Abstract
Breast tumor volume measured on MRI has been used to assess response to neoadjuvant chemotherapy. However, accurate and reproducible delineation of breast lesions can be challenging, since the lesions may have complicated topological structures and heterogeneous intensity distributions. In this article, the authors present an advanced computerized method to semiautomatically segment tumor volumes on T1-weighted, contrast-enhanced breast MRI. The method starts with manual selection of a region of interest (ROI) that contains the lesion to be segmented in a single image, followed by automated separation of the lesion volume from its surrounding breast parenchyma by using a unique combination of the image processing techniques including Gaussian mixture modeling and a marker-controlled watershed transform. Explicitly, the Gaussian mixture modeling is applied to an intensity histogram of the pixels inside the ROI to distinguish the tumor class from other tissues. Based on the ROI and the intensity distribution of the tumor, internal and external markers are determined and the tumor contour is delineated using the marker-controlled watershed transform. To obtain the tumor volume, the segmented tumor in one slice is propagated to the adjacent slice to form an ROI in that slice. The marker-controlled watershed segmentation is then used again to obtain a tumor contour in the propagated slice. This procedure is terminated when there is no lesion in an adjacent slice. To reduce measurement variations possibly caused by the manual selection of the ROI, the segmentation result is refined based on an automatically determined ROI based on the segmented volume. The algorithm was applied to 13 patients with breast cancer, prospectively accrued prior to beginning neoadjuvant chemotherapy. Each patient had two MRI scans, a baseline MRI examination prior to commencing neoadjuvant chemotherapy and a 1 week follow-up after receiving the first dose of neoadjuvant chemotherapy. Blinded to the computer segmentation results, two experienced radiologists manually delineated all tumors independently. The computer results were then compared with the manually generated results using the volume overlap ratio, defined as the intersection of the computer- and radiologist-generated tumor volumes divided by the union of the two. The algorithm reached overall overlap ratios of 62.6%±9.1% and 61.0%±11.3% in comparison to the two manual segmentation results, respectively. The overall overlap ratio between the two radiologists’ manual segmentations was 64.3%±10.4%. Preliminary results suggest that the proposed algorithm is a promising method for assisting in tumor volume measurement in contrast-enhanced breast MRI.
Keywords: breast cancer, contrast-enhanced MRI, segmentation, marker-controlled watershed, Gaussian mixture model
INTRODUCTION
Breast cancer is a major public health problem for women in the United States and many other parts of the world. The American Cancer Society estimates that among women in the United States, breast cancer will account for 26% of all new cancer cases and 40 480 deaths in 2008.1
Medical imaging plays a crucial role in breast cancer care, including detection, diagnosis, treatment planning, and therapy response assessment. Mammography is the imaging procedure most widely used to detect and diagnose breast cancer, although it has some limitations.2, 3 Other imaging modalities such as magnetic resonance imaging (MRI),4, 5 sonography,6, 7 and nuclear medicine imaging8, 9 may also be used in appropriate scenarios in conjunction with mammography in patients with known or suspected breast cancer.
Some studies have shown that MRI is superior to x-ray mammography and sonography for determining breast cancer tumor volume.10, 11, 12 Furthermore, contrast-enhanced MRI is gaining increased acceptance in breast tumor evaluation, especially in treatment response assessment.13, 14 Contrast-enhanced MRI has been found to more accurately reflect tumor presence and size after chemotherapy, and breast tumor volume measurements made with contrast-enhanced MRI have been used to assess pathologic response to neoadjuvant chemotherapy.15, 16
Tumor size can be measured manually on two-dimensional (2D) magnetic resonance (MR) images. However, three-dimensional (3D) delineation of tumor volume allows more accurate assessment of lesion extent. Manual measurement of 3D tumor volumes is less reproducible and extremely labor intensive, as it requires delineation of the tumor boundaries in all slices in which the tumor appears. Therefore, computerized techniques are being investigated to improve the accuracy, efficiency, objectivity, and consistency of tumor segmentation in breast images. Some computerized methods have been reported for tumor segmentation on x-ray mammography17, 18, 19 and sonography.20, 21, 22
Partridge et al.23 developed a semiautomated software algorithm for segmenting breast tumors from 3D MRI data using thresholding. A threshold based on the tumor enhancement ratio was applied on a voxel-by-voxel basis within the manually defined volume in their study. Arbach et al.24 also used thresholding to segment the enhancing region from the difference image (computed by subtracting the precontrast image from the postcontrast image). They evaluated three different threshold selections: A constant threshold, a threshold derived from a histogram, and a threshold defined as some percentage of the maximum value in the data. The voxel intensity of an MR image depends on the particular MRI instrumentation and contrast agent used, and thus threshold values must be selected carefully by the user. Moreover, the same tissues may not be enhanced uniformly by the contrast agent, and this also decreases the accuracy of threshold-based methods.
Chen et al.25 proposed a fuzzy c-means (FCM) clustering-based method for 3D lesion segmentation in dynamic contrast-enhanced MRI (DCE-MRI) data. They applied the FCM algorithm, an unsupervised learning technique in the pattern recognition field, to the enhancement kinetic curves of each voxel from five time points and partitioned the ROI voxels into two categories: Lesion and nonlesion. Wu et al.26 also analyzed the kinetic curves of voxels in dynamic MRI data and classified them as lesion or nonlesion regions based on the Bayesian theory and Markov random field model. Woods et al.27 used four-dimensional (3D spatial dimension and time dimension) co-occurrence-based texture analysis to segment malignant lesions from DCE-MR images. The textural features, gathered from statistical information regarding the change in voxel intensities over time, were used as the input of a model-free neural network-based classifier in their study. In these methods, complete series of dynamic data are required to obtain the voxel intensity changes over the entire DCE-MRI. If used in a preprocessing step, the registration of the serial images may improve the performance of the algorithms by lessening the effects of motion and breathing during acquisition of the serial MR data.25, 26
Without using serial MR data, Liu et al.28 proposed a dynamic-programming-based optimal edge detection method to segment the breast tumor from one volume of contrast-enhanced MRI data. Shi et al.29 also performed their algorithm on a single volume of postcontrast, T1-weighted data to segment breast tumors. Their method was based on 3D level set segmentation within a manually defined cuboid volume of interest. Both dynamic programming and level set methods achieve good segmentation results in isolated tumors with relatively smooth margins, but for scattered small foci of tumor (often seen in treated breast cancer) or spiculated tumors, these methods may be of limited value.
We present a fast and effective marker-controlled watershed segmentation method to semiautomatically delineate breast tumors in sequential contrast-enhanced MR slices. In contrast to the methods mentioned above, a watershed-based method can be applied effectively to spiculated and multifocal tumors. Watershed transform is formally defined in terms of flooding simulations30 and is widely used for image segmentation in various fields.31, 32, 33, 34 This method is intuitive, computationally efficient, and effective for completely dividing an image into separated regions.35, 36 The main problem with this method is that there are often a lot of local minima, which lead to oversegmentation of the image.36, 37 To overcome this problem, several strategies have been proposed, including region merging,38, 39 introducing a priori knowledge,37 and marker-controlled segmentation.40, 41, 42 In our study, we use a marker-controlled watershed segmentation approach because of its ability to detect a target boundary between markers inside and outside the tumor even if there are no clearly defined edges between two markers. The marker-controlled strategy was proposed by Meyer and Beucher40 and is based on the concept that the immersion of the gradient surface begins only from selected markers and not from all minima. If the markers are given, the image is segmented so that each segmented part contains a marker, and between any two markers a boundary line must exist. So the determination of the markers is crucial in the marker-controlled method.
In applying the watershed method, we focused mainly on determining internal markers that are completely within tumor tissue and external markers that are completely free of pixels containing tumor tissue. The rest of the marker-controlled watershed transform, i.e., modifying the gradient image and searching for the watershed lines through flooding simulation, follows the general strategy and is similar for all applications.40, 41, 42
PATIENT DATA
MR imaging was performed for patients who underwent neoadjuvant chemotherapy in a prospective study at our center. Thirteen patients ranging in age from 26 to 57 years (42.8±9.9 years, mean±stddev) were recruited to the study. All patients had needle biopsy-proven adenocarcinoma of the breast. MR scans were derived before beginning and 1 week after receiving the first dose of chemotherapy for all patients. MR imaging was done with the patient in the prone position in a 1.5 T MR system (Sigma, Lightening, or Excite, GE Medical Systems, Milwaukee, WI) using a dedicated surface breast coil. T1-weighted 3D fat-suppressed gradient-echo sequences (repetition time=5.420–7.536 ms, echo time=2.680–4.200 ms, flip angle=10°) were performed before and after a rapid bolus injection of 0.1 mmol∕kg gadopentetate dimeglumine (Magnevist, Berlex Laboratories, Wayne, NJ) delivered through an in-dwelling IV catheter. The scans were performed in the sagittal direction with 2–3 mm thick contiguous slices. The in-plane pixel spacing ranged from 0.7813 to 0.9375 mm, and each slice consisted of 256×256 pixels.
In our study, the baseline and the 1 week postchemotherapy MR imaging data from the above-mentioned study were used to develop and validate a segmentation algorithm for delineating breast tumors on sagittal T1-weighted, fat-suppressed, and contrast-enhanced images.
METHOD
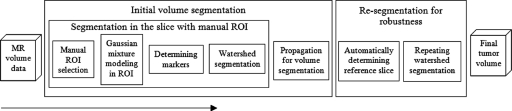
Tumor segmentation was implemented on a slice-by-slice basis. First, an ellipse-shaped region of interest (ROI) that included the entire tumor was manually drawn on a slice. A pair of thresholds was derived from this ROI using a Gaussian mixture model to generate internal and external markers. Then the tumor contour in the same slice was acquired from watershed segmentation. To automatically segment the tumor in other slices, this contour was then extended to form ROIs in the adjacent slices in both directions. The thresholds for markers were also passed to the adjacent slices and the tumor was segmented in these slices. This propagation process was only stopped when no tumor was found in the adjacent slice. After tumor contours were obtained in all slices, the slice with the largest tumor area was automatically chosen as a reference slice to recalculate the thresholds for markers. Finally, the watershed segmentation was automatically performed again in all slices using new thresholds, and the final 3D segmentation result was obtained. The purpose of segmenting tumor twice is to reduce the effect of manual initialization and improve the robustness of the algorithm. Details of these procedures are given in the following sections, and the flowchart of the overall implementation is shown in Fig. 1.
Figure 1.
Schematic diagram of the method.
ROI selection
A user interface developed in-house using IDL (Interactive Data Language, Boulder, CO) allowed a human operator to easily browse the MR volume data and draw an ellipse on any slice using the computer mouse. The inside of this ellipse served as the ROI in our work. For the initial delineation of the ROI, a slice was chosen in which the tumor appeared larger and contrasted to a greater degree with surrounding tissues than it did in other slices. This was to ensure that the ROI propagated to the next slice could cover the whole tumor in that slice. To help eliminate the surrounding nontumor tissue from the ROI, the ellipse was drawn so as to be as small as possible while encompassing the whole tumor. These guidelines for ROI delineation were used to avoid extreme instances of ROI selection. Figure 2a shows an example of a manually selected ROI. Different initial ROIs may produce different segmentation results, an issue that will be further addressed in this article. If a patient had multiple lesions and these lesions were far away from each other, an ROI was given for each lesion and the lesions were segmented separately.
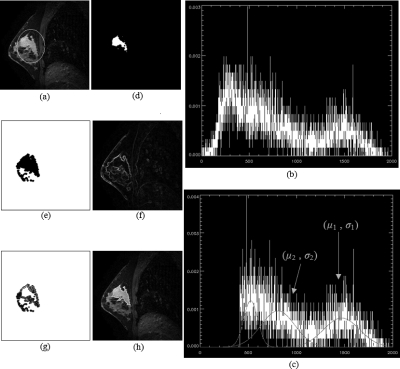
Figure 2.
Illustration of the marker-controlled watershed segmentation in a single slice where the ROI was manually drawn. (a) Original sagittal slice of MR image with manually drawn ROI (white ellipse) on it. (b) Image intensity histogram of the region inside ROI from (a). X axis is image intensity, and Y axis is normalized frequency of the intensity. (c) Truncated histogram of (b) and the three Gaussian curves representing three different Gaussian components from modeling. μ1 and σ1 are the mean value and standard deviation of the tumor distribution. μ2 and σ2 are the parameters of the Gaussian component next to the tumor class. (d) Extracted internal marker (white region) of image (a). (e) Extracted external marker (white region) of image (a). (f) Gradient image of (a). (g) Internal and external markers (white regions) from (d) and (e) are superimposed upon the gradient image from (f). (h) Tumor segmentation result of image (a). The white contour represents the segmented tumor boundary.
Internal and external markers
The way in which markers are determined has a strong impact on the marker-controlled method. The watershed transform automatically finds an optimal object boundary between two markers, and the markers define a search space for detection of the target boundary. If this search space (i.e., the space between markers) is too large, the algorithm may find a false edge. If the search space is too small, it means the markers are likely too large and may mask some parts of the true edge. In some applications of marker-controlled watershed methods in other fields, the markers were derived using thresholding41, 43 or distance transformation.32, 42 The distance transform is not suitable for use in breast cancer because of the irregular shapes of tumors, and fixed thresholding is inadequate for use in MR images because voxel intensities vary between scans.
In this paper, we applied a Gaussian mixture model to the histogram of the given ROI in the image to obtain an upper threshold Tupper and a lower threshold Tlower. Then, the internal markers were derived as the areas where the pixel intensities were larger than Tupper inside the ROI, and the external markers were the union of the whole region outside the ROI and lower intensity parts (lower than Tlower) inside the ROI. The process is described in detail below.
Gaussian mixture modeling
First, a histogram of the image inside the ROI was generated [Fig. 2b]. A certain portion of the histogram was assumed to be mainly contributed by tumor. Our goal was to use an algorithm to automatically find the tumor portion of this histogram to determine the dual thresholds Tupper and Tlower. Assuming that pixel intensities for each tissue accord with Gaussian distribution, we used a Gaussian mixture model to find the tumor portion of the histogram. Mixtures of distributions have been used in classifying brain MRI into different tissue types44, 45, 46 and extracting lung tumor volumes from positron emission tomography.47 The Gaussian mixture model assumes that the observed distribution, in our case the distribution of image intensities inside an ROI, can be expressed as a mixture of Gaussian distributions representing different tissues and also relies on an optimization technique such as expectation maximization (EM) to calculate maximum likelihood (ML) estimates of the parameters of each distribution component. A detailed analysis of mixture models can be found in the book by McLachlan and Peel,48 and the descriptions of the implementation of Gaussian mixture modeling are also available.45, 47 After applying this procedure, we could obtain the mean pixel value (μ) and standard deviation (σ) of each Gaussian distribution and also the distribution percentage of each one. (The distribution percentage represents how many of the pixels belong to the Gaussian component.)
To apply the Gaussian mixture model algorithm, it is necessary to know the number of Gaussian components in the mixed distribution. Liang et al.45 used the minimum description length (MDL) to select the optimal number of components for classifying brain images into different tissue types. However, the use of such information-based methods is time consuming and sometimes results in instability.47 Moreover, for our purposes it was only necessary to identify the tumor class from the histogram and not to precisely classify every type of tissue included in the ROI. Therefore, we used a fixed number of 3 to define the number of Gaussian distributions in the histogram. We chose this number because there are usually three different tissues—tumor, parenchyma, and muscle—in a given ROI. If there are less than three tissues in the ROI, for example only two, then the result of Gaussian mixture modeling using three classes will yield two main Gaussian distributions and one other Gaussian component with a very low distribution percentage, which can be easily removed from further processing. On the other hand, if the given ROI includes four different tissues, fat and the three tissues, three Gaussian components are not enough to describe the histogram. To deal with such a situation, we removed the fat portion from the histogram before Gaussian mixture modeling. The pixel intensity in fat is much lower than that in tumor on fat-suppressed and contrast-enhanced MR images, so it is easy to choose a threshold to truncate the histogram to remove the fat portion. It also would have been possible to use a fourth Gaussian component to account for fat, but this might have increased the computing time and decreased the reliability of the modeling algorithm. The threshold for truncating the histogram was calculated as follows:
| (1) |
where T is the truncating threshold, Imean 10% is the mean intensity value of 10% of pixels with the highest signal intensity within the ROI, and f is a scaling factor, whose value was fixed to 0.25 empirically in this study. Figure 2c shows the truncated histogram and the three Gaussian curves representing three different Gaussian components from modeling.
The Gaussian curve that represents the tumor can be identified easily because the tumor region is always the region with the highest intensity inside the ROI. Blood vessels may exhibit higher intensity in contrast-enhanced MR images, but it is easy to avoid including blood vessels in the selection of the ROI. Therefore the Gaussian distribution component with highest mean value was identified as tumor tissue in the histogram unless this component had a very low (lower than 10%) distribution percentage. If this Gaussian component had a distribution percentage lower than 10% (meaning that two components would be enough for the histogram), the Gaussian component of the next highest intensity was chosen to represent the tumor. The mean value and standard deviation of the Gaussian distribution for tumor were defined as μ1 and σ1, respectively. The Tupper and Tlower could be calculated easily from μ1 and σ1. However, in some cases, the pixel intensities of parenchyma or muscle may be very close to the pixel intensity of tumor. In such cases, another Gaussian curve must be considered, which is next to the tumor class and has a distribution percentage not lower than 10%. The mean value and standard deviation of this Gaussian curve were defined as μ2 and σ2, respectively [Fig. 2c]. Then the upper threshold Tupper was calculated as
| (2) |
Similarly, the lower threshold Tlower was
| (3) |
Determining markers
Finally, the internal and external markers were derived from the dual thresholds Tupper and Tlower. The two markers in fact constitute a binary mask of the image. The internal marker in our study was the area inside the ROI where the pixel intensities were higher than Tupper [Fig. 2d]. And the external marker was the union of the whole region outside the ROI and the area inside the ROI where the pixel intensities were lower than Tlower [Fig. 2e]. If the given ROI included a spiculated or multifocal tumor, then there might be multiple internal markers in one slice. If the tumor had holes inside it, then there might be multiple external markers.
Marker-controlled watershed segmentation
Once the internal and external markers are determined, the rest of the procedure is straightforward: A gradient image is obtained from the original image (in this paper, the Sobel operator was used on the smoothed image to acquire the gradient image) [Fig. 2f], and this gradient image is modified using a grayscale reconstruction algorithm41 in order to ensure the local minima only occur in the regions of the markers.41 Then, the watershed transform is applied to the modified gradient image to obtain the final contour in the belt between the markers [Fig. 2g]. No supervision is needed to perform the final segmentation, and the parameterization controlling the segmentation is concentrated in the marker construction step where it is easier to control. Figure 2h shows an example of segmented tumor on the slice in which the manual ROI was given. In this case, the intensity distribution of tumor was well separated from the surrounding tissues as shown in the histogram [Fig. 2c], so the searching space between internal and external markers could be very narrowed and was already quite close to the tumor boundary [Fig. 2g].
Volume segmentation by propagation
In the procedures described above, only one MR imaging slice is processed for tumor segmentation. To achieve complete volume delineation, the contours in all slices need to be acquired. In this study, we used a propagation strategy42 to segment the tumor in a slice using the segmentation result from its adjacent slice. Specifically, the segmented tumor region in one slice was morphologically dilated with a round element, and then this dilated region was used as the ROI in the next slice. This procedure was based on the assumption that tumor shape and size do not change dramatically between two adjacent slices. The radius of the round element used for dilation was proportional to the spacing of the adjacent two slices. The dual thresholds Tupper and Tlower of the current segmented slice were also passed to the adjacent slice without any change. Then in the adjacent slice, using the ROI and thresholds, internal and external markers were constructed in the same way as described above, and the watershed transform was performed to segment the tumor. The result in this adjacent slice was then applied to its adjacent slice, and this propagation procedure continued automatically in two directions until no tumor was found (i.e., no pixel in the ROI had intensity greater than Tupper).
Resegmentation for robustness of the algorithm
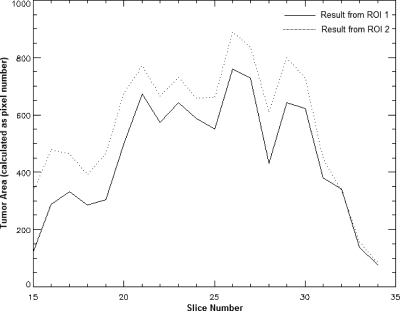
Although the whole volume of the tumor could be delineated by the procedures described above, manually selected ROIs from different slices may produce different segmentation results. For example, in the case shown in Fig. 3, the results were derived with two initial ROIs, ROI 1 and ROI 2 [Fig. 3a]. Although both ROIs generated similar and visually reasonable results [Figs. 3b, 3c], the overlap ratio (explained in Sec. 3F) between these two results was only 76%. This variation was mainly due to the fact that the dual thresholds Tupper and Tlower from different slices are not consistent (in the case shown in Fig. 3, Tupper was 1293.1 in ROI 1 and 1206.7 in ROI 2, and Tlower was 1007.1 in ROI 1 and 944.7 in ROI 2). The difference in thresholds causes variation in the internal and external markers and ultimately affects the automatically generated contour, especially in terms of degree of edge expansion and inclusion of small tumor pieces [Figs. 3b, 3c]. The addition of a layer as small as one voxel thick to the surface of an object may change its volume dramatically, especially if the object is small or has a large surface-to-volume ratio.
Figure 3.
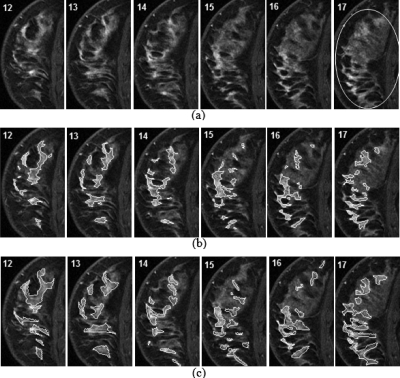
Illustration of the difference between results derived from ROIs in different slices. (a) Consecutive sagittal slices of original postcontrast MRI (from image numbers 20 to 31). Two ROIs were given on slice 22 (ROI 1) and slice 30 (ROI 2), respectively, to generate two different segmentation results. (b) Initial segmentation result from ROI 1. (c) Initial segmentation result from ROI 2.
To make the segmentation results derived from ROIs of different slices more consistent, a further processing step was included in our algorithm. We refer to the segmentation result acquired by previous steps as the “initial segmentation result.” From this initial result, we can find a reference slice, in which the segmented tumor has a larger area than on any other slice. For example, Fig. 4 shows the segmented tumor areas in each slice (calculated as the number of pixels inside the tumor contour) as a function of the slice number from the initial segmentation results in Figs. 3b, 3c. The solid line is from the initial result by ROI 1, and the dotted line is from the initial result by ROI 2. These two curves yielded the same reference slice, i.e., slice 26. Then, a new ROI was derived on this reference slice with a small amount of dilation of the initially segmented tumor area on this slice. The previously described Gaussian mixture modeling was then applied to the histogram of the image intensities inside the new ROI in the reference slice, setting the number of Gaussian components to be 1, because the initial segmentation result only included the tumor class. The final upper threshold and lower threshold for markers were then recalculated as
| (4) |
| (5) |
where μR and σR were the mean value and standard deviation of the Gaussian curve derived from the Gaussian mixture modeling in the new ROI in the reference slice. Because the different manually drawn ROIs led to the same reference slice eventually, the final dual thresholds Tupper_final and Tlower_final for different ROIs were very close and less affected by the selection of the slice for manual ROI delineation. For example, in the case shown in Fig. 3, Tupper_final was 1363.9 for ROI 1 and 1354.1 for ROI 2, and Tlower_final was 1082.5 for ROI 1 and 1070.5 for ROI 2. In each slice, the tumor was resegmented using a marker-controlled watershed transform with final thresholds and the ROI derived by dilation of the initial tumor segmentation. Because of the stability of the final dual thresholds, the internal marker and external marker in each slice were also stable, and therefore variation of the segmentation result, possibly caused by different ROIs, could be reduced. In the case of Fig. 3, the overlap ratio between the final segmentation results from ROI 1 and ROI 2 was 97%.
Figure 4.
Segmented tumor area in each slice (calculated as pixel number inside tumor contour) as a function of slice number. Solid line is from initial segmentation result in Fig. 3b. Dotted line is from initial segmentation result in Fig. 3c.
Postprocessing (e.g., filling of small holes and removal of blood vessels) was performed to further refine the segmentation result. Blood vessels also have high intensities on contrast-enhanced MR images. Although it could be intentionally excluded from the ROI in the slice where the manual operation was performed, that did not ensure that the vessels would be excluded from the ROIs automatically propagated in other slices. Considering that blood vessels have a thin linear shape, morphological opening was applied to the segmented tumor volume with a spherical element whose diameter was 3 mm.
Algorithm evaluation
In this work, two radiologists manually delineated all the tumors and the manual segmentation results were used as the reference to evaluate the algorithm. Blinded to the computer and the other radiologist’s results, each of the radiologists manually outlined the tumors slice by slice on the postcontrast-enhanced images using the computer mouse. The precontrast-enhanced image and the subtraction image (postcontrast image subtracted by precontrast image) were also used by the radiologists as additional information for determining tumor boundaries.
The performance of the proposed segmentation algorithm was assessed by comparing the computer segmentation result with each of the radiologists’ manual result. Besides visual evaluation of the agreement of these two contours, several quantitative measurements have been used in the literature19, 25, 29, 42 to evaluate segmentation. In this study, we used the overlap ratio to quantify how well the computer result and radiologist’s delineation agreed. For a particular scan (volume data), VC denoted the set of tumor voxels from the computer segmentation result, and VR denoted the set of tumor voxels in the radiologist’s segmentation. The volume overlap ratio (VOR) was defined as the intersection of VC and VR over the union of VC and VR, i.e.,
| (6) |
where ∩ is the logical AND operator and ∪ is the logical OR operator. VOR ranges from zero (for no overlap) to 1 (for exact overlap). The volume overlap ratio between the two radiologists’ manual results was also calculated to study the agreement between the two radiologists’ manual delineations.
RESULTS
Figures 56 show two examples of the segmentation results. Original images are overlapped with a manually drawn ROI [Fig. 5a], computer-generated tumor contours [Fig. 5b], and two radiologists-generated tumor contours [Figs. 5c, 5d]. Only six consecutive sagittal slices from the middle part of the volume data are shown in each case and only one radiologist’s manual result was shown in Fig. 6 because of space limitations. The original images with the manually drawn ROI, the segmentation result produced by our algorithm, and the radiologist’s manual result are presented for comparison. These examples visually demonstrate the ability of our algorithm to accurately delineate the tumor boundaries on T1-weighted and contrast-enhanced breast MR images. For the case shown in Fig. 5, the volume overlap ratios between the computer and the two radiologists’ results were 63.3% and 58.5%, respectively, and the VOR between the two radiologists’ results was 64.6%. For the case shown in Fig. 6, the corresponding three VORs were 51.7%, 51.8%, and 55.6%, respectively. Figure 7 shows 3D visualizations of the segmented tumor volumes of the other two cases. The quantitative evaluation results are provided in Table 1. For baseline MRI scans, the VOR was 60.9%±7.7% (mean±stddev) between computer and first radiologist’s segmentation results, 60.5%±11.4% between computer and second radiologist’s results, and 63.6%±10.1% between two radiologists’ segmentation results. There was no statistically significant difference between VOR of computer and first radiologist and VOR of the two radiologists (paired samples t test, p=0.1310). There was no statistically significant difference between VOR of computer and second radiologist and VOR of the two radiologists (p=0.1201). For the follow-up scans obtained 1 week after the first dose of chemotherapy, the VOR was 64.3%±10.4% between computer and first radiologist’s segmentation results, 61.5%±11.6% between computer and second radiologist’s results, and 65.0%±11.1% between the two radiologists’ segmentation results. Again, there was no significant difference between VOR of computer and first radiologist and VOR of the two radiologists (p=0.7598). There was no significant difference between VOR of computer and second radiologist and VOR of the two radiologists (p=0.0649). The overall VORs for a total of 26 MRI studies were 62.6%±9.1%, 61.0%±11.3%, and 64.3%±10.4% between computer and first radiologist, computer and second radiologist, and the two radiologists, respectively. There was no significant difference in the performance of the segmentation algorithm between baseline and 1 week postchemotherapy (paired samples t test, p=0.1107 for VORs between computer and first radiologist and p=0.5715 for VORs between computer and second radiologist). Once manual initialization of the algorithm was done, an average processing time for segmenting one tumor was about 4 s (PC of 3.2 GHz CPU and 3 Gbyte RAM). The computation time depended on a tumor’s size. The mean value and standard deviation of the tumor volumes from computer segmentation were 24.23±26.13 cm3 (mean±stddev, range: 4.42–81.67 cm3) for baseline and 23.05±30.01 cm3 (mean±stddev, range 1.46–102.68 cm3) for follow-up cases.
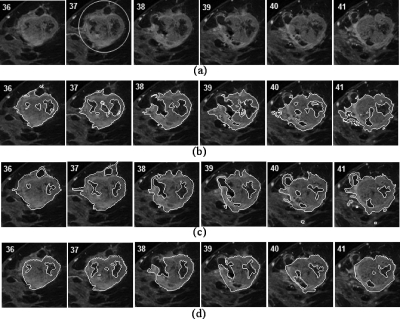
Figure 5.
Tumor segmentation result. (a) Original sequential images containing a malignant lesion. A manually selected ROI was shown in slice 37. (b) Computer-delineated tumor contours. (c) First radiologist’s manual segmentation result. VOR between (b) and (c) was 63.3%. (d) Second radiologist’s manual segmentation result. VOR between (b) and (d) was 58.5%. The VOR between (c) and (d) was 64.6%.
Figure 6.
Tumor segmentation result. (a) The original sequential images containing a malignant lesion. An initial ROI was given in slice 17. (b) Computer segmentation result. (c) First radiologist’s manual segmentation result. VOR between the computer and first manual results for this case was 51.7%. The VOR between first radiologist’s segmentation result and second radiologist’s result (not shown in here) was 55.6%.
Figure 7.
3D visualization of segmented tumor volumes from two cases (a) and (b). Computer segmentation result (upper row) viewed in three different angles and first radiologist’s manual result (lower row) viewed in the corresponding angles. The VORs were (a) 56.5% and (b) 69.9%.
Table 1.
Summary of the performance of the proposed tumor segmentation algorithm. (Com-R1: Comparison between computer and first radiologist’s segmentation results. Com-R2: Comparison between computer and second radiologist’s results. R1-R2: Comparison between the two radiologists’ results.)
| Cases | Comparison | Volume overlap ratio (%) | ||||
|---|---|---|---|---|---|---|
| Mean | Stddev | Max | Min | Median | ||
| Baseline | Com-R1 | 60.9 | 7.7 | 74.4 | 51.7 | 59.9 |
| Com-R2 | 60.5 | 11.4 | 75.4 | 42.0 | 58.5 | |
| R1-R2 | 63.6 | 10.1 | 79.5 | 46.6 | 62.9 | |
| Follow-up | Com-R1 | 64.3 | 10.4 | 78.8 | 48.2 | 66.0 |
| Com-R2 | 61.5 | 11.6 | 78.7 | 44.8 | 61.0 | |
| R1-R2 | 65.0 | 11.1 | 76.9 | 42.5 | 68.6 | |
| Overall | Com-R1 | 62.6 | 9.1 | 78.8 | 48.2 | 63.2 |
| Com-R2 | 61.0 | 11.3 | 78.7 | 42.0 | 59.6 | |
| R1-R2 | 64.3 | 10.4 | 79.5 | 42.5 | 63.7 | |
For a semiautomatic segmentation algorithm, results are often affected by manual selection of an initial ROI. The Gaussian mixture modeling and resegmentation strategy (Sec. 3E) used in our method can help reduce the variation in results caused by manually drawn ROIs. To validate the ability of our algorithm to cope with variations in manual initializations we chose four typical cases [Figs. 8a, 8b, 8c, 8d] as testing samples and manually drew three different ROIs for each case to initiate the segmentation. The testing cases included an isolated and a relatively large tumor [Fig. 8a], a spiculated medium-sized tumor [Fig. 8b, the same case shown in Fig. 3], a scattered multifocal tumor [Fig. 8c], and a tumor with low intensity contrast to the surrounding parenchyma [Fig. 8d]. For each case, we tested the consistency of the segmentation algorithm with three manually selected elliptical ROIs; two were made in the same slice and one in a different slice, all complying with the ROI drawing guidance (Sec. 3A). However, the center position, shape, and size of the ellipses were different. The degree of the differences of the two ROIs in the same slice was case depended. For example, if a tumor was quite isolated and there were no other tissues but fat surrounding the tumor, the variations of the ROI could be relatively large. The slice in which the third ROI was drawn was chosen randomly, but again, the ROI selection should comply with the requirements. We then obtained three segmentation results for each of the four cases. The three segmented volumes were compared with each other in terms of overlap ratio, yielding 3 comparison results for each case, and thus 12 in total. The mean value and standard deviation of the VOR from these 12 comparisons were 0.942 and 0.046, respectively. The high mean value and small standard deviation show that the algorithm is capable of yielding a robust segmentation result.
Figure 8.
Four different patterns of breast lesions for the evaluation of algorithm’s robustness.
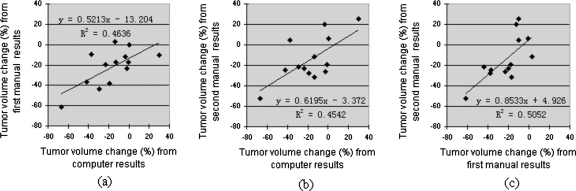
One of the major applications of the tumor volume segmentation is to assist in assessing therapy response by monitoring tumor volume change before and after treatment. In this study, tumor volume changes between baseline and 1 week follow-up studies were calculated using tumor’s baseline and 1 week follow-up volumes obtained by the computer algorithm and the two radiologists’ manual delineations, respectively. Correlations between computer and each of the two manual results were analyzed by using the least-squares regression method. Figure 9 shows the correlation results with scatter diagram, least-squares regression line, and Pearson’s correlation coefficient r (with p value). The correlation coefficient of computer and first radiologist’s results was 0.6809 (p=0.0104), and it was 0.6740 (p=0.0115) for computer and second radiologist. The tumor volume changes computed by the two radiologists’ segmentations were also compared and the correlation coefficient was 0.7107 (p=0.0065) (Fig. 9).
Figure 9.
Correlation between tumor volume changes [(follow-up−baseline)∕baseline∗100%] computed by computer segmentation results and manual segmentation results. Scatter diagram, least-squares regression line, and regression equation are provided. (a) Correlation between computer and first radiologist’s results. Correlation coefficient is r=(R2)1∕2=0.6809. The significance level is p=0.0104. (b) Correlation between computer and second radiologist’s results. Correlation coefficient is r=(R2)1∕2=0.6740. The significance level is p=0.0115. (c) Correlation between first radiologist’s results and second radiologist’s results. Correlation coefficient is r=(R2)1∕2=0.7107. The significance level is p=0.0065.
DISCUSSION
It is difficult to quantitatively compare the performance of our algorithm with the existing segmentation algorithms, because some of them did not provide quantitative results. Moreover, in our study the algorithm was performed and evaluated on T1-weighted, postcontrast-enhanced, and fat-suppressed sagittal MR images, and in other studies algorithms were tested on different types of images. However, the overlap ratio does provide a point of comparison with some of the studies. A study by Yuan et al.19 with digital mammograms and a study by Chen et al.25 with dynamic coronal MR slices both used an overlap ratio threshold of 0.4 to determine the correctness of the segmentation. In our study, all the tumor segmentation results had VOR above 0.4, which suggests that our algorithm performs at least comparably to the others. Shi et al.29 used a similar kind of MRI data that was used in our study and proposed a 3D level set-based method for tumor segmentation. They achieved a high VOR (between computer and radiologist’s manual results) of 0.72±0.08 for prechemotherapy tumors from ten patients and a relatively low VOR of 0.49±0.17 for the same tumors after chemotherapy. They suggested that their method performed worse in postchemotherapy cases because breast masses after therapy are topologically more complicated, whereas before treatment, breast masses have relatively smooth boundaries. Unlike their method, the marker-controlled watershed method used in our study can deal with spiculated and scattered tumors very well. This is illustrated by the fact that the VOR for baseline cases and the VOR for follow-up cases were similar in our study. Besides the segmentation accuracy and consistency, the computation time is a main concern of computer-aided diagnosis (CAD) procedure because a CAD system is intended to aid, not replace, the physician; thus an efficient human-computer interaction is essential. The segmentation speed of watershed-based algorithm presented in our study is superior to that of the level set method.
Although visually the segmentation results were well acceptable, the reason our VORs seem to be low is the spiculated shapes of the tumors. For such tumors, dilation or erosion of a single layer may cause a large change in volume value. To demonstrate this, we did a simple test using the case shown in Fig. 8c, in which the tumor had a large surface-to-volume ratio. We expanded the computer-derived tumor contour by only one pixel (0.78 mm) in each slice (i.e., we dilated the tumor area in each slice with a disk whose radius was equal to the width of one pixel) to obtain a new volume. The VOR between the original tumor volume and the expanded volume was 69.9%. When the tumor area was expanded by the width of two pixels, the VOR between the original volume and the expanded volume was only 54.3%. Voxels along the border of the tumor are likely to suffer from partial volume effects, which may increase the radiologist’s difficulty and uncertainty in manually delineating the tumor contour. This may also explain why the VOR between the two radiologists’ results is also not high. As seen in Table 1, segmentation agreement between our computer method and radiologists’ manual delineations is comparable with the interobserver agreement shown by the two radiologists’ results.
The resegmentation procedure (Sec. 3E) can reduce the effect of initial ROI selection on the segmentation result. But that does not mean that the initial ROI can be drawn in an arbitrary manner. On the contrary, the initial ROI should ensure that the initial segmentation result will be close to the tumor boundary. If there is a large leak or undersegmentation in the initial result, the automatically selected reference slice may not be reliable, and the final result may be skewed. Resegmentation does not necessarily improve the accuracy of our algorithm in a given case; rather, it improves the reproducibility and consistency of the algorithm. For example, some initial results may be closer to the “true volumes,” but the initial result is sensitive to the selection of the ROI.
To test the consistency of computer segmentation results due to different manual initiations of tumor ROIs (Sec. 4), it was difficult to have fixed and quantitative measurements for the variations of manually selected ROIs in terms of their position, shape, and size because there were large differences among different cases. In general, as presented in Sec. 3A, an initial ROI should be given to enclose the entire tumor in a reference slice and to possibly avoid other structures of high intensity. Furthermore, the reference slice should have a clear and relatively large dimension of tumor in it. In accordance with these prerequisites, we selected different initial ROIs to test the robustness of the algorithm.
Volumetric tumor segmentation from breast MRI data may be of clinical value in assessing treatment response and planning surgery in breast cancer patients treated with neoadjuvant chemotherapy. Manual outlining of tumors on 3D MRI data is time intensive and impractical. We describe a semiautomated computer method that shows excellent correlation with the radiologist’s visual segmentation. If the accuracy of our method is confirmed in studies with pathologic correlation, our technique may assist physicians in assessing tumor volume and thereby serve as an initial step in computerized image analysis.
CONCLUSION
We developed an effective, reproducible, and fast marker-controlled watershed algorithm to semiautomatically segment the tumors in contrast-enhanced T1-weighted MR images. The algorithm is initialized manually, with the use of a computer mouse to draw an elliptical ROI on a selected slice containing the target tumor. Gaussian mixture modeling is used to reliably and automatically determine the internal and external markers for watershed segmentation. A resegmentation strategy is included to stabilize the computer outcomes. The algorithm was applied to 13 patients’ data, each having a baseline and a 1 week follow-up MRI study, and received overall volume overlap ratios of 62.6%±9.1% and 61.0%±11.3% in comparison to the two manual segmentation results, respectively. The overall overlap ratio between the two radiologists’ manual segmentations was 64.3%±10.4%. Good segmentation results were achieved in tumors with irregular shapes. Our preliminary findings show that the proposed method is robust, efficient, and correlates well with the radiologist’s manual delineation of tumor volume. Further studies of this method in patients before and after completing neoadjuvant chemotherapy with pathologic correlation and long-term follow-up are necessary to confirm our method’s accuracy and to determine its value in predicting outcomes such as recurrence-free survival.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Avon-NCI Progress for Patients Supplement to P30 CA 00878-42 S5 and the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., and Thun M. J., “Cancer statistics, 2008,” Ca-Cancer J. Clin. 58, 71–96 (2008). 10.3322/CA.2007.0010 [DOI] [PubMed] [Google Scholar]
- Smith R. J., Cokinnides V., and Eyre H. J., “Cancer screening in the United States, 2007: A review of current guidelines, practices, and prospects,” Ca-Cancer J. Clin. 57, 90–104 (2007). 10.3322/canjclin.57.2.90 [DOI] [PubMed] [Google Scholar]
- Pisano E. D., Gatsonis C., Hendrick E., Yaffe M., Baum J. K., Acharyya S., Conant E. F., Fajardo L. L., Bassett L., D’Orsi C., Jong R., and Rebner M., “Digital mammographic imaging screening trial (DMIST) investigators group,” N. Engl. J. Med. 353, 1773–1783 (2005). 10.1056/NEJMoa052911 [DOI] [PubMed] [Google Scholar]
- Morris E. A., “Diagnostic breast MR imaging: Current status and future directions,” Radiol. Clin. North Am. 45, 863–880 (2007). 10.1016/j.rcl.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Saslow D., Boetes C., Burke W., Harms S., Leach M. O., Lehman C. D., Morris E., Pisano E., Schnall M., Sener S., Smith R. A., Warner E., Yaffe M., Andrews K. S., and Russell C. A., “American Cancer Society guidelines for breast cancer screening with MRI as an adjunct to mammography,” Ca-Cancer J. Clin. 57, 75–89 (2007). 10.3322/canjclin.57.2.75 [DOI] [PubMed] [Google Scholar]
- Berg W. A. and Gutierrez L., NessAiyer M. S., Carter W. B., Bhargavan M., Lewis R. S., and Ioffe O. B., “Diagnostic accuracy of mammography, clinical examination, US, and MR imaging in preoperative diagnosis of breast cancer,” Radiology 233, 830–849 (2004). 10.1148/radiol.2333031484 [DOI] [PubMed] [Google Scholar]
- Stavros A. T., Thickman D., Rapp C. L., Dennis M. A., Parker S. H., and Sisney G. A., “Solid breast nodules—Use of sonography to distinguish benign and malignant lesions,” Radiology 196, 123–134 (1995). [DOI] [PubMed] [Google Scholar]
- Berriolo-Riedinger A., Touzery C., Riedinger J. M., Toubeau M., Coudert B., Arnould L., Boichot C., Cochet A., Fumoleau P., and Brunotte F., “[F-18]FDG-PET predicts complete pathological response of breast cancer to neoadjuvant chemotherapy,” Eur. J. Nucl. Med. Mol. Imaging 34, 1915–1924 (2007). 10.1007/s00259-007-0459-5 [DOI] [PubMed] [Google Scholar]
- Berg W. A., Weinberg I. N., Narayanan D., Lobrano M. E., Ross E., Amodei L., Tafra L., Adler L. P., Uddo J., W.Stein3rd, and Levine E. A., “Positron emission mammography working group,” Breast J. 12, 309–323 (2006). 10.1111/j.1075-122X.2006.00269.x [DOI] [PubMed] [Google Scholar]
- Boetes C., Mus R. D. M., Holland R., Barentsz J. O., Strijk S. P., Wobbes T., Hendriks J., and Ruys S. H. J., “Breast-tumors—Comparative accuracy of MR-imaging relative to mammography and US for demonstrating extent,” Radiology 197, 743–747 (1995). [DOI] [PubMed] [Google Scholar]
- Esserman L., Hylton N., Yassa L., Barclay J., Frankel S., and Sickles E., “Utility of magnetic resonance imaging in the management of breast cancer: Evidence for improved preoperative staging,” J. Clin. Oncol. 17, 110–119 (1999). [DOI] [PubMed] [Google Scholar]
- Malur S., Wurdinger S., Moritz A., Michels W., and Schneider A., “Comparison of written reports of mammography, sonography and magnetic resonance mammography for preoperative evaluation of breast lesions, with special emphasis on magnetic resonance mammography,” Breast Cancer Res. 3, 55–60 (2001). 10.1186/bcr271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu-Maestro C., Chapellier C., Bleuse A., Chanalet I., Chauvel C., and Largillier R., “Imaging in evaluation of response to neoadjuvant breast cancer treatment benefits of MRI,” Breast Cancer Res. Treat. 72, 145–152 (2002). 10.1023/A:1014856713942 [DOI] [PubMed] [Google Scholar]
- Partridge S. C., Gibbs J. E., Lu Y., Esserman L. J., Sudilovsky D., and Hylton N. M., “Accuracy of MR imaging for revealing residual breast cancer in patients who have undergone neoadjuvant chemotherapy,” AJR, Am. J. Roentgenol. 179, 1193–1199 (2002). [DOI] [PubMed] [Google Scholar]
- Partridge S. C., Gibbs J. E., Lu Y., Esserman L. J., Tripathy D., Wolverton D. S., Rugo H. S., Hwang G. E., Ewing C. A., and Hylton N. M., “MRI measurements of breast tumor volume predict response to neoadjuvant chemotherapy and recurrence-free survival,” AJR, Am. J. Roentgenol. 184, 1774–1781 (2005). [DOI] [PubMed] [Google Scholar]
- Rosen E. L., Blackwell K. L., Baker J. A., Soo M. S., Bentley R. C., Yu D. H., Samulski T. V., and Dewhirst M. W., “Accuracy of MRI in the detection of residual breast cancer after neoadjuvant chemotherapy,” AJR, Am. J. Roentgenol. 181, 1275–1282 (2003). [DOI] [PubMed] [Google Scholar]
- Guliato D., Rangayyan R. M., Carnielli W. A., Zuffo J. A., and Desautels J. E. L., “Segmentation of breast tumors in mammograms using fuzzy sets,” J. Electron. Imaging 12, 369–378 (2003). 10.1117/1.1579017 [DOI] [Google Scholar]
- Dominguez A. R. and Nandi A. K., “Improved dynamic-programming-based algorithms for segmentation of masses in mammograms,” Med. Phys. 34, 4256–4269 (2007). 10.1118/1.2791034 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Giger M. L., Li H., Suzuki K., and Sennett C., “A dual-stage method for lesion segmentation on digital mammograms,” Med. Phys. 34, 4180–4193 (2007). 10.1118/1.2790837 [DOI] [PubMed] [Google Scholar]
- Horsch K., Giger M. L., Venta L. A., and Vyborny C. J., “Automatic segmentation of breast lesions on ultrasound,” Med. Phys. 28, 1652–1659 (2001). 10.1118/1.1386426 [DOI] [PubMed] [Google Scholar]
- Huang Y. L., Jiang Y. R., Chen D. R., and Moon W. K., “Level set contouring for breast tumor in sonography,” J. Digit. Imaging 20, 238–247 (2007). 10.1007/s10278-006-1041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi A. and Metaxas D. N., “Combining low-, high-level and empirical domain knowledge for automated segmentation of ultrasonic breast lesions,” IEEE Trans. Med. Imaging 22, 155–169 (2003). 10.1109/TMI.2002.808364 [DOI] [PubMed] [Google Scholar]
- Partridge S. C., Heumann E. J., and Hylton N. M., “Semi-automated analysis for MRI of breast tumors,” Stud. Health Technol. Inform. 62, 259–260 (1999). [PubMed] [Google Scholar]
- Arbach L., Stolpen A., and Reinhardt J. M., “Classification of breast MRI lesions using a backpropagation neural network (BNN),” in Biomedical Imaging: Nano to Macro, 2004. IEEE International Symposium on, Vol. 1 (2004), pp. 253–256.
- Chen W. J., Giger M. L., and Bick U., “A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images,” Acad. Radiol. 13, 63–72 (2006). 10.1016/j.acra.2005.08.035 [DOI] [PubMed] [Google Scholar]
- Wu Q., Salganicoff M., Krishnan A., Fussell D. S., and Markey M. K., “Interactive lesion segmentation on dynamic contrast enhanced breast MRI using a Markov model,” Proc. SPIE 6144, 61444M-1–61444M-8 (2006). 10.1117/12.654308 [DOI] [Google Scholar]
- Woods B. J., Clymer B. D., Kurc T., Heverhagen J. T., Stevens R., Orsdemir A., Bulan O., and Knopp M. V., “Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data,” J. Magn. Reson. Imaging 25, 495–501 (2007). 10.1002/jmri.20837 [DOI] [PubMed] [Google Scholar]
- Liu J., Ma W., and Lee S. -Y., “A segmentation method based on dynamic programming for breast mass in MRI images,” Medical Biometrics (Springer-Verlag, Berlin, 2008), pp. 307–313. [Google Scholar]
- Shi J., Sahiner B., Chan H. -P., Paramagul C., Hadjiiski L. M., Helvie M., Wu Y. -T., Ge J., Zhang Y., Zhou C., and Wei J., “Breast mass segmentation on dynamic contrast-enhanced magnetic resonance scans using the level set method,” Proc. SPIE 6915, 69152A-1–69152A-7 (2008). 10.1117/12.771390 [DOI] [Google Scholar]
- Vincent L. and Soille P., “Watersheds in digital spaces—An efficient algorithm based on immersion simulations,” IEEE Trans. Pattern Anal. Mach. Intell. 13, 583–598 (1991). 10.1109/34.87344 [DOI] [Google Scholar]
- Lin G., Adiga U., Olson K., Guzowski J. F., Barnes C. A., and Roysam B., “A hybrid 3D watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks,” Cytometry, Part A 56A, 23–36 (2003). 10.1002/cyto.a.10079 [DOI] [PubMed] [Google Scholar]
- Malpica N., deSolorzano C. O., Vaquero J. J., Santos A., Vallcorba I., GarciaSagredo J. M., and delPozo F., “Applying watershed algorithms to the segmentation of clustered nuclei,” Cytometry 28, 289–297 (1997). [DOI] [PubMed] [Google Scholar]
- Najman L. and Schmitt M., “Geodesic saliency of watershed contours and hierarchical segmentation,” IEEE Trans. Pattern Anal. Mach. Intell. 18, 1163–1173 (1996). 10.1109/34.546254 [DOI] [Google Scholar]
- Sijbers J., Scheunders P., Verhoye M., VanderLinden A., vanDyck D., and Raman E., “Watershed-based segmentation of 3D MR data for volume quantization,” Magn. Reson. Imaging 15, 679–688 (1997). 10.1016/S0730-725X(97)00033-7 [DOI] [PubMed] [Google Scholar]
- Jos B. T. M. R. and Arnold M., “The watershed transform: Definitions, algorithms and parallelization strategies,” Fund. Inform. 41, 187–228 (2000). [Google Scholar]
- Moga A. N. and Gabbouj M., “Parallel image component labeling with watershed transformation,” IEEE Trans. Pattern Anal. Mach. Intell. 19, 441–450 (1997). 10.1109/34.589204 [DOI] [Google Scholar]
- Grau V., Mewes A. U. J., Alcaniz M., Kikinis R., and Warfield S. K., “Improved watershed transform for medical image segmentation using prior information,” IEEE Trans. Med. Imaging 23, 447–458 (2004). 10.1109/TMI.2004.824224 [DOI] [PubMed] [Google Scholar]
- Bleau A. and Leon L. J., “Watershed-based segmentation and region merging,” Comput. Vis. Image Underst. 77, 317–370 (2000). 10.1006/cviu.1999.0822 [DOI] [Google Scholar]
- Haris K., Efstratiadis S. N., Maglaveras N., and Katsaggelos A. K., “Hybrid image segmentation using watersheds and fast region merging,” IEEE Trans. Image Process. 7, 1684–1699 (1998). 10.1109/83.730380 [DOI] [PubMed] [Google Scholar]
- Meyer F. and Beucher S., “Morphological segmentation,” J. Visual Commun. Image Represent. 1, 21–46 (1990). 10.1016/1047-3203(90)90014-M [DOI] [Google Scholar]
- Vincent L., “Morphological grayscale reconstruction in image analysis: Applications and efficient algorithms,” IEEE Trans. Image Process. 2, 176–201 (1993). 10.1109/83.217222 [DOI] [PubMed] [Google Scholar]
- Yan J. Y., Zhao B. S., Wang L., Zelenetz A., and Schwartz L. H., “Marker-controlled watershed for lymphoma segmentation in sequential CT images,” Med. Phys. 33, 2452–2460 (2006). 10.1118/1.2207133 [DOI] [PubMed] [Google Scholar]
- Nguyen H. T., Worring M., and van den Boomgaard R., “Watersnakes: Energy-driven watershed segmentation,” IEEE Trans. Pattern Anal. Mach. Intell. 25, 330–342 (2003). 10.1109/TPAMI.2003.1182096 [DOI] [Google Scholar]
- Choi H. S., Haynor D. R., and Kim Y. M., “Partial volume tissue classification of multichannel magnetic-resonance images—A mixel model,” IEEE Trans. Med. Imaging 10, 395–407 (1991). 10.1109/42.97590 [DOI] [PubMed] [Google Scholar]
- Liang Z., Jaszczak R. J., and Coleman R. E., “Parameter-estimation of finite mixtures using the EM algorithm and information criteria with application to medical image-processing,” IEEE Trans. Nucl. Sci. 39, 1126–1133 (1992). 10.1109/23.159772 [DOI] [Google Scholar]
- Sanjay-Gopel S. and Hebert T. J., “Bayesian pixel classification using spatially variant finite mixtures and the generalized EM algorithm,” IEEE Trans. Image Process. 7, 1014–1028 (1998). 10.1109/83.701161 [DOI] [PubMed] [Google Scholar]
- Aristophanous M., Penney B. C., Martel M. K., and Pelizzari C. A., “A Gaussian mixture model for definition of lung tumor volumes in positron emission tomography,” Med. Phys. 34, 4223–4235 (2007). 10.1118/1.2791035 [DOI] [PubMed] [Google Scholar]
- McLachlan G. and Peel D., Finite Mixture Models, 1st ed. (Wiley, New York, 2000). [Google Scholar]