Abstract
BACKGROUND
The genes underlying the risk of stroke in the general population remain undetermined.
METHODS
We carried out an analysis of genomewide association data generated from four large cohorts composing the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium, including 19,602 white persons (mean [±SD] age, 63±8 years) in whom 1544 incident strokes (1164 ischemic strokes) developed over an average follow-up of 11 years. We tested the markers most strongly associated with stroke in a replication cohort of 2430 black persons with 215 incident strokes (191 ischemic strokes), another cohort of 574 black persons with 85 incident strokes (68 ischemic strokes), and 652 Dutch persons with ischemic stroke and 3613 unaffected persons.
RESULTS
Two intergenic single-nucleotide polymorphisms on chromosome 12p13 and within 11 kb of the gene NINJ2 were associated with stroke (P<5×10−8). NINJ2 encodes an adhesion molecule expressed in glia and shows increased expression after nerve injury. Direct genotyping showed that rs12425791 was associated with an increased risk of total (i.e., all types) and ischemic stroke, with hazard ratios of 1.30 (95% confidence interval [CI], 1.19 to 1.42) and 1.33 (95% CI, 1.21 to 1.47), respectively, yielding population attributable risks of 11% and 12% in the discovery cohorts. Corresponding hazard ratios were 1.35 (95% CI, 1.01 to 1.79; P = 0.04) and 1.42 (95% CI, 1.06 to 1.91; P=0.02) in the large cohort of black persons and 1.17 (95% CI, 1.01 to 1.37; P = 0.03) and 1.19 (95% CI, 1.01 to 1.41; P = 0.04) in the Dutch sample; the results of an underpowered analysis of the smaller black cohort were nonsignificant.
CONCLUSIONS
A genetic locus on chromosome 12p13 is associated with an increased risk of stroke.
Stroke is the leading neurologic cause of death and disability.1 Twin and familial aggregation studies suggest that the risk of stroke has a substantial genetic component,2–4 but the genes underlying this risk in the general population remain undetermined. Studies of candidate genes or studies that use classical linkage approaches have yielded inconsistent findings.5
Genomewide association studies have uncovered previously unsuspected common variants underlying the risk of complex diseases such as diabetes6 and coronary disease.7,8 Two previous genomewide association studies of stroke were limited by a case–control design that is more susceptible to survival and selection biases than the design of prospective cohort studies.9,10 We combined data derived from four large, prospective, population-based cohorts consisting predominantly of white persons: the Atherosclerosis Risk in Communities (ARIC) cohort,11 the Cardiovascular Health Study cohort,12 the Framingham Heart Study cohort,13,14 and the Rotterdam Study cohort.15 The four cohorts are part of a consortium, the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE),16 formed to generate a discovery sample of 19,602 participants. We also present the findings from three replication samples, a prospectively evaluated cohort of 2430 black participants in the ARIC study, a second small, prospectively evaluated cohort of 574 black participants in the Cardiovascular Health Study, and a case–control sample of 4265 self-reported Caucasian (hereafter called white) Dutch persons.
METHODS
STUDY DESIGN AND SAMPLES
Details of the cohort selection and risk-factor assessment in the four studies11–15 are described in Section 2 in the Supplementary Appendix, available with the full text of this article at NEJM.org. The institutional review board of each study approved the study design. All participants gave written informed consent for study participation, including genetic research.
Stroke-free participants entered the current study on the date of the blood draw used for their genotyping, and they were followed prospectively for incident stroke. Almost all participants in the Framingham Heart Study and Rotterdam Study described themselves as white (Framingham Heart Study) or Caucasian (Rotterdam Study); thus, participants in the ARIC and Cardiovascular Health Study who were black according to self-report were excluded in our discovery analyses. Details of how ancestry was determined in the individual cohorts are provided in Section 2 in the Supplementary Appendix. Participants were also excluded if they did not provide informed consent or were not successfully genotyped. The analysis included 7686 participants from the ARIC study, 2022 from the Cardiovascular Health Study, 4131 from the Framingham Heart Study, and 5763 from the Rotterdam Study. Baseline demographic and clinical characteristics of the samples are shown in Table 1.
Table 1.
Characteristics of the Study Population in the Discovery Cohorts for Analysis of Incident Total Stroke and Incident Ischemic Stroke.*
| Variable | ARIC (N = 7686) | CHS (N = 2022) | FHS (N = 4131) | Rotterdam (N = 5763) |
|---|---|---|---|---|
| Female sex (%) | 53 | 55 | 55 | 59 |
| Mean follow-up (yr) | 15 | 11 | 6 | 10 |
| Mean age (yr) | ||||
| At DNA draw | 54±6 | 73±6 | 66±12 | 69±9 |
| At incident stroke | 66±7 | 81±6 | 80±10 | 80±8 |
| Strokes (no.) | ||||
| Prevalent | 12 | 0‡ | 135 | 170 |
| Incident total | 312 | 459 | 156 | 617 |
| Incident ischemic | 277 | 389 | 131 | 367 |
| Incident atherothrombotic | 243 | 264 | 82 | 296 |
| Cardiovascular risk factors at baseline† | ||||
| Systolic blood pressure (mm Hg) | 118±17 | 138±22 | 131±20 | 139±22 |
| Diastolic blood pressure (mm Hg) | 72±10 | 71±11 | 74±10 | 74±12 |
| Hypertension (%) | 27 | 61 | 52 | 61 |
| Diabetes mellitus (%) | 6 | 14 | 12 | 10 |
| Current smoker (%) | 25 | 11 | 14 | 23 |
| Prevalent cardiovascular disease other than stroke (%) | 5 | 0‡ | 16 | 10 |
Plus–minus values are means ±SD. Sample numbers include only genotyped persons who also provided consent for these analyses and had high-quality genotyping (i.e., that met quality-control criteria). ARIC denotes Atherosclerosis Risk in Communities, CHS Cardiovascular Health Study, and FHS Framingham Heart Study.
The definition of baseline risk factors was uniform in all four studies. Hypertension was defined with the use of criteria from the seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: systolic blood pressure of 140 mm Hg or more, diastolic blood pressure of 90 mm Hg or more, or the use of an antihypertensive agent. Diabetes mellitus was defined as a blood glucose level of 200 mg per deciliter (11 mmol per liter) or more in a random or 2-hour postprandial specimen, a fasting blood glucose level of 126 mg per deciliter (7 mmol per liter) or more, or the use of insulin or oral hypoglycemic agents. Cardiovascular disease was defined as the presence of congestive heart failure, coronary heart disease, or intermittent claudication.
In the CHS, persons with prevalent stroke or other cardiovascular disease were not genotyped.
STROKE DEFINITION, SURVEILLANCE, AND CLASSIFICATION
Stroke was defined as a focal neurologic deficit of presumed vascular cause with a sudden onset and lasting for at least 24 hours or until death if the participant died less than 24 hours after the onset of symptoms. Details of stroke surveillance and diagnostic criteria for stroke and stroke types in the four studies have been published17–24 and are summarized in Section 3 in the Supplementary Appendix. Strokes were classified as ischemic, hemorrhagic, or of “unknown” type on the basis of clinical and imaging criteria. Ischemic strokes were further subdivided into atherothrombotic and cardioembolic subtypes. We included ischemic strokes, hemorrhagic strokes, and strokes of unknown type in our analyses but excluded sub-arachnoid hemorrhages.
GENOTYPING
The consortium was formed after the individual studies had finalized their genomewide association study platforms. In the ARIC study, genotyping was performed with the GeneChip SNP Array 6.0 (Affymetrix); in the Cardiovascular Health Study, the HumanCNV370-Duo (Illumina) was used; in the Framingham Heart Study, the Gene-Chip Human Mapping 500K Array Set and 50K Human Gene Focused Panel (Affymetrix) were used; and in the Rotterdam Study, version 3.0 of the Infinium HumanHap550 chip (Illumina) was used. All studies used their genotype data to impute the 2.5 million autosomal single-nucleotide polymorphisms (SNPs) using HapMap CEU as the reference population. Imputation methods and quality-control measures are described in Section 4 in the Supplementary Appendix. We used a TaqMan assay (Applied Biosystems) to directly genotype the SNPs reaching genomewide significance in the combined analysis in each cohort in which the SNP had originally been imputed.
STATISTICAL ANALYSIS
Study-Specific Analysis
Cox proportional-hazards models were used in the individual studies to evaluate time to first stroke; participants were excluded at death or at the time of their last follow-up examination or health status update when they were known to be stroke-free. For the analyses of ischemic stroke, persons were also excluded when they had an alternative type of stroke (e.g., hemorrhagic or unknown). Each study fit an additive genetic model relating the genotype dose (0 to 2 copies of the minor allele) to the outcome (total stroke or ischemic stroke). Primary analyses were adjusted for age and sex. In addition, the ARIC study and the Cardiovascular Health Study adjusted the analysis for study site, and the Framingham Heart Study adjusted the analysis for familial structure and for whether the DNA samples had been subject to whole-genome amplification. We also adjusted the analyses yielding the most significant associations in order to account for baseline systolic blood pressure, hypertension (defined as systolic blood pressure ≥140 mm Hg, diastolic pressure ≥90 mm Hg, or the use of antihypertensive medications),25 and other risk factors for stroke described in the stroke risk profile of the Framingham Heart Study: diabetes, current smoking, and atrial fibrillation.26 Finally, we examined the association of the two SNPs that reached genome-wide significance in our initial genomewide association study with the subtype of atherothrombotic stroke. All studies screened for genetic variations in each population; these variations were negligible. Additional details concerning the statistical analyses are available in Section 5 in the Supplementary Appendix.
Combined Analysis of Study-Specific Data
We combined the results from the four cohorts using inverse-variance weighting (fixed-effects) analysis. After verification of DNA strands across studies and quality control, filtering, and imputation within each study, we restricted our analysis to the 2,194,468 autosomal SNPs that were common to all studies. Details of the analytic strategy are available in Section 6 in the Supplementary Appendix. We decided a priori on a genomewide significance threshold of 5×10−8, which corresponds to a target α level (P value) of 0.05 with a Bonferroni correction for 1 million independent tests. The linkage-disequilibrium pattern seen in ongoing detailed sequencing of the genome within European populations also provides support for the use of this threshold.27 SNPs with 5×10−8 ≤P <1×10−5 were considered to show a highly suggestive association; SNPs with 1×10−5 ≤P<1×10−4 were considered to show a moderately suggestive association.
Replication
The first replication sample comprised the black participants in the ARIC study.11 We genotyped 2430 persons (889 men; mean [±SD] age, 53±6 years) who were initially stroke-free and consented to genotyping. The stroke definition and surveillance methods were identical to those used for the white participants in the ARIC study.20 We genotyped rs12425791 with the use of the Affy-metrix GeneChip SNP Array 6.0 and rs11833579 (which was absent from the Affymetrix chip) by means of a TaqMan assay.
The Cardiovascular Health Study cohort included 574 black persons; genotyping of rs11833579 and rs12425791 in these persons was undertaken at the same time as in the white persons (with the use of a TaqMan assay). Further details on selection of the replication sample, genotyping quality-control filters, and analyses are outlined in Section 7 in the Supplementary Appendix.
We carried out a third test of replication in a sample of 652 white Dutch persons with prevalent stroke (313 women; mean age, 77±7 years; 501 ischemic strokes, of which 400 were atherothrombotic) and age-matched controls. We genotyped rs12425791 and rs11833579 in persons with prevalent stroke in the Rotterdam Study at baseline, in a clinical series from three hospitals in the Netherlands, and in controls selected from among Rotterdam Study participants who were stroke-free at baseline and within the same age range as the persons with stroke. We genotyped both SNPs with the use of the TaqMan system. Inclusion and exclusion criteria for the replication samples were the same as these used for the discovery samples.
RESULTS
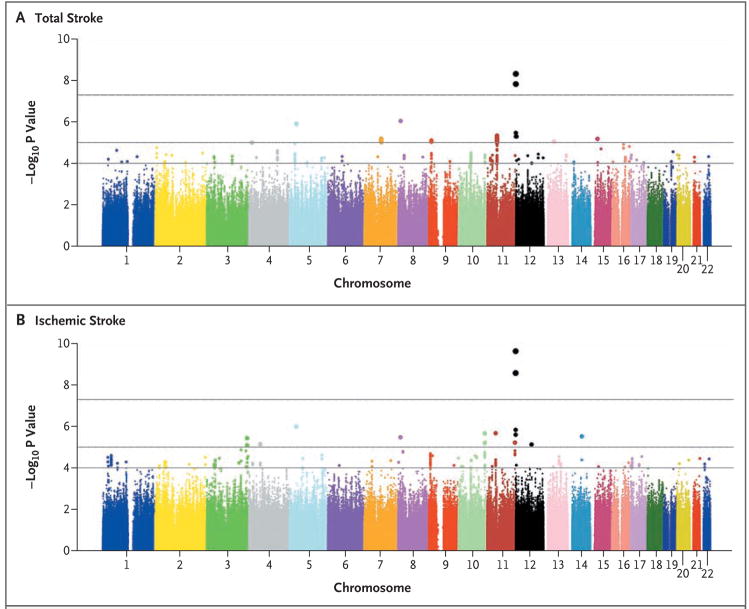
We observed 1544 incident strokes (1164 ischemic strokes) among 19,602 persons followed for an average of 11 years (Table 1). Figures 1A and 1B show the results of the genomewide association study for total and ischemic stroke. For highly suggestive loci with P<1×10−5, the hazard ratios and population attributable risks associated with the minor allele are shown in Table 2; results for all associated SNPs, including moderately suggestive loci with P<1×10−4, are shown in Tables 1 and 2 in the Supplementary Appendix. The full set of results can be obtained at the European Genotype Archive (www.ebi.ac.uk/ega; accession number, EGA00000000060).
Figure 1. Results of Tests for the Association between Stroke and Each SNP Measured in the Genomewide Association Study.
P values (based on the fixed-effects model) are shown in signal-intensity (Manhattan) plots relative to their genomic position for total stroke (Panel A) and ischemic stroke (Panel B). Within each chromosome, the results are plotted left to right from the p-terminal end. The top horizontal line indicates the chosen threshold for genomewide significance, P = 5×10−8; the middle line indicates the threshold for P = 10−5; and the bottom line indicates the threshold for P = 10−4.
Table 2.
Most Significant Associations between Single-Nucleotide Polymorphisms (SNPs) and Stroke Phenotype.*
| SNP | SNP Function | Minor Allele† | MAF | Chromosome: Position | Hazard Ratio (95% CI) | P Value | PAR | Closest Gene‡ | Second Closest Gene‡ | Additional SNPs at P<10−5 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Distance | Name | Distance | |||||||||
| Total stroke | ||||||||||||
| rs11833579 | Upstream | A | 0.23 | 12:645460 | 1.32 (1.20–1.44)§ | 4.8×10−9 | 0.13 | NINJ2 | 2.4 | WNK1 | 87.0 | 3 |
| rs12425791 | Intergenic | A | 0.19 | 12:653745 | 1.31 (1.19–1.44)§ | 1.5×10−8 | 0.11 | NINJ2 | 10.7 | WNK1 | 78.7 | 3 |
| rs713536 | Intergenic | T | 0.47 | 8:12943177 | 1.23 (1.13–1.33) | 9.1×10−7 | 0.18 | C8orf79 | 11.5 | DLC1 | 42.1 | |
| rs4867131 | Intergenic | A | 0.11 | 5:33047743 | 1.41 (1.23–1.62) | 1.2×10−6 | 0.09 | C5orf23 | 220.1 | NPR3 | 224.7 | |
| rs10734548 | Intronic | T | 0.22 | 11:46744149 | 1.23 (1.13–1.35) | 4.4×10−6 | 0.10 | CKAP5 | WG | F2 | 26.5 | 49¶ |
| rs11609145 | Intergenic | G | 0.20 | 12:3885559 | 1.31 (1.17–1.48) | 4.9×10−6 | 0.12 | PARP11 | 32.7 | EFCAB4B | 153.0 | |
| rs3211928 | Intronic | G | 0.45 | 7:79942371 | 0.83 (0.77–0.90) | 6.4×10−6 | 0.21|| | CD36 | WG | SEMA3C | 74.1 | 14 |
| rs877087 | Intronic | T | 0.45 | 15:31661567 | 1.22 (1.12–1.33) | 6.5×10−6 | 0.17 | RYR3 | WG | AVEN | 284.1 | |
| rs7853368 | Intergenic | G | 0.39 | 9:13447920 | 1.20 (1.11–1.29) | 7.8×10−6 | 0.14 | MPDZ | 207.5 | NFIB | 623.9 | 1 |
| rs4151467 | Intronic | C | 0.06 | 13:47817924 | 1.44 (1.23–1.69) | 8.7×10−6 | 0.05 | RB1 | WG | P2RY5 | 65.2 | |
| rs6449093 | Intergenic | G | 0.09 | 4:14638765 | 1.32 (1.27–1.49) | 9.8×10−6 | 0.06 | CPEB2 | 43.0 | C1QTNF7 | 379.1 | |
| Ischemic stroke | ||||||||||||
| rs11833579 | Upstream | A | 0.23 | 12:645460 | 1.41 (1.27–1.56)§ | 2.3×10−10 | 0.17 | NINJ2 | 2.4 | WNK1 | 87.0 | 3 |
| rs12425791 | Intergenic | A | 0.19 | 12:653745 | 1.39 (1.25–1.54)§ | 2.6×10−9 | 0.14 | NINJ2 | 10.7 | WNK1 | 78.7 | 3 |
| rs4867131 | Intergenic | A | 0.11 | 5:33047743 | 1.49 (1.27–1.75) | 9.9×10−7 | 0.10 | C5orf23 | 220.1 | NPR3 | 224.7 | |
| rs10837576 | Intergenic | A | 0.29 | 11:41103075 | 0.78 (0.70–0.85) | 2.1×10−6 | 0.27|| | LRRC4C | 830.8 | API5 | 218.7 | |
| rs10794579 | Downstream | T | 0.43 | 10:124676646 | 1.24 (1.13–1.35) | 2.1×10−6 | 0.18 | C10orf88 | 3.7 | FAM24A | 14.0 | 3 |
| rs2318308 | Intergenic | A | 0.27 | 14:65460852 | 0.75 (0.66–0.85) | 3.0×10−6 | 0.29|| | FUT8 | 181.1 | GPHN | 583.0 | |
| rs713536 | Intergenic | T | 0.47 | 8:12943177 | 1.26 (1.14–1.38) | 3.3×10−6 | 0.20 | C8orf79 | 11.5 | DLC1 | 42.0 | |
| rs17429019 | Intergenic | G | 0.06 | 3:190727847 | 1.49 (1.26–1.76) | 3.6×10−6 | 0.05 | TP63 | 104.1 | TPRG1 | 203.8 | 1 |
| rs12786704 | Intronic | G | 0.16 | 11:131684538 | 1.36 (1.19–1.55) | 6.0×10−6 | 0.10 | HNT | WG | OPCML | 105.5 | |
| rs6820391 | Intronic | A | 0.28 | 4:54255624 | 1.24 (1.13–1.36) | 6.9×10−6 | 0.12 | LNX1 | WG | FIP1L1 | 88.8 | 2 |
| rs11615969 | Intergenic | C | 0.09 | 12:75229404 | 1.53 (1.27–1.84) | 7.4×10−6 | 0.09 | BBS10 | 11.3 | OSBPL8 | 18.6 | |
P values, hazard ratios, and 95% confidence intervals (CIs) are based on a fixed-effects (inverse-variance–weighted) analysis. Each row specifically identifies only the SNP–phenotype association with the lowest P value for that locus, except for the two associations reaching genomewide significance that are both shown here. Four SNPs (rs11833579, rs12425791, rs713536, and rs4867131) were common to both total and ischemic stroke. The last column shows the number of additional SNPs at the same locus, within 250 kb of the specified SNP, that were also associated with the phenotype with a P value of less than 10−5. At the NINJ2 locus, these additional SNPs include rs728096 and rs7297967. MAF denotes minor-allele frequency, PAR population attributable risk, and WG within gene.
Alleles were identified on the basis of the plus strand of the National Center for Biotechnology Information (NCBI) build 36. The minor allele was also the coded allele. The MAF is based on allele frequency in the combined sample.
The closest gene and second closest gene show the Human Gene Organization (HUGO) Gene Nomenclature System symbols for the two genes located closest to each SNP and the distance of the associated SNP from the 5 end (start) of the gene. Standardized gene annotations for all SNP results were derived programmatically from the University of California, Santa Cruz, Genome Browser RefSeq gene track (hg18). Distances to genes are expressed in kilobase pairs, based on NCBI build 36.
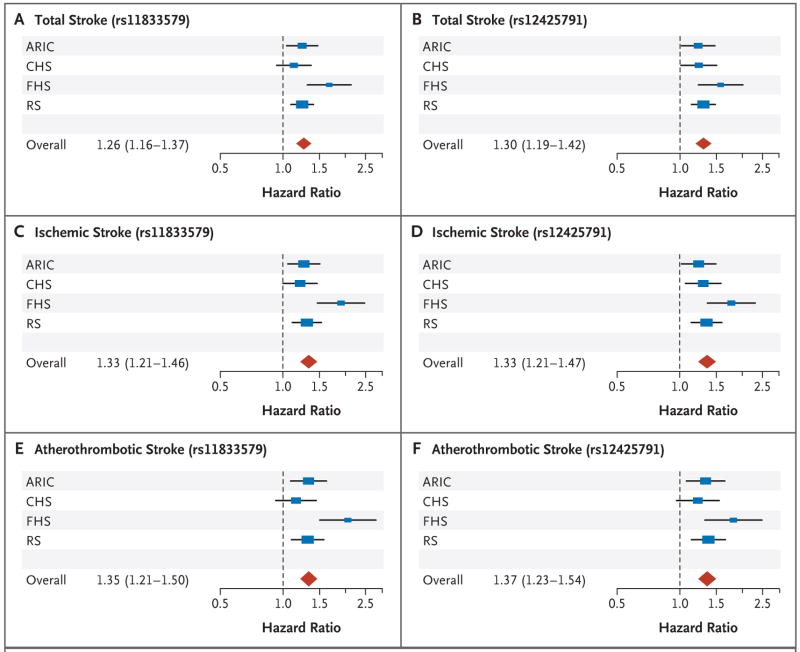
The hazard ratios, which were derived by combining genotyped data from the various genomewide association study platforms and from the TaqMan assay (for cohorts wherein the SNP was initially imputed), were as follows: 1.26 (95% CI, 1.16 to 1.37) for the association of rs11833579 with incident total stroke, 1.33 (95% CI, 1.21 to 1.47) with incident ischemic stroke, and 1.35 (95% CI, 1.21 to 1.50) with incident atherothrombotic stroke; for rs12425791, the corresponding hazard ratios were 1.30 (95% CI, 1.19 to 1.42), 1.33 (95% CI, 1.21 to 1.47), and 1.37 (95% CI, 1.23 to 1.54).
SNPs at this locus included intragenic SNPs within the following seven genes: CKAP5, F2, LRP4, ARFGAP2, C11orf49, DDB2, and PACSIN3. Functional annotation of all SNPs with P<1×10−6 was undertaken with the use of the FASTSNP program available online.28 We looked for SNPs that were intragenic nonsynonymous coding SNPs or intronic splice-site variants in at least moderate linkage disequilibrium (r2>0.5) with any of the significant or highly suggestive SNPs, with the use of the SNAP program (www.broad.mit.edu/mpg/snap/). We observed that rs107345486 and five other highly significant SNPs at this locus were in linkage disequilibrium with rs3816614, a nonsynonymous coding SNP within the LRP4 gene; rs10734548 was also in linkage disequilibrium with rs2070852, an intronic splice-site variant within the F2 gene.
Population attributable risks reported for these three SNPs were calculated with the use of the major allele as the risk allele, since the minor alleles were protective (i.e., the minor alleles had an inverse association with stroke risk).
P values for two SNPs located on chromosome 12p13 (rs11833579 and rs12425791) surpassed our preset threshold (P = 5×10−8) for genomewide significance both for total stroke and for ischemic stroke (Table 2, and Tables 1 and 2 in the Supplementary Appendix). The hazard ratios were larger and the P values were smaller for the association of these two SNPs with the more specific phenotype of ischemic stroke than for the association with total stroke, despite a smaller number of ischemic stroke events. Results of the initial genomewide association study suggested that each copy of a minor allele at these two loci increased the hazard ratio for total stroke by 1.31 to 1.32 (95% confidence interval [CI], 1.19 to 1.44) and for ischemic stroke by 1.39 to 1.41 (95% CI. 1.27 to 1.56). The corresponding population attributable risks were 11 to 13% for total stroke and 14 to 17% for ischemic stroke. We found no association between rs11833579 and rs12425791 and nonischemic stroke (hazard ratio, 1.13; 95% CI, 0.94 to 1.36; P = 0.20 for each SNP). The risk estimates for both SNPs were similar across the four studies, as shown by a Forest plot (Fig. 2 in the Supplementary Appendix). We also observed an association of rs11833579 and rs12425791 with ischemic stroke when we used genotypes obtained by means of a TaqMan assay.
We tested for an association between the two implicated SNPs and the atherothrombotic subtype of ischemic stroke, and we observed associations that were stronger than those of the two SNPs with all ischemic strokes. Thus, for rs11833579 hazard ratios were 1.26 (95% CI, 1.16 to 1.37) for total stroke, 1.33 (95% CI, 1.21 to 1.47) for ischemic stroke, and 1.35 (95% CI, 1.21 to 1.50) for atherothrombotic stroke. Hazard ratios for rs12425791 were 1.30 (95% CI, 1.19 to 1.42) for total stroke, 1.33 (95% CI, 1.21 to 1.47) for ischemic stroke, and 1.37 (95% CI, 1.23 to 1.54) for atherothrombotic stroke (Fig. 2).
Figure 2. Forest Plots Showing Associations between Single-Nucleotide Polymorphisms and Total, Ischemic, and Atherothrombotic Strokes.
The associations between rs11833579 and total stroke (Panel A), ischemic stroke (Panel C), and atherothrombotic stroke (Panel E) and between rs12425791 and total stroke (Panel B), ischemic stroke (Panel D), and atherothrombotic stroke (Panel F) are based on directly genotyped data. Individual studies (blue boxes) are plotted against the individual effect sizes (hazard ratios). The red diamonds indicate the overall hazard ratios. The size of the blue box is inversely proportional to the variance. Horizontal lines indicate 95% confidence intervals. The dashed vertical line in each panel shows the value for no effect (hazard ratio = 1.0). ARIC denotes Atherosclerosis Risk in Communities study, CHS Cardiovascular Health Study, FHS Framingham Heart Study, and RS Rotterdam Study.
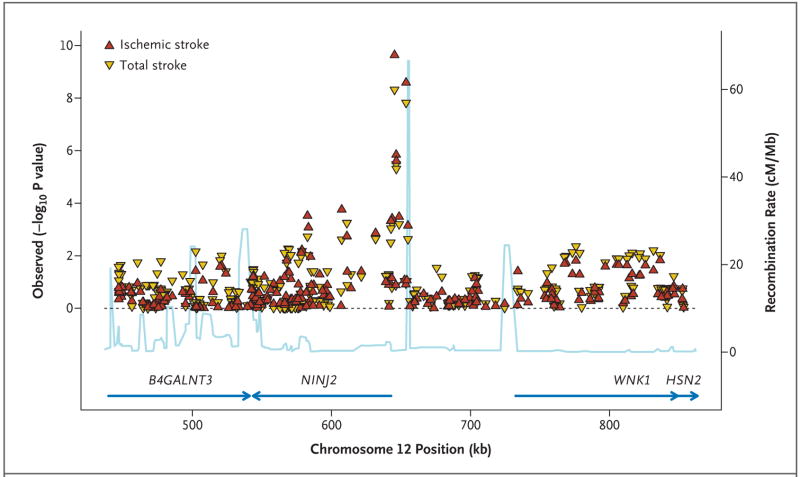
Both SNPs were in close proximity to NINJ2, which encodes ninjurin2, and they were in significant linkage disequilibrium with each other (r2 = 0.73 based on HapMap CEU data, National Center for Biotechnology Information [NCBI] build 36) and with SNPs in the 5′ untranslated region of NINJ2 (Fig. 3 in the Supplementary Appendix). Figure 3 shows all SNPs within a 200-kb region on either side of these two SNPs, together with the recombination rates and the known genes in that region. Two other SNPs that were also close to the 5′ end of NINJ2 (rs7298096 and rs7297967) showed a strong, albeit not significant, association with both total and ischemic stroke at P values of less than 5×10−5 (Table 2, and Tables 1 and 2 in the Supplementary Appendix). In addition, several other SNPs within NINJ2 showed a modest association (P<0.01) with both phenotypes (13 SNPs showed a modest association with total stroke, and 10 SNPs with ischemic stroke).
Figure 3. Associations in the Region Centered on rs11833579 and Containing NINJ2.
All single-nucleotide polymorphisms are plotted with their P values (on combined analysis) against their genomic position. P values for total stroke and ischemic stroke are shown. The light blue line represents the estimated recombination rates. Blue arrows indicate gene annotations.
The second closest gene to rs12425791 and rs11833579 is WNK1, although it is separated from these SNPs by a putative recombination hot spot. This gene encodes the lysine-deficient protein kinase 1. WNK1 has been related to blood-pressure levels and to the severity of hypertension in a general population.29,30 Adjusting the analyses for systolic blood pressure, hypertension, diabetes, atrial fibrillation, and current smoking individually or together had negligible effects on the observed associations (Table 3 in the Supplementary Appendix).
We observed a replication of the association between rs12425791 and stroke in the black participants in the ARIC study. Over a follow-up period of 15 years, 215 persons had an incident stroke (191 strokes were ischemic; of these, 153 were atherothrombotic). With a minor-allele frequency of 10% in this sample, rs12425791 was associated with incident total stroke (hazard ratio, 1.35; 95% CI, 1.01 to 1.79; P = 0.04), ischemic stroke (hazard ratio, 1.42; 95% CI, 1.06 to 1.92; P = 0.02), and atherothrombotic stroke (hazard ratio, 1.41; 95% CI, 1.01 to 1.95; P = 0.04). We observed no association between rs11833579 and stroke, probably because of the low linkage disequilibrium (r2 = 0.35) between the two SNPs in this sample. Our tests of association in the small sample of black persons in the Cardiovascular Heart Study, which included only 68 persons with incident ischemic strokes, did not provide evidence of replication. This sample had 21% power to detect a 30% increase in risk at an alpha level of 0.05.
In the Dutch case–control sample, we observed replication of the association of rs12425791 with total stroke (odds ratio, 1.17; 95% CI, 1.01 to 1.37; P = 0.03), ischemic stroke (odds ratio, 1.19; 95% CI, 1.01 to 1.41; P = 0.04), and atherothrombotic stroke (odds ratio, 1.29; 95% CI, 1.08 to 1.54; P = 0.005) and replication of the association of rs11833579 with the prevalence of atherothrombotic stroke (odds ratio,1.19; 95% CI, 1.00 to 1.40; P = 0.05).
A combined analysis of the discovery and replication samples of white subjects yielded, for rs12425791, an overall hazard ratio for the risk of ischemic stroke of 1.29 (95% CI, 1.19 to 1.41; P=1.1×10−9) (Table 5 in the Supplementary Appendix). We observed highly suggestive associations (which were not significant at a genome-wide level) with both the total stroke and ischemic stroke phenotypes (in 4 SNPs), with total stroke only (in 71 SNPs) and with ischemic stroke only (in 13 SNPs) (Table 2). We also examined associations with candidate SNPs previously reported to be significantly associated with either total or ischemic stroke (Section 8 and Table 6 in the Supplementary Appendix).
DISCUSSION
In this combined analysis of genomewide association study data from four large cohort studies of incident stroke, two previously unsuspected common SNPs on chromosome 12p13 were consistently associated with total, ischemic, and atherothrombotic stroke in white persons. We observed a replication of one of these SNPs in two independent samples: North American black persons and Dutch white persons. We did not observe a replication of the association of either SNP with stroke in a second, smaller sample of North American blacks.
We had a priori considered total stroke, a heterogeneous phenotype, as our primary outcome. However, the stronger association with ischemic and atherothrombotic strokes (as compared with total stroke) and the absence of any association with nonischemic stroke indicate that rs11833579 and rs12425791 were specifically associated with ischemic stroke and, in particular, the atherothrombotic stroke subtype. The effect sizes were similar when we tested for an association using directly genotyped (rather than imputed) SNPs, and the estimated hazard ratios for incident stroke were similar in all four discovery cohorts and in the black replication sample from the ARIC cohort.
Ninjurin2 is one of two transmembrane proteins in the ninjurin, or “nerve-injury-induced protein” family, and it has invertebrate homologues in the genomes of anopheles and drosophila species. It is a homophilic cell–cell adhesion molecule that interacts with matrix metalloproteinases.31 In the peripheral nervous system of male Sprague-Dawley rats, ninjurin is inducible in Schwann cells and dorsal-root ganglia through nerve injury, is transported from the perikaryon to the axon, and promotes neurite extension resulting in nerve regeneration.32 In the central nervous system, it is constitutively expressed at low levels by radial glia.32 Microarray studies have shown differences in the expression of ninjurin2 within the spinal cord of Nogo-A–specific knockout mice (in which neurites are increased by a factor of two to four after injury) as compared with wild-type controls.33 It is possible that the level of expression of ninjurin2 affects how the brain tolerates ischemic insults.
Wnk1 is expressed in the developing nervous system and in the aorta and brain vasculature of the mouse.34 WNK1 regulates the transport of sodium, potassium, and chloride ions across the plasma membrane.35,36 It phosphorylates the protein synaptotagmin, which results in an increased calcium requirement for synaptotagmin to bind to phospholipid vesicles, a process facilitating vesicle fusion.37 WNK1 mutations have been linked to familial hyperkalemic hypertension,35 to elevated ambulatory blood pressures in a community sample,29 and to the severity of essential hypertension.30 Although hypertension is a major risk factor for stroke,38 we did not observe a change in the strength of association between SNPs (rs11833579 or rs12425791) adjacent to WNK1 and stroke, after adjusting for systolic blood pressure or hypertension, suggesting that the role of WNK1 in regulating blood pressure does not explain the genetic associations we report here.
A recent genomewide association study of stroke cases and controls did not detect the NINJ2 signal.10 However, this study used a platform that does not genotype either rs11833579 or rs12425791 and has no proxy SNPs in moderate or significant linkage disequilibrium (r2≥0.4) with these two SNPs.
Our study has limitations. The identified intergenic SNPs are probably not the causal variants; it is more likely that they are in linkage disequilibrium with the causal variants. Further exploration of this genomic region with more detailed genotyping, expression, and translational studies will be required. There were too few events to study genotype–phenotype associations underlying individual subtypes of atherothrombotic stroke, such as cortical or lacunar ischemic strokes. Finally, we had limited power to detect associations with small effect sizes and associations with rare variants. Nonetheless, our analysis of genomewide association study data from four large community-based studies has identified a previously unsuspected association with incident stroke.
Supplementary Material
Acknowledgments
Supported by grants or contracts from the National Heart, Lung, and Blood Institute (N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022 R01HL087641, R01HL59367, and R01HL086694), the Na- tional Human Genome Research Institute (U01HG004402), and the National Institutes of Health (NIH) (HHSN268200625226C) and the NIH Roadmap for Medical Research (UL1RR025005) to the Atherosclerosis Risk in Communities study; grants from the National Heart, Lung, and Blood Institute (N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01-HC-15103, N01-HC-55222, N01-HC-75150, N01-HC-45133, U01HL080295, and R01HL087652), the National Institute of Neurological Disorders and Stroke (NINDS), the National Center for Research Resources (M01RR00069, to the Cedars–Sinai General Clinical Research Center Genotyping core), and the National Institute of Diabetes and Digestive and Kidney Diseases (DK063491, to the Southern California Diabetes Endocrinology Research Center) to the Cardiovascular Health Study; grants from the National Heart, Lung, and Blood Institute (N01-HC-25195) and its contract with Affymetrix for genotyping services (N02-HL-6-4278), the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center for the use of the Linux Cluster for Genetic Analysis; and by grants from the NINDS (NS17950) and the National Institute of Aging (AG08122, AG16495, and AG033193 to the Framingham Heart Study), the Nether-lands Organization of Scientific Research (175.010.2005.011), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Netherlands Consortium for Healthy Ageing (050-060-810), the Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands Organization for Health Research and Development, the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture, and Science, the Ministry for Health, Welfare, and Sports, the European Commission, and the Municipality of Rotterdam to the Rotterdam Study.
Dr. Lopez reports receiving consulting fees from Pfizer and Eisai and Neurochem and lecture fees from Novartis; and Dr. Kase, consulting fees from Sanofi-Aventis and Organon. No other potential conflict of interest relevant to this article was reported.
We thank the staff and participants of the Atherosclerosis Risk in Communities study, the Cardiovascular Heart Study, the Framingham Heart Study, and the Rotterdam Study for their important contributions; the investigators participating in the SNP Health Association Resource (SHARe) project of the Framingham Heart Study for developing tools used in the analysis; and Dr. Michael Moorhouse, Pascal Arp, Jeannette Vergeer, Dr. Marie Josee van Rijn, and Mila Jhamai for their help in creating the Rotterdam database.
APPENDIX
The authors’ affiliations are as follows: Rotterdam Study — the Departments of Epidemiology (M.A.I., Y.S.A., M.J.B., M.S., A.H., B.A.O., A.G.U., C.M.D., M.M.B.B.), Neurology (E.G.H., M.J.B., P.J.K., L.M.L.), Forensic Molecular Biology (M.S.), Internal Medicine (F.R., A.G.U.), and Clinical Chemistry (A.G.U.), the Erasmus MC University Medical Center, Rotterdam; Department of Neurology, St. Elisabeth Hospital, Tilburg (G.R., P.L.M.K.); and Department of Neurology, TweeSteden Hospital, Tilburg (G.R., P.L.M.K.) — all in the Netherlands; Framingham Heart Study — Department of Neurology, Boston University School of Medicine (S.S., A.L.D., S.D., A.B., Y.D., M.K.-H., C.S.K., P.A.W.) and Department of Biostatistics, Boston University School of Public Health (A.L.D., A.B., Y.D., Q.Y.) — both in Boston; Department of Neurology and Center for Neuroscience, University of California at Davis, Sacramento (C.D.); Cardiovascular Health Study — the Departments of Medicine (J.C.B., N.L.G., B.M.P., W.T.L.), Biostatistics (T.L., K.R.), Epidemiology (S.R.H., N.L.S., B.M.P., W.T.L.), Health Services (B.M.P.), and Neurology (W.T.L.), University of Washington; the Center for Health Studies, Group Health (S.R.H., B.M.P.), and the Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (N.L.S.) — all in Seattle; Brown Foundation Institute of Molecular Medicine and Graduate School of Biomedical Sciences, University of Texas Health Science Center at Houston, Houston (M.F.); the Department of Medicine and Pathology, University of Vermont, Burlington, (M.C.); the Departments of Neurology and Psychiatry, University of Pittsburgh School of Medicine, Pittsburgh (O.L.L.); the Medical Genetics Institute, Cedars–Sinai Medical Center, Los Angeles (T.H., K.D.T., J.I.R.); Atherosclerosis Risk in Communities Study — Brown Foundation Institute of Molecular Medicine and Human Genetics Center, University of Texas Health Science Center at Houston, Houston (M.F., E.B.); Division of Epidemiology and Community Health School of Public Health, University of Minnesota, Minneapolis (A.R.F.); Division of Epidemiology and Biostatistics, Mel and Enid Zuckerman College of Public Health, University of Arizona, Tucson (E.S.); Department of Epidemiology, University of North Carolina, Chapel Hill (W.D.R.); Department of Epidemiology and Welch Center for Prevention, Johns Hopkins Bloomberg School of Public Health, Johns Hopkins University, Baltimore (J.C.); Department of Medicine (Geriatrics), University of Mississippi Medical Center, Jackson (T.H.M.); Aging Gene-Environment Susceptibility-Reykjavik Study — Intramural Research Program, National Institute of Aging, Washington, DC (L.J.L.).Z
References
- 1.Rosamond W, Flegal K, Furie K, et al. Heart disease and stroke statistics — 2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117(4):e25–e146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 2.Bak S, Gaist D, Sindrup SH, Skytthe A, Christensen K. Genetic liability in stroke: a long-term follow-up study of Danish twins. Stroke. 2002;33:769–74. doi: 10.1161/hs0302.103619. [DOI] [PubMed] [Google Scholar]
- 3.Liao D, Myers R, Hunt S, et al. Familial history of stroke and stroke risk: the Family Heart Study. Stroke. 1997;28:1908–12. doi: 10.1161/01.str.28.10.1908. [DOI] [PubMed] [Google Scholar]
- 4.Jousilahti P, Rastenyte D, Tuomilehto J, Sarti C, Vartiainen E. Parental history of cardiovascular disease and risk of stroke: a prospective follow-up of 14371 middle-aged men and women in Finland. Stroke. 1997;28:1361–6. doi: 10.1161/01.str.28.7.1361. [DOI] [PubMed] [Google Scholar]
- 5.Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6:149–61. doi: 10.1016/S1474-4422(07)70028-5. [DOI] [PubMed] [Google Scholar]
- 6.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matarín M, Brown WM, Scholz S, et al. A genome-wide genotyping study in patients with ischaemic stroke: initial analysis and data release. Lancet Neurol. 2007;6:414–20. doi: 10.1016/S1474-4422(07)70081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gretarsdottir S, Thorleifsson G, Manolescu A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–9. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 11.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 12.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–76. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 13.Dawber TR, Kannel WB. The Framingham Study: an epidemiological approach to coronary heart disease. Circulation. 1966;34:553–5. doi: 10.1161/01.cir.34.4.553. [DOI] [PubMed] [Google Scholar]
- 14.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study: design and preliminary data. Prev Med. 1975;4:518–25. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 15.Hofman A, Breteler MM, van Duijn CM, et al. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22:819–29. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carandang R, Seshadri S, Beiser A, et al. Trends in incidence, lifetime risk, severity, and 30-day mortality of stroke over the past 50 years. JAMA. 2006;296:2939–46. doi: 10.1001/jama.296.24.2939. [DOI] [PubMed] [Google Scholar]
- 18.Hollander M, Koudstaal PJ, Bots ML, Grobbee DE, Hofman A, Breteler MM. Incidence, risk, and case fatality of first ever stroke in the elderly population: the Rotterdam Study. J Neurol Neurosurg Psychiatry. 2003;74:317–21. doi: 10.1136/jnnp.74.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longstreth WT, Jr, Bernick C, Fitzpatrick A, et al. Frequency and predictors of stroke death in 5,888 participants in the Cardiovascular Health Study. Neurology. 2001;56:368–75. doi: 10.1212/wnl.56.3.368. [DOI] [PubMed] [Google Scholar]
- 20.Rosamond WD, Folsom AR, Chambless LE, et al. Stroke incidence and survival among middle-aged adults: 9-year follow-up of the Atherosclerosis Risk in Communities (ARIC) cohort. Stroke. 1999;30:736–43. doi: 10.1161/01.str.30.4.736. [DOI] [PubMed] [Google Scholar]
- 21.Bots ML, Looman SJ, Koudstaal PJ, Hofman A, Hoes AW, Grobbee DE. Prevalence of stroke in the general population: the Rotterdam Study. Stroke. 1996;27:1499–501. doi: 10.1161/01.str.27.9.1499. [DOI] [PubMed] [Google Scholar]
- 22.Price TR, Psaty B, O’Leary D, Burke G, Gardin J. Assessment of cerebrovascular disease in the Cardiovascular Health Study. Ann Epidemiol. 1993;3:504–7. doi: 10.1016/1047-2797(93)90105-d. [DOI] [PubMed] [Google Scholar]
- 23.Seshadri S, Beiser A, Kelly-Hayes M, et al. The lifetime risk of stroke: estimates from the Framingham Study. Stroke. 2006;37:345–50. doi: 10.1161/01.STR.0000199613.38911.b2. [DOI] [PubMed] [Google Scholar]
- 24.Wolf PA, Kannel WB, Dawber TR. Prospective investigations: the Framingham Study and the epidemiology of stroke. Adv Neurol. 1978;19:107–20. [PubMed] [Google Scholar]
- 25.Chobanian AV, Bakris GL, Black HR, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA. 2003;289:2560–72. doi: 10.1001/jama.289.19.2560. [Erratum, JAMA 2003; 290:197.] [DOI] [PubMed] [Google Scholar]
- 26.Wolf PA, D’Agostino RB, Belanger AJ, Kannel WB. Probability of stroke: a risk profile from the Framingham Study. Stroke. 1991;22:312–8. doi: 10.1161/01.str.22.3.312. [DOI] [PubMed] [Google Scholar]
- 27.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179–85. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 28.Yuan HY, Chiou JJ, Tseng WH, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34(Web Server Issue):W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tobin MD, Raleigh SM, Newhouse S, et al. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation. 2005;112:3423–9. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 30.Newhouse SJ, Wallace C, Dobson R, et al. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Hum Mol Genet. 2005;14:1805–14. doi: 10.1093/hmg/ddi187. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Dailey GM, Kwan E, Glasheen BM, Sroga GE, Page-McCaw A. An MMP liberates the Ninjurin A ectodomain to signal a loss of cell adhesion. Genes Dev. 2006;20:1899–910. doi: 10.1101/gad.1426906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Araki T, Milbrandt J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J Neurosci. 2000;20:187–95. doi: 10.1523/JNEUROSCI.20-01-00187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimou L, Schnell L, Montani L, et al. Nogo-A-deficient mice reveal strain-dependent differences in axonal regeneration. J Neurosci. 2006;26:5591–603. doi: 10.1523/JNEUROSCI.1103-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaloy C, Hadchouel J, Imbert-Teboul M, Clemessy M, Houot AM, Jeunemaitre X. Cardiovascular expression of the mouse WNK1 gene during development and adulthood revealed by a BAC reporter assay. Am J Pathol. 2006;169:105–18. doi: 10.2353/ajpath.2006.051290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson FH, Disse-Nicodème S, Choate KA, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–12. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 36.Eladari D, Chambrey R. WNKs: new concepts in the regulation of NaCl and K+ balance. J Nephrol. 2007;20:260–4. [PubMed] [Google Scholar]
- 37.Shekarabi M, Girard N, Rivière JB, et al. Mutations in the nervous system–specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J Clin Invest. 2008;118:2496–505. doi: 10.1172/JCI34088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokes J, III, Kannel WB, Wolf PA, D’Agostino RB, Cupples LA. Blood pressure as a risk factor for cardiovascular diseased: the Framingham Study — 30 years of follow-up. Hypertension. 1989;13(Suppl):I-13–I-18. doi: 10.1161/01.hyp.13.5_suppl.i13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.