Abstract
Osteosarcomas are the most prevalent primary bone tumors found in pediatric patients. To understand their molecular etiology, cell culture models are used to define disease mechanisms under controlled conditions. Many osteosarcoma cell lines (e.g., SAOS-2, U2OS, MG63) are derived from Caucasian patients. However, patients exhibit individual and ethnic differences in their responsiveness to irradiation and chemotherapy. This motivated the establishment of osteosarcoma cell lines (OS1, OS2, OS3) from three ethnically Chinese patients. OS1 cells, derived from a pre-chemotherapeutic tumor in the femur of a 6-year-old female, were examined for molecular markers characteristic for osteoblasts, stem cells and cell cycle control by immunohistochemistry, reverse transcriptase-PCR, western blotting and flow cytometry. OS1 have aberrant G-banded karyotypes, possibly reflecting chromosomal abnormalities related to p53 deficiency. OS1 had ossification profiles similar to human fetal osteoblasts rather than SAOS-2 which ossifies ab initio (p<0.05). Absence of p53 correlates with increased Runx2 expression, while the slow proliferation of OS1 cells is perhaps attenuated by pRB retention. OS1 express mesenchymal stem cell markers (CD44, CD105) and differ in relative expression of CD29, CD63 and CD71 to SAOS-2. (p<0.05). Cell cycle synchronization with nocodazole did not affect Runx2 and CDK1 levels but decreased cyclin-E and increased cyclin-A (p<0.05). Xenotransplantion of OS1 in SCID mice yields spontaneous tumors that were larger and grew faster than SAOS-2 transplants. Hence, OS1 is a new osteosarcoma cell culture model derived from a pre-chemotherapeutic ethnic Chinese patient, for mechanistic studies and development of therapeutic strategies to counteract metastasis and deregulation of mesenchymal development.
Keywords: osteosarcoma, osteoblast, bone, cancer, proliferation, differentiation, chemotherapy, genetic differences, ethnicity, metastasis, cell cycle, DNA damage
Osteosarcomas are the most common primary malignant bone tumors in adolescents and young adults. Multi-agent chemotherapy primarily based on cisplatin and adriamycin and perhaps high-dose methotrexate has greatly improved the 5-year survival of osteosarcoma patients (Taylor et al., 1978; Beijamin et al., 1992; Bacci et al., 1993; Link et al., 1991; Bielack et al., 2002; Wilkins et al., 2003). Nevertheless, local recurrence (4% to 8%) and post-operative pulmonary metastasis (about 30%) are frequent (Grimer et al., 2002; Picci et al., 1997). This indicates a need to identify prognostically poor individuals as only a modest fraction of relapsed cases remains disease free with poor long term survival (30%) (Meyers and Gorlick, 1997; Nathan et al., 2006; Coffin et al., 2005). To improve the prognosis and treatment of osteosarcomas, it is necessary to examine the growth phenotypic properties, cellular regulatory pathways, and biochemical mechanisms that are altered in osteosarcomas using cell culture models. Culture models also permit controlled investigation of in vivo parameters related to osteosarcoma progression including hydrostatic pressure and angiogenesis (Nathan et al., 2008; Nathan et al., 2005). However, there are only a limited number of human osteosarcoma cell lines available that permit examination of mechanisms contributing to the etiology of this bone cancer.
Osteosarcomas may arise from dedifferentiated osteoblasts or from pre-osteoblastic mesenchymal progenitors that exhibit deregulated growth potential. Human tumors frequently are deficient for classic tumor suppressors such as p53 and pRB (Hatakeyama and Weinberg, 1995; Vogelstein et al., 2000). Genetic loss of either p53 or pRB in human patients increases the osteosarcoma incidence (Fuchs and Pritchard, 2002; Deshpande and Hinds, 2006), while recently developed p53 and/or pRB deficient mouse models develop bone-specific lesions and are highly prone to osteosarcoma (Lengner et al., 2006; Wang et al., 2006; Walkley et al., 2008; Berman et al., 2008). Both p53 and pRB have been biologically linked to normal control of osteogenesis (Lengner et al., 2006; Wang et al., 2006; Thomas et al., 2001; Thomas et al., 2004). Both tumor suppressors have also been linked to the expression and/or activity of the Runt-related transcription factor Runx2, which is a bone-specific cell fate determining factor that controls osteoblast growth (Galindo et al., 2005; Pratap et al., 2003; Teplyuk et al., 2008; Zaidi et al., 2007; Young et al., 2007a; Young et al., 2007b; Lian et al., 2004). For example, calvarial osteoprogenitors from mice with a bone-specific p53 null mutation have elevated levels of Runx2 (Lengner et al., 2006). Runx2 levels are modulated in human osteosarcoma cells deficient for pRB and/or p53 (Thomas et al., 2004; Nathan et al., 2009), and Runx2 interacts directly with pRB which controls its transcriptional activity (Thomas et al., 2001; Thomas et al., 2004; Berman et al., 2008). Runx2 is a multi-faceted gene regulator that integrates cell signaling at transcriptional subnuclear domains and maintains phenotypic identity in osteoprogenitors, osteoblasts and osteosarcomas through epigenetic mechanisms (Young et al., 2007a; Young et al., 2007b; Lian et al., 2004). Expression of Runx2 correlates with that of the CDK inhibitor (CKI) p27 (Thomas et al., 2004; Galindo et al., 2005). Furthermore, Runx2 expression is inversely related to the levels of the p53 responsive CDK inhibitor p21/Kip1/CDKN1A (Pratap et al., 2003; Westendorf et al., 2002). Both p27 (Cip1/CDKN1B) and p21 control the CDK dependent release of pRB from E2F (Hatakeyama and Weinberg, 1995). Thus, pRB, p53 and Runx2 form a bone-specific regulatory network that controls normal cell cycle progression in osteoblasts and that is deregulated in osteosarcoma cells.
Clinical treatment of osteosarcoma involves surgery, as well as radiation and chemotherapy that cause DNA damage at the cellular level (Nathan et al., 2006). Genetic differences among individuals influence cancer disposition and response to therapy, and these differences may be more pronounced in ethnically diverse populations (Gao et al., 2008; Lal et al., 2007; Tadokoro et al., 2003). Indeed, there are individual (Haslam et al., 1998; Gilman et al., 1985) and ethnic (Parkin et al., 1993; Stiller et al., 2000; Settakorn et al., 2006; Mirabello et al., 2009) differences in the occurrence of osteosarcomas that may be directly relevant to differences in the therapeutic responses of bone cancer patients. Hence, it is necessary to develop distinct cell culture models for osteosarcomas from different skeletal sites that reflect the pluriformity of the human population and to account for possible skeletal site-specific, ethno-geographic and/or genetic differences in responses of osteosarcomas to radiation and chemo-therapy.
The most frequently studied osteosarcoma cell lines (e.g., SAOS-2, U2OS and MG63) were derived from Caucasian patients. In this study, we established a novel osteosarcoma cell culture model (OS1) based on a tumor from a pediatric patient of Chinese descent in Singapore. For this patient-derived cell line, the interesting findings are that OS1 cells which exhibit high levels of Runx2 have a relatively long population doubling time (~5 days) atypical to the aggressive nature of osteosarcoma cell lines commercially available where the population doubling time averages between 1 and 3 days. Yet OS1 cells xenografted in mice with severe combined immune deficiency (SCID) formed larger tumors that grew twice as fast as tumors from SAOS-2 cells. We also show data indicating that this slow population doubling time is consistent with abnormalities in molecular parameters of cell cycle control and DNA damage response pathways, and differences in cell surface expression.
MATERIALS AND METHODS
Derivation of new osteosarcoma cell lines
The study was approved by the Institutional Review Board of the National University Hospital (NHG DSRB B/00/301) in Singapore and informed consent for experimental use of the tissues was obtained. Three osteosarcoma cell lines (OS1, OS2 and OS3) were obtained prior to chemotherapy from bone tumor biopsies from patients of, respectively, Singaporean Chinese (OS1) or Indonesian Chinese (OS2 and OS3) descent. Preliminary data on OS1 have been presented earlier in a brief report (Nathan et al., 2009). Osteosarcoma biopsies were rinsed in phosphate-buffered saline (PBS), minced into small fragments, and initially propagated as outgrowth cultures. Tissue fragments were incubated at 37°C with 5% CO2 (Sanyo MCO-15A incubator, Sanyo Electric Co Ltd, Japan) in mixed culture medium (referred to as ‘15%RD’) containing (9:1[v/v]) of Roswell Park Memorial Institute 1640 (RPMI 1640) medium and Dulbecco's modified Eagle's medium (DMEM; Gibco Industries, Inc. OK, USA) supplemented with 15% fetal bovine serum (HyClone Laboratories, Inc. UT, USA) in plastic culture flasks (NUNC A/S, Roskilde, Denmark). Maintenance of the incubator was in line with institutional guidelines and included regular testing of cultures for bacterial contamination and we discarded samples with any appearance of contamination. Cells from outgrowth cultures were dispersed in PBS containing 0.25% trypsin (JRH Biosciences, Inc. KS, USA) and the resulting single-cell suspension was then cultured in 15%RD medium. Uniform colonies were morphologically selected by removing contaminating fibroblast-like and highly proliferative cells with a TPP Cell Scraper (Techno Plastic Products AG, Switzerland) under an Olympus IX70 inverse microscope (Olympus, Japan). The cell morphology of osteosarcoma cells was observed after long term culture (>50 passages) to ensure the maintenance of the phenotype. Isolated osteosarcoma cells were obtained by removing surrounding cells with a TPP cell scraper and their morphology was recorded once every 24 hours for 5 days using a digital camera (Media Cybernetics, Inc. MD, USA).
Determination of cell population doubling time
Cells were seeded at 103 cells per well in twelve 96-well plates, incubated at 37°C in 5% CO2 in 15%RD medium and half of the medium was replenished every day. Cell population growth was measured at 24h intervals over twelve days using MTT assays (Mosmann, 1983). Used culture medium was replaced with 200 μl of fresh pre-warmed medium containing 2% MTT (Sigma, St. Louis, MI, USA; Catalog# M-5655) and cells were incubated for 2 hours. The medium was then replaced with 100 μl 20% SDS (Pharmacia Biosciences, Catalog# 17.1313-01) in a mixture of N,N-Dimethylformamide (Sigma, D4551) and distilled water (1:1 (v/v)) and treated for another 45 min at 37°C . The optical density at 570 nm (OD570) was measured with the Tecan Sunrise ELISA Reader (Scimed Pte Ltd, Singapore). The colony doubling time was calculated by regression analysis of the OD570 over the observed period.
Karyotype Analysis
Karyotype analysis was performed as described previously (Polito et al., 2003) to investigate the chromosomal stability of the OS1 cell line. Cultured cells at different passages (15, 21, 31 and 59) were treated for 16 hours with the metaphase blocker Demecolcine (Sigma) at a final concentration of 0.4 μg/ml. Cells were then dispersed by trypsinization for 20 min, collected by low-speed centrifugation (1600 rpm for 6 minutes), rinsed in 0.075M KCl solution and fixed overnight in 5 ml of a 3:1 (v/v) mixture of absolute methanol/glacial acetic acid at 4°C. Cells were then rinsed three times in with the same solution, transferred to slides and stained by immersion in freshly prepared Giemsa staining solution (Sigma, G9641) for 7 minutes. Excess staining solution was removed by rinsing with distilled water. Chromosomes were examined by microscopy and analyzed on an enlarged picture format.
Alizarin red staining was used to further establish the osteoblast-like character of OS1 cells. Near-confluent (~80%) cells were cultured for 4 weeks in differentiation medium (50 μg/ml ascorbic acid-2-phosphate, 10 mM β-glycerophosphate and 100 nM dexamethasone)(Sigma). Cultures were fixed with cold methanol (−20 °C), rinsed with distilled water and stained with 0.01% Alizarin Red S (pH 5.5) for 30 min. Staining of the cultures was documented on an Epson Perfection photo scanner (Seiko Epson, Japan).
Examination of osteogenic markers
Immuno-histochemical staining of OS1 cell cultures was carried out with antibodies for Osteocalcin, Osteonectin and Collagen type I. Cells were seeded in a LAB-TEK chamber slide (Nalgene/NUNC Inc, USA) at a concentration of 2 × 104 cells in a 200 μl volume per well. Near-confluent cells (~80%) were washed with PBS, fixed with pre-cooled 75% ethanol for 30 minutes and stained using the Dako LSABTM + system (DakoCytomation, Carpinteria, CA). Slides were washed with PBS, incubated in 3% hydrogen peroxide for 5 minutes followed by immuno-staining for 30 min at 37°C with mouse monoclonal antibodies for Collagen type I, Osteocalcin and Osteonectin (Biodesign Inc, USA). PBS without primary antibodies was included as a negative control in slides prepared in parallel. Upon rinsing with PBS, slides were incubated with a biotinylated secondary antibody followed addition of Avidin-Biotin Complex reagents. Antigen-antibody complexes were detected using the DAB substrate chromogen system (DakoCytomation) and immuno-histochemical results were evaluated using a light microscope attached to a digital camera system (Sony SSC-DC58AP).
Conventional RT-PCR was carried out with OS1, OS2 and OS3 cells, as well as human 143.98.2 osteosarcoma cells (ATCC, CRL-11226) to assess commitment of OS1 cells to the osteogenic lineage based on Runx2 gene expression. Total RNA was extracted from 2 × 106 cells using the RNA Blood Mini Kit (QIAGEN, Valencia, CA, USA). Cells were dissolved in lysis buffer containing guanidine isothiocyanate and homogenized through a QIAshredder column (QIAGEN Inc, CA). RNA was purified using QIAamp spin columns and total RNA was recovered in 30 μl RNase-free water. RNA concentration was determined using a DU 530 UV/Vis Spectrophotometer (Beckman). RNA quality was assessed by observing the integrity of 18S and 28S rRNAs upon separation by 1% agarose gel electrophoresis in 0.5 × TBE buffer and visualization using a Wealtec UV transilluminator (Wealtec Corp. NV, USA). Total RNA (1 μg) was reverse transcribed into cDNA using the Access RT-PCR Kit (Promega, Madison, WI, USA). Briefly, the reverse transcription reactions and PCR were carried out in one 50 μl reaction system (0.5 mg/mL of oligo-dT primer, AMV/Tf1 5 × Buffer, 10 mmol/L dNTP Mix, AMV Reverse Transcriptase and Tf1 DNA Polymerase). Total reactions were reverse transcribed at 45°C for 45 minutes. Samples were denatured for 2 minutes at 94°C and amplified by 30 cycles at 94°C for 30 s, 57°C for 1 min and 68°C for 2 min, and final cycle at 68°C for 7 min. The primer sequences for human Runx2 (GenBank NM_004348) were 5′- cag ttc cca agc att tca tcc -3′ and 5′- tca ata tgg tcg cca aac ag -3′ (Nakayama et al., 2004). The size of the PCR product was 443bp. The internal control β-actin (GenBank X00351) was amplified using the forward primer 5′- cac act gtg ccc atc tac gag g -3′ and the reverse primer 5′- agt ttc gtg gat gcc aca gga -3′ (Bodo et al., 2002). The size of the β-actin product is 350 bp. Aliquots (2.5 μl) of the PCR reactions were subjected to electrophoresis in a 1.5% agarose gel with ethidium bromide (final concentration of 0.25 μg/ml in 0.5 × TBE electrophoresis buffer. We examined OS1 cells in parallel with the two other osteosarcoma cell lines (OS2 and OS3), as well as fibroblast-like cells (fb) from the original tumor tissue used to derive OS1 and a commercial osteosarcoma human osteosarcoma cell line (143.98.2, CRL-11226, ATCC).
Analysis of mRNA and protein expression of cell growth markers
Reverse transcriptase quantitative real time PCR (RT-qPCR) analysis was applied to analyze mRNA levels of cell growth related genes in different cell types (HFOB, SAOS-2, OS1, OS2 and/or OS3). RNA was extracted with TRIzol (Invitrogen) and total RNA (1 μg) was reverse transcribed into cDNA using a commercial kit (Superscript III, Invitrogen). Real time PCR was performed on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems) using SYBR Green PCR Master Mix (Applied BIosystems) in triplicates. PCR samples were denatured for 10 min at 95°C followed by 40 PCR cycles for 15 sec at 95°C and 1 min at 60°C. Primer sequences are available on request. The results are calculated as relative expression using ΔCT values.
Western blot analysis was used to determine protein expression in our panel of cell lines. Cells were rinsed three times in ice-cold PBS and proteins samples were obtained by suspending cells in lysis buffer (5 mM MgCl2, 150 mM NaCl, 0.05% Nonidet P-40, 0.1%SDS, 1% Triton X100, pH7.5) and protease inhibitor cocktail (Calbiochem, UK) at 4°C for 10-30 minutes. The BCA protein assay kit (Pierce, USA) was used to determine protein concentrations and cell lysates (20-30 μg) were resolved by 10% SDS-PAGE. Upon transfer of proteins to nitrocellulose membranes, membranes were blocked with 5% low fat milk in Tris-buffered saline (TBS) for 1 h and then incubated with the indicated primary antibodies (1:1,000 dilution). Mouse monoclonal antibodies for Runx2 (gift from Dr. Yoshiaki Ito), pRB (Biodesign, USA), p53 and p27 (Cell signal), as well as rabbit polyclonal Cyclin D antibody (Santa Cruz Biotechnology, USA) were obtained from the indicated suppliers. Following incubation with peroxide-conjugated goat anti-mouse/rabbit secondary antibodies (Santa Cruz) for 1 h, the immuno-reactive bands were visualized with a Western Lightening Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Inc., Waltham, MA) and the autoradiograms of the blot were captured by a scanner. The relative quantification of each protein blot was analyzed with Gel-Pro Analyzer software (Media Cybernetics, Inc, Bethesda, MD).
Immunofluorescence microscopy
Immunofluorescence microscopy was used to establish in situ correlations between Runx2 and the proliferation marker Ki67. Cells were fixed for 30 min in cold 3.7% paraformaldehyde (Sigma), permeabilized for 15 min with 0.1% Triton-X100 in PBS (T-PBS) and non-specific reactivity was suppressed by a 30 min incubation with 5% goat serum for 30 min. Cells were incubated overnight at 4°C with monoclonal antibodies for Runx2 (1:10,000 dilution) and Ki67, followed by a 30 min incubation at room temperature with Alexa Fluor 594 chicken anti-mouse secondary antibody (Molecular Probes, USA) for 30 min. DAPI staining was used to visualize nuclei and micrographs were obtained with an Olympus fluorescence microscope (Olympus, Tokyo Japan).
Flow cytometry
Flow cytometry was performed using a panel of established cell surface markers: CD29, CD34, CD38, CD44, CD63, CD71, CD105, CD117 and CD135 (BD Biosciences). These antigens reflect distinct cell fates and stages in the mesenchymal and/or hematopoietic lineages. OS1 and SAOS-2 cells were removed from culture using a non-enzymatic cell dissociation solution (CellStripper; Mediatech, Manassas, VA) washed twice in PBS and then aliquoted into a 96-well plate. Cells were pelleted at 450g for 5 minutes, resuspended in FCS/PBS solution and incubated on ice for 20 min. Cells were, then washed twice in 2% FCS/PBS before resuspension in 2% FCS/PBS and analyzed on a Guava PCA-96 bench top flow cytometer (Guava Technologies, Hayward,CA) by fluorescence activated cell sorting. All samples were measured in triplicates for each cell line.
Xenotransplantation assays in SCID mice
Xenotransplantion assays with 6-week old locally-bred immuno-compromised SCID mice were performed to estimate the rate of growth of OS1-derived tumors. One day before transplantation, OS1 and SAOS-2 cells were recovered from tissue culture flasks using PBS buffer containing 5 mM EDTA. For each cell line, suspensions of ~ 1 × 107 cells were prepared in serum-free RPMI-1640 medium, washed with PBS then mixed with normal saline before being introduced subcutaneously on the left flank of different mice. All experiments were conducted under sterile conditions in a laminar flow hood. After the procedure, all animals were maintained in a pathogen-free environment. The implants were allowed to develop for up to thirty weeks with the animals monitored daily. The volume of the tumor, once palpable under the skin, was estimated by measuring the average diameter of the mass weekly with a vernier caliper.
Statistics
Standard descriptive statistics were used: mean, standard deviation, and percentage changes were determined with respect to a control (SPSS, SPSS, Inc., Chicago, Illinois). A paired T-test was used where appropriate to compare paired data sets. If three of more treatment groups were assessed, a One-way Analysis Of Variance (ANOVA) was used to test if treatment groups were homogenous. If a significant difference was found, the Scheffe post-hoc multiple comparison test was used to determine the paired difference between treatment groups by comparing each set of data. Statistical significance was accepted at P < 0.05.
RESULTS
Establishment of a slow growing pediatric osteosarcoma cell line, OS1, that remains capable of differentiating into mature osteoblasts
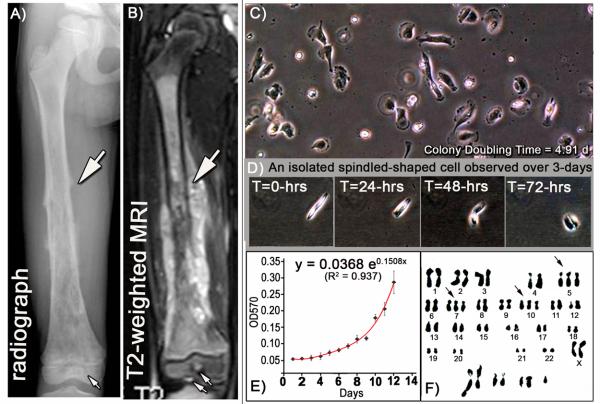
The tumor biopsy for OS1 was obtained from a 6 year old female Singaporean pediatric patient who had a rapidly progressing high-grade neoplastic tumor in the right femur (Figs. 1A and 1B) and early lung cancer metastasis. The biopsy was diagnosed as an osteosarcoma based on pathological analysis that revealed osteoid material containing pleiomorphic malignant cells, including many mitotic cells and some cells with atypical mitotic spindles. Outgrowth cultures from the biopsy specimen were propagated for over 30 passages resulting in a cell line (OS1) (Fig. 1C). Upon plating, OS1 cells were rounded in appearance just after attachment and eventually became more polygonal or cuboidal. Cells elongate and become wider and spindled-shaped, and later appear thinner as they begin to migrate (Fig 1D). The OS1 cell line exhibited stable proliferative properties after 21 months (76 passages). Daily cultures were sampled in triplicate for 12 days and used in MTT Assays and OD570 measurements were graphed as a function of time. Line plots were fitted with an exponential regression curve (y= 0.0368e0.1508x, R2 = 0.937, Fig. 1E). Based on regression analysis, the estimated mean colony doubling time for OS1 cells at passage 50 was estimated as 4.91 days. Karyotype analysis of metaphase-arrested cells with demecolcine (Petkovic and Cepulic, 1996) showed that OS1 cells have complex chromosomal abnormalities that vary from cell to cell. This chromosomal complexity persisted for the duration of observations (from passage 19 to 59). A representative metaphase spread of OS1 cells at passage 31, exhibited trisomy for chromosomes 5, 7 and 10; as well as a set of marker chromosomes with structural deletions and chromosomal aberrations including translocations (Fig. 1F). The karyotypic abnormalities of OS1 cells are consistent with their derivation from a high grade osteosarcoma.
Figure 1. Derivation of OS1 cells from an aggressive bone tumor.
OS1 cells were derived from a 6-year-old girl of Singaporean-Chinese descent who presented with pain and swelling of the right thigh which rapidly developed over one week. A) Antero-posterior radiograph of the right femur, and B) MRI scans showed a tumor extending the entire length of the femur shaft (block white arrows), from the sub-trochanteric to the physis with permeation into the epiphysis (small arrows). C) A primary culture was established (OS1) from the tumor biopsy, and cells maintained for 30-passages demonstrated distinct cell morphology. D) A representative isolated cell exhibited morphological changes related to cell attachment, cell migration and cell division over a period of 72-hours (Original magnification, 100X). E) After 50 passages, the observed growth curve for OS1 was fitted with an exponential regression function and the mean colony doubling time was estimated to be 4.91 days. F) Karyotype analysis of a representative metaphase from the OS1 cell line at passage 31, demonstrates a variety of chromosomal abnormalities and aberrations, with no consistencies noted between metaphases. In this metaphase, trisomy was noted in chromosomes 5, 7 and 10, and genetic abnormalities also included translocations and structural deletions.
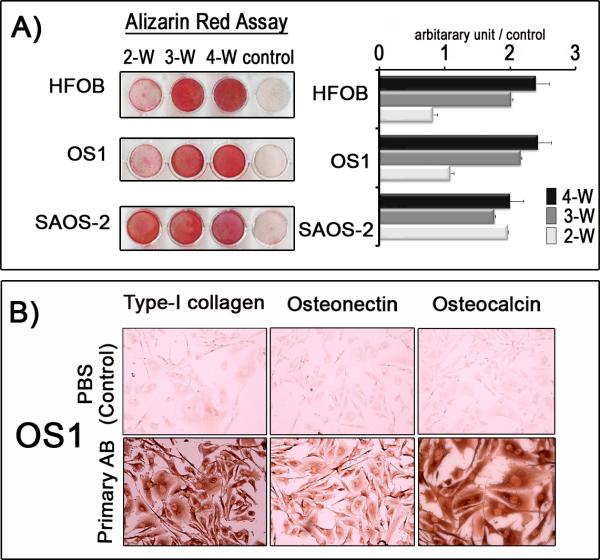
The relative ability of OS1 cells (passage 50) to differentiate into mature osteoblasts was investigated by Alizarin red staining. OS1 cells were maintained in osteogenic media in long term cultures (up to four weeks) and compared to SAOS-2 osteosarcoma cells and immortalized human fetal osteoblasts (HFOB) (Fig. 2A). SAOS-2 cells stained prominently with Alizarin red after 2 weeks. However, like HFOB cells, OS1 cells displayed limited Alizarin red staining at this point, presumably because both cells grew slower and reached confluence at a later stage. All three cell lines exhibited robust Alizarin red staining by 3 weeks in culture under osteogenic conditions. Thus, OS1 represents a mineralizing osteoblastic cell line, like SAOS-2 and HFOB cells.
Figure 2. OS1 cells are immature osteoblastic cells capable of differentiating into mature osteoblasts.
A) Alizarin-red assays were performed to assess mineralization of human fetal osteoblasts (HFOB), OS1 and SAOS-2 cells cultured in triplicate in osteogenic media for 2, 3 and 4 weeks. Quantification of the Alizarin-red assay was based on the pixel intensities of the staining and presented as a proportion of the control in arbitrary units. By Week 2 (2-W), OS1 cells showed initial mineralization (reflecting osteogenic induction) that doubled by Week 3 (3-W) and decelerated by Week 4 (4-W) (ANOVA, p<0.05). A similar trend was observed for HFOB over the same period (ANOVA, P<0.05). For SAOS-2 cells, mineralization accelerated twice as fast as OS1 and HFOB during the same period, with no significant changes during 4-W (ANOVA, P>0.05); B) Immunohistochemistry staining (DAKO LSAB+ system) show that OS1 cells are positive for osteogenic markers (Type-1 collagen, osteonectin and osteocalcin) (bottom panel). Phosphate-buffered solution (PBS) was substituted for primary antibodies as a negative control (top panel) (Original magnification, X20).
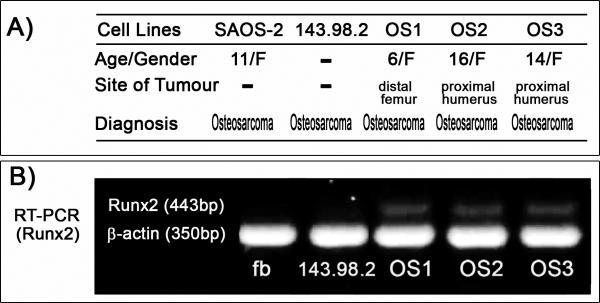
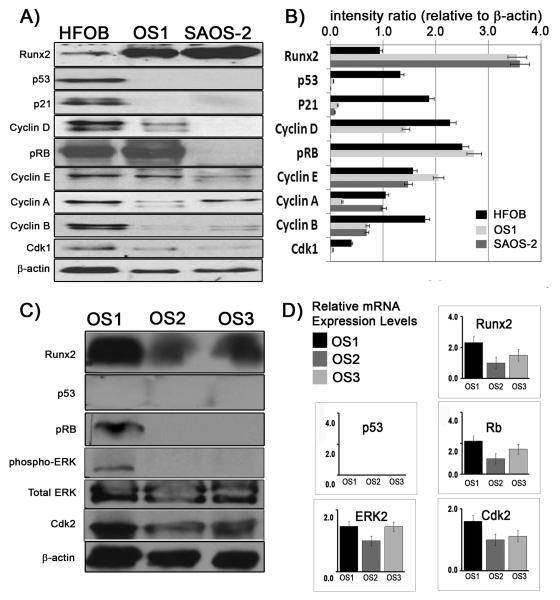
Similar differentiating cultures at passage 50 were also subjected to immuno-histochemistry using a panel of osteoblast specific markers (Osteocalcin, Osteonectin, Collagen type I, BMP4). Differentiated OS1 cells stained strongly for Osteocalcin and Collagen type I and less clearly for Osteonectin (Fig. 2B), while no staining was observed for BMP4 (data not shown). Furthermore, OS1 cells and two other osteosarcoma cell lines (OS2 and OS3) that were derived from two other pediatric patients of Chinese descent, expressed the osteogenic lineage-commitment marker Runx2 as demonstrated by conventional RT-PCR (Fig. 3A and B). Runx2 gene expression was below detection levels for human 143.98.2 osteosarcoma cells (ATCC) and for fibroblasts derived from the original OS1-generating tumor tissue (Fig. 3B). All three cell lines (OS1, OS2 and OS3) were confirmed by qPCR to have increased Runx2 gene expression with corresponding p53 deletion and pRB retention (Fig 3c). The clear detection of the bone-phenotypic markers Runx2, Osteocalcin and Collagen type I, as well as Alizarin red staining (Figs. 2 to 3) together indicate that OS1 cells are bona fide immortalized osteogenic cells capable of differentiating into mature osteoblasts.
Figure 3. Expression of the osteogenic marker and master regulator Runx2 in OS1 cells.
A) Brief summary of data on the origin of osteosarcoma cell lines. Three different Singaporean patients diagnosed with osteosarcoma were used to establish primary cultures and eventually cell lines from bone biopsies (respectively, OS1, OS2 and OS3). The other osteosarcoma cells lines (SAOS-2, 143.98.2) were obtained from ATCC. B) Conventional RT-PCR demonstrates the expression of Runx2 gene in OS1, OS2 and OS3 cell lines. The human osteosarcoma cell line (143.98.2) and human fibroblasts (fb) obtained from the original OS1 tumor tissue were used for comparison and do not exhibit Runx2 expression.
OS1 cells represent de-differentiated osteoblasts that express stem cell markers
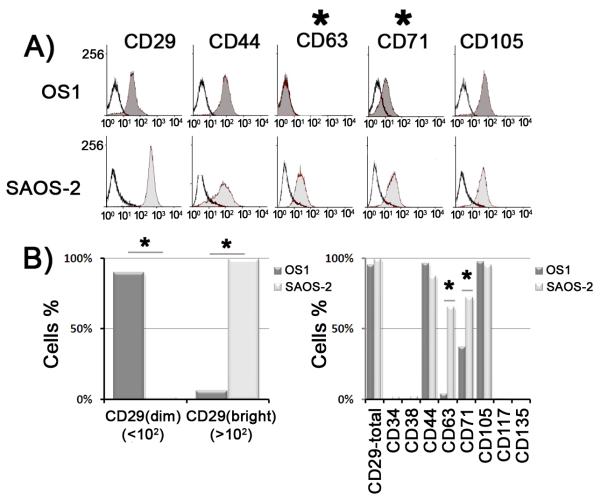
Osteosarcoma is a pediatric disease that may originate from somatic mutations in differentiated osteoblasts that revert into earlier lineage stages, or from the failure of mesenchymal stem cells and/or osteoprogenitors to differentiate into mature osteoblasts. To examine the phenotypic state of OS1 cells from a mesenchymal stem cell perspective, we generated a profile of cell surface markers by flow cytometry. We compared expression of a standard panel of antigens (mesenchymal lineage: CD29, CD44, CD63, CD71, CD105; hematopoietic lineage: CD34, CD38, CD117 and CD135) relative to an isotype-matched control antibody in both OS1 and SAOS-2 cells (Fig. 4). Both cell types express at least four of the five mesenchymal stem cell markers we tested (i.e., CD29, CD44, CD71, CD105) (Fig 4A). Thus, OS1 and SAOS-2 cells are both bona fide cells of the mesenchymal lineage. One clear difference is that, unlike SAOS-2 cells, OS1 do not express detectable levels of CD63, yet both OS1 and SAOS-2 cells express CD44 and CD105 at comparable levels (Fig. 4B). OS1 cells also differ from SAOS-2 cells, because they express lower levels of CD29 (β1-integrin), which activates signaling pathways through cell-cell and cell matrix interactions, and CD71. As expected, no expression was observed for the hematopoiesis markers CD34, CD38, CD117 and CD135 (Fig 4B). Taken together, the differences in cell surface expression between OS1 and SAOS-2 may be related to the differences in the biological properties of these mesenchymal-derived tumor cell lines (see Discussion).
Figure 4. OS1 cells represent osteoblastic cells committed to the mesenchymal lineage.
A) Flow-cytometric profile for mesenchymal lineage antigens was obtained in which isotype-matched control (non-colored profile) antibody was compared with antibodies for CD29, CD44, CD63, CD71 and CD105 (colored profile) for OS1 (top panel) and SAOS-2 (bottom panel) cells. B) CD29 (β1-integrin) which is involved in the cell-cell and cell matrix interactions is significantly different between OS1 (dark grey) and SAOS-2 (light grey). A large fraction of OS1 cells had low fluorescence intensity [‘CD29(dim)’], and large fraction of SAOS-2 had high fluorescence intensity [‘CD29(bright)’]. Significant differences between OS1 and SAOS-2 were also noted for CD63, and CD71 (* - P<0.05), while no significant differences were noted for CD44 and CD105. No expression was detected for the hematopoietic lineage markers CD34, CD38, CD117 and CD135.
OS1 exhibit abnormalities in molecular parameters of cell growth control
To understand the molecular basis of the cell growth defect in OS1 cells, we performed western blot analysis with antibodies for transcriptional regulators of cell growth (p53, pRB and Runx2), the CDK inhibitor p21 (target gene of p53), cyclins D1 and E that are linked to pRB activation, as well as cyclins A and B as markers of events related to late S and G2 phases or mitosis (Fig. 5A). Compared to HFOB, both OS1 and SAOS-2 cells express high levels of Runx2 protein. This may be biologically linked to the observed absence of p53 and p21 protein expression. Similar observations were obtained from OS2 and OS3 osteosarcoma cells, who do not express p53 (Fig 5C). These results are consistent with previous data obtained with calvarial osteoblasts from p53 null mice (Lengner et al., 2006) and a brief preliminary report on OS1 cells (Nathan et al., 2009). Unlike SAOS-2 OS2 and OS3 cells, OS1 retains expression of pRB at levels similar to those in HFOB cells, while both OS1 and HFOB cells express higher levels of cyclins D1 and E than SAOS-2 cells. Interestingly, OS1 cells but not OS2 and OS3 cells expressed detectable levels of phospho-ERK, which is documented to promote cell growth and survival (Fig. 5C). The remaining results show that cyclin A expression in OS1 is lower than that in SAOS-2 and HFOB, whereas both cyclin B and CDK1 levels are reduced in both OS1 and SAOS-2 cells. The western blot data are corroborated by real time RT-qPCR data showing that RNA expression parallels protein levels to a large extent (Figs. 5B and D). For example, Runx2 transcripts levels in OS1 and SAOS-2 cells are, approximately 2.5 times higher than in HFOB cells. Cyclin D1 mRNA levels in HFOB and OS-1 were >100 times higher than those in SAOS-2 cells. Taken together, our results indicate that OS1 cells are defective in the p53/p21 pathway, which is a component of the DNA damage response, but retention of pRB and an activated MAPK/phosphoERK pathway indicates that cyclin/CDK dependent mechanisms remain capable of controlling the periodic inactivation of pRB during the cell cycle in OS1 cells.
Figure 5. The molecular pathology of OS1 cells involves loss of p53, elevated Runx2 and retention of pRB.
Western blot analyses for transcriptional regulators of cell growth (p53, pRB and Runx2), as well as the indicated cell cycle markers (e.g., cyclins D1, E, B and A), was carried out with lysates from three different cell types. A) A representative protein expression profile and B) quantification of the expression levels (intensity levels as a ratio of β-actin) comparing HFOB, OS1 and SAOS-2 cells. C) Runx2 protein and cell cycle markers analysis in OS1, OS2 and OS3 osteosarcoma cells. D) Relative gene expression levels by qPCR demonstrate Runx2 mRNA is present in all three cell lines (OS1, OS2 and OS3). None of these cell lines expressed p53, but pRB mRNA is present. The three cell lines have varying expression levels Erk2 and Cdk2.
Elevated Runx2 expression in proliferating OS1 cells
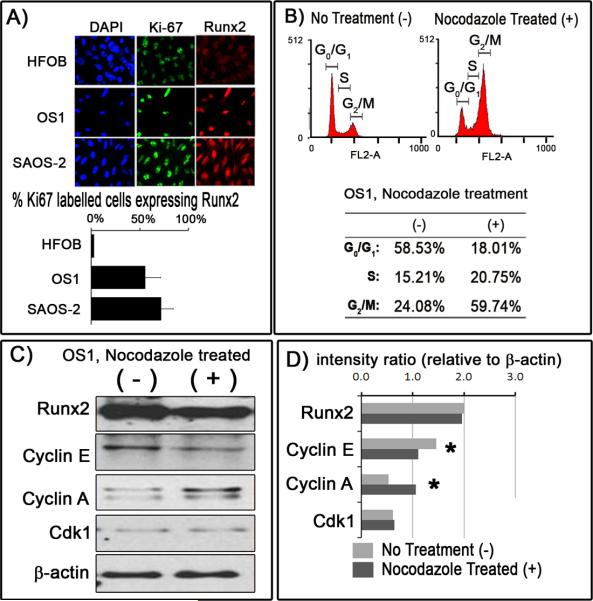
Previous studies from our group have shown that Runx2 levels are low in actively proliferating cells and elevated in quiescent or differentiated osteoblasts (Galindo et al., 2005). The high levels of Runx2 protein observed in proliferating OS1 and SAOS-2 cells compared to HFOB cells based on western blot analysis (Fig. 5A) suggests that regulation of Runx2 protein levels are uncoupled from cell cycle progression in osteosarcoma cells. Therefore, we evaluated whether there is a correlation at the single cell level between expression of Runx2 and Ki67, which represents a proliferation-specific marker that resides in nucleoli. Immunofluorescence microscopy revealed that nucleolar Ki67 signals and nuclear localization of Runx2 are evident in all three cell types. Runx2 signals are very weak in HFOB cells with only a limited number of cells displaying significantly positive staining, while the majority of OS1 and SAOS-2 cells (>70%) showed strong Runx2 signals. Although there are few cells with strong signals for both Runx2 and Ki67 in HFOB cells, high levels of each of these proteins clearly co-exist in OS1 and SAOS-2 cells (Fig. 6A). Thus, unlike HFOB cells, actively proliferating Ki67-positive OS1 and SAOS-2 cells are permissive for high levels of Runx2.
Figure 6. Runx2 levels remain constitutively elevated in actively proliferating OS1 cells.
A) Immunoflorescence staining was carried out with Ki67 and Runx2 antibodies in HFOB, OS1 and SAOS-2 cells. The fraction of Ki67-positive and Runx2-positive cells were quantified and correlated. B) In a separate experiment, OS1 cells were treated with nocodazole (+) to inhibit cells exiting from the cell cycle. Flow-cytometric profile of the nocodazole-treated OS1 cells (+) and untreated OS1 cells showed a percentage increase in cells in the G2/M phase from 24.08% to 59.74%. C&D) Western blot analyses of the cell cycle regulators comparing between nocodazole-treated OS1 cells (dark grey bar) and untreated nocodazole (light grey bar) demonstrated a significant decrease in Cyclin E levels and a corresponding increase in Cyclin A levels when nocodazole was introduced, consistent with a cell cycle arrest in G2/M. No significant differences were noted for Runx2 and Cdk1 levels (Note: * - t-test, P<0.05).
To assess whether OS1 cells can be synchronized in mitosis, we treated cells with nocodazole and examined the cell cycle distribution by FACS analysis. Flow cytometry showed that nocodazole effectively inhibits exit from mitosis and increases the 4N population, reflecting cells with a double amount of DNA in G2 and M phases (Fig. 6B). Western blot analysis of the same samples indicates that expression of cyclin E was reduced and cyclin A was increased relative to asynchronous cells, consistent with a G2/M block (Figs. 6C and 6D). Thus, OS1 cells can be used for cell cycle synchronization studies using nocodazole. Interestingly, Runx2 protein and mRNA were maintained at high levels during G2/M (Figs. 6C and 6D, which is consistent with the elevated expression of this factor in asynchronous cells.
Xenografted OS1 cells form faster growing tumors than SAOS-2 cell implants in vivo
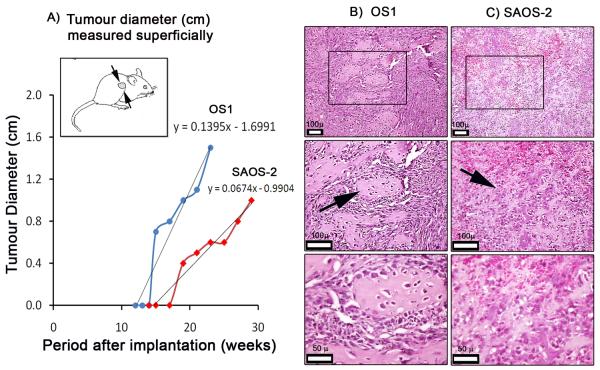
OS1 cells were isolated from a patient with high grade metastatic primary osteosarcoma and early lung metastasis, yet these cells proliferate slowly in culture. To assess the growth rate of OS1 cells in vivo, we injected these cells in the flanks of immuno-deficient (SCID) mice. In parallel, we injected SAOS-2 cells, which grow faster in culture (doubling time of 24-32 hr, data not shown) for external comparison. Mice with xenografts were analyzed for spontaneously arising palpable tumors at weekly intervals and tumor size was measured with calipers that were applied superficially (Fig 7A). OS1 cell implants presented with flank tumors after about 14 weeks, while tumors from SAOS-2 cells lagged behind by about 4 weeks. Linear regression analysis of tumor size over time indicates a trend that OS1 tumors grow faster than SAOS-2 cells in vivo. Histology of the xenografted mass for OS1 (Fig 7B) and SAOS-2 (Fig 7C) in SCID mice are consistent with recurrent osteosarcoma. The xenografted OS1 mass demonstrated the presence of osteoids and a pleiomorphic arrangement. The osteoid showed osteoclastic activities and with well-distributed osteocytes. The xenografted SAOS-2 mass however had more vascular channels, with small and irregular shaped acellular osteoid formation. Thus, while OS1 cells have a slow doubling time in culture compared to SAOS-2 cells, they grow faster in a tumor environment in vivo.
Figure 7. Xenotransplantation studies reveal that OS1 forms faster growing tumors than SAOS-2.
A) OS1 and SAOS-2 cells were injected in the left flank of SCID mice (~ 1 ×107 cells). The xenografts for OS1 and SAOS-2 yielded tumor masses that became palpable at twelve and fifteen weeks after implantation, respectively. Linear regression analysis revealed that OS1 derived tumors grew twice as fast as SAOS-2 derived tumors (respectively, 1.4 and 0.7 mm in diameter per week). Histology (H&E) of xenografted B) OS1 and C) SAOS-2 mass collected at the end point both demonstrated highly pleiomorphic cells. OS1 showed a typical pleiomorphic arrangement with well-defined osteoid formation, with osteocytes present. Osteoclast activity on the osteoid matrix was also noted. The xenografted SAOS-2 tumor mass was more vascular and with ill-defined osteoids that appeared acellular. (Low magnification, 10x; high magnification, 20x and 40x respectively)
DISCUSSION
In this study, we have established a panel of osteosarcoma cell lines (OS1, OS2 and OS3) from three different pediatric patients admitted to the National University Hospital in Singapore. As compared to SAOS-2 cells, OS1 grow slow in culture but form earlier and larger palpable tumors in vivo. It is possible that SAOS-2 cells, which have been in use for more than three decades, have adapted more to growth in culture at the expense of their in vivo growth properties. As OS1 cells are from a patient with early lung metastasis, this cell line will be useful for examining osteosarcoma metastasis to lung. Osteosarcomas form a highly vascularized tissue, and angiogenesis and vascular invasion may be promoted by tumor cells. Angiogenesis is promoted by paracrine VEGF production and because expression of VEGF is controlled by Runx2 (Zelzer et al., 2001), the elevated levels of Runx2 present in OS1 cells may promote blood vessel growth in osteosarcoma cells to support tumor growth and increase its metastatic potential. However, in vivo the xenografted OS1 mass did not have as many vascular channels. From a practical perspective, it would be interesting to investigate the interrelationships between local angiogenesis and osteosarcomas in both the in vitro and in vivo OS1 models. For in vitro modeling, it is necessary to perform co-culture experiments. However, human vascular epithelial cells (HuVECs) have a slow doubling time. Considering the slow population doubling time of OS1 cells in vitro, these cells would have the potential for studying paracrine interactions in co-culture systems with HuVECs or other cells with similar doubling times (e.g. HuVECs, osteoblasts, osteoclasts, fibroblasts).
Established osteosarcoma cell lines (e.g., SAOS-2, U2OS and MG63) are deficient for expression of p53 and/or pRB. Specifically, SAOS-2 cells are p53−/pRB−, U2OS are p53+/pRB− and MG63 cells are p53−/pRB+ (Chandar et al., 1992). Our study shows that OS1 cells have a p53−/RB+ expression phenotype, similar to MG63 cells. While p53 deficiency in OS1 cells may account for the immortalized phenotype (>76 passages) and abnormal karyotypes (e.g. partial trisomy in multiple chromosomes and other chromosomal abnormalities) due to increased genomic instability, retention of pRB expression may render these cells at least in part growth factor dependent.
Previous studies have established that Runx2 levels are low in proliferating osteoblasts and elevated in quiescent osteoblasts (Galindo et al., 2005). In situ expression of the nucleolar proliferation marker Ki67 is expected to be inversely correlated with Runx2 protein levels. Indeed, our immuno-fluorescence microscopy studies show that non-tumorigenic immortalized HFOB cells exhibit limited co-expression between Ki67 and Runx2 with the majority of cells being Ki67(+)/Runx2(dim). However, both OS1 and SAOS-2 osteosarcoma cells show deregulation of this in situ relationship and representative cell populations exhibit abundant Ki67(+)/Runx2(bright) double positive cells. SAOS-2 cells have constitutively elevated Runx2 levels during the cell cycle (Galindo et al., 2005). The similarities in the correlation between Ki67 and Runx2 levels between SAOS-2 and OS1 cells suggest that regulation of Runx2 levels are also uncoupled from cell growth in OS1 cells.
Flow cytometry for cell surface antigens characteristic for mesenchymal stem cells show that OS1 and SAOS-2 have the following phenotype: OS1 cells are CD29(dim)/ CD63(−)/ CD71(dim)/ CD44(+)/ CD105(+), while SAOS-2 cells are CD29(bright)/ CD63(+)/ CD71(bright)/ CD44(+)/ CD105(+). Both SAOS-2 and OS1 cells express CD44, a transmembrane glycoprotein that functions as the cell surface receptor for its ligand hyaluronan and is also involved in cell adhesion/homing and to tumor growth and metastasis. Likewise, both osteosarcoma cell types express CD105 (ENG, endoglin), a proliferation-related marker that is a component of the transforming growth factor-β receptor complex. CD105 is expressed on the cell surface of endothelial cells, but also represents a putative mesenchymal stem cell marker that is associated with self-renewal (Haasters et al., 2009). The observation that OS1 and SAOS-2 qualitatively share surface expression of CD44 and CD105 markers, as well as CD29 and CD71 (see below) indicates that OS1 and SAOS-2 cells are derived from cells committed to the mesenchymal lineage.
CD29 is identical to integrin β1 (ITGB1, fibronectin receptor), a 140 kDa subunit of heterodimeric integrin receptors that is expressed in many cell types including osteoblasts to mediate cell signaling in response to cell adhesion. Reduced surface expression of CD29 in OS1 cells suggests that OS1 cells may be less responsive to substrate adherence and cell-cell contact than SAOS-2 cells. CD71 (TFRC, transferrin receptor, p90) is involved in iron metabolism and is a positive marker for tumor cell growth. The reduced levels of CD71 in cultured OS1 cells relative to SAOS-2 cells are consistent with the slower in vitro proliferative rates of OS1 cells. CD63 is a cell surface receptor (Tetraspanin-30,TSPAN30) that is linked with integrin-β1 signaling and its expression reduces metastatic potential by suppressing cell motility. The absence of CD63 expression in OS1 cells is consistent with the motile nature of OS1 cells as observed microscopically. Interestingly, CD63 supports homing of hematopoietic stem cells (HSCs) to osteoblasts in bone marrow (Gillette et al., 2009). CD63 deficiency in OS1 cells indicates that they lack this HSC homing function. Differences in the surface expression between OS1 and SAOS-2 are consistent with possible biological distinctions in their migratory and adhesive properties.
In conclusion, we have established three new osteosarcoma cell lines (OS1, OS2 and OS3) and have characterized OS1 cells as a culture model for in vitro and in vivo growth of osteosarcoma cells. All three lines are derived from pediatric patients of Asian descent, thus increasing the repertoire of osteosarcoma cell lines. Our study reveals that these lines have phenotypic properties similar to previously established osteosarcoma cell lines, and may be useful for future studies that examine molecular mechanisms that abrogate cell growth control in osteosarcoma cells.
ACKNOWLEDGMENTS
This study was co-supported by funding from NIH (grant R01AR049069 to AvW and grant P01 CA082834 to GS) and Singapore Cancer Syndicate, A*STAR (grants MN-005 and MN-077, to MST) and NUS ARF/Lee Kuan Yew Fellowship (grant R-364-000-089-112 to DL). We thank the members of the laboratories participating in the Singapore Osteobiology group for stimulating discussions. In particular, we thank Ren Xia Fei, Richie Soong, Wong Hee Kit, Lee Eng Hin, Nadiya Teplyuk, Kakoli Das, Christian Dombrowski, Dietmar Hutmacher, Victor Nurcombe, Shazib Pervais, Yoshiaki Ito, Motomi Osato, Jane Lian and Janet Stein for comments and support.
These studies were supported in part by NIH grants (AR49069 and CA082834), as well as the Lee Kuan Yew post-doctoral fellowship awarded to David Leong.
Footnotes
Conflict of interest statement: No conflict
Ethical Board Review statement : Approved by the National University Hospital Singapore Institutional Review Board (NHG DSRB B/00/301).
Literature Cited
- Bacci G, Picci P, Ferrari S, Ruggieri P, Casadei R, Tienghi A, Brach del Prever A, Gherlinzoni F, Mercuri M, Monti C. Primary chemotherapy and delayed surgery for nonmetastatic osteosarcoma of the extremities. Results in 164 patients preoperatively treated with high doses of methotrexate followed by cisplatin and doxorubicin. Cancer. 1993;72:3227–3238. doi: 10.1002/1097-0142(19931201)72:11<3227::aid-cncr2820721116>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Beijamin R, Chawla S, Carrasco H, Raymond A, Murray J, Armen T, Patel S, Wallace S, Ayala A, Papadopoulos N. Preoperative chemotherapy for osteosarcoma with intravenous Adriamycin and intoa-arterial cis-platinum. Ann Oncol suppl. 1992;3:3–6. doi: 10.1093/annonc/3.suppl_2.s3. [DOI] [PubMed] [Google Scholar]
- Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, Mukherjee S, Lees JA. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008;6:1440–1451. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, Zoubek A, Jürgens H, Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- Bodo M, Lilli C, Bellucci C, Carinci P, Calvitti M, Pezzetti F, Stabellini G, Bellocchio S, Balducci C, Carinci F, Baroni T. Basic fibroblast growth factor autocrine loop controls human osteosarcoma phenotyping and differentiation. Mol Med. 2002;8:393–404. [PMC free article] [PubMed] [Google Scholar]
- Chandar N, Billig B, McMaster J, Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br J Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin CM, Lowichik A, Zhou H. Treatment effects in pediatric soft tissue and bone tumors: practical considerations for the pathologist. Am J Clin Pathol. 2005;123:75–90. doi: 10.1309/h0d4vd760nh6n1r6. [DOI] [PubMed] [Google Scholar]
- Deshpande A, Hinds PW. The retinoblastoma protein in osteoblast differentiation and osteosarcoma. Curr Mol Med. 2006;6:809–817. doi: 10.2174/1566524010606070809. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop Relat Res. 2002:40–52. doi: 10.1097/00003086-200204000-00007. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Price DK, Sissung T, Reed E, Figg WD. Ethnic disparities in Americans of European descent versus Americans of African descent related to polymorphic ERCC1, ERCC2, XRCC1, and PARP1. Mol Cancer Ther. 2008;7:1246–1250. doi: 10.1158/1535-7163.MCT-07-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette JM, Larochelle A, Dunbar CE, Lippincott-Schwartz J. Intercellular transfer to signalling endosomes regulates an ex vivo bone marrow niche. Nat Cell Biol. 2009;11:303–311. doi: 10.1038/ncb1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman PA, Wang N, Fan SF, Reede J, Khan A, Leventhal BG. Familial osteosarcoma associated with 13;14 chromosomal rearrangement. Cancer Genet Cytogenet. 1985;17:123–132. doi: 10.1016/0165-4608(85)90022-6. [DOI] [PubMed] [Google Scholar]
- Grimer RJ, Taminiau AM, Cannon SR, Surgical Subcommitte of the European Osteosarcoma Intergroup Surgical outcomes in osteosarcoma. J Bone Joint Surg Br. 2002;84:395–400. doi: 10.1302/0301-620x.84b3.12019. [DOI] [PubMed] [Google Scholar]
- Haasters F, Prall WC, Anz D, Bourquin C, Pautke C, Endres S, Mutschler W, Docheva D, Schieker M. Morphological and immunocytochemical characteristics indicate the yield of early progenitors and represent a quality control for human mesenchymal stem cell culturing. J Anat. 2009;214:759–767. doi: 10.1111/j.1469-7580.2009.01065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam SI, Van Hul W, Morales-Piga A, Balemans W, San-Millan JL, Nakatsuka K, Willems P, Haites NE, Ralston SH. Paget's disease of bone: evidence for a susceptibility locus on chromosome 18q and for genetic heterogeneity. J Bone Miner Res. 1998;13:911–917. doi: 10.1359/jbmr.1998.13.6.911. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M, Weinberg RA. The role of RB in cell cycle control. Prog Cell Cycle Res. 1995;1:9–19. doi: 10.1007/978-1-4615-1809-9_2. [DOI] [PubMed] [Google Scholar]
- Lal S, Wong ZW, Jada SR, Xiang X, Chen Shu X, Ang PCS, Figg WD, Lee EJ, Chowbay B. Novel SLC22A16 polymorphisms and influence on doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics. 2007;8:567–575. doi: 10.2217/14622416.8.6.567. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- Link MP, Goorin AM, Horowitz M, Meyer WH, Belasco J, Baker A, Ayala A, Shuster J. Adjuvant chemotherapy of high-grade osteosarcoma of the extremity. Updated results of the Multi-Institutional Osteosarcoma Study. Clin Orthop Relat Res. 1991;270:8–14. [PubMed] [Google Scholar]
- Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44:973–989. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Kato N, Nakajima Y, Shimizu E, Ogata Y. Effect of TNF-alpha on human osteosarcoma cell line Saos2--TNF-alpha regulation of bone sialoprotein gene expression in Saos2 osteoblast-like cells. Cell Biol Int. 2004;28:653–660. doi: 10.1016/j.cellbi.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Nathan SS, DiResta GR, Casas-Ganem JE, Hoang BH, Sowers R, Yang R, Huvos AG, Gorlick R, Healey JH. Elevated physiologic tumor pressure promotes proliferation and chemosensitivity in human osteosarcoma. Clin Cancer Res. 2005;11:2389–2397. doi: 10.1158/1078-0432.CCR-04-2048. [DOI] [PubMed] [Google Scholar]
- Nathan SS, Gorlick R, Bukata S, Chou A, Morris CD, Boland PJ, Huvos AG, Meyers PA, Healey JH. Treatment algorithm for locally recurrent osteosarcoma based on local disease-free interval and the presence of lung metastasis. Cancer. 2006;107:1607–1616. doi: 10.1002/cncr.22197. [DOI] [PubMed] [Google Scholar]
- Nathan SS, Huvos AG, Casas-Ganem JE, Yang R, Linkov I, Sowers R, DiResta GR, Gorlick R, Healey JH. Tumor interstitial fluid pressure may regulate angiogenic factors in osteosarcoma. J Orthop Res. 2008;26:1520–1525. doi: 10.1002/jor.20633. [DOI] [PubMed] [Google Scholar]
- Nathan SS, Pereira BP, Zhou YF, Gupta A, Dombrowski C, Soong R, Pho RWH, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Elevated expression of Runx2 as a key parameter in the etiology of osteosarcoma. Mol Biol Rep. 2009;36:153–158. doi: 10.1007/s11033-008-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM, Stiller CA, Nectoux J. International variations in the incidence of childhood bone tumours. Int J Cancer. 1993;53:371–376. doi: 10.1002/ijc.2910530305. [DOI] [PubMed] [Google Scholar]
- Petkovic I, Cepulic M. Chromosome aberrations in two osteosarcoma. Cancer Genetics and Cytogenetics. 1996;158:91. [Google Scholar]
- Picci P, Sangiorgi L, Bahamonde L, Aluigi P, Bibiloni J, Zavatta M, Mercuri M, Briccoli A, Campanacci M. Risk factors for local recurrences after limb-salvage surgery for high-grade osteosarcoma of the extremities. Ann Oncol. 1997;8:899–903. doi: 10.1023/a:1008230801849. [DOI] [PubMed] [Google Scholar]
- Polito P, Cin P, Debiec-Rychter M, et al. Human solid tumors: cytogenetic techniques. In: Swansbery J, editor. Cancer cytogeneticsmethods and protocols. Humana Press; Totowa, New Jersey: 2003. pp. 135–50. [DOI] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- Settakorn J, Lekawanvijit S, Arpornchayanon O, Rangdaeng S, Vanitanakom P, Kongkarnka S, Cheepsattayakorn R, Ya-In C, Thorner PS. Spectrum of bone tumors in Chiang Mai University Hospital, Thailand according to WHO classification 2002: A study of 1,001 cases. J Med Assoc Thai. 2006;89:780–787. [PubMed] [Google Scholar]
- Stiller CA, Bunch KJ, Lewis IJ. Ethnic group and survival from childhood cancer: report from the UK Children's Cancer Study Group. Br J Cancer. 2000;82:1339–1343. doi: 10.1054/bjoc.1999.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Kobayashi N, Zmudzka BZ, Ito S, Wakamatsu K, Yamaguchi Y, Korossy KS, Miller SA, Beer JZ, Hearing VJ. UV-induced DNA damage and melanin content in human skin differing in racial/ethnic origin. FASEB J. 2003;17:1177–1179. doi: 10.1096/fj.02-0865fje. [DOI] [PubMed] [Google Scholar]
- Taylor WF, Ivins JC, Dahlin DC, Edmonson JH, Pritchard DJ. Trends and variability in survival from osteosarcoma. Mayo Clin Proc. 1978;53:695–700. [PubMed] [Google Scholar]
- Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J Biol Chem. 2008;283:27585–27597. doi: 10.1074/jbc.M802453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Thomas DM, Johnson SA, Sims NA, Trivett MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR, Holloway AJ, Diyagama D, Grim JE, Clurman BE, Bowtell DDL, Lee JS, Gutierrez GM, Piscopo DM, Carty SA, Hinds PW. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, Alt FW, McMahon AP, Orkin SH. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, Li B. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins RM, Cullen JW, Odom L, Jamroz BA, Cullen PM, Fink K, Peck SD, Stevens SL, Kelly CM, Camozzi AB. Superior survival in treatment of primary nonmetastatic pediatric osteosarcoma of the extremity. Ann Surg Oncol. 2003;10:498–507. doi: 10.1245/aso.2003.03.061. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E, Glotzer DJ, Hartmann C, Thomas D, Fukai N, Soker S, Olsen BR. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech Dev. 2001;106:97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]