Abstract
In 1918 the Spanish influenza pandemic, caused by an avian H1N1 virus, resulted in over 50 million deaths worldwide. Several outbreaks of H7 influenza A viruses have resulted in human cases, including one fatal case. Since 1997, the outbreaks of highly pathogenic avian influenza (HPAI) of the H5N1 subtype have affected a wide variety of mammals in addition to poultry and wild birds. Here, we give an overview of the current knowledge of the determinants of pathogenicity of these three subtypes of avian influenza A virus in mammals. Common mechanisms for acquisition of virulence and replication of these avian influenza viruses in mammals are becoming apparent. Therefore, monitoring these and additional genetic changes upon zoonotic infections is important. Identification of genetic changes responsible for transmission between mammals will be an important task for the near future.
Keywords: influenza A virus, HPAI, pathogenesis
Introduction
Influenza A virus is a member of the Orthomyxoviridae family, comprising enveloped, negative strand RNA viruses with a segmented genome. Influenza A viruses are characterized based on the envelope glycoproteins hemagglutinin (HA) and neuraminidase (NA). To date, 16 subtypes of HA and 9 subtypes of NA have been found and almost all subtype combinations (Fouchier et al., 2005; Olsen et al., 2006; Webster et al., 1992).
Although influenza A viruses are best known for the annual epidemics in humans, wild birds form their natural reservoir (Olsen et al., 2006; Webster et al., 1992). Whereas only 3 influenza A virus subtypes circulated in humans in the past century, all known subtypes have been isolated from wild birds. Numerous subtypes have also been detected in poultry, generally causing mild disease, or no disease at all. Upon introduction in poultry, viruses of the subtypes H5 and H7 may cause outbreaks of highly pathogenic avian influenza (HPAI). The transition of a low pathogenic avian influenza (LPAI) virus to a HPAI virus generally results from the introduction of multiple basic amino acids at the HA cleavage site, facilitating systemic virus replication and a mortality of up to 100% in poultry.
Occasionally, a new subtype of influenza A virus is introduced into the human population that is able to spread efficiently from human to human, causing a pandemic. This process is called antigenic shift and may be the result of a reassortment event of avian and human influenza A viruses (Scholtissek et al., 1978), or of adaptation of a fully avian virus to humans (Claas et al., 1998; Tumpey et al., 2005a). During the past century, three such pandemics occurred. The introduction of influenza A virus of the H1N1 subtype in 1918 is probably the best-known example of a pandemic. An estimated 50 million people died as a result of the so-called ‘Spanish influenza’ (Patterson et al., 1991). Introduction of the H2N2 and H3N2 subtypes caused relatively mild pandemics in 1957 and 1968 respectively. Not all introductions of a new influenza A virus subtype in the human population lead to a pandemic. Often, transmission is limited because the virus does not spread easily from one person to the other. The knowledge of the pathogenicity of avian influenza viruses in mammals has increased considerably in recent years, sparked by the increased number of transmissions of avian influenza viruses to humans. Here, we present the current knowledge of the determinants of pathogenicity of three important avian influenza viruses that have caused infections in humans: the 1918 Spanish influenza virus, HPAI H5N1 viruses and H7 viruses.
1918 Spanish influenza
The cause of the hitherto unparalleled morbidity and mortality during the Spanish influenza pandemic in 1918 has long remained a mystery, since the influenza A virus was only discovered more than 10 years after this pandemic. The recent reconstruction of a 1918 virus based on archival and frozen tissue samples has shed some light on its pathogenic properties (Basler et al., 2001; Geiss et al., 2002; Kobasa et al., 2007; Kobasa et al., 2004; Taubenberger et al., 2005; Tumpey et al., 2005a; Tumpey et al., 2005b; Tumpey et al., 2007).
Several studies in mammals have shown that this is a very pathogenic human influenza A virus. When ferrets were infected with 106 pfu of this virus, 2/3 animals succumbed to the infection (Tumpey et al., 2007). Macaques infected with 7×106 pfu started to show symptoms of disease within 24h post infection and all succumbed to the infection by day 8 (Kobasa et al., 2007).
Multiple gene segments are involved in the pathogenicity of the 1918 virus. The complete 1918 virus was 100 times more lethal for mice than a reassortant 1918 virus with the polymerases (PB1, PB2, PA) of a human H3N2 virus (Tumpey et al., 2005a). Only 10 amino acids in the 1918 polymerase complex distinguish it from the consensus avian polymerase complex. Among these 10 amino acid substitutions is a E627K substitution in PB2 (Taubenberger et al., 2005). This residue has been described as a determinant of pathogenicity of HPAI H5N1 and H7N7 viruses (see below) and may thus also play a role in the pathogenicity of the 1918 spanish influenza virus.
HA and NA are also important determinants of pathogenicity of the 1918 virus, although the mechanism through which they exert their effect is not yet known (Kobasa et al., 2004; Tumpey et al., 2005a; Tumpey et al., 2005b). NA of the 1918 virus is unusual in its’ ability to enable virus replication in the absence of trypsin in tissue culture (Tumpey et al., 2005a).
In vitro, NS1 of the 1918 virus is a potent inhibitor of the IFN pathway (Geiss et al., 2002). It may thus also contribute to the virulence of the 1918 virus in humans. However, the role of NS1 in the pathogenicity of the 1918 virus could not be confirmed in mice; rather, viruses with NS1 of the 1918 virus seemed attenuated in these animals (Basler et al., 2001). Whether NS1 is not involved in pathogenesis of the 1918 virus or whether the mouse model is not a suitable animal model remains to be determined. Experimental infection of macaques suggests that the 1918 virus induces an antiviral response different from that induced by a contemporary human H1N1 virus (Kobasa et al., 2007). This difference may be due to NS1 of the 1918 virus.
HPAI H7 viruses
Several outbreaks of LPAI and HPAI H7 viruses in poultry have resulted in transmission of these viruses to humans, indicating the unusual zoonotic properties of these viruses. During an experimental infection of seals with influenza virus A/Seal/Massachusetts/1/80, one of the investigators developed conjunctivitis after being sneezed on by an infected seal (Webster et al., 1981). In 1996, a LPAI H7N7 virus, A/England/268/96, was transmitted from ducks to a 43-year-old woman, resulting in a case of conjunctivitis (Kurtz et al., 1996). During a large outbreak of HPAI H7N7 virus in the Netherlands in 2003, 89 humans became infected. Although most of these developed conjunctivitis and in some cases mild respiratory symptoms, one patient died as a result of severe pneumonia and related complications (Fouchier et al., 2004; Koopmans et al., 2004). In 2004, an outbreak of HPAI H7N3 virus in Canada resulted in conjunctivitis and mild influenza-like illness in two men that had been in contact with infected poultry (Tweed et al., 2004). An outbreak of LPAI H7N3 virus in the UK in 2006 resulted in a case of conjunctivitis in a poultry worker; virus was also detected in a throat swab collected from this person (Nguyen-Van-Tam et al., 2006). Finally, several people were infected with a low pathogenic H7N2 virus during an outbreak in the UK in 2007 (ProMed, 2007). Viruses of the H7 subtype display an unusual tissue tropism compared to other subtypes, since cases of conjunctivitis have occurred in many outbreaks of LPAI and HPAI H7 viruses, but have rarely been reported with other subtypes.
When A/Seal/Massachusetts/1/80 (H7N7) was serially passaged in chicken embryo cells it obtained a multibasic cleavage site in HA, rendering it highly pathogenic in chickens. Subsequently, this virus was passaged in mouse lungs and became highly pathogenic to mice. Two substitutions in PB2, D701N and S714R were shown to be important determinants of virulence in mice, which was related to an increased polymerase activity (Gabriel et al., 2005).
The pathogenicity of a conjunctivitis virus isolated during the Dutch HPAI H7N7 outbreak and the virus isolated from the patient with the fatal outcome of H7N7 infection was compared in a mouse model. The fatal case virus was intrinsically more pathogenic than the conjunctivitis virus. A E627K substitution in PB2 was the main determinant of pathogenicity. HA of the fatal case virus, containing 3 substitutions, including a substitution introducing a potential glycosylation site near the receptor binding site, increased virus titers in the lungs of infected mice and distribution of virus to different organs. The effect of HA on pathogenicity may be related to subtle differences in the attachment pattern of the conjunctivitis virus and the fatal case virus to the lower respiratory tract, as shown in virus-binding studies (Munster et al., 2007). An E627K substitution in PB2 is often found as a determinant of pathogenicity of HPAI viruses in mice. Adaptation of A/Equine/London/1416/73 (H7N7) to mice resulted, amongst others, in a E627K substitution in PB2, leading to a 1000x increased virulence of this virus (Shinya et al., 2007). Moreover, out of three viruses isolated during the outbreak of HPAI H7N1 from 1999–2000 in Italy, the virus with lysine at position 627 of PB2 was the most pathogenic in mice (Rigoni et al., 2007).
HPAI H5N1 viruses
During an outbreak of HPAI H5N1 in poultry markets in Hong Kong in 1997, 18 people were infected, six of whom died. This was the first recorded direct transmission of an avian influenza virus to humans causing fatality (de Jong et al., 1997).
Two viruses isolated from humans during this 1997 outbreak showed a different pathogenicity in mice: A/Hong Kong/483/97, isolated from a patient with a fatal infection, caused a lethal systemic infection in mice, whereas A/Hong Kong/486/97, isolated from a patient with relatively mild disease, caused a non-lethal respiratory infection. This difference in pathogenicity was mainly determined by a E627K substitution in PB2. An I227S substitution in HA also increased the virulence of this virus in mice (Hatta et al., 2001).
The HPAI H5N1 viruses isolated during the 1997 outbreak were able to interfere with the innate immune response of the host in mice and pigs. Isolates from the 1997 outbreak were more pathogenic in mice than HPAI H5 viruses isolated during previous outbreaks in the UK, Italy and Mexico. In contrast to other HPAI H5 strains, the 1997 strains did not lead to increased production of the proinflammatory cytokine TGF-β in serum of infected mice (Dybing et al., 2000). Also, the 1997 virus was insensitive to IFN-α, IFN-γ and TNF-α in vitro, due to a glutamic acid at position 92 of NS1 (Seo et al., 2002). This residue was also a determinant of pathogenicity in pigs infected with a reassortant A/PR/8/34 virus with NS of A/HK/156/97 (Seo et al., 2002). It was shown that HPAI H5N1 viruses that are highly pathogenic to mice induced much higher levels of inflammatory cytokines in mouse lungs (Lipatov et al., 2005). Finally, in vitro assays using human monocyte-derived macrophages showed that HPAI H5N1 virus from 1997 induced higher levels of proinflammatory cytokines (Cheung et al., 2002).
HPAI H5N1 viruses were isolated from healthy ducks in southern China from 1999 onwards. These viruses differed remarkably in their pathogenicity in mice. The main determinant of pathogenicity was shown to be a D701N substitution in PB2 (Li et al., 2005).
In 2003, the HPAI H5N1 virus reemerged in Hong Kong and has spread to poultry in other parts of South East Asia since. Since the outbreak in wild migratory birds in Qinghai Lake in 2005, the HPAI H5N1 virus has surfaced across Asia, Europe, the Middle East and Africa. Since then, the HPAI H5N1 virus caused disease in several mammalian species such as tigers, leopards (Keawcharoen et al., 2004), dogs (Butler, 2006) and humans (Beigel et al., 2005). In humans, over 300 cases of HPAI H5N1 infection were detected so far, with a fatal outcome in ~2/3 of these (WHO, 2007). Based on a cohort of 18 hospitalized patients infected with H5N1 virus of whom 13 died, it was shown that lethal outcome of infection correlated with a high viral load and a high production of cytokines and chemokines. Plasma levels of IP-10, MCP-1, IL-8, IL-6 and IL-10 correlated with pharyngeal viral load, suggesting that the overproduction of cytokines is a result of increased virus replication (de Jong et al., 2006).
The pathogenicity of an avian HPAI H5N1 virus, A/Chicken/Vietnam/C58/04, and a human HPAI H5N1 virus, A/Vietnam/1203/04, was compared in mouse and ferret models. In both species, the chicken isolate was non-lethal, whereas the human isolate caused considerable mortality (Salomon et al., 2006). The polymerase complex contributed to the pathogenicity of A/Vietam/1203/04. Viruses with PB2 or PB1 of the chicken virus in the background of A/Vietnam/1203/04 were attenuated in both species, indicating the importance of adaptation of the avian polymerase complex to the mammalian host. The polymerase activity of A/Vietnam/1203/04 was significantly higher than that of the chicken virus polymerase complex in human cells, again indicating that increased virus replication is related to pathogenicity. In ferrets, but not in mice, NS was an important determinant of pathogenicity (Salomon et al., 2006).
Whereas human influenza A viruses use α-2,6-linked sialic acids (SA) as a receptor, avian viruses use α-2,3-linked SA. This difference in receptor binding is, in part, related to the different pathogenicity of these viruses in humans. Attachment studies using HPAI H5N1 virus showed that this virus rarely attached to the trachea, in contrast to a human H3N2 virus that attached abundantly to the trachea. In the lower respiratory tract, H5N1 virus attached predominantly to type II pneumocytes, alveolar macrophages and nonciliated cuboidal epithelial cells in terminal bronchioles (van Riel et al., 2006). This observation corresponded with the presence of α-2,3-linked SA on nonciliated cuboidal bronchiolar cells and type II pneumocytes and the virtual absence of these receptors in the upper respiratory tract (Shinya et al., 2006). The attachment pattern of H5N1 virus to these cells was in concordance with the diffuse alveolar damage that is observed in human cases of HPAI H5N1 virus infection and could very well be related to the pathogenicity of these viruses (van Riel et al., 2006).
Common determinants of pathogenicity
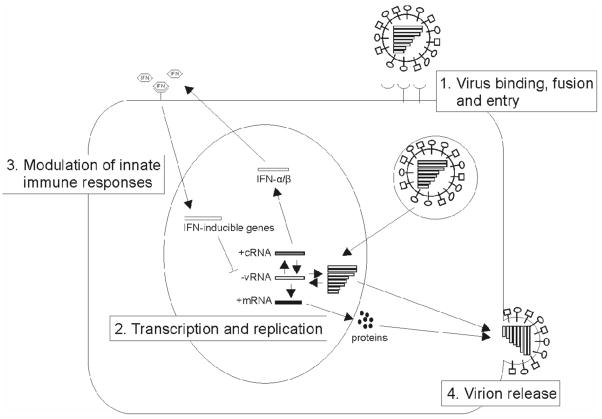
From the data above, common patterns for the acquistition of virulence of avian influenza viruses for mammals are becoming apparent (Fig. 1). Obviously, the pathogenicity of avian influenza viruses in mammals is a polygenic trait. Changes in the polymerase complex have been observed in the 1918 Spanish influenza and H5 and H7 avian influenza viruses. Thus it seems that adaptation of the avian virus polymerase complex is an important prerequisite for replication in mammals. One of the most commonly described substitutions is the E627K substitution in PB2. This residue is a glutamic acid in avian viruses and a lysine in human viruses and was described as a determinant of host range in vitro (Subbarao et al., 1993). Avian viruses lacking this E627K substitution may acquire it spontaneously upon a single passage in mice (Li et al., 2005; Mase et al., 2006). Although the importance of this residue in pathogenesis was described in mice for different influenza A virus subtypes, there was no correlation between the presence of a lysine at position 627 of PB2 and the outcome of disease in HPAI H5N1 virus infected humans (de Jong et al., 2006). However, all viruses in this cohort contained other substitutions in the polymerase complex involved in adaptation to efficient replication in mammals. This suggests that the ability to replicate in humans is not determined by the E627K substitution in PB2 alone. Sequence comparison of avian and human influenza A viruses identified 52 species-associated positions in the influenza A virus genome. Of these 52 positions, 35 are in the polymerase complex (Chen, 2006).
Figure 1.
Determinants of avian influenza virus pathogenicity in mammals. 1. HA of influenza A virus is important for receptor binding, fusion and entry into the host cell. HA of avian viruses preferentially bind to α-2,3-linked SA, whereas human viruses preferentially bind to α-2,6-linked SA. 2. The influenza virus polymerase complex undergoes changes to facilitate replication in the mammalian host cell. 3. Virus replication results in an innate immune response to block virus replication. The NS1 protein is able to antagonize this response and modulate production of proinflammatory cytokines and chemokines. 4. NA is essential for the release of virus particles from the infected cell and thus for spread of the virus by destroying α-2,3-and α-2,6-linked SA receptors.
Specific substitutions in HA and NA can increase virulence, for instance through changes in or near the receptor binding site (Hatta et al., 2001; Munster et al., 2007; Yamada et al., 2006) or a substitution in NA that enables replication in the absence of trypsin (Tumpey et al., 2005a). Moreover, avian influenza A viruses have a different receptor specificity than human influenza A viruses and thus infect different cell types, which may be related to the pathogenesis of these viruses (Munster et al., 2007; van Riel et al., 2006).
NS1 is also an important determinant of pathogenesis through its ability to antagonize the innate immune response. The fact that this has not been described in vivo as often as e.g. changes in the polymerase complex may be related to the fact that many studies were performed in mice. The inbred mouse strains used for these studies lack a functional Mx1 gene (Staeheli et al., 1988), while the Mx1 gene is an important factor in bringing cells in antiviral state (Staeheli et al., 1986). This and other host-specific factors may contribute to the fact that the role of NS1 has not been described often in vivo.
Human-to-human transmission
Now that common traits are becoming apparent of how avian influenza viruses acquire pathogenicity, the minimal requirements for efficient human-to-human transmission need to be elucidated. The pandemics of the past century have shown that there are two ways of pandemic viruses arising: adaptation of an avian virus by mutation, as was probably the case with Spanish influenza, or reassortment of an avian and a human influenza virus, as was the case with the pandemics of 1957 and 1968. There are two minimal requirements for sustained human-to-human transmission: efficient replication in the human host and replication in the upper respiratory tract to enable virus transmission via the airways, e.g. by coughing and sneezing.
Receptors for avian influenza viruses, α-2,3-linked SA, are abundantly present only in the lower respiratory tract of humans (Shinya et al., 2006; van Riel et al., 2006). It was suggested that this explains why HPAI H5N1 virus is not transmitted efficiently from human to human. If this were true, adaptation of HA to recognize α-2,6-linked SA in the upper respiratory tract would suffice for human-to-human transmission. However, HPAI H5N1 viruses that have dual receptor specificity for both α-2,3- and α-2,6-linked SA are currently circulating (Yamada et al., 2006), but so far this has not resulted in efficient human-to-human transmission. This suggests that efficient human-to-human transmission is not solely determined by receptor recognition. Several transmission experiments have been performed in ferrets using the 1918 virus and HPAI H5N1 virus. Firstly, after introduction of two amino acid changes in HA of the 1918 virus, thereby changing the receptor specificity from α-2,6 to α-2,3-linked SA, the virus was no longer transmitted to contact ferrets (Tumpey et al., 2007). Secondly, reassortant viruses consisting of the internal genes of a human H3N2 virus and HA and NA of HPAI H5N1 virus did not transmit from ferret to ferret; neither did reassortant viruses consisting of HA, NA, MA and NS of H5N1 virus and the polymerases and NP of a human H3N2 virus, despite the fact that virus was present in nasal washes of infected animals and sneezing was observed in some animals (Maines et al., 2006).
A third transmission study showed inefficient transmission of 2 HPAI H5N1 viruses. Transmission was not related to the ability of some of these viruses to recognize both α-2,3 and α-2,6-linked SA, since only one out of 2 viruses with affinity for α-2,6-linked SA as well as α-2,3-linked SA was transmitted. Again, virus was present in the nasal washes of all ferrets, independent of whether the virus was transmitted or not (Yen et al., 2007). Taken together, it seems that although recognition of α-2,6-linked SA is an important determinant of transmission, mere presence of virus in the upper respiratory tract is not the only factor of importance. So far, all transmission experiments have been performed in ferrets; a recently described transmission model in guinea pigs (Lowen et al., 2006) may increase our knowledge of the determinants of transmission in mammals further.
Now that common determinants of pathogenicity of avian influenza viruses in mammals are becoming clear, the requirements for efficient transmission in mammalian model systems need to be elucidated. The data summarized here were almost all generated in vitro or in animal models, and care should be taken when extrapolating these data to humans. More (post mortem) data from humans are needed to establish with certainty the determinants of pathogenesis and efficient human-to-human transmission.
Acknowledgments
We thank Vincent Munster for helpful discussions and other members of the Department of Virology for continuous support.
Abbreviations
- HPAI
highly pathogenic avian influenza
- LPAI
low pathogenic avian influenza
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, Garcia-Sastre A, Palese P, Taubenberger JK. Sequence of the 1918 pandemic influenza virus nonstructural gene (ns) segment and characterization of recombinant viruses bearing the 1918 ns genes. Proc Natl Acad Sci U S A. 2001;98:2746–2751. doi: 10.1073/pnas.031575198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY. Avian influenza a (h5n1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- Butler D. Thai dogs carry bird-flu virus, but will they spread it? Nature. 2006;439:773. doi: 10.1038/439773a. [DOI] [PubMed] [Google Scholar]
- Chen G-W, Chang S-C, Mok C-K, Lo Y-L, Kung Y-N, Huang J-H, Shih Y-H, Wang J-Y, Chiang C, Chen C-J, Shih S-R. Genomic signatures of human versus avian influenza a viruses. Emerg Infect Dis. 2006;12:1353–1360. doi: 10.3201/eid1209.060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, Gordon S, Guan Y, Peiris JS. Induction of proinflammatory cytokines in human macrophages by influenza a (h5n1) viruses: A mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- Claas EC, Osterhaus AD. New clues to the emergence of flu pandemics. Nat Med. 1998;4:1122–1123. doi: 10.1038/2617. [DOI] [PubMed] [Google Scholar]
- de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza a (h5n1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybing JK, Schultz-Cherry S, Swayne DE, Suarez DL, Perdue ML. Distinct pathogenesis of hong kong-origin h5n1 viruses in mice compared to that of other highly pathogenic h5 avian influenza viruses. J Virol. 2000;74:1443–1450. doi: 10.1128/jvi.74.3.1443-1450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza a virus hemagglutinin subtype (h16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. Avian influenza a virus (h7n7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. Cellular transcriptional profiling in influenza a virus-infected lung epithelial cells: The role of the nonstructural ns1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of hong kong h5n1 influenza a viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Keawcharoen J, Oraveerakul K, Kuiken T, Fouchier RA, Amonsin A, Payungporn S, Noppornpanth S, Wattanodorn S, Theambooniers A, Tantilertcharoen R, Pattanarangsan R, Arya N, Ratanakorn P, Osterhaus DM, Poovorawan Y. Avian influenza h5n1 in tigers and leopards. Emerg Infect Dis. 2004;10:2189–2191. doi: 10.3201/eid1012.040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, Suzuki H, Nishimura H, Mitamura K, Sugaya N, Usui T, Murata T, Maeda Y, Watanabe S, Suresh M, Suzuki T, Suzuki Y, Feldmann H, Kawaoka Y. Enhanced virulence of influenza a viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. Transmission of h7n7 avian influenza a virus to human beings during a large outbreak in commercial poultry farms in the netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Kurtz J, Manvell RJ, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet. 1996;348:901–902. doi: 10.1016/S0140-6736(05)64783-6. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K. Molecular basis of replication of duck h5n1 influenza viruses in a mammalian mouse model. J Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatov AS, Andreansky S, Webby RJ, Hulse DJ, Rehg JE, Krauss S, Perez DR, Doherty PC, Webster RG, Sangster MY. Pathogenesis of hong kong h5n1 influenza virus ns gene reassortants in mice: The role of cytokines and b- and t-cell responses. J Gen Virol. 2005;86:1121–1130. doi: 10.1099/vir.0.80663-0. [DOI] [PubMed] [Google Scholar]
- Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A. 2006;103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM. Lack of transmission of h5n1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mase M, Tanimura N, Imada T, Okamatsu M, Tsukamoto K, Yamaguchi S. Recent h5n1 avian influenza a virus increases rapidly in virulence to mice after a single passage in mice. J Gen Virol. 2006;87:3655–3659. doi: 10.1099/vir.0.81843-0. [DOI] [PubMed] [Google Scholar]
- Munster VJ, de Wit E, van Riel D, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Kuiken T, Fouchier RA. The molecular basis of the pathogenicity of the dutch highly pathogenic human influenza a h7n7 viruses. J Infect Dis. 2007;196:258–265. doi: 10.1086/518792. [DOI] [PubMed] [Google Scholar]
- Nguyen-Van-Tam JS, Nair P, Acheson P, Baker A, Barker M, Bracebridge S, Croft J, Ellis J, Gelletlie R, Gent N, Ibbotson S, Joseph C, Mahgoub H, Monk P, Reghitt TW, Sundkvist T, Sellwood C, Simpson J, Smith J, Watson JM, Zambon M, Lightfoot N. Outbreak of low pathogenicity h7n3 avian influenza in uk, including associated case of human conjunctivitis. Euro Surveill. 2006;11:E060504–060502. doi: 10.2807/esw.11.18.02952-en. [DOI] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Patterson KD, Pyle GF. The geography and mortality of the 1918 influenza pandemic. Bull Hist Med. 1991;65:4–21. [PubMed] [Google Scholar]
- ProMed. Avian influenza h7n2, human - united kingdom (wales) (08) 2007 Available from http://www.promedmail.org [archive no. 20070606.1830]
- Rigoni M, Shinya K, Toffan A, Milani A, Bettini F, Kawaoka Y, Cattoli G, Capua I. Pneumo- and neurotropism of avian origin italian highly pathogenic avian influenza h7n1 isolates in experimentally infected mice. Virology. 2007;364:28–35. doi: 10.1016/j.virol.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, Webster RG, Hoffmann E. The polymerase complex genes contribute to the high virulence of the human h5n1 influenza virus isolate a/vietnam/1203/04. J Exp Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes h2n2 and h3n2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- Seo SH, Hoffmann E, Webster RG. Lethal h5n1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- Shinya K, Watanabe S, Ito T, Kasai N, Kawaoka Y. Adaptation of an h7n7 equine influenza a virus in mice. J Gen Virol. 2007;88:547–553. doi: 10.1099/vir.0.82411-0. [DOI] [PubMed] [Google Scholar]
- Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. Influenza virus-susceptible mice carry mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: Constitutive expression in 3t3 cells transformed with cloned mx cdna confers selective resistance to influenza virus. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the pb2 gene of influenza a virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. Characterization of the reconstructed 1918 spanish influenza pandemic virus. Science. 2005a;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Garcia-Sastre A, Taubenberger JK, Palese P, Swayne DE, Pantin-Jackwood MJ, Schultz-Cherry S, Solorzano A, Van Rooijen N, Katz JM, Basler CF. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: Functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J Virol. 2005b;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, Garcia-Sastre A. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. Human illness from avian influenza h7n3, british columbia. Emerg Infect Dis. 2004;10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5n1 virus attachment to lower respiratory tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza a viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Geraci J, Petursson G, Skirnisson K. Conjunctivitis in human beings caused by influenza a virus of seals. N Engl J Med. 1981;304:911. doi: 10.1056/NEJM198104093041515. [DOI] [PubMed] [Google Scholar]
- WHO. Cumulative number of confirmed human cases of avian influenza a/(h5n1) reported to who. 2007 Available from http://www.who.int/csr/disease/avian_influenza/country/cases_table_2007_06_15/en/index.html.
- Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, Shinya K, Sawada T, Usui T, Murata T, Lin Y, Hay A, Haire LF, Stevens DJ, Russell RJ, Gamblin SJ, Skehel JJ, Kawaoka Y. Haemagglutinin mutations responsible for the binding of h5n1 influenza a viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- Yen HL, Lipatov AS, Ilyushina NA, Govorkova EA, Franks J, Yilmaz N, Douglas A, Hay A, Krauss S, Rehg JE, Hoffmann E, Webster RG. Inefficient transmission of h5n1 influenza viruses in a ferret contact model. J Virol. 2007;81:6890–6898. doi: 10.1128/JVI.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]