Abstract
OBJECTIVE
To examine the role of aldo-keto reductases (AKRs) in the cardiovascular metabolism of the precursors of advanced glycation end products (AGEs).
RESEARCH DESIGN AND METHODS
Steady-state kinetic parameters of AKRs with AGE precursors were determined using recombinant proteins expressed in bacteria. Metabolism of methylglyoxal and AGE accumulation were studied in human umbilical vein endothelial cells (HUVECs) and C57 wild-type, akr1b3 (aldose reductase)-null, cardiospecific-akr1b4 (rat aldose reductase), and akr1b8 (FR-1)-transgenic mice. AGE accumulation and atherosclerotic lesions were studied 12 weeks after streptozotocin treatment of C57, akr1b3-null, and apoE- and akr1b3-apoE–null mice.
RESULTS
Higher levels of AGEs were generated in the cytosol than at the external surface of HUVECs cultured in high glucose, indicating that intracellular metabolism may be an important regulator of AGE accumulation and toxicity. In vitro, AKR 1A and 1B catalyzed the reduction of AGE precursors, whereas AKR1C, AKR6, and AKR7 were relatively ineffective. Highest catalytic efficiency was observed with AKR1B1. Acetol formation in methylglyoxal-treated HUVECs was prevented by the aldose reductase inhibitor sorbinil. Acetol was generated in hearts perfused with methylglyoxal, and its formation was increased in akr1b4- or akr1b8-transgenic mice. Reduction of AGE precursors was diminished in hearts from akr1b3-null mice. Diabetic akr1b3-null mice accumulated more AGEs in the plasma and the heart than wild-type mice, and deletion of akr1b3 increased AGE accumulation and atherosclerotic lesion formation in apoE-null mice.
CONCLUSIONS
Aldose reductase–catalyzed reduction is an important pathway in the endothelial and cardiac metabolism of AGE precursors, and it prevents AGE accumulation and atherosclerotic lesion formation.
Nonenzymatic glycation and oxidation of proteins and lipids results in the formation of advanced glycation end products (AGEs) (1,2). The AGEs are formed when α-dicarbonyl and oxoaldehydes react with amines (Maillard reaction). These reactions generate multiple products that lead to Amadori modified proteins or aminophospholipids. AGEs are generated normally as tissues age, but they are formed at an accelerated rate during diabetes (3,4). Excessive AGE formation has been linked to cross-linking of matrix molecules such as collagen, vitronectin, and laminin (3,5,6). In addition binding of AGEs to receptor of AGE (RAGE) results in the activation of mitogen-activated protein (MAP) kinases, nuclear factor (NF)-κB, and cAMP response element binding (CREB) (7,8). These events stimulate the production of reactive oxygen species and lead ultimately to an increase in vascular permeability and inflammation. The pathological significance of such receptor-mediated events is underscored by studies showing that RAGE blockage suppresses accelerated atherosclerotic lesion formation in diabetic mice (9), decreases neointimal expansion (10), and restores diabetic deficits in wound healing (11). Pharmacological blockage or genetic deletion of RAGE has been shown also to decrease albuminuria and mesangial expansion/glomerulosclerosis in mouse models of type 1 diabetes (12). Taken together, these studies suggest that the AGEs are significant mediators of hyperglycemic injury.
In most tissues, AGEs are derived from products generated from the auto-oxidation of glucose and fructose. These include deoxyglucosone, methylglyoxal, and glyoxal (1,2). Methylglyoxal is generated nonenzymatically from the oxidation and spontaneous dismutation of intermediates in the glycolysis pathway or enzymatic oxidation reaction catalyzed by peroxidases, whereas other AGE precursors such as deoxyglucosone are generated from fructose or from nonenzymatic degradation of Amadori rearrangement compounds (13). Previous studies show that the AGE precursor methylglyoxal is metabolized and detoxified by the glyoxalase system consisting of glyoxalase-I and -II (14,15). Nevertheless, the participation of other enzymes in the metabolism of methylglyoxal and other AGE precursors remains unclear. In vitro studies show that members of the aldo-keto reductase (AKR) superfamily catalyze methylglyoxal reduction (16,17), and in bacteria these enzymes participate in the conversion of methylglyoxal to acetol (18,19). Nevertheless, the in vivo role of AKRs in mammalian metabolism of AGE precursors has not been studied. Accordingly, we tested the efficacy of several AKRs in catalyzing the reduction of AGE precursors. Our results show that reduction catalyzed by aldose reductase (AKR1B) is a significant metabolic fate of AGE precursors in endothelial cells and in the heart and that this metabolism prevents AGE accumulation during diabetes and atherosclerosis.
RESEARCH DESIGN AND METHODS
See the online supplement available at http://diabetes.diabetesjournals.org/cgi/full/db09-0375/DC1.
RESULTS
High glucose–induced AGE formation.
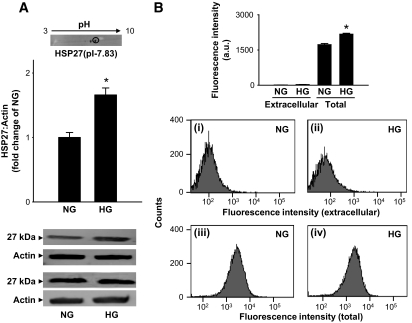
AGEs are generated from nonenzymatic processes both in the cytosol and at the external matrix. To examine AGE abundance in these compartments, human umbilical vein endothelial cells (HUVECs) were cultured in media containing normal (5.5 mmol/l) and high (30 mmol/l) glucose for 7 days, and AGE formation was measured. Western analysis of the total cell lysates using the anti-argpyrimidine antibody showed that cells cultured in high glucose accumulated more AGEs than those grown in normal medium (Fig. 1A). Upon two-dimensional gel electrophoresis and matrix-assisted laser description ionization–mass spectrometry (MALDI-MS) analysis, the major immunopositive protein was identified to be HSP27 (Fig. 1A, inset), indicating that intracellular proteins are most likely to participate in AGE formation. These observations are in agreement with the studies by Schalkwijk et al. (20), showing that high glucose selectively increases argpyrimidine adduct formation (with HSP27) in endothelial cells. However, to examine the extent of formation of extracellular AGEs, intact cells were stained with the anti-argpyrimidine antibody and cell fluorescence was measured by fluorescence-activated cell sorter. To measure intracellular AGEs, the cells were permeabilized and then treated with anti-argpyrimidine antibody. Although minimal fluorescence was detected in nonpermeabilized cells (Fig. 1B[i, ii]), permeabilization increased fluorescence in both normal and high glucose–treated cells (Fig. 1B[iii, iv]). Relative fluorescence was higher in cells cultured in high glucose than in normal glucose medium. These data indicate that high glucose promotes AGE formation and that most AGEs are generated in intracellular proteins such as HSP27. It follows then that accumulation and toxicity of AGEs is likely to be regulated not only by processes that generate AGE forming species but also by intracellular pathways that metabolize AGE precursors.
FIG. 1.
High glucose–induced AGE formation. A: Western blot analysis of total cell lysates prepared from HUVECs cultured in the presence of normal (NG; 5.5 mmol/l) or high (HG; 30 mmol/l) glucose for 7 days and probed with anti-argpyrimidine antibodies. Western blots from two representative experiments are shown. Inset shows the major immunopositive spot resolved by two-dimensional gel analysis of the cell lysates. The spot, corresponding to a molecular weight of 27 kDa and a pI of 7.83, was identified to be HSP27 by MALDI/MS analysis. B: Fluorescence-activated cell sorter data obtained from HUVECs cultured in normal (i, iii) and high (ii, iv) glucose. Extracellular AGEs were detected by labeling the cells with the anti-argpyrimidine antibody and phycoerythrin-conjugated secondary antibody. To detect total (extracellular + intracellular) AGEs, the cells were permeabilized before antibody treatment. Relative mean fluorescence was calculated by subtracting the fluorescence obtained from the isotype-matched antibody control. Group data are presented as means ± SE. *P < 0.05 vs. normal glucose; n = 3–4.
Relative efficacy of AKRs in AGE reduction.
Although methylglyoxal and glyoxal are known to be metabolized by glyoxalases (14,15), the metabolism of other AGE precursors remains unknown. It is also unclear whether methylglyoxal and glyoxal are metabolized by pathways other than glyoxalases. Hence, we measured the catalytic efficiencies of the human AKRs (AKR1, AKR6, and AKR7 families), some of which have been shown to catalyze methylglyoxal reduction in vitro (16). For these experiments, the AKR coding sequences containing His-tag were expressed in Escherichia coli and the proteins were purified on a Ni-affinity column. Using glyoxal or methylglyoxal as substrates, highest catalytic efficiency was observed with AKR1B1 (human aldose reductase). For both AKR1B1 and AKR1B3 (mouse aldose reductase), methylglyoxal was a better substrate than glyoxal. AKR1A4 (aldehyde reductase) was also an efficient catalyst for methylglyoxal reduction, although its catalytic activity with glyoxal was much less than that of AKR1B1. Members of the AKR1C and AKR7A5 family displayed low catalytic efficiencies, mostly because of their high Km values (Table 1).
TABLE 1.
Steady-state kinetic parameters for the reduction of AGE precursors by AKRs
| Substrates | Genbank accession number | Proteins | Km (μM) | kcat (min−1) | kcat/Km (min−1/μM−1) |
|---|---|---|---|---|---|
| Glyoxal | NM_021473 | AKR1A4 | 3,229 ± 904 | 45.8 ± 4.2 | 0.014 |
| NM_001628 | AKR1B1 | 350 ± 20 | 29.5 ± 2.02 | 0.082 | |
| NM_009658 | AKR1B3 | 333 ± 62 | 12.1 ± 0.36 | 0.036 | |
| NM_009731 | AKR1B7 | 7,450 ± 1,500 | 5.6 ± 0.35 | 0.001 | |
| NM_008012 | AKR1B8 | 635 ± 81 | 20 ± 0.52 | 0.031 | |
| NM_020299 | AKR1B10 | 7,800 ± 359 | 8.87 ± 0.63 | 0.002 | |
| NM_030611 | AKR1C3 | NDA | — | — | |
| NM_013778 | AKR1C6 | 12,073 ± 1,031 | 12.8 ± 0.67 | 0.001 | |
| NM_134066 | AKR1C18 | NDA | — | — | |
| NM_025337 | AKR7A5 | 5,931 ± 1,393 | 12.87 ± 0.27 | 0.002 | |
| Methylglyoxal | AKR1A4 | 874 ± 91 | 483 ± 0.27 | 0.55 | |
| AKR1B1 | 22 ± 2 | 37.2 ± 2.21 | 1.698 | ||
| AKR1B3 | 35 ± 4 | 19.5 ± 2.3 | 0.56 | ||
| AKR1B7 | 1,996 ± 274 | 8.9 ± 0.31 | 0.004 | ||
| AKR1B8 | 184 ± 15 | 18.2 ± 1.21 | 0.098 | ||
| AKR1B10 | 1,250 ± 89 | 26.82 ± 5 | 0.389 | ||
| AKR1C3 | NDA | — | — | ||
| AKR1C6 | 139 ± 35 | 7.65 ± 0.7 | 0.054 | ||
| AKR1C18 | NDA | — | — | ||
| AKR7A5 | 9,075 ± 1,250 | 22 ± 0.7 | 0.002 | ||
| Furfural | AKR1A4 | 5,918 ± 1,379 | 17.4 ± 0.78 | 0.002 | |
| AKR1B1 | 316 ± 34 | 30.86 ± 1.91 | 0.097 | ||
| AKR1B3 | 69 ± 10 | 13.36 ± 0.93 | 1.91 | ||
| AKR1B7 | NDA | — | — | ||
| AKR1B8 | 622 ± 100 | 2.6 ± 0.27 | 0.001 | ||
| AKR1B10 | 1,617 ± 287 | 178 ± 15 | 0.11 | ||
| AKR1C3 | 156 ± 35 | 3.09 ± 0.33 | 0.019 | ||
| Hydroxymethyl Furfural | AKR1A4 | 6,428 ± 1,479 | 24.05 ± 4 | 0.003 | |
| AKR1B1 | 482 ± 92 | 14.55 ± 2 | 0.03 | ||
| AKR1B3 | 98 ± 15 | 4.15 ± 0.3 | 0.042 | ||
| AKR1B7 | NDA | — | — | ||
| AKR1B8 | 1,197 ± 117 | 7.82 ± 0.05 | 0.016 | ||
| AKR1B10 | 1,117 ± 135 | 100 ± 15 | 0.085 | ||
| AKR1C3 | 1,939 ± 425 | 167 ± 0.02 | 0.008 | ||
| Deoxyglucosone | AKR1A4 | 2,067 ± 178 | 777 ± 25 | 0.376 | |
| AKR1B1 | 112 ± 5 | 38.9 ± 2 | 0.345 | ||
| AKR1B3 | 89 ± 5 | 11.6 ± 1 | 0.13 | ||
| AKR1B10 | 3,293 ± 118 | 14.36 ± 1.9 | 0.004 | ||
| AKR1B10 | 3,293 ± 118 | 14.36 ± 1.9 | 0.004 |
Enzyme activity was measured in 0.1 mol/l potassium phosphate (pH 7.0) using the indicated substrates and 0.15 mmol/l NADPH at room temperature. Proteins were reduced with dithiothreitol before assay. NDA, no detectable activity.
AKR1B1 was also efficient at catalyzing the reduction of furfural and hydroxymethylfurfural, which are the major AGE precursors found in food (21). Daily human consumption of hydroxymethylfurfural is about 50 mg/kg per day (22). Furfural was reduced by AKR1B1, AKR1B3, and AKR1B10 with a catalytic efficiency that was higher than glyoxal. In contrast, hydroxymethylfurfural was poorly reduced by any of the AKRs. AKR1B1 and AKR1B3 were also efficient with the AGE precursor 3-deoxyglucosone, which was reduced efficiently by AKR1A4 as well (Table 1). Collectively, these data indicate that AKR1B1 (aldose reductase) is the most efficient enzyme involved in the reduction of several naturally occurring AGE precursors. The role of other AKRs, however, could not be discounted. Some of these enzymes displayed high kcat values with the AGE precursors, and their contribution to AGE metabolism may depend upon their relative abundance in specific tissues and the presence of other pathways of subsidiary metabolism.
Reductive metabolism of AGE precursors.
Given that our results indicated that AKRs catalyze the reduction of AGE precursors and that AGEs accumulate intracellularly, we tested whether in cells methylglyoxal undergoes reductive transformation. For this, HUVECs were incubated with methylglyoxal, and their lysates were used to measure acetol formation. Upon gas-liquid chromatography analysis, reagent methylglyoxal (Rt = 13.1; Fig. 2A[i]) was well separated from acetol (Rt = 7.5; Fig. 2A [ii]). Mass analysis of acetol revealed characteristic fragments with m/z 326 and 285 (Fig. 2A[ii]). Similar fragments were obtained with 13C3-acetol (m/z 329 and 286) with expected shift in mass. Using the 13C3-labeled internal standard, the abundance of acetol was measured from cells that were either left untreated or treated with the AKR1B inhibitors sorbinil or tolrestat. Ions with m/z 285 and 326 were used to quantify the abundance of natural acetol. The concentration of these ions was calculated using m/z 286 and 329 ions of the 13C-internal standard. In cells incubated with methylglyoxal, the average concentration of acetol was 6.2 ± 0.7 (n = 4) nmoles/mg protein. Treatment with sorbinil or tolrestat led to a significant decrease in the intensity of these ions (Fig. 2A[iii]). These results demonstrate that in HUVECs, AKR1B catalyzes the reduction of methylglyoxal to acetol.
FIG. 2.
Methylglyoxal metabolism in endothelial cells. A: GC-MS analysis of acetol formation in HUVECs. For acetol quantification, 13C3-methylglyoxal was synthesized from 13C3-acetone and 13C3-acetol was prepared by incubating 13C3-methylglyoxal with AKR1B1 and 0.15 mmol/l NADPH. i: Natural and 13C3-methylglyoxal were derivatized using O-(2,3,4,5,6 pentaflourobenzyl)-hydroxylamine hydrochloride (PFBHA) extracted in hexane and separated by gas chromatography. The 1,2 dioxime methylglyoxal eluted with a retention time of 13.1 min (inset). On MS analysis, the fragmentation pattern of 12C-1,2, dioxime methylglyoxal showed a parent ion with m/z 462. Ions with m/z 432 and 265 represented the loss of nitric oxide (NO) [M-30] and OC7H3F5 [M-197] groups, respectively, from 1,2 dioxime methylglyoxal. Corresponding ions with m/z 465, 435, and 268 are because of the 13C3-methylglyoxal. ii: Acetol was derivatized using PFBHA and N,O-bis (trimethylsilyl) trifluoroacetamide with trimethylchlorosilane (BSTFA). Derivatized acetol eluted with a retention time of 7.5 min (inset). Ions with m/z 326 and 285 were assigned to TMS-2-oxime acetol suffering a loss of CH3 [M-15] or C3H4O [M-56], respectively. Corresponding ions with m/z 329 and 286 are because of 13C3-acetol. iii: Acetol formation in HUVECs, cultured in media containing 1 mmol/l methylglyoxal for 24 h in the absence and presence of the AKR1B inhibitors sorbinil (50 μmol/l) or tolrestat (25 μmol/l). After treatment, 13C3-acetol was added to cell lysates and lysates were derivatized and analyzed by GC-MS. *P < 0.05 vs. control. B: Western analysis of lysates prepared from HUVECs cultured in normal (NG) or high (HG) glucose, with or without sorbinil (50 μmol/l), probed with anti-argpyrimidine and anti-HSP27 antibodies. Intensity of immunopositive band (argpyrimidine) was normalized to HSP27. Data are presented as means ± SE. *P < 0.05 vs. normal glucose (n = 4) and #P < 0.05 vs. high glucose without sorbinil (n = 4).
To examine the role of AKR1B in regulating AGE formation, HUVECs were cultured in high glucose (30 mmol/l) media and steady-state levels of the most abundant AGE protein (Fig. 2B) AGE-HSP27 were measured. As before, incubation with glucose increased the levels of antiarpyrimidine reactive HSP27, but the abundance of AGE-HSP27 was increased further in cells treated with sorbinil. No changes in the total protein levels of HSP27 were observed. In addition to the anti-argpyrimidine antibody, which recognizes arginine adducts of methylglyoxal, we also examined the formation of adducts between glyoxal and lysine using the anti-carboxymethyl lysine (CML) antibody. Treatment with high glucose, however, did not increase anti-CML reactivity, and this reactivity was not affected by sorbinil (data not shown), indicating that under the experimental conditions studied, arginine adducts were more abundant. Overall, these results indicated that in HUVECs inhibition of AKR1B (aldose reductase) increases the formation and accumulation of argpyrimidine containing HSP27.
Role of AKR1B in methylglyoxal metabolism.
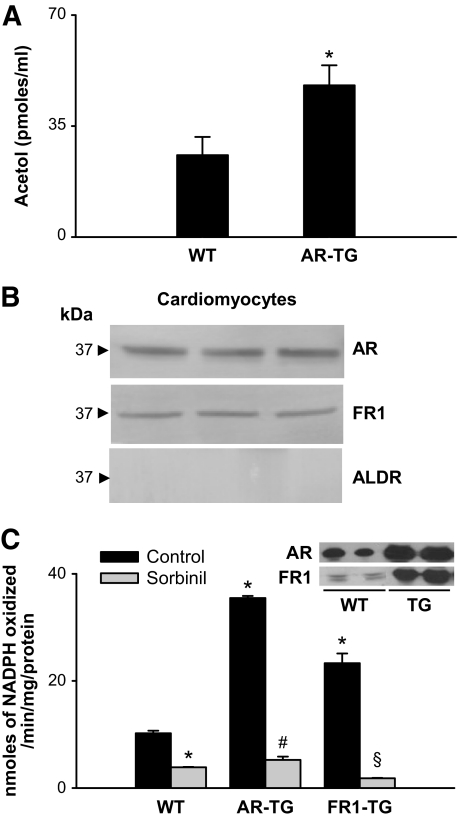
To determine whether methylglyoxal is metabolized to acetol in the heart, wild-type and AKR1B4 (rat aldose reductase) transgenic mouse hearts were continuously perfused with methylglyoxal, and the perfusate was collected. The expression of the transgene was driven by the α-myosin heavy chain promoter, leading to cardiomyocyte-restricted increase in gene expression; thus changes in methylglyoxal metabolism could be attributed to cardiomyocyte-specific metabolism. Gas chromotography–mass spectrometry (GC-MS) analysis of the perfusate revealed clearly resolved ions with m/z 285 and 326, which were ascribed to acetol. The concentration of acetol in effluents of aldose reductase–transgenic hearts was significantly higher (45 ± 4 pmoles/ml) than that collected from wild-type hearts (25 ± 3 pmoles/ml; Fig. 3A), indicating that an increase in aldose reductase in the heart increases the metabolism of methylglyoxal to acetol.
FIG. 3.
AKR-catalyzed reduction of methylglyoxal in mouse heart. A: Acetol generated in effluents of isolated wild-type (C57) or aldose reductase–transgenic hearts perfused with 20 μmol/l methylglyoxal. Acetol concentration was measured by GC-MS after derivatization with PFBHA and BSTFA. 13C3 Acetol was used as an internal standard. Data are presented as means ± SE. *P < 0.01 vs. wild type (n = 4). B: Western blot analysis of cardiac myocytes isolated from adult male C57 mice probed with antibodies raised against AKR1B1 (aldose reductase), AKR1B8 (FR-1), and AKR1A4 (ALDR). Figure shows bands from three different mice. C: Rate of methylglyoxal reduction in homogenates prepared from hearts of wild-type mice (n = 6) or mice with cardiac myocyte–specific transgene expressing AKR1B4 (rat aldose reductase; n = 6) or AKR1B8 (FR-1; n = 6). The enzyme activity was determined with 1 mmol/l methylglyoxal and 0.15 mmol/l NADPH, with or without 1 μmol/l sorbinil. Inset shows Western blots from wild-type and transgenic hearts developed with anti–aldose reductase and anti–FR-1 antibodies. *P < 0.01 vs. wild type (control), #P < 0.01 vs. aldose reductase–transgenic (control), and §P < 0.01 vs. FR1–transgenic (control). AR-TG, aldose reductase–transgenic; WT, wild type.
Myocytes isolated from mouse hearts expressed AKR1B3 (aldose reductase), AKR1B8 (FR-1), but not AKR1A3 (Fig. 3B). To establish the contribution of these enzymes, reduction of methylglyoxal was measured in cardiac homogenates of wild-type, AKR1B4-transgenic, and AKR1B8-transgenic hearts, with cardiomyocyte-restricted expression of the transgene. Significant rates of methylglyoxal reduction were observed in wild-type cardiac homogenates. Methylglyoxal reduction was inhibited by sorbinil, indicating that the reaction was catalyzed by AKR1B (Fig. 3C). That AKR1B reduces methylglyoxal in the heart is further supported by data showing a significant increase in reduction rates in hearts of AKR1B4 or AKR1B8-transgenic mice (Fig. 3C). In agreement with in vitro data (Table 1), the methylglyoxal reductase activity in AKR1B4-transgenic mice was 35% higher than methylglyoxal reductase activity in the AKR1B8-transgenic hearts, indicating that AKR1B1/3/4 is a better methylglyoxal reductase than AKR1B8. Based on these observations, we conclude that aldose reductase (AKR1B1/3/4) plays a major role in the reductive metabolism of methylglyoxal; however, AKR1B8 can also catalyze methylglyoxal reduction.
Because the purified AKR1B proteins were found to catalyze the reduction of several different AGE precursors, we also measured the reduction of major AGE precursors in cardiac homogenates of wild-type and AKR1B3-null mice. Methylglyoxal and deoxyglucosone reductase activities (Fig. 4A[ii, iii]) in wild-type mice were similar in magnitude to the DL-glyceraldehyde reductase activity in these hearts (Fig. 4A[i]), whereas, glyoxal reduction (Fig. 4A[iv]) was twofold lower than the DL-glyceraldehyde reductase activity. Reduction of AGE precursors was significantly inhibited by sorbinil in wild type but not in AKR1B3-null, consistent with the involvement of AKR1B3 in this reduction.
FIG. 4.
Genetic ablation of AKR1B3 (aldose reductase) diminishes the reduction of AGE precursors. A: Rate of reduction of glyceraldehyde (i), methylglyoxal (ii), 3-deoxyglucosone (iii), and glyoxal (iv) in cardiac homogenates prepared from wild-type and akr1b3-null mice. The enzyme activity was determined with glyceraldehyde (10 mmol/l), glyoxal (1 mmol/l), methylglyoxal (1 mmol/l), or deoxyglucosone (1 mmol/l) and 0.15 mmol/l NADPH, with or without 1 μmol/l sorbinil. Values are presented as means ± SE. *P < 0.05 vs. wild type (n = 6). Inset shows the expression of the proteins in wild-type and aldose reductase–knockout mice. B: Rate of formation of S-d-lactoylglutathione (i) and S-glycolylglutathione (ii) in homogenates prepared from wild-type and akr1b3-null hearts. Glyoxalase I activity was measured with methylglyoxal (1 mmol/l) or glyoxal (1 mmol/l) and GSH (1 mmol/l) in the absence or presence of glyoxalase I inhibitor BBGC (0.2 mmol/l). Inset to panel i shows Western blots developed from wild-type and akr1b3-null (knockout) hearts using the anti–glyoxalase-I antibody. Data are means ± SE (n = 6). *P < 0.01 vs. wild-type (methylglyoxal or glyoxal) and #P < 0.01 vs. aldose reductase–null mice (methylglyoxal or glyoxal). Inset shows the expression of glyoxalase I in wild-type and aldose reductase–knockout mice. C: Computer simulations for the relative contributions of aldose reductase and glyoxalase I in the metabolism of glyoxal (i) and methylglyoxal (ii). Relative contribution of the enzymes was calculated on the basis of measurement of aldose reductase and glyoxalase I enzyme activities assuming that the concentration of AGE precursors is in steady state achieved between the processes of formation and those of elimination. AR, aldose reductase; WT, wild type.
In addition to AKRs, glyoxalase I is also involved in the metabolism of methylglyoxal. To determine the relative contribution of the two enzymes to the overall methylglyoxal metabolism, we measured the expression and activity of glyoxalase I in cardiac homogenates of wild-type and akr1b3-null mice. Glyoxalase I protein was abundant in the heart, and its abundance was not affected by deletion of the akr1b3 gene. Our kinetic measurements indicated that 100 ± 8 nmoles · min−1 · mg−1 protein of S-d-lactoylglutathione (a product of glyoxalase I) were generated in the heart. S-d-lactoylglutathione formation was significantly inhibited by glyoxalase I inhibitor S-p-bromobenzylglutathione cyclopentyl diester (BBGC) (Fig. 4B[i]), indicating that it is primarily derived from glyoxalase I. Approximately, 3–4 nmoles · min−1 · mg−1 protein of S-glycolylglutathione were formed by glyoxalase I when glyoxal was used as substrate (Fig. 4B[ii]). The values of the kinetic parameter of glyoxalase I were similar to those reported by Allen et al. (23). No glyoxalase I activity was observed with deoxyglucosone. Metabolism of methylglyoxal by glyoxalase I was ∼10-fold higher than the methylglyoxal reductase activity by AKR1B3: 10 ± 2 nmoles acetol · min−1 · mg−1 protein; (Fig. 4A[i]). Formation of S-glycolylglutathione by glyoxalase I from glyoxal was, however, approximately the same as the reduction of glyoxal by AKR1B3 (Fig. 4A[iv]). To assess the relative contribution of aldose reductase and glyoxalase I in the metabolism of AGE precursors in the heart, we performed simulation experiments using the steady-state kinetic parameters listed in supplemental Table 1. These calculations suggest that at low concentrations (glyoxal 1–100 μmol/l and methylglyoxal 1–20 μmol/l) aldose reductase–catalyzed reduction accounts for ∼85% of glyoxal and ∼40% of methylglyoxal metabolism, whereas glyoxalase I contributes to ∼15% glyoxal and ∼60% methylglyoxal metabolism. At higher concentrations, the relative contribution of glyoxalase I increases as a function of substrate concentration. This analysis is in agreement with the kinetic model of methylglyoxal metabolism in yeast (24). Together, these analyses suggest that both aldose reductase and glyoxalase I play significant and nonredundant roles in the metabolism of AGE precursors.
Regulation of AGE formation by AKR1B3.
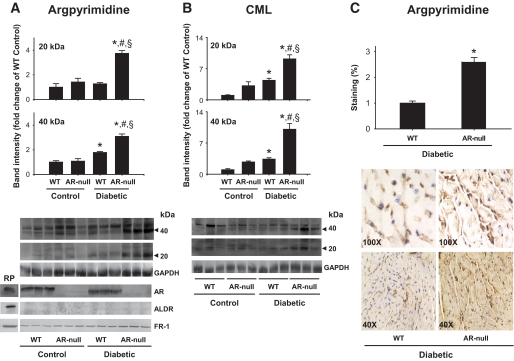
To examine how AKR1B3 regulates AGE formation in diabetes, wild-type and akr1b3-null mice were made diabetic by injecting streptozotocin (STZ). Control mice were treated with the vehicle alone. Induction of diabetes led to an increase in A1C levels from 5.4 ± 1.1% to 7.9 ± 1.1% after 8 weeks of diabetes. The STZ-injected mice showed marked hyperglycemia, and the levels of plasma glucose, when measured after 12 weeks, were slightly higher in the aldose reductase–null than wild-type mice (supplemental Table 2). Detectable levels of multiple argpyrimidine-derived AGEs were present in murine plasma. Quantification of clearly resolved band at 27 kDa showed that the intensity of this band was higher in diabetic mice (Fig. 5A). The intensity of this band was even greater in akr1b3-null diabetic mice, indicating that deletion of this gene increases the formation of 27 kDa argpyrimidine AGE in the plasma of diabetic mice. Similar changes were observed when AGE formation was detected using the anti-CML antibody. As shown in Fig. 5B, increased accumulation of CML AGEs (2- to 12-fold) p27, p40, and p100 kDa was detected in the plasma of diabetic and nondiabetic akr1b3-null mice. Overall, the akr1b3-null diabetic plasma showed a greater plasma AGE accumulation than plasma from wild-type mice. These data suggest that deletion of the akr1b3 gene increases AGE accumulation in the plasma.
FIG. 5.
Increased accumulation of plasma AGEs in the aldose reductase–null mice. Western blots of plasma from nondiabetic and diabetic wild- type and akr1b3-null (aldose reductase–null) mice, probed with anti-argpyrimidine (A) and anti-CML (B) antibodies. Inset shows positive recognition of glyoxlyic acid–treated BSA. Bar graphs show the intensity of indicated anti-argpyrimidine- or anti-CML–positive bands normalized to Amido-Black–stained blots. Data are presented as means ± SE. *P < 0.01 vs. wild type (control), #P < 0.01 vs. aldose reductase–null (control), and §P < 0.01 vs. wild-type diabetic plasma. AR, aldose reductase; WT, wild type.
To evaluate changes in tissue levels, AGE formation was quantified in the heart. In hearts of 20-week-old akr1b3-null diabetic mice, AGEs of 20 and 40 kDa were detected by anti-argpyrimidine and anti-CML antibodies (Fig. 6A and B). Changes in AGE formation in the hearts of wild-type and akr1b3-null mice were similar to those observed in the plasma. Induction of diabetes was associated with greater AGE accumulation in wild-type hearts largely because of the increase in p20 and p40 AGEs. In contrast, in hearts of diabetic akr1b3-null mice, higher accumulation of p20 and p40 AGEs was observed. In each case, two- to threefold increase in the abundance of these AGEs was observed in akr1b3-null than in wild-type hearts (Fig. 6B). However, the intensity of some of the anti-AGE antibody–positive bands was decreased in the diabetic aldose reductase–null mice. The reasons for this decrease are unclear, but it is unlikely to be because of nonspecific reactivity of the antibody, in which case the intensity would remain the same. Because the 45 and 40 kDa proteins show reciprocal relationship, we speculate that the 45 and 40 kDa proteins may be the same protein and that because of multiple AGE formation, its apparent molecular weight shifts from 45 to 40 kDa. Further experiments are, however, required to identify changes in individual bands. Nevertheless, overall these data support the view that AKR1B3 prevents AGE formation and that metabolism of AGE precursors by this enzyme prevents AGE accumulation.
FIG. 6.
Increased AGE accumulation in the hearts of aldose reductase–null mice. A: Western blots of heart homogenates from diabetic and nondiabetic wild-type and aldose reductase–null mice were probed with anti-argpyrimidine (A) and anti-CML (B) antibodies. Nondiabetic wild-type and aldose reductase–null hearts served as respective controls. The expression of aldose reductase, FR-1, and ALDR in the hearts of these mice was examined by Western blots developed using anti–aldose reductase, FR-1, and ALDR antibodies. Recombinant proteins were used as positive controls. Bar graphs show the intensity of the indicated anti-argpyrimidine- or anti-CML–positive bands normalized to GAPDH. Data are presented as means ± SE. *P < 0.01 vs. wild type (control), #P < 0.01 vs. aldose reductase null (control), and §P < 0.01 vs. wild-type diabetic. C: Immunohistochemical analyses of AGE accumulation in hearts of diabetic wild-type and aldose reductase–null mice. Sections were stained with anti-argpyrimidine antibody, and staining was quantified by image analysis. Group data shows the extent of staining quantified using the MetaMorph imaging software. Data are presented as means ± SE. *P < 0.01 vs. wild type (diabetic). AR, aldose reductase; RP, recombinant proteins; WT, wild type. (A high-quality color digital representation of this figure is available in the online issue.)
To examine AGE localization, sections of the diabetic hearts were stained with the anti-argpyrimidine antibody. Low levels of positive reactivity with the antibody were observed in wild-type diabetic hearts. This staining was associated strongly with blood vessels, but the cardiac myocytes showed diffuse staining as well (Fig. 6C). Staining of both myocytes and blood vessel was two- to threefold higher in the diabetic akr1b3-null than wild-type hearts (Fig. 6C). These results indicate that deletion of akr1b3 increases cardiovascular AGE accumulation in diabetes.
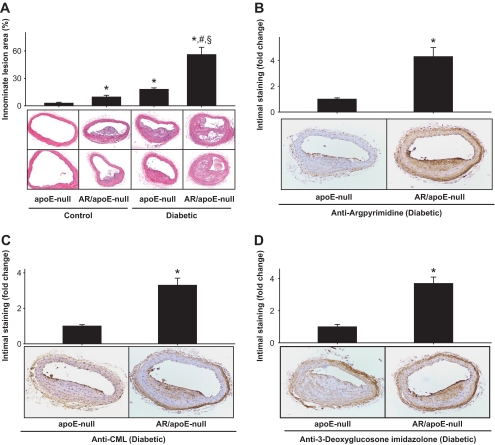
To assess how AKR1B3 regulates AGE formation during atherosclerosis, we generated akr1b3-apoE–null mice. Diabetes was induced by STZ, and lesion formation was examined 12 weeks after the induction of diabetes. The levels of blood glucose, cholesterol, and triglycerides in akr1b3-apoE–null mice were similar to those in apoE-null mice (supplemental Table 2). Image analysis of hematoxylin-eosin–stained sections of innominate arteries showed that lesion sizes in diabetic akr1b3-apoE–null mice were two- to threefold higher than diabetic apoE-null mice (Fig. 7A). These data support the notion that deletion of the akr1b3 gene increases atherosclerotic lesion formation in apoE-null mice.
FIG. 7.
Genetic ablation of aldose reductase exacerbates diabetic lesion formation and AGE accumulation. A: Photomicrographs of cross sections of innominate arteries of 20-week-old nondiabetic (control) and diabetic apoE-null and akr1b3-apoE–null mice. Sections were stained with hematoxylin and eosin, and the lesion area was quantified by image analysis. Data are presented as means ± SE. *P < 0.01 vs. apoE-null (control), #P < 0.01 vs. aldose reductase/apoE–null (control) and §P < 0.01 vs. apoE-null (diabetic). Arterial sections of diabetic apoE-null and akr1b3-apoE–null mice stained with anti-argpyrimidine (B), anti-CML (C), and anti–3-deoxyglucosone imidazolone (D) antibodies. The extent of staining was quantified by image analysis. Data are presented as means ± SE. *P < 0.05 vs. apoE-null (diabetic). AR, aldose reductase. (A high-quality color digital representation of this figure is available in the online issue.)
Increase in lesion formation in the akr1b3-apoE–null mice was accompanied by an increase in AGE accumulation. For quantifying AGE formation, lesions of comparable sizes were stained with three different anti-AGE antibodies. As shown in Fig. 7A, low levels of positive staining were observed in apoE-null mice; however, the extent of staining was three- to fourfold higher in the akr1b3-apoE–null mice. Lesions of the akr1b3-apoE–null mice showed increases in their reactivity to anti-argpyrimidine, anti-CML, and anti–3-deoxyglucosone imidazolone antibodies. Anti-argpyrimidine–positive immunostaining was more predominant in the endothelial layer and the highly proliferative areas of the lesions as well as smooth muscle cells (Fig. 7B). Staining for CML was particularly localized to atherosclerotic plaques (Fig. 7C), whereas the staining for 3-deoxyglucosone imidazolone increased both in smooth muscle cells and endothelial layer. Intense staining was also observed in the adventitia (Fig. 7D). Collectively, these data indicate that in diabetic apoE-null mice, deletion of akr1b3 increases the formation of several structurally different AGEs.
DISCUSSION
The major findings of this study are that aldose reductase (AKR1B1/3/4)-catalyzed reduction is a significant pathway for the cardiovascular metabolism of AGE precursors and that the reduction of AGE precursors by aldose reductase diminishes the AGE formation and accumulation. Our observations that aldose reductase reduces a variety of AGE precursors and that pharmacological inhibition or genetic deletion of the enzyme increases AGE accumulation both in cells exposed to high glucose and in diabetic mice provide direct evidence supporting the notion that aldose reductase prevents AGE accumulation in vivo by detoxifying AGE precursors. These findings provide a new view of the role of this enzyme in the development of secondary diabetic complications.
Formation and accumulation of AGEs has been linked to the development of several diabetic complications (1,25–27). The AGEs are formed by reactions of proteins with glucose-derived carbonyls and could be generated either inside cells or on the extracellular surface, leading to collagen cross-linking (3). Our results, however, show that most AGEs, at least during short-term hyperglycemia, are generated within cells, indicating that metabolism of AGE precursors could affect AGE formation. Intracellular formation of AGEs suggests that their formation could be regulated by the metabolism of AGE precursors. Indeed the AGE precursors methylglyoxal and glyoxal have been shown to be metabolized by glyoxalases, and overexpression of glyoxalase I prevents AGE formation in endothelial cells cultured in high glucose (14). Nonetheless, the in vivo role of glyoxalase I in preventing AGE formation in diabetic animals has not been tested. Moreover, given the limited substrate specificity of glyoxalase I, it is not clear how AGE precursors other than methylglyoxal (e.g., deoxyglucosone, furfural, hydroxymethylfurfural) are metabolized. Therefore, we studied the AKR superfamily. Many members of this family catalyze the reduction of keto-aldehydes (28). We found significant activity with AKR1A4, AKR1B1, AKR1B3, AKR1B7, and AKR1B8. No activity was observed with the AKR1C family. Even with AKRs, family- wide variations in catalytic activities were observed with different AGE precursors, indicating that these enzymes may have a tissue-specific role such that in tissues in which they are expressed in highest abundance, they may be capable of metabolizing AGE precursors. Consistently high activity was, however, observed with AKR1B1/3 (aldose reductase), indicating that this enzyme is likely to play the most general and significant role in the reduction of AGE precursors.
Previous work shows that aldose reductase is a broad-specificity aldehyde reductase. It catalyzes the reduction of several endogenously generated aldehydes including glucose, products of lipid peroxidation such as hydroxynonenal (28–32), as well as AGE precursors such as methylglyoxal (16,33). Overexpression of aldose reductase in kidney tubules decreases carbonyl content (34), and the deletion of the aldose reductase gene in Saccharomyces cervisiae enhances the accumulation of argpyrimidine adducts (24). Studies from our lab show that aldose reductase prevents ischemia injury (35) and mediates ischemic preconditioning (36), indicating that the enzyme may be involved in removing reactive aldehydes. The results of the current study, however, establish for the first time the in vivo cardiovascular role of aldose reductase in the metabolism of methylglyoxal and other AGE precursors. Several lines of evidence indicate that aldose reductase is a significant route of methylglyoxal metabolism. These include data showing that 1) acetol is generated in hearts perfused with methylglyoxal; 2) in endothelial cells aldose reductase inhibition increases AGE formation; and 3) reduction of methylglyoxal is increased in aldose reductase-transgenic hearts and decreased in aldose reductase–null hearts. A significant, nonredundant role of aldose reductase in the metabolism of AGE precursors is also supported by the observation that higher AGE levels were detected in aldose reductase–null than in wild-type mice. In comparison with tissues of wild-type mice, aldose reductase–null mouse heart and plasma showed higher abundance of several AGEs that reacted with the anti-argpyrimidine, anti-CML, and anti–3-deoxyglucosone imidazolone antibodies. The difference between wild-type and aldose reductase–null mouse tissues was evident even in the absence of diabetes, indicating that AGEs are formed during normal metabolism and that aldose reductase metabolizes AGE precursors even under basal conditions.
A significant role of aldose reductase in preventing AGE accumulation, indicated by our data, is in contrast to the current view that the polyol pathway generates AGEs (8,37). In the polyol pathway glucose is reduced to sorbitol by aldose reductase, and then sorbitol is converted to fructose by sorbitol dehydrogenase. In the lens, fructose-3-phosphate is then generated either from the phosphorylation of fructose by 3-phosphokinase or the reduction of sorbitol-3-phosphate (38). Fructose-3-phosphate then spontaneously breaks down into 3-deoxyglucosone (39), which generates CML or imidazolone adducts (37). In agreement with this view, it has been shown that levels of 3-deoxyglucosone are increased in erythrocytes of diabetic subjects and inhibition of aldose reductase decreases the levels of CML adducts in the erythrocytes (37). Nonetheless, even in the lens, fructose-3-phosphate is not generated upon incubation with high glucose (only with high fructose) (40). In cardiovascular tissues, it is not clear whether the low levels of sorbitol dehydrogenase (41) are sufficient to support significant formation of fructose from glucose. Moreover, as shown in current study, aldose reductase directly catalyzes the reduction of 3deoxyglucosone, and in contrast to changes seen in erythrocytes (37), genetic deletion of aldose reductase increased the abundance of CML adducts in diabetic heart and 3-deoxyglucosone–derived imidazolone adducts in atherosclerotic lesions of diabetic mice. This disagreement may be because of differences in the tissue-specific roles of the aldose reductase or sorbitol dehydrogenase expression or metabolic conditions that either favor the removal or the production of deoxyglucosone and other AGEs by aldose reductase. Nonetheless, the observation that lack of aldose reductase permits greater accumulation of several structurally diverse AGEs suggests that the role of this enzyme is more complex than previously thought and that at least in cardiovascular tissues, aldose reductase prevents metabolites of glucose from forming AGEs.
Our results also show that atherosclerotic lesion formation in apoE-null mice was enhanced by deletion of the aldose reductase gene. The increase in lesion formation in ar-null mice was accompanied by greater accumulation of AGEs. Although STZ-treated mice displayed slightly (20–30%) higher levels of glucose, greater accumulation of AGEs in these mice could not be directly attributed to high glucose alone because deletion of aldose reductase increased AGE accumulation in apoE-null mice without exacerbating hyperglycemia. Furthermore, diabetic mice accumulated AGEs that displayed positive reactivity with anti-argpyrimidine, anti-CML, as well as anti–3-deoxyglucosone imidazolone antibodies, suggesting that the lack of aldose reductase permits greater accumulation of several structurally diverse AGEs (Fig. 7), consistent with the broad specificity of the enzyme observed in kinetic studies (Table 1). Several previous studies, however, suggest that aldose reductase is the underlying cause of secondary diabetic complications. This view is based on several decades of work showing that inhibition of aldose reductase delays, prevents, or even reverses cataractogenesis and neuropathy in diabetic rats (42). In addition, it has been recently reported that general overexpression of aldose reductase in all tissues exaggerates motor nerve conduction velocity defect (43) and atherosclerosis in diabetic mice (44). However, significance of the effects of such nonspecific increase in aldose reductase, even in tissues where it is not basally expressed (e.g., smooth muscle cells, liver etc), is not clear. Our results showing that cardiac-specific overexpression of aldose reductase increases methylglyoxal and that deletion of aldose reductase in the tissues in which it is expressed increases AGE formation and lesion formation in mice indicates that aldose reductase promotes the removal of AGE precursors and increases atherosclerotic lesion formation in a tissue-specific manner. It is also likely that the effects of aldose reductase may also depend on the state of the disease and the total tissue carbonyl load. Indeed, results from our own laboratory show that inhibition of aldose reductase prevents smooth muscle growth and diminishes restenosis in diabetic rats (42) and also prevents high glucose–induced inflammatory signaling and the release of TNF-α from vascular smooth muscle cells (45), indicating that inhibition of the enzyme could prevent some of the harmful effects of high glucose. Hence, in different metabolic scenarios, aldose reductase may be cytoprotective by removing toxic aldehydes or harmful because it depletes NADPH and thereby reduces the concentration of reducing equivalents. In this regard, aldose reductase may be similar to NF-κB or reactive oxygen species, both of which are regulated by aldose reductase and both of which could have protective or deleterious effects depending upon the metabolic context. Further studies are required to fully understand the metabolic dependence of aldose reductase action. Nevertheless, the data presented here provide direct support to the notion that reduction of methylglyoxal and related AGE precursors is a significant metabolic activity that could be ascribed to aldose reductase and that chronic deficiency of this enzyme could increase AGE accumulation and the formation of atherosclerotic lesions.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants ES-17260, HL-89380, HL-8930-0281, HL-55477, HL-59378, and RR-024489.
No potential conflicts of interest relevant to this article were reported.
The authors thank Dan Riggs, David Young, Erica Werkman, and Barbara Bishop for their technical assistance.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Brownlee M: Advanced protein glycosylation in diabetes and aging. Annu Rev Med 1995;46:223–234 [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Barden A, Mori T, Beilin L: Advanced glycation end-products: a review. Diabetologia 2001;44:129–146 [DOI] [PubMed] [Google Scholar]
- 3.Horie K, Miyata T, Maeda K, Miyata S, Sugiyama S, Sakai H, van Ypersole de SC, Monnier VM, Witztum JL, Kurokawa K: Immunohistochemical colocalization of glycoxidation products and lipid peroxidation products in diabetic renal glomerular lesions. Implication for glycoxidative stress in the pathogenesis of diabetic nephropathy. J Clin Invest 1997;100:2995–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schleicher ED, Wagner E, Nerlich AG: Increased accumulation of the glycoxidation product N(epsilon)-(carboxymethyl)lysine in human tissues in diabetes and aging. J Clin Invest 1997;99:457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammes HP, Weiss A, Hess S, Araki N, Horiuchi S, Brownlee M, Preissner KT: Modification of vitronectin by advanced glycation alters functional properties in vitro and in the diabetic retina. Lab Invest 1996;75:325–338 [PubMed] [Google Scholar]
- 6.Schmidt AM, Hori O, Chen JX, Li JF, Crandall J, Zhang J, Cao R, Yan SD, Brett J, Stern D: Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest 1995;96:1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wautier JL, Schmidt AM: Protein glycation: a firm link to endothelial cell dysfunction. Circ Res 2004;95:233–238 [DOI] [PubMed] [Google Scholar]
- 8.Yan SF, Ramasamy R, Naka Y, Schmidt AM: Glycation, inflammation, and RAGE: a scaffold for the macrovascular complications of diabetes and beyond. Circ Res 2003;93:1159–1169 [DOI] [PubMed] [Google Scholar]
- 9.Park L, Raman KG, Lee KJ, Lu Y, Ferran LJ, Jr, Chow WS, Stern D, Schmidt AM: Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat Med 1998;4:1025–1031 [DOI] [PubMed] [Google Scholar]
- 10.Sakaguchi T, Yan SF, Yan SD, Belov D, Rong LL, Sousa M, Andrassy M, Marso SP, Duda S, Arnold B, Liliensiek B, Nawroth PP, Stern DM, Schmidt AM, Naka Y: Central role of RAGE-dependent neointimal expansion in arterial restenosis. J Clin Invest 2003;111:959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goova MT, Li J, Kislinger T, Qu W, Lu Y, Bucciarelli LG, Nowygrod S, Wolf BM, Caliste X, Yan SF, Stern DM, Schmidt AM: Blockade of receptor for advanced glycation end-products restores effective wound healing in diabetic mice. Am J Pathol 2001;159:513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wendt TM, Tanji N, Guo J, Kislinger TR, Qu W, Lu Y, Bucciarelli LG, Rong LL, Moser B, Markowitz GS, Stein G, Bierhaus A, Liliensiek B, Arnold B, Nawroth PP, Stern DM, D'Agati VD, Schmidt AM: RAGE drives the development of glomerulosclerosis and implicates podocyte activation in the pathogenesis of diabetic nephropathy. Am J Pathol 2003;162:1123–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conklin D, Prough R, Bhatanagar A: Aldehyde metabolism in the cardiovascular system. Mol Biosyst 2007;3:136–150 [DOI] [PubMed] [Google Scholar]
- 14.Shinohara M, Thornalley PJ, Giardino I, Beisswenger P, Thorpe SR, Onorato J, Brownlee M: Overexpression of glyoxalase-I in bovine endothelial cells inhibits intracellular advanced glycation endproduct formation and prevents hyperglycemia-induced increases in macromolecular endocytosis. J Clin Invest 1998;101:1142–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thornalley PJ: Glyoxalase I–structure, function and a critical role in the enzymatic defense against glycation. Biochem Soc Trans 2003;31:1343–1348 [DOI] [PubMed] [Google Scholar]
- 16.Vander Jagt DL, Robinson B, Taylor KK, Hunsaker LA: Reduction of trioses by NADPH-dependent aldo-keto reductases. Aldose reductase, methylglyoxal, and diabetic complications. J Biol Chem 1992;267:4364–4369 [PubMed] [Google Scholar]
- 17.Wermuth B: Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J Biol Chem 1981;256:1206–1213 [PubMed] [Google Scholar]
- 18.Ko J, Kim I, Yoo S, Min B, Kim K, Park C: Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J Bacteriol 2005;187:5782–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misra K, Banerjee AB, Ray S, Ray M: Reduction of methylglyoxal in Escherichia coli K12 by an aldehyde reductase and alcohol dehydrogenase. Mol Cell Biochem 1996;156:117–124 [DOI] [PubMed] [Google Scholar]
- 20.Schalkwijk CG, van BJ, van der Schors RC, Uchida K, Stehouwer CD, van Hinsbergh VW: Heat-shock protein 27 is a major methylglyoxal-modified protein in endothelial cells. FEBS Lett 2006;580:1565–1570 [DOI] [PubMed] [Google Scholar]
- 21.Feron VJ, Til HP, de Vrijer F, Woutersen RA, Cassee FR, van Bladeren PJ: Aldehydes: occurrence, carcinogenic potential, mechanism of action and risk assessment. Mutat Res 1991;259:363–385 [DOI] [PubMed] [Google Scholar]
- 22.Ulbricht RJ, Northup SJ, Thomas JA: A review of 5-hydroxymethylfurfural (HMF) in parenteral solutions. Fundam Appl Toxicol 1984;4:843–853 [DOI] [PubMed] [Google Scholar]
- 23.Allen RE, Lo TW, Thornalley PJ: A simplified method for the purification of human red blood cell glyoxalase. Characteristics I., immunoblotting, and inhibitor studies. J Protein Chem 1993;12:111–119 [DOI] [PubMed] [Google Scholar]
- 24.Gomes RA, Sousa SM, Vicente MH, Ferreira AE, Cordeiro CA, Freire AP: Protein glycation in Saccharomyces cerevisiae. Argpyrimidine formation and methylglyoxal catabolism. FEBS J 2005;272:4521–4531 [DOI] [PubMed] [Google Scholar]
- 25.Ahmed N, Thornalley PJ: Advanced glycation endproducts: what is their relevance to diabetic complications? Diabetes Obes Metab 2007;9:233–245 [DOI] [PubMed] [Google Scholar]
- 26.Goh SY, Cooper ME: Clinical review: the role of advanced glycation end products in progression and complications of diabetes. J Clin Endocrinol Metab 2008;93:1143–1152 [DOI] [PubMed] [Google Scholar]
- 27.Goldin A, Beckman JA, Schmidt AM, Creager MA: Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation 2006;114:597–605 [DOI] [PubMed] [Google Scholar]
- 28.Srivastava SK, Ramana KV, Bhatnagar A: Role of aldose reductase and oxidative damage in diabetes and the consequent potential for therapeutic options. Endocr Rev 2005;26:380–392 [DOI] [PubMed] [Google Scholar]
- 29.Spite M, Baba SP, Ahmed Y, Barski OA, Nijhawan K, Petrash JM, Bhatnagar A, Srivastava S: Substrate specificity and catalytic efficiency of aldo-keto reductases with phospholipid aldehydes. Biochem J 2007;405:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava S, Watowich SJ, Petrash JM, Srivastava SK, Bhatnagar A: Structural and kinetic determinants of aldehyde reduction by aldose reductase. Biochemistry 1999;38:42–54 [DOI] [PubMed] [Google Scholar]
- 31.Srivastava S, Spite M, Trent JO, West MB, Ahmed Y, Bhatnagar A: Aldose reductase-catalyzed reduction of aldehyde phospholipids. J Biol Chem 2004;279:53395–53406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Jagt DL, Kolb NS, Vander Jagt TJ, Chino J, Martinez FJ, Hunsaker LA, Royer RE: Substrate specificity of human aldose reductase: identification of 4-hydroxynonenal as an endogenous substrate. Biochim Biophys Acta 1995;1249:117–126 [DOI] [PubMed] [Google Scholar]
- 33.Vander Jagt DL, Hunsaker LA: Methylglyoxal metabolism and diabetic complications: roles of aldose reductase, glyoxalase-I, betaine aldehyde dehydrogenase and 2-oxoaldehyde dehydrogenase. Chem Biol Interact 2003;143–144:341–351 [DOI] [PubMed] [Google Scholar]
- 34.Dunlop M: Aldose reductase and the role of the polyol pathway in diabetic nephropathy. Kidney Int 2000;58:S3–S12 [DOI] [PubMed] [Google Scholar]
- 35.Kaiserova K, Tang XL, Srivastava S, Bhatnagar A: Role of nitric oxide in regulating aldose reductase activation in the ischemic heart. J Biol Chem 2008;283:9101–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinmura K, Bolli R, Liu SQ, Tang XL, Kodani E, Xuan YT, Srivastava S, Bhatnagar A: Aldose reductase is an obligatory mediator of the late phase of ischemic preconditioning. Circ Res 2002;91:240–246 [DOI] [PubMed] [Google Scholar]
- 37.Niwa T, Tsukushi S: 3-deoxyglucosone and AGEs in uremic complications: inactivation of glutathione peroxidase by 3-deoxyglucosone. Kidney Int Suppl 2001;78:S37–S41 [DOI] [PubMed] [Google Scholar]
- 38.Szwergold BS, Kappler F, Brown TR: Identification of fructose 3-phosphate in the lens of diabetic rats. Science 1990;247:451–454 [DOI] [PubMed] [Google Scholar]
- 39.Hamada Y, Araki N, Koh N, Nakamura J, Horiuchi S, Hotta N: Rapid formation of advanced glycation end products by intermediate metabolites of glycolytic pathway and polyol pathway. Biochem Biophys Res Commun 1996;228:539–543 [DOI] [PubMed] [Google Scholar]
- 40.Lal S, Szwergold BS, Kappler F, Brown T: Detection of fructose-3-phosphokinase activity in intact mammalian lenses by 31P NMR spectroscopy. J Biol Chem 1993;268:7763–7767 [PubMed] [Google Scholar]
- 41.Ramana KV, Friedrich B, Tammali R, West MB, Bhatnagar A, Srivastava SK: Requirement of aldose reductase for the hyperglycemic activation of protein kinase C and formation of diacylglycerol in vascular smooth muscle cells. Diabetes 2005;54:818–829 [DOI] [PubMed] [Google Scholar]
- 42.Srivastava S, Ramana KV, Tammali R, Srivastava SK, Bhatnagar A: Contribution of aldose reductase to diabetic hyperproliferation of vascular smooth muscle cells. Diabetes 2006;55:901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yagihashi S, Yamagishi SI, Wada RR, Baba M, Hohman TC, Yabe-Nishimura C, Kokai Y: Neuropathy in diabetic mice overexpressing human aldose reductase and effects of aldose reductase inhibitor. Brain 2001;124:2448–2458 [DOI] [PubMed] [Google Scholar]
- 44.Vikramadithyan RK, Hu Y, Noh HL, Liang CP, Hallam K, Tall AR, Ramasamy R, Goldberg IJ: Human aldose reductase expression accelerates diabetic atherosclerosis in transgenic mice. J Clin Invest 2005;115:2434–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramana KV, Tammali R, Reddy AB, Bhatnagar A, Srivastava SK: Aldose reductase-regulated tumor necrosis factor-α production is essential for high glucose-induced vascular smooth muscle cell growth. Endocrinology 2007;148:4371–4384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.