Abstract
Noonan syndrome (NS) is a genetically heterogeneous disorder caused most commonly by activating mutations in PTPN11. We report a patient with hypotonia, developmental delay and clinical features suggestive of NS. High-resolution chromosome analysis was normal, and sequence analyses of PTPN11, SOS1, KRAS, BRAF, RAF1, MEK, and MEK2 were also normal. Array CGH revealed a single copy gain of 9 BAC clones at 12q24.11q24.21 (8.98 Mb in size), which encompassed the PTPN11 locus at 12q24.13 and was confirmed by FISH analysis. Shchelochkov et al., [2008] reported a similar case and speculated that such duplications might account for 15–30% of NS cases with no detectable mutation in NS genes. We screened more than 250 NS cases without mutation in known NS disease-causing genes by quantitative PCR, and none of these studies produced results in the duplicated range. We also explored the possibility that de novo changes affecting the untranslated region (UTR) of the PTPN11 transcript might represent an alternative event involved in SHP2 enhanced expression. DHPLC analysis and direct sequencing of the entire 3' UTR in 36 NS patients without mutation in known genes did not show any disease-associated variant. These findings indicate that duplications of PTPN11 represent an uncommon cause of NS, and functionally relevant variations within the 3'UTR of the gene do not appear to play a major role in NS. However, recurrent observations of NS in individuals with duplications involving the PTPN11 locus suggest that increased dosage of SHP2 may have dysregulating effects on intracellular signaling.
Keywords: Noonan syndrome, PTPN11, TBX3, TBX5, Gene Duplication PTPN11, Array CGH, Duplication 12q24.3, Dosage Sensitive Gene
INTRODUCTION
Noonan syndrome (NS) is a relatively common autosomal dominant condition, which may be sporadic or inherited [Noonan, 2006; Allanson 2007]. It consists of characteristic facial features, short stature, heart defects, particularly pulmonic stenosis, developmental delay in about one third of cases (usually preceded by failure to thrive), blood clotting disorders, variable ectodermal involvement, and susceptibility to certain cancers, particularly juvenile monomyelocytic leukemia (JMML). Dermatologic findings may include café-au-lait spots, keratosis pilaris, lentigines, and nevi. NS may be caused by gain of function mutations in PTPN11 (50%), which encodes SHP2, a protein tyrosine phosphatase with positive modulatory role in RAS-MAPK signaling, or functional dysregulation of other signal transducers participating in this signal cascade, including, SOS1, KRAS, RAF1 or BRAF [Tartaglia et. al. 2001; Zenker et al.,, 2004; Schubbert et al 2006; Roberts et al 2007; Tartaglia et al., 2007; Nava et al.,, 2007; Razzaque et al 2007; Pandit et al 2007; Sarkozy et al.,, 2009]. Overall, mutations in identified genes account for approximately 70–75% of affected cases, indicating that other disease-causing events remain to be identified.
We report on the identification of a de novo duplication of 8.98 Mb at 12q24.11q24.21, encompassing PTPN11, in a 4-year-old male with a phenotype fitting NS, apparently normal chromosomes and no mutation in known disease genes. Quantitative PCR analysis and denaturing high-performance liquid chromatography (DHPLC) screening performed to explore prevalence of PTPN11 copy number gain and functionally relevant changes in the 3' untranslated region (UTR) of PTPN11 support the view that duplications of PTPN11 are uncommon as a cause of NS and that functionally relevant 3'UTR variants do not appear to be associated significantly with this disorder.
CLINICAL REPORT
The patient was the AGA term product of an uncomplicated pregnancy, labor and delivery, born to a 28-year-old primigravida mother and a 27-year-old father. After her second pregnancy, in retrospect, his mother noted that the fetal movements of her first pregnancy were less vigorous than in her second pregnancy. His delivery was via cesarean for failure to progress. Family history was non-contributory, and he was born weighing 3.72 kg, and measuring 53 cm in length, with a head circumference of 36 cm. Pyloric stenosis was detected and repaired at age 4 weeks, and he underwent inguinal hernia repair with orchiopexy at age 4 months. Feeding difficulties remained evident after birth, and a lingual frenulum was clipped. He continued to have poor postnatal growth about 2 SD below mean for age with preservation of his head size at the 50thcentile. He was a very slow feeder, with a weak suck, and he seldom took more than 100 cc of feeding at a time. He was delayed in transitioning to solid foods. Motor milestones were delayed during infancy, and he was referred for early intervention to developmental services at age 1 year because he was not crawling. A neurologist noted generalized hypotonia, and thyroid studies and pediatric eye evaluation were normal. He had recurrent ear infections requiring insertion of pressure-equalizing tubes and adenoidectomy. Echocardiogram revealed only mild mitral valve regurgitation. His coagulation parameters were normal.
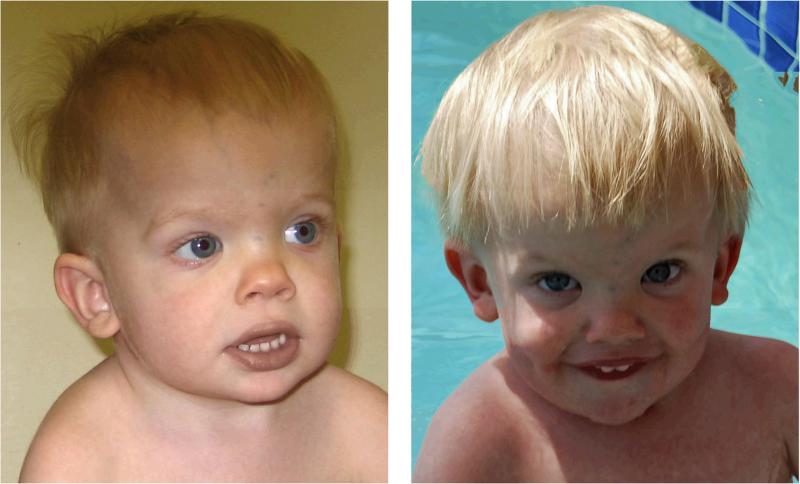
His skin appeared thin and smooth with easy bruising and several faint café au lait macules. His hair was thin and fine, and he had a prominent forehead with down-slanting palpebral fissures, bilateral epicanthal folds, and a flat nasal bridge (Fig 1). He had full lips, an upturned nasal tip, and a short, wide neck. He had hypoplastic nipples and mild pectus deformity with superior pectus carinatum and inferior pectus excavatum. He had bilateral single transverse palmar flexion creases and flat feet, and his joints were very loose, especially in his hands and feet. He had generalized hypotonia with normal hearing, with mildly delayed social skills, fine and gross motor skills and expressive language skills. Formal developmental assessment at age 29 months placed his cognitive skills at 23–25 months, social skills at 25 months, fine motor skills at 18–23 months, and self-help skills at 29 months. At 33 months, his gross motor skills were at 22–26 months, and at age 4 years his auditory comprehension skills were at 3.1 years, with expressive communication skills at 2.7 years. His height was 84.5 cm (just under the 3rd centile), weight 11.6 kg (3rd centile), head circumference 50.5 cm (50th centile).
Figure 1.
Clinical features of the patient at ages 25 months and 45 months (top left and right) illustrating facial features for Noonan syndrome along with typical pectus deformity.
METHODS
Patient cohorts
As a collaborative multicentric effort, 250 subjects with NS or a phenotype suggestive of this condition were screened to explore prevalence of PTPN11 copy number gains. Clinical features for 124 of these subjects satisfied diagnostic criteria reported by van der Burgt et al. [1994], while another 53 cases were more atypical, most of them displaying variable cognitive deficits and developmental delay as well as additional anomalies compatible with NS. These patients had been evaluated by experienced clinicians either personally or by reviewing history, documented clinical findings and photographs. The remaining 73 subjects of this cohort were referred for mutation analysis by clinicians who suspected a diagnosis of NS. None of the subjects carried a mutation in the PTPN11, SOS, KRAS, or RAF1 gene based on scanning of the coding exons by denaturing high-performance liquid chromatography (DHPLC) analysis and/or direct sequencing.
Scanning of the 3' UTR of the PTPN11 gene was performed on a subgroup of 36 subjects with clinical features that satisfied NS diagnostic criteria (see above). None of the subjects carried a mutation in the PTPN11, SOS, KRAS, RAF1, MEK1 or MEK2 genes.
Array CGH Analysis and FISH Analysis
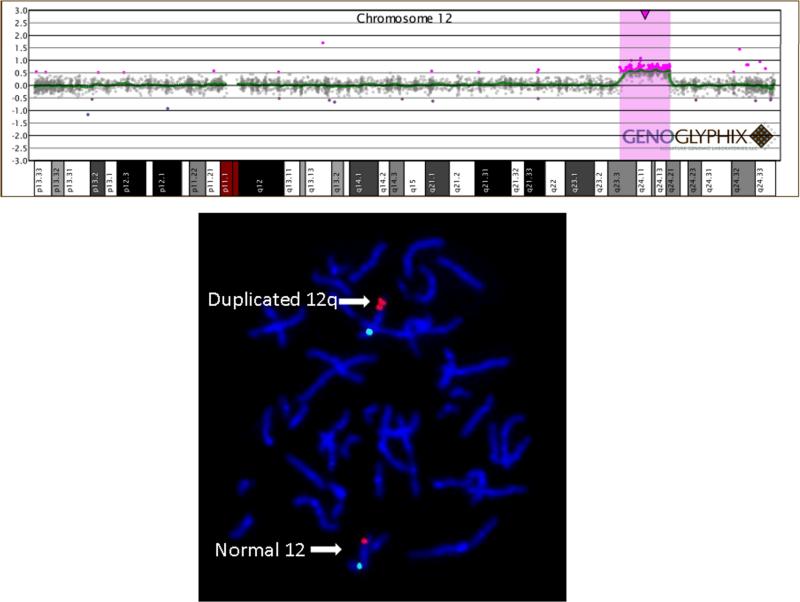
Array CGH was performed with a BAC microarray (the SignatureChip®-www.signaturegenomics.com) that was developed for the detection of microdeletions, microduplications, aneuploidy, unbalanced translocations, and subtelomeric and pericentromeric copy-number alterations [Bejjani et al., 2005]. As shown in Figure 2, microarray analysis was performed and analyzed as described [Bejjani et al., 2005], and with an oligonucleotide array (the SignatureChipOS®) as described [Ballif et al., 2008]. All abnormalities detected by array CGH were confirmed by fluorescence in situ hybridization (FISH) as published [Shaffer et al., 1994] using a BAC clone from the PTPN11 locus (RP11–9P8), revealing a duplication at 12q24.13 visible on metaphase chromosomes. The centromere probe to chromosome 12 was use as a control and showed a normal hybridization pattern.
Figure 2.
PTPN11 single copy gain in the patient with clinical features fitting NS. Array CGH analysis demonstrated a single-copy gain of nine BAC clones at 12q24.11q24.21 (centromeric RP1-46F2, RP11-124I12, RP11-767G4, RP11-421J14, RP11-9P8, RP5-1048I22, RP11-438N16, RP11-269C10, and RP11-100F15 telomeric) encompassing the PTPN11 locus at 12q24.13 (top). FISH analysis confirmed the duplication of the PTPN11 locus (bottom). Clone RP11-9P8 from the PTPN11 locus is labeled in red, and 12q centromere probe D12Z3 is labeled in green as a control. The normal chromosome 12 shows one green and one red signal, while the chromosome with the duplication shows one green and two red signals.
Quantitative PCR
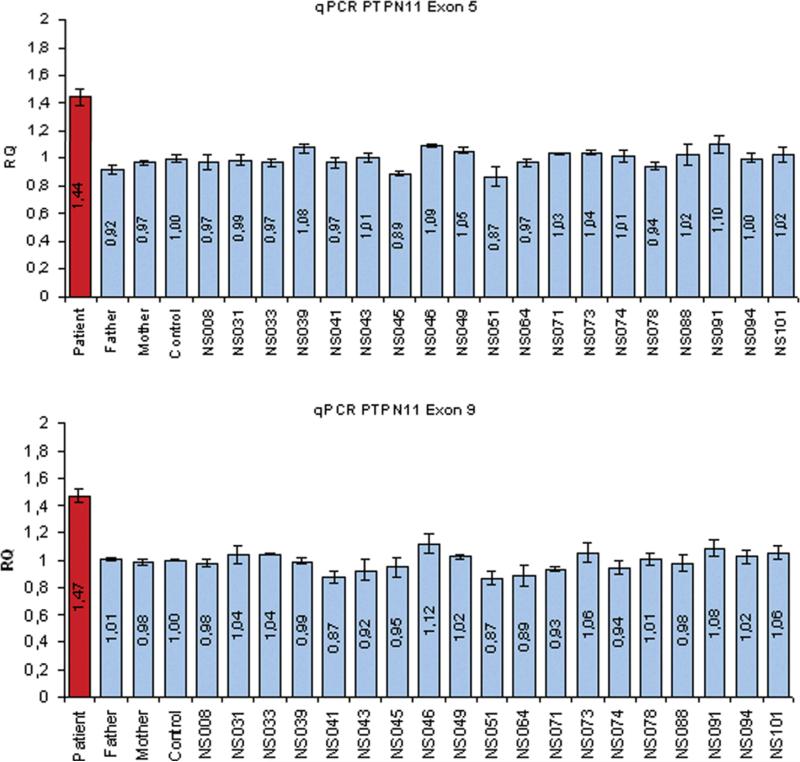
Quantitative real-time PCR analysis for PTPN11 exon 5 and exon 9 was performed using custom designed primers and TaqMan minor-groove binder probes according to the manufacturer's recommendations with the PrimerExpress 2.0 Software (Applied Biosystems, Foster City, USA) from the Genbank sequence NM_002834 (PTPN11 exon 5: forward primer 5'-GTGGAGGAGAACGGTTTGATTC-3', reverse primer 5'-CCAATGTTTCCACCATAGGATTC-3', probe 6FAM-AGATCTTGTGGAACATTATA. PTPN11 exon 9: forward primer: 5'-GAATTTGAAACCAAGTGCAACAAT-3', 5'-CAGCCTTGTGTGGCAATGTAA-3', probe 6FAM-CAAAGCCCAAAAAG). The assay for each sample was performed in 384-well plates with a final volume of 20 μl on an ABI 7900HT in a multiplex reaction with the endogenous control of albumin exon 12 as described previously [Thiel et al., 2003]. Copy number levels were calculated using the ΔΔCt method and normalized to the expression levels of healthy control samples.
Mutation analysis
The entire coding sequence and flanking intronic portions of the PTPN11 gene were screened by direct sequencing using the ABI BigDye terminator Sequencing Kit v.1.1 (Applied Biosystem, Foster City, CA) and an ABI 3700 Capillary Array Sequencer). Exclusion of mutations within the affected exons of SOS1, KRAS, BRAF, RAF1, MEK1, and MEK2 as well as of disease-associated sequence variants within the 3' UTR of the PTPN11 gene was based on DHPLC (3100 and 3500HT WAVE DNA fragment analysis systems, Transgenomic) scanning. Amplimers having abnormal denaturing profiles were purified (Microcon PCR, Millipore) and sequenced bi-directionally. Primer pairs, and PCR and DHPLC analysis settings are available upon request.
RESULTS
Initial high resolution chromosome analysis revealed a normal male karyotype. After PTPN11 was sequenced and found to be normal, array CGH was performed because of the child's hypotonia and developmental delay. This assay demonstrated a single copy gain of 9 BAC clones at 12q24.11q24.21 (Figure 2), which included the PTPN11 locus, as well as TBX3 and TBX5. This finding was confirmed by FISH analysis (Fig 2). Informed consent was obtained from the family to analyze the other NS causative genes on a research basis. An additional array CGH analysis was performed on an oligonucleotide array and further defined the extent of duplication to be 8.98 Mb. The genomic coordinates of the duplication on 12q24.3 were determined to be: 104,641,698– 113,603,100. The child's DNA was also used as reference to check for changes in PTPN11 copy number in a cohort of 250 subjects with NS or a phenotype suggestive of this disorder without mutations in known NS disease-causing genes by qPCR. We did not observe any duplication, indicating that PTPN11 gene duplication is not a frequent event among subjects with this disorder. We explored the possibility that de novo changes affecting the 3' untranslated sequence of the PTPN11 transcript might represent an alternative mechanism involved in SHP2 dysregulated expression. DHPLC screening and direct sequencing performed on 36 subjects with NS without mutation in known genes did not reveal any disease-associated variant.
DISCUSSION
NS is a genetically heterogeneous disorder that may be caused by gain of function mutations in PTPN11 (50%) or other genes coding for proteins participating in the RAS-MAPK pathway, including SOS1 (15–20%), KRAS (2%), or RAF1 (5–10%) or BRAF (2%) [Sarkozy et al., 2009; Tartaglia and Gelb, 2009]. The molecular cause underlying this disorder is still unknown in approximately 25–30% of affected individuals. NS-causing PTPN11 mutations generally promote upregulation of SHP-2 function and enhanced signal flow through RAS [Tartaglia and Gelb, 2009]. Most mutations affect amino acid residues participating in stabilization of the N-SH2/PTP domain interaction that maintains SHP-2 in its catalytically active conformation, but other mechanisms appear to be also involved [Keilhack et al., 2005; Tartaglia et al., 2006; Martinelli et al., 2008]. While it has been established that a different group of PTPN11 mutations is acquired as a somatic event in childhood acute leukemias and myeloproliferative or myelodysplastic disorders [Tartaglia et al., 2003; 2004], a study documented an alternative perturbing effect of SHP-2 on signaling and leukemogenesis via enhanced espression [Xu et al., 2005]. This observation raised the hypothesis that PTPN11 gene duplication/amplification might affect developmental processes via gene dosage effect. Consistently, a dosage effect for a mutated PTPN11 gene has been documented in mice [Araki et al., 2004] and humans [Becker et al., 2007].
In our patient with NS clinical features, no mutation was found in the PTPN11, KRAS, SOS1, RAF1, BRAF, MEK1 and MEK2 genes, while array-CGH demonstrated an 8.98 Mb duplication at 12q24.3, which included the PTPN11 locus. A similar finding was recently documented by Shchelochkov et al. [2008] who reported a 3-year-old girl with a similar duplication that also included PTPN11. This patient had hypotonia, coarctation of the aorta, ASD and VSD (closed spontaneously), velopharyngeal incompetence, hypertelorism, ptosis, epicanthal folds, cupped ears, microcephaly, pectus excavatum and short stature (10th centile) with developmental delay. Based on this finding and available records, these authors suggested that PTPN11 duplication might be a common cause for NS due to increased gene dosage.
There are several other reports of patients with duplications of 12q24 encompassing PTPN11 [Harrod et al., 1980; Dixon et al., 1993; Doco-Fenzy et al., 2006; Cappellacci et al., 2007]. Only the patient reported herein and the patients reported by Doco-Fenzy et al. [2006] and Shchelochkov et al. [2008] were initially suspected of having NS, and these other two reported patients had microcephaly and aortic anomalies. Many of their other manifestations are seen in NS, such as failure to thrive, developmental delay, short neck, wide spaced nipples, chest deformity, and minor facial anomalies such as hypertelorism, small chin, and down-turned corners of the mouth. The duplication observed in all 3 of these patients who were initially suspected of having NS also includes TBX3 and TBX5, whose action may be dosage-sensitive. Overexpression of TBX5 is known to have an impact on cardiogenesis [Hatcher et al., 2004]. While these data suggest that duplications of the region containing PTPN11 might result in an enhanced expression level of SHP2 that could have activating effects on intracellular signaling similar to a mutant SHP2 protein with dysregulated function and may affect development, our findings support the view that PTPN11 duplication does not represent a common cause of this disorder. Since our qPCR screening was performed using two intragenic fragment only (exons 5 and 11), we however cannot rule out the hypothesis that smaller intragenic in-frame duplications or deletions affecting PTPN11 might upregulate SHP2 function and cause NS or a related phenotype.
Supplementary Material
Figure 3.
PTPN11 gene duplication does not represent a major event in NS. Quantitative real-time PCR analysis was used to check for changes in PTPN11 copy number in a cohort of 250 NS cases without a mutation in known NS disease-causing genes. Two coding regions of the gene (exon 5 and exon 9) were analyzed, and none of the cases produced results in the duplicated range.
ACKNOWLEDGMENTS
We appreciate support from SHARE's Childhood Disability Center, the Steven Spielberg Pediatric Research Center, and the NIH/NICHD Program Project Grant (HD22657 to JMG), Medical Genetics NIH/NIGMS Training Program Grant (5-T32-GM08243 to JMG), Telethon-Italy Grant (GGP07115 to MT), “Collaborazione Italia-USA/malattie rare 2007/2009” Grant (to MT), and the German Research Foundation (DFG; ZE 524/4–1 to MZ). We also appreciate assistance from the Cedars-Sinai General Clinical Research Center Grant (M01-RR00425) for samples collected under CSMC IRB Protocols 0463 and 4232.
REFERENCES
- Allanson JE. Noonan syndrome. Am J Med Genet Part C Semin Med Genet. 2007;145C:274–279. doi: 10.1002/ajmg.c.30138. [DOI] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, McDonald M, Campbell L, Ming JE, Zackai EH, Bejjani BA, Shaffer LG. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. 1. Vol. 28. 2008. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Hughes H, Howard K, Armstrong M, Roberts D, Lazda EJ, Short JP, Shaw A, Patton MA, Tartaglia M. Early fetal death associated with compound heterozygosity for Noonan syndrome-causative PTPN11 mutations. Am J Med Genet Part A. 2007;143A:1249–1252. doi: 10.1002/ajmg.a.31738. [DOI] [PubMed] [Google Scholar]
- Bejjani BA, Saleki R, Ballif BC, Rorem EA, Sundin K, Theisen A, Kashork CD, Shaffer LG. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: Is less more? Am J Med Genet Part A. 2005;134A:259–267. doi: 10.1002/ajmg.a.30621. [DOI] [PubMed] [Google Scholar]
- Cappellacci S, Martinelli S, Rinaldi R, Martinelli E, Parisi P, Mancini B, Pescosolido R, Grammatico P. De novo pure 12q22q24.33 duplication: first report of a case with mental retardation, ADHD, and Dandy-Walker malformation. Am J Med Genet Part A. 2007;140A:1203–1207. doi: 10.1002/ajmg.a.31219. [DOI] [PubMed] [Google Scholar]
- Dixon JW, Costa T, Teshima IE. Mosaicism for duplication 12q (12q13a`q24.2) in a dysmorphic male infant. J Med Genet. 1993;30:70–72. doi: 10.1136/jmg.30.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doco-Fenzy M, Mauran P, Lebrun JM, Bock S, Bednarek N, Struski S, Albuisson J, Ardalan A, Collot N, Schneider A, Dastot-LeMoal F, Gaillard D, Goossens M. Pure direct duplication (12)(q24.1! q24.2) in a child with Marcus-Gunn phenomenon and multiple congenital anomalies. Am J Med Genet Part A. 2006;140A:212–221. doi: 10.1002/ajmg.a.31057. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Tartaglia M. Noonan syndrome and related disorders: Dysregulated RAS-mitogen activated protein kinase signal transduction. Hum Mol Genet. 2006;15:R220–R226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- Harrod MJE, Byrne JB, Dev VG, Francke U. Duplication 12q mosaicism in two unrelated patients with a similar syndrome. Am J Med Genet. 1980;7:123–129. doi: 10.1002/ajmg.1320070206. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Diman NY, Kim MS, Pennisi D, Song Y, Goldstein MM, Mikawa T, Basson CT. Role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol Genomics. 2004;18:129–140. doi: 10.1152/physiolgenomics.00060.2004. [DOI] [PubMed] [Google Scholar]
- Jones KL. Noonan syndrome. In: Jones KL, editor. Smith's Recognizable Patterns of Human Malformations. 6th edition Elsevier, Inc.; Philadelphia, PA: 2006. pp. 124–126. [Google Scholar]
- Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- Martinelli S, Torreri P, Tinti M, Stella L, Bocchinfuso G, Flex E, Grottesi A, Ceccarini M, Palleschi A, Cesareni G, Castagnoli L, Petrucci TC, Gelb BD, Tartaglia M. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Hum Mol Genet. 2008;17:2018–2029. doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava C, Hanna N, Michot C, Pereira S, Pouvreau N, Niihori T, Aoki Y, Matsubara Y, Arveiler B, Lacombe D, Pasmant E, Parfait B, Baumann C, He`ron D, Sigaudy S, Toutain A, Rio M, Goldenberg A, Leheup B, Verloes A, Cave` H. Cardiofacio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: Genotype-phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JA. Noonan syndrome and related disorders: alterations in growth and puberty. Rev Endocr Metab Disord. 2006;7:251–255. doi: 10.1007/s11154-006-9021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, Lo`pez Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M, Fujiwara Y, Matsushima M, Mizuno K, Tokuyama M, Hirota H, Muneuchi J, Higashinakagawa T, Matsuoka R. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- Sarkozy A, Carta C, Moretti S, Zampino G, Digilio M, Pantaleoni F, Scioletti A, Esposito G, Cordeddu V, Lepri F, Petrangeli V, Dentici M, Mancini G, Selicorni A, Rossi C, Mazzanti L, Marino B, Ferrero GB, Silengo M, Memo L, Stanzial F, Faravelli F, Stuppia L, Gelb B, Dallapiccola B, Tartaglia M. Germline BRAF mutations in Noonan, LEOPARD and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat. 2009;30:695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, McCaskill C, Han JY, Choo KH, Cutillo DM, Donnenfeld AE, Weiss L, Van Dyke DL. Molecular characterization of de novo secondary trisomy 13. Am J Hum Genet. 1994;55:968–974. [PMC free article] [PubMed] [Google Scholar]
- Shchelochkov OA, Patel A, Weissenberger GM, Chinault AC, Wiszniewska J, Fernandes PH, Eng C, Kukolich MK, Sutton VR. Amer. J. Med Genet. Part A. 2008;146A:1042–1048. doi: 10.1002/ajmg.a.32215. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD. Germ line and somatic PTPN11 mutations in human disease. Eur J Med Genet. 2005;48:81–96. doi: 10.1016/j.ejmg.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD. Molecular genetics of Noonan syndrome. In: Zenker M, editor. Monographs in Human Genetics - Vol. 17. Noonan syndrome and related disorders: A matter of deregulated RAS signaling. Karger Press; Basel: 2009. pp. 20–39. [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase, SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Kalidas K, Shaw A, Song X, Musat DL, van der Burgt I, Brunner HG, Bertola DR, Crosby A, Ion A, Kucherlapati RS, Jeffery S, Patton MA, Gelb BD. PTPN11 mutations in Noonan syndrome: Molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hählen K, Hasle H, Licht JD, Gelb BD. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, Palmi C, Carta C, Pession A, Aricò M, Masera G, Basso G, Sorcini M, Gelb BD, Biondi A. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104:307–313. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Martinelli S, Stella L, Bocchinfuso G, Flex E, Cordeddu V, Zampino G, Burgt I, Palleschi A, Petrucci TC, Sorcini M, Schoch C, Foa R, Emanuel PD, Gelb BD. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, Ustaszewska A, Martin J, Bristow J, Carta C, Lepri F, Neri C, Vasta I, Gibson K, Curry CJ, Siguero JP, Digilio MC, Zampino G, Dallapiccola B, Bar-Sagi D, Gelb BD. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- Thiel CT, Kraus C, Rauch A, Ekici AB, Rautenstrauss B, Reis A. A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2–12 duplications and deletions leading to HMSN/HNPP. Eur J Hum Genet. 2003;11:170–8. doi: 10.1038/sj.ejhg.5200920. [DOI] [PubMed] [Google Scholar]
- Van der Burgt I, Berends E, Lommen E, van Beersum S, Hamel B, Mariman E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet. 1994;53:187–191. doi: 10.1002/ajmg.1320530213. [DOI] [PubMed] [Google Scholar]
- Xu R, Yu Y, Zheng S, Zhao X, Dong Q, He Z, Liang Y, Lu Q, Fang Y, Gan X, Xu X, Zhang S, Dong Q, Zhang X, Feng GS. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood. 2005;106:3142–3149. doi: 10.1182/blood-2004-10-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker M, Buheitel G, Rauch R, Koenig R, Boose K, Kress W, Tietze HU, Doerr HG, Hofbeck M, Singer, Reis A, Rauch A. Genotype-phenotype correlations in Noonan syndrome. J Pediatr. 2004;144:368–374. doi: 10.1016/j.jpeds.2003.11.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.