oulu ethics committee approved the study. The NFBC1966 samples used were also included in the genome-wide association studies that identified common genetic variation in and around G6PC2 and MTNR1B as determinants of glucose levels in European populations (7,9).
Genotyping.
Genotyping using the Illumina Hap610 BeadChip was carried out according to standard methodology. In brief, each sample was whole genome amplified, fragmented, precipitated, and resuspended in appropriate hybridization buffer. Denatured samples were hybridized on prepared HumanHap610 BeadChips for a minimum of 16 h at 48°C. After hybridization, the BeadChips were processed for the single-base extension reaction as well as staining and imaging on an Illumina Bead Array Reader. Normalized bead intensity data obtained for each sample were loaded into the Illumina Beadstudio 2.0 software, which converted fluorescence intensities into SNP genotypes. Based on 17 duplicate scans, mean genotyping concordance rate was 99.9987% (range 99.9930–99.9998%). For the 582,539 autosomal SNPs included on the Hap610 BeadChip, average SNP call rate was 99.7%, with call rates >95% for 99.7% of SNPs. Call rate was >95% for 99.1% of people; 46 people were excluded for call rates <95%. We also excluded SNPs with minor allele frequency <0.01, call rate <0.95%, or Hardy-Weinberg equilibrium of P < 10−6. This left 544,390 autosomal SNPs for the genome-wide association analysis. Methods and quality control for genotyping carried out using the Illumina Hap300 BeadChip in Indian Asians and the Illumina Hap370 BeadChip in European Caucasians have been described previously (11,13).
Statistics.
Single SNP marker tests were carried out for association with fasting glucose levels in nondiabetic Indian Asians under an additive genetic model with adjustment for age and sex. Principal components analysis was used to characterize population substructure in Indian Asians genotyped on the Hap610 and Hap300 BeadChips, and the top four components were included in models (15). Analysis of QQ plots for association over all SNPs showed good adherence to null expectations, indicating that the approach was sufficient to allow for any inflation due to population substructure (see supplemental Fig. S1, available in the online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1805/DC1). λ values for the genome-wide analyses were 1.01 in both the Illumina Hap610 and the Illumina Hap300 samples (compared with 1.07 and 1.02, respectively, for analyses without principal components). The Hap610 sample size provided 80% power to identify SNPs associated with 0.8% of population variation in glucose at genome-wide significance (P < 5 × 10−8 [16]).
We then carried out a meta-analysis of results from the Illumina Hap610 and the Illumina Hap300 samples. Imputation was done using a hidden Markov model algorithm implemented in MACH software and pooled phased haplotypes for the CEU, CHB/JPT, and YRI samples from HapMap build35, dbSNP build 125 (17). Imputed SNPs with minor allele frequency <0.01 or low-quality score (r2 < 0.30) were removed. This generated ∼1.9 million directly genotyped or imputed autosomal SNPs per participant with data available in both samples. Meta-analysis was carried out using z scores weighted by square root of sample size, implemented in the software package METAL (www.sph.umich.edu/csg/abecasis/metal). QQ plots showed good adherence to null expectations (λ for meta-analyzed data = 1.047; supplemental Fig. S1).
The associations of SNPs with type 2 diabetes and other phenotypic traits, and of risk allele score with glucose levels, were tested in the Illumina Hap300 data using regression analysis and an additive genetic model. Analyses were adjusted for age and sex in Indian Asians as well as for sex among European Caucasians (NFBC1966 participants are all 31 years of age). Heterogeneity of effect between Indian Asians and European Caucasians was tested by joint analysis of data for Indian Asians and European Caucasians, with incorporation of ethnicity and genotype-ethnicity interaction terms into the regression models. Two SNP regression models were used to identify whether SNPs from the same genetic locus have separate relationships with glucose levels. Glucose risk allele scores were also compared between Indian Asians and European Caucasians by independent samples t test.
RESULTS
The characteristics of Indian Asian and European Caucasian participants are summarized in Table 1.
TABLE 1.
Characteristics of Indian Asians and European Caucasians studied
| Indian Asians Hap610 sample | Indian Asian Hap300 sample |
European Caucasians | ||
|---|---|---|---|---|
| Nondiabetic | Diabetic | |||
| n | 5,089 | 1,687 | 698 | 4,462 |
| Age (years) | 53.9 ± 10.6 | 48.4 ± 10.6 | 56.0 ± 10.1 | 31 |
| Sex (% male) | 85 | 100 | 100 | 47.6 |
| Waist-to-hip ratio | 0.95 ± 0.07 | 0.96 ± 0.07 | 0.99 ± 0.07 | 0.86 ± 0.09 |
| BMI (kg/m2) | 26.8 ± 4.2 | 26.7 ± 4.2 | 27.6 ± 4.4 | 24.6 ± 4.2 |
| Known hypertension | 35 | 21 | 55 | 19.8 |
| Systolic blood pressure (mmHg) | 133.4 ± 18.9 | 132.4 ± 20.3 | 139.9 ± 20.5 | 124.7 ± 13.5 |
| Diastolic blood pressure (mmHg) | 82.3 ± 10.7 | 82.5 ± 12.1 | 82.9 ± 11.8 | 77.2 ± 11.4 |
| Type 2 diabetes | 0 | 0 | 100 | 0 |
| Glucose (mmol/l) | 5.2 ± 0.62 | 5.1 ± 0.6 | 8.8 ± 2.9 | 5.0 ± 0.5 |
| Insulin (IU/l) | 11.7 ± 8.8 | 12.1 ± 8.6 | 15.4 ± 13.7 | 8.5 ± 4.3 |
| HOMA-B | 142 ± 124 | 157 ± 122 | 68 ± 224 | 100 ± 29 |
| HOMA-S | 0.57 ± 1.48 | 0.55 ± 1.33 | 0.45 ± 1.56 | 1.05 ± 0.37 |
| Total cholesterol (mmol/l) | 5.21 ± 1.12 | 5.41 ± 1.05 | 4.73 ± 1.07 | 5.09 ± 1.01 |
| HDL cholesterol (mmol/l) | 1.22 ± 0.30 | 1.24 ± 0.31 | 1.14 ± 0.27 | 1.57 ± 0.38 |
| Triglycerides (mmol/l) | 1.77 ± 1.11 | 1.64 ± 0.81 | 1.78 ± 0.86 | 1.19 ± 0.74 |
Data are means ± SD for continuous and percent for categorical traits.
Genome-wide association in the Hap610 sample.
We identified three SNPs associated with glucose levels in Indian Asians at a genome-wide level of significance (P < 5 × 10−8 [16]) in the Hap610 sample (Table 2; supplemental Fig. S2). All were located near melatonin receptor MTNR1B and in high linkage disequilibrium (LD) with each other (pairwise r2 > 0.5; Fig. 1, supplemental Table S1). The most closely associated SNP was rs2166706; when conditioned on rs2166706, there was no evidence for a separate effect of rs3847554 or rs1387153 on glucose levels. The associations of rs2166706, rs3847554, and rs1387153 with glucose levels were confirmed among Indian Asians in the Hap300 sample, in whom risk allele of rs2166706 was associated with ∼0.05 mmol/l higher glucose levels than that for the alternate allele (Table 2).
TABLE 2.
Genomic context and association test results for the three SNPs associated with fasting blood glucose levels among Indian Asians in the Hap610 sample at P < 5 × 10−8
| Genomic context |
Alleles |
Risk allele frequency |
Indian Asians |
European Caucasians |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap610 sample |
Hap300 sample |
Combined analysis | |||||||||||
| SNP | Chr | Gene locus | Alt | Risk | IA | EC | Effect | P | Effect | P | P | Effect | P |
| rs2166706 | 11 | MTNR1B | A | G | 46.2 | 45.0 | 0.07 (0.04–0.09) | 1.6 × 10−8 | 0.05 (0.01–0.08) | 0.006 | 2.1 × 10−9 | 0.05 (0.03–0.07) | 4.9 × 10−7 |
| rs3847554 | 11 | MTNR1B | G | A | 49.8 | 49.4 | 0.06 (0.04–0.09) | 1.9 × 10−8 | 0.04 (0.01–0.08) | 0.01 | 3.9 × 10−9 | 0.06 (0.04–0.08) | 2.5 × 10−8 |
| rs1387153 | 11 | MTNR1B | G | A | 38.1 | 35.3 | 0.06 (0.04–0.09) | 3.5 × 10−8 | 0.05 (0.01–0.08) | 0.01 | 1.5 × 10−8 | 0.05 (0.03–0.07) | 2.6 × 10−7 |
Data are effect size (95% CI) for fasting glucose in millimoles per liter for each copy of the risk allele. All three SNPs were directly genotyped, with call rates >99% in the three study samples. Alt, alternate allele; Chr, chromosome; EC, European Caucasian; IA, Indian Asian.
FIG. 1.
Genomic context of melatonin receptor MTNR1B region showing pairwise LD between SNPs with minor allele frequency ≥0.05 directly genotyped in both Indian Asians (A) and European Caucasians (B) using Haploview's standard color scheme.
Further testing of rs2166706 in the Hap300 sample.
The risk allele of SNP rs2166706 was associated with elevated A1C and reduced HOMA-B among nondiabetic Indian Asians (Table 3) in the Hap300 sample. In contrast, there was no association of rs2166706 with weight, BMI, insulin levels, or HOMA-S (Table 3). Risk allele frequency of rs2166706 was higher among Indian Asians with type 2 diabetes than among nondiabetic Indian Asians in the Hap300 sample. The age- and sex-adjusted odds ratio (OR) for type 2 diabetes was 1.21 (95% CI 1.06–1.38) per copy of risk allele of rs2166706 among Indian Asians (P = 0.006). After additional adjustment for HOMA-B, OR for type 2 diabetes was reduced and no longer statistically significant (1.12 [0.97–1.30]; P = 0.14).
TABLE 3.
Associations of rs2166706 with secondary phenotypes among Indian Asians in the Hap300 sample and European Caucasians
| Indian Asians | P | European Caucasians | P | Phetero | |
|---|---|---|---|---|---|
| Type 2 diabetes† | 1.21 (1.06–1.38) | 0.006 | — | — | — |
| A1C (%)* | 0.12 (0.04–0.19) | 0.003 | — | — | — |
| HOMA-B* | −7.1 (−11.0 to −3.0) | 0.001 | −1.9 (−3.6 to −0.2) | 0.03 | 0.02 |
| HOMA-S* | 5.4 (−4.5 to 16.2) | 0.29 | 3.5 (−0.7 to 7.9) | 0.10 | 0.74 |
| BMI (kg/m2)* | −0.17 (−0.40 to 0.06) | 0.14 | −0.36 (−0.80 to 0.08) | 0.11 | 0.45 |
| Waist-to-hip ratio* | −0.001 (−0.005 to 0.003) | 0.64 | −0.005 (−0.013 to 0.002) | 0.16 | 0.36 |
| Hypertension† | 0.93 (0.82–1.05) | 0.22 | 1.00 (0.95–1.06) | 0.72 | 0.28 |
| Systolic blood pressure (mmHg)* | −0.9 (−1.9 to 0.12) | 0.08 | −0.2 (−1.6 to 1.2) | 0.79 | 0.43 |
| Diastolic blood pressure (mmHg)* | −0.6 (−1.3 to 0.0) | 0.05 | 0.0 (−1.2 to 1.2) | 0.99 | 0.39 |
| Cholesterol (mmol/l)* | 0.00 (−0.05 to 0.06) | 0.88 | 0.02 (−0.09 to 0.13) | 0.69 | 0.75 |
| HDL cholesterol (mmol/l)* | −0.2 (−1.5 to 1.1) | 0.78 | 1.4 (−1.1 to 3.9) | 0.28 | 0.27 |
| Triglycerides (mmol/l)* | 5.1 (−1.0 to 11.7) | 0.10 | −2.1 (−7.0 to 3.1) | 0.43 | 0.08 |
Effects are *unit change (initial to final quantity) or †odds ratio (95% CI) per allele copy, under additive genetic model, adjusted for sex (and age among Indian Asians).
Meta-analysis of genome-wide association data in Indian Asians.
In meta-analysis of the Indian Asian Hap610 and Hap300 samples, 29 SNPs reached P < 5 × 10−8. These were distributed in the MTNR1B, GCK, and G6PC2 loci (supplemental Table S2 and Fig. S2). The LD structure of the GCK and G6PC2 loci were similar among Indian Asians and European Caucasians (supplemental Figs. S3 and S4). Genetic variants near GCKR that have been associated with glucose levels in European populations (9,10) were also associated with fasting glucose levels among Indian Asians, although not at a genome-wide significance level (GCKR: rs1260326, P = 3.3 × 10−4).
Comparisons with European Caucasians.
SNP rs2166706 was associated with glucose levels in European Caucasians (Table 2). Based on data from the present study and on the HapMap CEU population (HapMap PhaseIII/Rel#1 Sept 2008, NCBI B36 assembly, dbSNP b126), SNP rs2166706 is in moderate LD with both rs1387153 and rs10830963 (supplemental Table S1), SNPs that have been reported to be associated with glucose in European Caucasians (7,8). Frequencies for risk allele of rs2166706 were similar among Indian Asians and European Caucasians (46.2 vs. 45.0%, respectively; P = 0.44) with no evidence for heterogeneity of effect on glucose (P = 0.84; Table 2). Results for association of rs2166706 with secondary phenotypes were similar in Indian Asians and European Caucasians (Table 3) except for HOMA-B, where the relationship with rs2166706 was weaker and less statistically significant among European Caucasians.
Combined effects of genetic loci influencing glucose levels.
Risk allele frequencies of rs1260326 (GCKR) and rs560887 (G6PC2) were higher, and of rs2166706 (MTNR1B) and rs4607517 (GCK) similar, among Indian Asians and European Caucasians (Table 2 and supplemental Table S3). Effect sizes on glucose levels at the MTNR1B, GCKR, G6PC2, and GCK loci were similar among Indian Asians and European Caucasians, with no evidence for heterogeneity between the populations (Table 2 and supplemental Table S3).
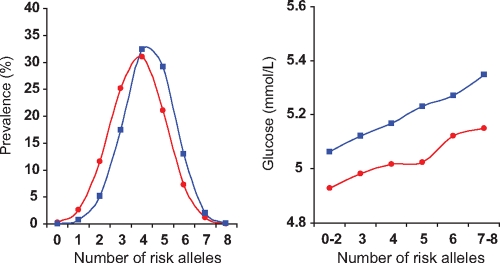
A score comprising number of risk alleles at these four loci was closely related to glucose levels in both Indian Asians and European Caucasians (change in glucose per unit of allele score: Indian Asians 0.06 ± 0.01 mmol/l, P = 1.1 × 10−16; European Caucasians 0.04 ± 0.01 mmol/l, P = 4.6 × 10−15; Fig. 2). Risk allele score was greater among Indian Asians than European Caucasians (4.3 ± 1.2 vs. 3.8 ± 1.3, respectively; P = 2.3 × 10−85; Fig. 2) and accounted for 1.2% of population variance in glucose levels in both populations. However, at each level of risk allele score, Indian Asians had higher glucose levels than European Caucasians (P = 3.3 × 10−33; Fig. 2).
FIG. 2.
Distribution of risk allele score (number of risk alleles at the MTNR1B, GCKR, GCK, and G6PC2 loci) among Indian Asians (blue squares) and European Caucasians (red circles) as well as the relationship of risk allele score to glucose levels in the two populations.
DISCUSSION
We have identified and replicated an association of SNP rs2166706 near MTNR1B with plasma glucose levels and show that rs2166706 is also associated with increased risk of type 2 diabetes among Indian Asians. We also replicate associations of the GCK, GCKR, and G6PC2 loci with glucose levels in Indian Asians.
Our findings extend the results of recent studies in European Caucasian populations that identify genetic variation near MTNR1B as an important determinant of glucose levels (7,8). MTNR1B encodes a high-affinity receptor for melatonin, a circulating hormone released by the pineal gland (18). Secretion of melatonin is tightly regulated by the hypothalamic suprachiasmatic nucleus, the anatomic center of the mammalian circadian clock (19,20). Melatonin release is one of the key mechanisms by which the circadian clock maintains synchronization and regulates the biologic activities of peripheral tissues (18). Melatonin receptors (including MTNR1B) are highly expressed in the hypothalamus, where they contribute to melatonin-induced negative feedback and phase timing of circadian activity (21). MTNR1B is also expressed in human pancreatic islet cells and may mediate the effects of melatonin on basal and glucose-induced insulin release (22).
Our findings of an association between SNP rs2166706 near MTNR1B and raised glucose, reduced pancreatic β-cell function, and increased risk of type 2 diabetes are consistent with accumulating evidence that circadian clocks play a key role in the regulation of carbohydrate and energy metabolism. Clock/Clock null mice develop hyperglycemia, and in humans sleep loss and depression are associated with circadian desynchronization and increased risk of type 2 diabetes (23–25). Although further functional studies will be required to clarify the contribution of peripheral and central melatonin signaling pathways to glucose metabolism and type 2 diabetes, our observations suggest that MTNR1B signaling may be a therapeutic target for the development of agents to improve glucose regulation and to prevent or treat type 2 diabetes.
We also confirm association of GCK, GCKR, and G6PC2 loci with glucose levels in Indian Asians. Effect sizes were similar among Indian Asians and European Caucasians, and a risk allele score was associated with glucose similarly in the two populations. However, risk allele frequencies were higher among Indian Asians than European Caucasians. These findings suggest that genetic variation at these loci makes an important contribution to raised glucose levels among Indian Asians.
These are the first data identifying genetic variants that influence glucose metabolism among Indian Asians, a population at high risk for type 2 diabetes and related metabolic disorders. The 21% of Indian Asians homozygous for the G allele of rs2166706 of MTNR1B have ∼0.1 mmol/l higher fasting glucose levels and ∼45% higher odds of type 2 diabetes compared with people with AA genotype. Our findings give new insight into the genetic mechanisms underlying abnormalities of glucose metabolism among high-risk Indian Asians and provide the basis for future studies to investigate the role of circadian rhythms and melatonin signaling in the etiology of type 2 diabetes.
Supplementary Material
Acknowledgments
The LOLIPOP study was supported by the British Heart Foundation (SP/04/002) and the Wellcome Trust. NFBC1966 study was supported by the Academy of Finland (104781), the Medical Research Council (G0500539), University Hospital Oulu, Biocenter, University of Oulu, Finland, National Heart, Lung and Blood Institute Grant 5R01HL087679-02 through the SNP Typing for Association with Multiple Phenotypes from Existing Epidemiological Data (STAMPEED) program (1RL1MH083268-01) and the Wellcome Trust (Project Grant GR069224). The DNA extractions, sample quality control subjects, biobank upkeeping, and aliquotting were performed in the national Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland (120315) and Biocentrum Helsinki. For NFBC1966 the wide genotyping was conducted by the Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, Massachusetts.
No potential conflicts of interest relevant to this article were reported.
We thank Nelson Freimer, Leena Peltonen, and the research teams involved in LOLIPOP and NFBC1966.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.King H, Aubert RE, Herman WH: Global burden of diabetes, 1995–2025: prevalence, numerical estimates, and projections Diabetes Care 1998;21:1414–1431 [DOI] [PubMed] [Google Scholar]
- 2.McKeigue PM, Shah B, Marmot MG: Relation of central obesity and insulin resistance with high diabetes prevalence and cardiovascular risk in South Asians Lancet 1991;337:382–386 [DOI] [PubMed] [Google Scholar]
- 3.Chambers JC, Obeid OA, Refsum H, et al. : Plasma homocysteine concentrations and risk of coronary heart disease in UK Indian Asian and European men Lancet 2000;355:523–527 [DOI] [PubMed] [Google Scholar]
- 4.Mohan V, Deepa M, Deepa R, et al. : Secular trends in the prevalence of diabetes and impaired glucose tolerance in urban South India–the Chennai Urban Rural Epidemiology Study (CURES-17) Diabetologia 2006;49:1175–1178 [DOI] [PubMed] [Google Scholar]
- 5.Ramachandran A, Snehalatha C, Baskar AD, et al. : Temporal changes in prevalence of diabetes and impaired glucose tolerance associated with lifestyle transition occurring in the rural population in India Diabetologia 2004;47:860–865 [DOI] [PubMed] [Google Scholar]
- 6.Kahn SE, Hull RL, Utzschneider KM: Mechanisms linking obesity to insulin resistance and type 2 diabetes Nature 2006;444:840–846 [DOI] [PubMed] [Google Scholar]
- 7.Prokopenko I, Langenberg C, Florez JC, et al. : Variants in MTNRIB influence fasting glucose levels Nat Genet 2009;41:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, et al. : A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk Nat Genet 2009;41:89–94 [DOI] [PubMed] [Google Scholar]
- 9.Bouatia-Naji N, Rocheleau G, Van Lommel L, et al. : A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels Science 2008;320:1085–1088 [DOI] [PubMed] [Google Scholar]
- 10.Vaxillaire M, Cavalcanti-Proença C, Dechaume A, et al. : The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population Diabetes 2008;57:2253–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabatti C, Service SK, Hartikainen AL, et al. : Genome-wide association analysis of metabolic traits in a birth cohort from a founder population Nat Genet 2009;41:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Diabetes Association. American Diabetes Association. Diagnosis and classification of diabetes mellitus Diabetes Care 2006;29(Suppl. 1):S43–S48 [PubMed] [Google Scholar]
- 13.Chambers JC, Elliott P, Zabaneh D, et al. : Common genetic variation near MC4R is associated with waist circumference and insulin resistance Nat Genet 2008;40:716–718 [DOI] [PubMed] [Google Scholar]
- 14.Wallace TM, Levy JC, Matthews DR: Use and abuse of HOMA modeling Diabetes Care 2004;27:1487–1495 [DOI] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, et al. : Principal components analysis corrects for stratification in genome-wide association studies Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 16.Pe'er I, Yelensky R, Altshuler D, et al. : Estimation of the multiple testing burden for genomewide association studies of nearly all common variants Genet Epidemiol 2008;32:381–385 [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Li Y, Singleton AB, et al. : Genotype-imputation accuracy across worldwide human populations Am J Hum Genet 2009;84:235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brzezinski A: Melatonin in humans N Engl J Med 1997;336:186–195 [DOI] [PubMed] [Google Scholar]
- 19.Saper CB, Scammell TE, Lu J: Hypothalamic regulation of sleep and circadian rhythms Nature 2005;437:1257–1263 [DOI] [PubMed] [Google Scholar]
- 20.Reppert SM, Weaver DR: Coordination of circadian timing in mammals Nature 2002;418:935–941 [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Weaver DR, Jin X, et al. : Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock Neuron 1997;19:91–102 [DOI] [PubMed] [Google Scholar]
- 22.Ramracheya RD, Muller DS, Squires PE, et al. : Function and expression of melatonin receptors on human pancreatic islets J Pineal Res 2008;44:273–279 [DOI] [PubMed] [Google Scholar]
- 23.Turek FW, Joshu C, Kohsaka A, et al. : Obesity and metabolic syndrome in circadian Clock mutant mice Science 2005;308:1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golden SH, Lazo M, Carnethon M, et al. : Examining a bidirectional association between depressive symptoms and diabetes JAMA 2008;299:2751–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knutson KL, Van Cauter E: Associations between sleep loss and increased risk of obesity and diabetes Ann N Y Acad Sci 2008;1129:287–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.