Abstract
The stringent regulation of cell cycle progression helps maintain genetic stability in cells. MicroRNAs (miRNAs) are critical regulators of gene expression in diverse cellular pathways, including developmental patterning, hematopoietic differentiation, and antiviral defense. Here we show that two c-Myc-regulated miRNAs, miR-17 and miR-20a, govern the transition through G1 in normal diploid human cells. Inhibition of these miRNAs leads to a G1 checkpoint due to an accumulation of DNA double strand breaks, resulting from premature temporal accumulation of the E2F1 transcription factor. Surprisingly, gross changes in E2F1 levels were not required to initiate the DNA damage response and checkpoint, as these responses could occur with a less than two-fold change in E2F1 protein levels. Instead, our findings indicate that the precise timing of E2F1 expression dictates S phase entry and that accurate timing of E2F1 accumulation requires converging signals from the Rb/E2F pathway and the c-Myc-regulated miR-17 and miR-20a miRNAs to circumvent a G1 checkpoint arising from the untimely accumulation of E2F1. These data provide a mechanistic view of miRNA-based regulation of E2F1 in the context of the emerging model that miRNAs coordinate the timing of cell cycle progression.
Keywords: checkpoint, DNA damage, E2F1, miRNA
Strictly monitoring cell cycle progression is essential for maintaining genetic integrity in cells. One critical regulatory step in G1, the restriction point (R), commits cells to undergo replication. Major events associated with R include hyperphosphorylation of the retinoblastoma (Rb) protein and derepression of the E2F transcription factors (Dimova & Dyson, 2005). E2F1, E2F2, and E2F3a act as transcriptional activators on promoters of gene products required for DNA replication, like DNA polymerase α and thymidine kinase (DeGregori et al., 1995). These proteins are required for G1 to S phase progression as demonstrated by the observation that mouse embryo fibroblasts deleted for all three genes are defective in S phase entry (Wu et al., 2001). Genetic stability of cells is also maintained by checkpoints, which halt cell cycle progression following an encountered stressor, like DNA damage. Data have shown that inappropriate oncogene activation leads to checkpoint activation by signaling a DNA damage response (DDR) (Halazonetis et al., 2008). Accordingly, Rb inactivation or ectopic expression of one of the activator E2F proteins, E2F1, activates a G1 checkpoint (Frame et al., 2006; Lomazzi et al., 2002) by generating a DDR (Bartkova et al., 2005; Hong et al., 2008; Pickering & Kowalik, 2006; Powers et al., 2004; Rogoff et al., 2004).
Rb regulated growth control is crucial to controlling cell proliferation, which may explain why it is disrupted in most, if not all, solid tumors. Emerging data indicates that mis-expressed microRNAs (miRNAs) can promote cancer progression by functioning as tumor suppressors or oncogenes (Calin & Croce, 2006). One commonly overexpressed miRNA cluster is the miR-17-92 cluster, which encodes six mature miRNAs (Mendell, 2008), and is directly transactivated by c-Myc, a transcription factor that accumulates early in G1 (O’Donnell et al., 2005). Although the timing of miR-17-92 expression suggests that miRNAs generated from this cluster may function in G1, the physiological role of these miRNAs in G1 is unclear.
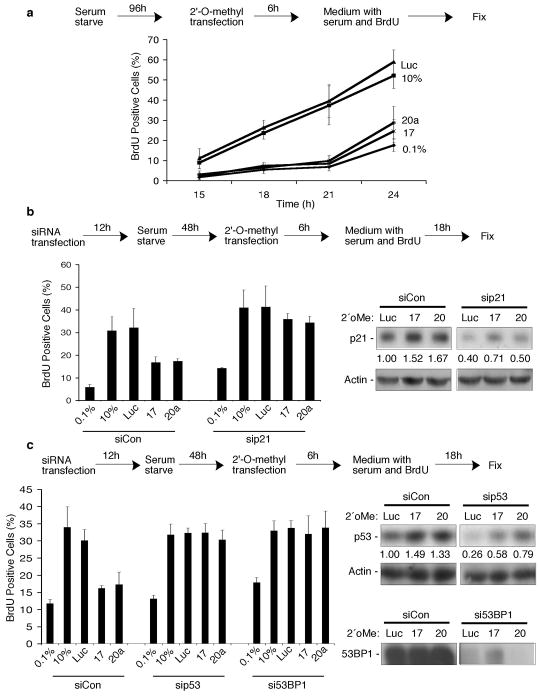
Given that c-Myc accumulates early in G1 and is then degraded as cells pass through R (Persson et al., 1985; Rabbitts et al., 1985) and that expression of miR-17-92 temporally correlates with c-Myc expression (O’Donnell et al., 2005), we determined whether two mature miRNAs derived from that cluster, miR-17 and miR-20a, play physiological roles in G1 cell cycle progression. Serum-starved normal human fibroblasts were transfected with 2′-O-methyl oligonucleotides complementary to either miRNA sequence (2′oMe-miR17; 2′oMe-miR20a) (O’Donnell et al., 2005) or a control 2′-O-methyl oligonucleotide (2′oMe-Luc) (Hutvagner et al., 2004). Following transfection, growth medium containing serum and BrdU was added to cells and BrdU incorporation was used to measure the ability of the cells to progress from G1 to S. While the BrdU-positive population of untransfected (denoted as 10%) or 2′oMe-Luc transfected cells increased over time, inhibition of miR-17 or miR-20a resulted in reduced levels of BrdU incorporation, similar to levels seen in control cells maintained in medium with reduced serum (Fig 1a; 0.1%). These results suggest that miR-17 and miR-20a regulate the G1 to S phase transition of the cell cycle.
Figure 1. Inhibition of miR-17 or miR-20a results in a G1 checkpoint.
(a) Mid-passage normal human embryonic lung fibroblasts were serum-starved in media containing 0.1% serum for 96h and then mock transfected (0.1%, 10%) or transfected with 2′-O-methyl oligonucleotides complementary to miR-17 (17), miR-20a (20a), or luciferase (luc) using Oligofectamine (Invitrogen, Carlsbad, CA, USA) at a final concentration of 20 nM. After 6h, transfection media was replaced with media containing 0.1% serum and 10uM BrdU (0.1%), or media containing 10% serum and 10uM BrdU (10%, Luc, 17, or 20a). Cells were fixed and immunostained for BrdU incorporation at the indicated time points and the percentage of BrdU positive cells is graphed. At least 200 cells per treatment were counted positive if they had 3 or more foci per cell. Error bars indicate the s.d. for at least three independent experiments. (b–c) Fibroblasts were transfected with the indicated siRNAs using Lipofectamine 2000 (Invitrogen) at a final concentration of 60 nM prior to serum-starvation for 48h. During the Oligofectamine: RNA complex reaction, serum-starved cells were trypsinized and soybean trypsin inhibitor was added to inactivate the trypsin. Cells were pelleted and resuspended in DMEM containing 0.1% FBS. The transfection complex was mixed with cells and plated. After 6h, transfection media was replaced with medium containing BrdU until the indicated time points. Protein depletion by siRNA transfection is quantified using Multianalyst software (BioRad, Hercules, CA) of immunoblot analysis.
We then asked whether the reduction in BrdU incorporation upon miR-17 or miR-20a inhibition was due to activation of a G1 checkpoint. To ask this question, we depleted cells of key mediators of G1 arrest (Frame et al., 2006) and assessed whether cell cycle progression was still impeded by inhibition of miR-17 or miR-20a. Inhibiting the miRNAs resulted in the expected reduction of BrdU positive cells when cells were transfected with a control siRNA (siCon). However, siRNA-induced depletion of the cyclin-dependent kinase inhibitor, p21 (sip21; Fig. 1b), alleviated the block in cell cycle progression observed when miR-17 or miR-20a was inhibited to levels similar to untransfected (10%) or 2′oMe-Luc transfected cells (Fig. 1b). We also examined the potential contribution of the p53 transcription factor and the DNA damage sensing and checkpoint signaling protein, 53BP1, to the block in cell cycle progression initiated by miR-17 or miR-20a inhibition. Similar to depleting p21, transfecting sip53 or si53BP1 (Fig. 1c) increased the percentage of cells entering S phase when miR-17 or miR-20a was inhibited (Fig. 1c). Because of the large molecular weight of 53BP1, an actin loading control was not feasible. For this reason, transfected cells were also immunostained for 53BP1 (SI Fig. 1). Taken together, these results suggest that miR-17 and miR-20a govern G1 cell cycle progression, and that disrupting this regulation leads to activation of a G1 checkpoint involving p21, p53, and 53BP1.
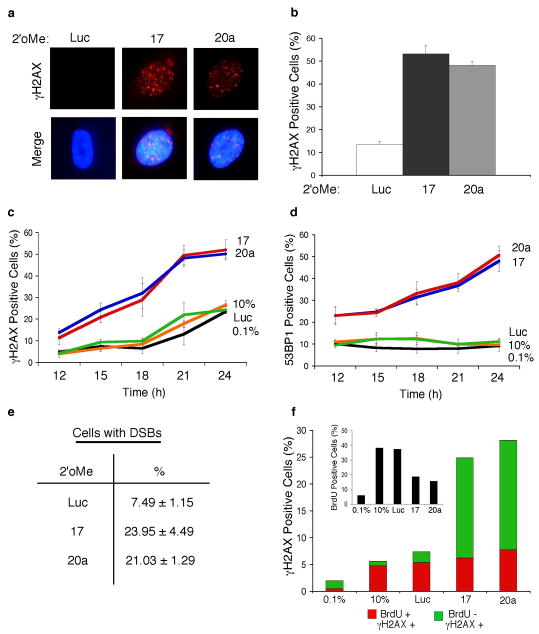
Data have shown that disrupting the control of oncoproteins involved in regulating G1 progression can lead to a cell cycle checkpoint (Dimri et al., 2000; Frame et al., 2006; Lomazzi et al., 2002) due to an accumulation of DNA double strand breaks (DSBs) (Bartkova et al., 2005; Pickering & Kowalik, 2006). Hallmarks of DNA damage are the phosphorylation of the H2AX histone protein, termed γH2AX (Rogakou et al., 1998), and localization of γH2AX adjacent to sites of DNA breaks, which can be visualized as discrete foci by immunostaining (Rogakou et al., 1999). We initially determined whether inhibition of miR-17 or miR-20a resulted in γH2AX foci formation in cycling cells. Transfecting 2′oMe-miR17 or 2′oMe-miR20a into cycling cells resulted in a 4–5 fold increase in the number of cells containing γH2AX foci (Fig. 2a–b). To determine whether γH2AX foci formation correlated with the reduction in BrdU incorporation, cells were treated as in Fig 1a, and then immunostained for markers of DNA damage. Similar to what was observed in cycling cells, the percentage of cells containing γH2AX foci was higher during the duration of serum-stimulation when miR-17 or miR-20a was inhibited (Fig. 2c). Given that 53BP1 localizes at DSB sites (Schultz et al., 2000) and that we found a role for it in the G1 checkpoint associated with inhibiting miR-17 or miR-20a, we examined 53BP1 foci formation following miRNA inhibition. The percentage of cells containing 53BP1 foci increased following serum and BrdU addition when miR-17 or miR-20a was inhibited (Fig. 2d; SI Fig. 1). We then asked whether inhibiting the miRNAs leads to DSB accumulation by neutral comet assay, a direct measure of DSBs (Olive et al., 1991; Olive et al., 1992). Indeed, an ~3 fold increase in DSB-positive cells was observed following inhibition of miR-17 or miR-20a (Fig. 2e). To determine whether progression into S phase was required to generate the DDR when the miRNAs were inhibited, we treated cells as in Fig. 1a and then co-immunostained cells for γH2AX and BrdU. Of the γH2AX foci containing cells, the majority (75.0% for 2′oMe-miR17; 72.4% for 2′oMe-miR20a) did not costain with BrdU (Fig. 2f and SI Fig. 2). As expected, untransfected or 2′oMe-Luc transfected cells, had low levels of γH2AX foci, and most (84.6% for 10%; 73.3% for Luc) of these γH2AX foci containing cells costained with BrdU. Taken together, these data show that inhibiting miR-17 or miR-20a results in a DDR that correlates with the activation of a G1 checkpoint.
Figure 2. Inhibition of miR-17 or miR-20a leads to a DNA damage response.
(a) Cycling fibroblasts were transfected with 2′oMe-miR17, 2′oMe-miR20a or 2′oMe-Luc and immunostained for γH2AX foci (Pickering & Kowalik, 2006). (b) Quantification of panel a. (c–f) Cells were treated as in Fig 1a and the percentage of cells positive for γH2AX foci (c) 53BP1 foci (Frame et al., 2006; Pickering & Kowalik, 2006) (d) neutral comet assay tail formation (Pickering & Kowalik, 2006) (e) is shown. (f) The total percentage of cells positive for γH2AX foci is shown at 21h post serum addition. The percentage of cells that costain (red) and do not costain with BrdU (green) is shown. The percentage of cells in S phase is shown on the inset graph.
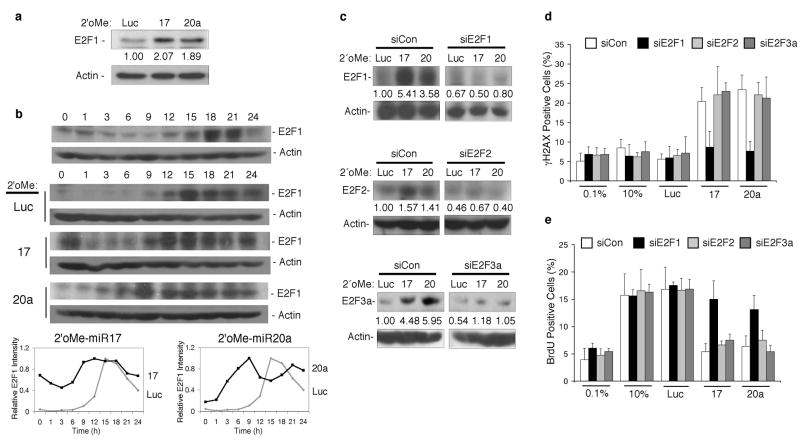
One of the demonstrated targets of miR-17 and miR-20a is the E2F1 transcript (O’Donnell et al., 2005) (Fig 3a). Given that E2F1 protein is expressed late in G1 despite E2F1 mRNA accumulation earlier in G1 (O’Donnell et al., 2005), we asked whether miR-17 and miR-20a regulate the precise timing of E2F1 expression as cells progress through R. The temporal accumulation of E2F1 levels was examined in extracts from serum-stimulated fibroblasts. In untransfected or 2′oMe-Luc transfected cells (Fig 3b), E2F1 accumulation peaked 15–18h following serum-stimulation. However, inhibiting miR-17 or miR-20a resulted in earlier (9–12h) E2F1 accumulation, following serum-stimulation (Fig. 3b). Despite the temporal shift in E2F1 accumulation, E2F1 levels were not appreciably higher when miR-17 or miR-20a was inhibited compared to 2′oMe-Luc transfected control cells (Fig. 3b). However, the timing of E2F1 accumulation suggests that it may play a role in the DDR and G1 checkpoint associated with inhibiting miR-17 or miR-20a.
Figure 3. Inhibition of miR-17 or miR-20a leads to an E2F1-associated DDR and G1 checkpoint.
(a) The levels of E2F1 protein in random cycling fibroblasts 24 h post-transfection with 2'oMe-miR17, 2'oMe-miR20a or 2'oMe-Luc. The fold change in E2F1 levels is indicated. (b) E2F1 levels were examined by immunoblot analysis of serum starved, G0 fibroblasts that were released into complete media (top) and of cells treated as shown in Figure 1a (middle). The relative intensity of E2F1 signal in 2'oMe-Luc-transfected cells compared to 2'oMe-miR17 (bottom left) or 2'oMe-miR20a (bottom right) is shown. (c) Fibroblasts were synchronized and treated as shown in Figure 1b. At 18 h, cells were harvested and the level of protein depletion is shown for the indicated protein. (d and e) Cells were treated as shown in Figure 1b, immunostained, and the percentage of cells positive for (d) gH2AX foci or (e) BrdU incorporation is shown.
Overexpression data from our laboratory and others have shown that E2F1 deregulation leads to a DDR (Bartkova et al., 2005; Hong et al., 2008; Pickering & Kowalik, 2006; Powers et al., 2004; Rogoff et al., 2004) and G1 checkpoint (Frame et al., 2006; Lomazzi et al., 2002). Therefore, we asked whether the DDR and G1 checkpoint that we observed following inhibition of miR-17 or miR-20a were due to deregulation of physiological levels of E2F1. To assess whether E2F1 was involved in signaling a DDR and G1 checkpoint following miR-17 or miR-20a inhibition, the percentage of γH2AX or BrdU positive cells was determined in cells depleted for E2F family members (Fig. 3c). Although the E2F family consists of eight genes, we focused on E2F1, E2F2, and E2F3a because collectively they mediate the G1 to S transition (Dimova & Dyson, 2005) and because miR-17 and miR-20a regulate their protein levels (O’Donnell et al., 2005; Sylvestre et al., 2007). Depleting E2F2 or E2F3a did not reduce γH2AX foci formation following inhibition of miR-17 or miR-20a beyond controls (Fig. 3d). However, depleting E2F1 diminished the percentage of γH2AX positive cells resulting from miR-17 or miR-20a inhibition to levels in untransfected or 2′oMe-Luc transfected controls (Fig. 3d). Depleting E2F1 also alleviated the G1 checkpoint associated with inhibiting miR-17 or miR-20a, whereas depleting E2F2 or E2F3a had little effect (Fig. 3e), despite the increase in E2F2 and E2F3a levels (Sylvestre et al., 2007) (Fig. 3c). These data implicate these miRNAs in delaying the accumulation of E2F1 to prevent an E2F1-associated DDR that leads to a G1 arrest.
Here we show that temporal regulation of cell cycle progression is exerted by two miRNAs, miR-17 and miR-20a. Disrupting miR-17 or miR-20a during G1 progression resulted in premature E2F1 accumulation, leading to a DNA damage-induced G1 checkpoint. These miRNAs maintain low E2F1 protein levels through G1 until increased E2F1 levels are tolerated by the cell at the G1/S boundary. It will be interesting to determine what environmental change(s) occurs during the temporal progression from G1 to S that allows cells to tolerate increased E2F1 levels. Our results are consistent with recent data indicating that E2F activation is “all-or-none” through R (Yao et al., 2008) and suggests a miRNA-mediated mechanism as a regulator of this activation. We and others have previously shown that marked changes in E2F1 levels by overexpression or Rb inactivation leads to a DDR (Bartkova et al., 2005; Pickering & Kowalik, 2006; Powers et al., 2004; Rogoff et al., 2004) and subsequent G1 arrest (Frame et al., 2006; Lomazzi et al., 2002). Interestingly, inactivation of the miRNAs in random cycling cells led to only a ~2-fold induction in E2F1 (Fig. 3a), but this inappropriately timed increase in E2F1 was sufficient to induce a DDR-induced checkpoint (Fig 2a, b, e). Given that E2F1 levels accumulate during normal progression into S (Fig. 3b) and that inhibition of the miRNAs did not lead to gross changes in E2F1 levels, but rather, disrupted the timing of E2F1 accumulation, we suggest that the strict timing of E2F1 accumulation may serve as a sensitive gauge for a cellular alarm system, alerting the cell to potentially oncogenic changes that culminate in E2F1 accumulation at a prohibited time.
It remains to be determined what the long-term consequence(s) of inhibiting these miRNAs is to an untransformed cell. Data have suggested that inhibiting the miRNAs in cancer cells results in apoptosis (He et al., 2005; Matsubara et al., 2007; Sylvestre et al., 2007). Another possibility is that long-term inhibition of these miRNAs in normal cells with intact checkpoint functions may prolong G1 arrest, and could lead to senescence, given that E2F1 overexpression can generate this phenotype (Dimri et al., 2000).
Consistent with the idea that there are multiple levels of regulation during cell cycle transitions, a recent study shows that a related miRNA, miR-106b, promotes G1 exit by directly down-regulating the p21 cyclin-dependent kinase inhibitor (Ivanovska et al., 2008). Additionally, E2F1 and E2F3 contribute to the regulation of the miR-17-92 cluster (Woods et al., 2007), although this likely occurs following R, when E2F1 and E2F3 are transcriptionally active. These observations, together with our results, highlight the importance of stringently regulating G1 progression on (at least) three different levels and raise the possibility that miRNAs may regulate other cell cycle transitions. Our findings fit the emerging concept that miRNAs “fine-tune” gene expression to precisely regulate the temporal expression of target genes and provide a novel mechanism whereby miRNAs contribute to a cellular alarm system, consisting of a DDR and G1 checkpoint, that becomes activated upon errant timing of oncoprotein expression.
Supplementary Material
Acknowledgments
Supported in parts by research grants from the March of Dimes Foundation (6-FY06-344) and the NIH (AI0766189 and 5 P30 DK32520). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental Information is available at Oncogene’s website.
References
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. E2F-1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Mol Cell Biol. 1995;15:4215–24. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–26. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20:273–85. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene. 2006;25:3258–66. doi: 10.1038/sj.onc.1209352. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Paulson QX, Johnson DG. E2F1 and E2F3 activate ATM through distinct mechanisms to promote E1A-induced apoptosis. Cell Cycle. 2008;7:391–400. doi: 10.4161/cc.7.3.5286. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomazzi M, Moroni MC, Jensen MR, Frittoli E, Helin K. Suppression of the p53- or pRB-mediated G1 checkpoint is required for E2F-induced S-phase entry. Nat Genet. 2002;31:190–4. doi: 10.1038/ng891. [DOI] [PubMed] [Google Scholar]
- Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Olive PL, Wlodek D, Banath JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 1991;51:4671–6. [PubMed] [Google Scholar]
- Olive PL, Wlodek D, Durand RE, Banath JP. Factors influencing DNA migration from individual cells subjected to gel electrophoresis. Exp Cell Res. 1992;198:259–67. doi: 10.1016/0014-4827(92)90378-l. [DOI] [PubMed] [Google Scholar]
- Persson H, Gray HE, Godeau F. Growth-dependent synthesis of c-myc-encoded proteins: early stimulation by serum factors in synchronized mouse 3T3 cells. Mol Cell Biol. 1985;5:2903–12. doi: 10.1128/mcb.5.11.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–55. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG. E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res. 2004;2:203–14. [PubMed] [Google Scholar]
- Rabbitts PH, Watson JV, Lamond A, Forster A, Stinson MA, Evan G, et al. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. Embo J. 1985;4:2009–15. doi: 10.1002/j.1460-2075.1985.tb03885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rogoff HA, Pickering MT, Frame FM, Debatis ME, Sanchez Y, Jones S, et al. Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol. 2004;24:2968–77. doi: 10.1128/MCB.24.7.2968-2977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–90. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–62. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–82. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.