Abstract
In this article we consider a semiparametric generalized mixed-effects model, and propose combining local linear regression, and penalized quasilikelihood and local quasilikelihood techniques to estimate both population and individual parameters and nonparametric curves. The proposed estimators take into account the local correlation structure of the longitudinal data. We establish normality for the estimators of the parameter and asymptotic expansion for the estimators of the nonparametric part. For practical implementation, we propose an appropriate algorithm. We also consider the measurement error problem in covariates in our model, and suggest a strategy for adjusting the effects of measurement errors. We apply the proposed models and methods to study the relation between virologic and immunologic responses in AIDS clinical trials, in which virologic response is classified into binary variables. A dataset from an AIDS clinical study is analyzed.
Key words and phrases: AIDS clinical trial, generalized linear mixed-effects models, linear mixed-effects model, local linear, local quasilikelihood, longitudinal data, measurement error, penalized quasilikelihood
1 Introduction
In recent years, the relation between viral load and CD4 cell counts has been well studied (Wu et al., 1999; Wu, 2002; Liang, Wu, and Carroll, 2003). These studies have investigated the concordance and discordance between virologic and immunologic variables and may help clinicians more deeply understand AIDS pathogenesis and improve therapy regimes. Analysis of these AIDS data poses statistical challenges. A common feature of the markers, such as viral load measurements and CD4+ cell counts, used to evaluate antiviral therapy is that their measurement produces longitudinal data-that is, a series of independent and dependent variables obtained by taking repeated measurements, over time, from any one subject. Consideration of the features of longitudinal data, such as within-subject and between-subject variations and the correlation structure, is of practical interest.
Antiretroviral therapy for HID-1 infected patients has greatly improved in recent years. Administration of drug cocktails consisting of three or more drugs may reduce and maintain virus load copies below the detection limit in many patients, although it is unlikely that combination therapies alone can eradicate HID in infected patients because long-lived infected cells and sites within the body exist where drugs may not be effective. Because of the success of highly active antiretroviral therapy for HID infection, viral load in AIDS patients is suppressed and reaches to magnitudes that are below the limit of quantification, and AIDS becomes a chronic disease. Clinicians and patients are therefore sometimes only interested in maintaining the viral load below the detection limit, and in how the immune system (measured by CD4 cell counts) relates to this suppression.
Our primary goal in this paper is to propose a generalized partially linear mixed-effects model (GPLMeM) to study the relation between binary viral load measurement and CD4 cell count. Our focus is not only on the population characteristic but also on individual diversities. This is particularly important in AIDS research because that there is generally a large between-subject variation, which indicates the importance of estimating each individual’s parameters for the individualization of treatment management and care of patients with AIDS. We propose a new method to explore the population and individual characteristics by combining local linear regression (Fan and Gijbels, 1996), penalized quasilikelihood (PQL, Breslow and Clayton, 1993), and local quasilikelihood (Severini and Staniswalis, 1994) techniques.
For longitudinal binary data analysis, generalized linear mixed-effects models (GLMeM) have been proposed to incorporate the between-subject and within-subject variations. See, for example, Breslow and Clayton (1993) and Schall (1991) for the PQL method, Zeger and Karim (1991) for the Gibbs sampler. Wang et al. (1998) considered generalized linear mixed models when one of the covariates is measured with error, and they proposed a simulation extrapolation (SIMEX, Stefanski and Cook, 1995) estimation method.
To weaken model assumption for possible misspecification and to avoid the curse of dimensionality of fully nonparametric regression in the presence of several predictor variables, semipara-metric models have been studied and used for longitudinal data analysis in literature (Severini and Staniswalis, 1994). Lin and Carroll (2001) studied marginal semiparametric mixed-effects models and used the generalized estimating equation to estimate the parameter of interest. Wu and Zhang (2002) and Ke and Wang (2001) proposed semiparametric nonlinear mixed-effects models. Lin and Zhang (1999), Zhang (2004), and Zhang et al. (1998) used the smoothing spline to fit semi-parametric mixed-effects models. These authors considered between-subject and within-subject variations in their estimation methods, but they mainly aimed at delineating population features, while individual characteristics are ignored.
The article is organized as follows. In Section 2 we formally introduce the model’s framework, propose an estimation algorithm, and develop asymptotic results; extend to the case that X is measured with error; and discuss practical implementation of the methods. We illustrate the methods by conducting a small simulation experiment and extensively analyzing a data set from an AIDS study in Section 3 and provide a discussion in Section 4. The proof of the theoretical results is given in the Appendix.
2 Generalized Partially Linear Mixed-effects Models
2.1 Model
Suppose that data are obtained from n independent subjects with outcome variables Yij, linear covariates Xij(p × 1) and Aij(q × 1), and nonlinear scalar Zij, where i = 1, …, n indicates the subjects; j = 1, …, ni indicates the observation of the ith subject. Given the covariates Xij, Zij, Aij, and unobserved q × 1 random effects vectors bi and unobserved random effects function ci(·), the observations Yij are assumed to be independent with means μij and variance ϕωijV(μij), where ωij is a known weight and V(·) is a known variance function. The GPLMeM of Y given X, Z is specified by
| (2.1) |
where g(·) is a known monotonic differentiable link function, β is a p × 1 vector, and θ(·) and ci(·) are unknown smoothing functions. ci(·) indicates the random-effects, which can be regarded as a realization of a stochastic process with mean zero. The random effects bi are independent of ci(·) and are independently distributed N(0, Σb), where the covariance matrix Σb is determined by finite parameters. Given the covariates Xij, Aij and Zij, and bi and ci(·), the observations Yij and Yik are independent; i.e., E(Yij|Xij, Zij) = E{Yij|Xij, Zij, (Xik, Zik)k≠j}. In longitudinal data, Zij are often the treatment times, and Xij, Aij, and Yij are the observations of the ith subject at time Zij, for example, in the AIDS data set studied later.
2.2 Estimation
To estimate the parametric and nonparametric parts β, bi, θ(·), and ci(·), we combine local linear regression, the PQL technique, and the quasilikelihood principle. We estimate these parameters by considering local PQL, then we update the estimate of β and bi, using all data points, by relying on the global PQL. In local PQL, we approximate θ(·) and ci(·) by linear functions:
Denote α = (α0, α1)T, νi = (νi0, νi1)T, . In a neighborhood of z0, model (2.1) can be approximated by a GLMeM and described as
where νi ~ N(0, Σν) are random effects. In a conventional GLMeM setting, we can estimate α, β, bi, and νi by using the PQL principle; i.e., maximize
with respect to α, β, bi, and νi, where and .
To consider the localization, we propose a locally PQL approach. The motivation of doing so is essentially the same as localization in conventional nonparametric regression.
Denote ρk(t) = {dg−1)(t)/dt}k/[σ2V{g−1(t)}] and qk(t,y) = ∂kQ{g−1(t), y}/∂tk for k = 1, 2, 3. Let kj = ∫ ujK(u)du, μj = ∫ ujK2(u)du for j = 1, 2, , and .
Step 1
For each fixed z0, we consider the maximization
| (2.2) |
with respect to α, β, bi, and νi. Kh(·) = K(·/h)/h, K(·) is a kernel function, and h is a bandwidth.
Take the differential of (2.2) on α, β, bi, and νi. Step 1 is equivalent to solving the equations
| (2.3) |
and, for each i,
| (2.4) |
Denote , Zi = (Zi1,…Zini)T, and Ai similarly. Let Kih = diag{Kh(Zi1 − z0), …, Kh(Zini − z0)}, Ri = diag(Ri1, …, Rini) with Rij = {g′(μ̃ij)}2V(μ̃ij).
Equations (2.3) and (2.4) are approximately equivalent to iteratively fitting the following linear mixed-effects (Laird and Ware, 1982) model:
| (2.5) |
where εi ~ (0, Ri) and Ji = (Λi1, ···, (Λini)T.
Consequently, the estimates defined by step 1 are approximately given in closed-form expressions (Davidian and Giltinan, 1995):
where with . &thetas;(z0) and ci(z0) are estimated by θ̂(z0) = α̂0 and ĉi(z0) = νi0.
Note that β and bi are global parameters and their estimates given in step 1 are based only on local information. We update their estimate, using all data, by considering a global penalized quasilikelihood procedure:
Step 2
Update estimates of β and bi by maximizing the object function
| (2.6) |
with respect to β and bi, where .
The expressions containing matrices Σb, Σν, and Ri, which are generally unknown and need to be estimated. To estimate these matrices, one may use the maximum and the restricted maximum likelihood methods to estimate the unknown components of Σb, Σν, and Ri under the normality assumption. To implement these non-trivial methods, the EM-algorithm and New-Raphson methods have been proposed(Laird and Ware, 1982; Davidian and Giltinan, 1995). The standard statistical software packages such as SAS and Splus/R provide user-friendly functions to implement these methods( lme Splus/R function and the SAS procedure SAS MIXED). After obtaining the point estimates of the unknown components, we have Σ̂b, Σ̂ν, and R̂i, and then Σ̂i and Ω̂i. The estimators of α, β, bi, and μi thus can be obtained by substitution of Σ̂i and Ω̂i in steps 1 and 2.
2.3 Asymptotic Results
We claim the following condition, which is standard in the literature describing the generalized partially linear models.
Condition
The function q2(t, y) < 0 for t ∈ R and y in the range of the response variable.
The random-effects functions ci(z) are iid and have zero mean gaussian marginal distribution, and have the same distribution as a random-effects curve c(z), a two times continuously differentiable function.
The density function f(z) of Z is positive and continuous at the point z0.
The functions θ(·) and θ(2)(·) are continuous at the point z0.
With R = XTβ+ATb+θ(Z)+c(Z), , and are twice differentiable in z.
-
and , for some δ > 2.
Mh/N → λ, a finite nonnegative constant.
Let γy(z1, z2) be the covariance of [Y − g−1{XTβ + ATb + θ(Z) + c(Z)}]ρ1{XTβ + ATb+ θ (Z) + c(Z)} for Z = z1 and z2. γy(z1, z2) is continuous on z1 and z2.
The functions V″(·) and g‴(·) are continuous.
Theorem 1
Consider the estimator θ̂(z0) given in step 1. Then, as n → ∞, h → 0 and Nh → ∞, and under the specified condition, we have the asymptotic expansion
| (2.7) |
and hence
| (2.8) |
where
and
Theorem 2
Let β̂ be the estimate given in step 2. Under the condition, as n → ∞, Nh4 → 0 and Nh2/log(1/h) → ∞,
| (2.9) |
where B = E[ρ2(R)XXT] and .
As in generalized partially linear models for cross-sectional data, theorem 2 indicates that in order to estimate β at the root–n rate, one must undersmooth the nonparametric part θ(·). This undersmoothing request is standard in the kernel literature, although ordinary bandwidth rates are permissible in partially linear models.
2.4 Implementation
To obtain the estimates by fitting (2.5), we give an initial value for μ̃ij, say , and then have the working response variable
For a given bandwidth h, we fit model (2.5) and obtain estimates α̂, β̂, b̂i, and ν̂i. Update and g′(μ̂ij). Correspondingly, Ỹij can be updated as g(μ̂ij) + (yij − μ̂ij)g′(μ̂ij), and fit (2.5) again. Repeat these procedures until convergence, and collect the final estimates θ̂ (z0) = α̂0, ĉi(z0) = ν̂i0.
During the process of the implementation, one should keep in mind that the bandwidth needs to be selected in the estimation procedure involved in step 1. Theorem 2 indicates that undersmoothing the nonparametric part is necessary to guarantee to estimate β at the root-n rate. Because only the rates of convergence for the bandwidth h are necessary for the same limiting distribution for the estimators of β, we select h by using the empirical bias bandwidth selection method (Ruppert, 1997) in our data analysis. In our numerical analysis below, we select initial values by fitting the data by a generalized linear mixed-effects model. Because iteration is needed, we set the maximum number of iteration to be 30. Computation results for our numerical analysis are obtained within 18 iterations. This adoption leads to potentially burdensome computation, which may be attenuated with a powerful computation platform.
2.5 Measurement errors in covariates
In practice, the covariates may not be exactly observable, and like CD4 cell counts in our AIDS dataset analyzed later, are measured with substantial errors. The estimators and inference may be biased if one ignores these measurement errors. We need to adjust the resulting bias. In this special case, we denote Xij by Xi(Zij), the true covariates of subject i, and Wij by Wi(Zij), the observed values of Xij at time Zij. Suppose we have the measurement error model,
| (2.10) |
where Wi(z) is the observed value and Xi(z) is the underlying true value for the ith patient at treatment time z. The error ui(z) represents measurement error in the CD4+ cell counts. We assume that ui(z) is a mean zero process, and {Xi(z), ui(z)} are mutually independent. Fuller (1987) and Carroll, Ruppert, and Stefanski (1995) provided a good survey of measurement error models. Buonaccorsi, Demidenko, and Tosteson (2000) studied measurement errors in linear mixed-effects models. Higgins, Davidian, and Giltinan (1997) proposed a two-step approach to deal with measurement errors in nonlinear mixed-effects models. Wang et al. (1998) studied generalized linear mixed measurement error models. Liang and Ren (2005) investigated generalized partially linear models with errors-in-variables and proposed a measurement error calibration based on a SIMEX algorithm. The computation burden in a similar platform is tremendously expensive, although its implementation is straightforward. In this paper we adjust measurement error by using a mixed-effects regression spline model, as done by Higgins et al. (1997); a similar idea was used by Liang, Wu, and Carroll (2003).
The data points repeatedly measured along time z are similar to replications if we can assume that the measured variable is a smooth function of z. Shi, Weiss, and Taylor (1996) and Rice and Wu (2001) have modeled the natural history of the CD4+ cell process in untreated HID-infected patients by using a mixed-effects regression spline model. Model the CD4+ cell counts as follows:
| (2.11) |
where Ψi(z) = {ψ0(z), ψ1(z), ···, ψp(z)} and Φi(z) = {φ0(z), φ1(z), ···, φq(z)} are basis functions such as cubic B-spline basis, ξ = (ξ0, ξ1, ···, ξp)T is a fixed-effect parameter vector, and ηi = (ηi0, ηi1, ···, ηiq)T is a random effect vector with mean zero and covariance matrix Ση (Ση may be unstructured or can be specified with a special structure). The selection of the number (p and q) and locations of knots for regression splines can be achieved by using cross-validation (Eubank, 1999). Rice and Wu (2001) suggest setting p = q and ψk(z) = φk(z). Model (2.11) is a standard linear mixed-effects model, which can be fitted by using the LME function of Splus (Pinheiro and Bates, 2000).
Higgins et al. (1997) have proposed a two-step approach to deal with measurement errors in time-dependent covariates in nonlinear mixed-effects models. The first step is to estimate the covariate function by fitting an appropriate model for covariate processes, and then fit the nonlinear mixed-effects model by plugging in the estimates of the covariates in the second step. This is essentially similar to the regression calibration idea (Carroll et al. 1995). By applying a similar idea to our model for measurement error in CD4+ cell counts, we fit a LME model,
| (2.12) |
and obtain an estimate Ŵij of Wij, which is regarded as Xij.
3 Numerical Illustration
In this section, we first conduct a small simulation study for an illustration. We then use the proposed methods to analyze a data set from an AIDS study. In our numerical analysis, we calculate the naive estimates, i.e., those obtained by ignoring measurement errors, the proposed estimates. We use the quartic kernel k(u) = 15/16(1 − u2)2I(|u|≤1) for nonparametric regression.
3.1 A Small Simulation Experiment
We performed a small simulation experiment and the data were generated from a logistic model, whose log(odd) can be expressed as
where Xij is independent uniform (0, 1) components, Zij is uniformly distributed U(0, 1), and Ui is normally distributed . The parameter β is equal to 1.85, and the nonparametric function is θi(z) = (1 + c1i) cos(2πz) + (2 + c2i) sin(2πz) with (c1i, c2i) being 2-dimensional normal distribution .
We consider a combination of (n, m) = (20, 30), (40, 50), (60, 70), σu = 0.1, 0.25, and σb = 0.1. In each of configurations, we run 500 replications. We calculate the proposed estimates and the naive estimates (i.e., we ignore the measurement errors). The estimated values of β are summarized in Table 1.
Table 1.
The estimates with standard errors (s.e.) of the parameter β for the simulated data. “Mean”: the means of the estimated values; “s.e.”: sample standard errors based on 500 replications.
| n | m | σu | Proposed | Naive | ||
|---|---|---|---|---|---|---|
| mean | s.e. | mean | s.e. | |||
| 20 | 30 | 0.1 | 1.876 | 0.355 | 1.784 | 0.352 |
| 40 | 1.873 | 0.256 | 1.759 | 0.243 | ||
| 60 | 1.887 | 0.305 | 1.742 | 0.299 | ||
| 20 | 50 | 1.831 | 0.338 | 1.65 | 0.337 | |
| 40 | 1.822 | 0.227 | 1.67 | 0.266 | ||
| 60 | 1.848 | 0.18 | 1.706 | 0.201 | ||
| 20 | 70 | 1.824 | 0.238 | 1.672 | 0.238 | |
| 40 | 1.796 | 0.214 | 1.627 | 0.208 | ||
| 60 | 1.813 | 0.18 | 1.628 | 0.185 | ||
| 20 | 30 | 0.25 | 1.89 | 0.343 | 1.298 | 0.354 |
| 40 | 1.885 | 0.317 | 1.257 | 0.326 | ||
| 60 | 1.891 | 0.264 | 1.253 | 0.264 | ||
| 20 | 50 | 1.822 | 0.258 | 1.128 | 0.254 | |
| 40 | 1.799 | 0.237 | 1.1 | 0.248 | ||
| 60 | 1.832 | 0.207 | 1.179 | 0.206 | ||
| 20 | 70 | 1.821 | 0.239 | 1.127 | 0.219 | |
| 40 | 1.79 | 0.197 | 1.128 | 0.194 | ||
| 60 | 1.809 | 0.192 | 1.089 | 0.195 | ||
The results are in accord with the theory for measurement error models. The simulation results based on the proposed method indicate that the naive estimator of β is markedly biased, while the estimated values based on the proposed method is much closer to the true value than the naïve estimate of β.
3.2 Data Analysis
Let Y be binary viral load, X be CD4 cell counts and Z be the treatment time. An ordinary logistic mixed-effects model says that the logit of Y = 1 satisfies
| (3.1) |
where b1i and b2i reflect the individual variations. The merit of this model includes its computational convenience and easy interpretation of the model parameter. However, this model may not be able to capture some curvature, as we shall see later because drug resistance, and noncompliance probably contribute nonlinearly along the treatment time. An alternative is a partially logistic mixed-effects model, described as
| (3.2) |
where θ(z) and ci(z) are unknown smoothing functions, which describe the population characteristic and individual diversities.
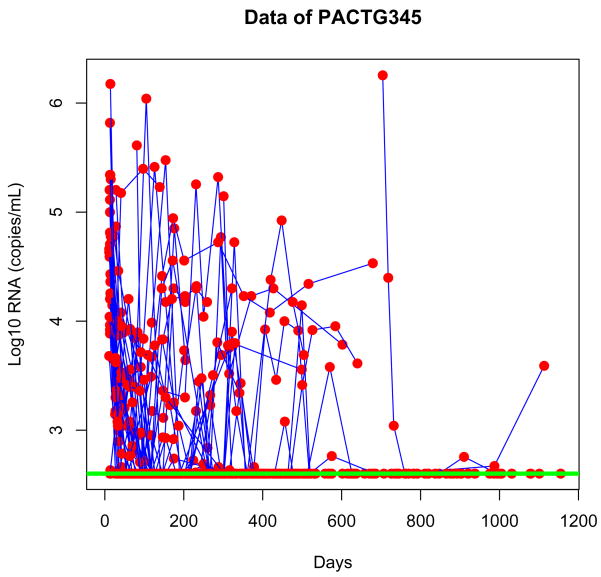
In this section, we present the results of analysis of an AIDS clinical study conducted by the Pediatric AIDS Clinical Trials Group (PACTG 345, Scott et al., 2001). In this study, 33 patients were enrolled as cohort II. Specimens were obtained on days 0, 1, 3, 7, 14, 28, 56, …, 1115, and 559 observations were obtained with 256 HID-1 RNA measurements below the detection limit of 400 copies/mL: 45% viral load observations were therefore suppressed below the detection limit. Figure 1 presents the individual observations of plasma HID RNA concentration (viral load) after initial antiretroviral treatments. A main objective of the treatment is to suppress the viral load below the limit of detection.
Figure 1.
Individual viral load measurements of plasma HID RNA concentration in the PACTG 345 study. The detection limit of 400 copies of HID RNA per mL of plasma is indicated by the horizontal line.
The purpose of this analysis focuses on whether the viral load is suppressed and how the of CD4 cell counts change during the treatment. We apply the model and estimation method described in section 2 to explore this data set and address the concerns mentioned in Section 1 by modeling the dynamic relation between the binary response (with or without viral suppression) and CD4 cell counts during the treatment period of about 3 years.
Let Yij = 1 if the viral load of the ith subject at time Zij is below the detection limit, and 0 otherwise. In this analysis, we consider 4 scenarios: (i) model (3.1) and ignoring consideration of measurement error in CD4+ cell counts; (ii) model (3.1) with consideration of measurement error in CD4+ cell counts; (iii) model (3.2), ignoring measurement error in CD4+ cell counts; and (iv) model (3.2) with consideration of measurement error in CD4+ cell counts. When considering measurement errors in CD4+ cell counts and using the method proposed in subsection 2.5 to produce the “true” CD4+ cell counts, the smoothing parameters (the number of knots) are selected by the model selection criterion BIC and the location of knots is selected at the quantiles of the data (Eubank, 1999). We obtain p = q = 3. To stabilize the variance and computational algorithms, we centered the covariate CD4+ cell counts and took a log-transformation for time in the model fitting. We fitted the model by using the LME function of Splus.
The population estimated values of the parameter β for these 4 scenarios are presented in Table 2. Comparing the estimates of β under (i) scenarios to (iii) and (ii) to (iv), we saw that models (3.1) and (3.2) derive remarkably different results. We also compared the estimates obtained with or without considering measurement error in the CD4+ cell counts. The estimates attenuated toward zero regardless of the models (3.1) or (3.2) used when the measurement error in the covariate is not considered. This finding is similar to that in standard linear or nonlinear regression models with measurement error (Carroll et al. 1995).
Table 2.
Estimates of the parameters in scenarios (i)–(iv), based on PACTG 345 data
| (i) | (ii) | (iii) | (iv) | |
|---|---|---|---|---|
| β̂ | −0.272 | −0.358 | −0.692 | −1.046 |
| SE | 0.10 | 0.143 | 0.134 | 0.223 |
| Pvalue | 0.007 | 0.0014 | < 10−4 | < 10−4 |
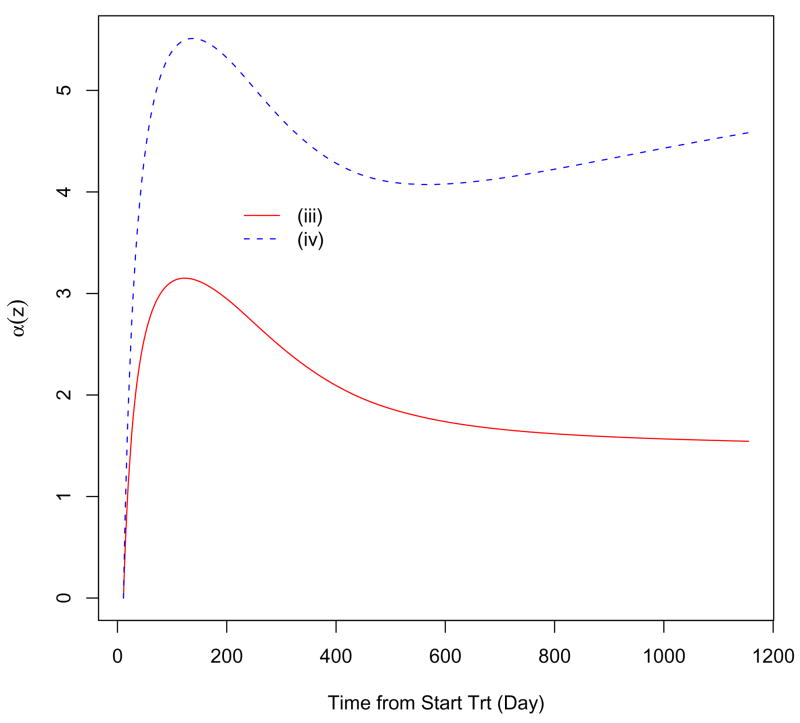
The population estimates of θ(z) for scenarios (iii) and (iv) are shown in Figure 2. Two population curves have similar patterns; that is, the curves rise rapidly at first and then maintain a steady decline from day 56 to the end of treatment. However, a consideration of measurement error reflects a sharper increase at the beginning.
Figure 2.
Population curve estimates of θ(z) considering measurement errors (solid line) and ignoring measurement errors (dashed line).
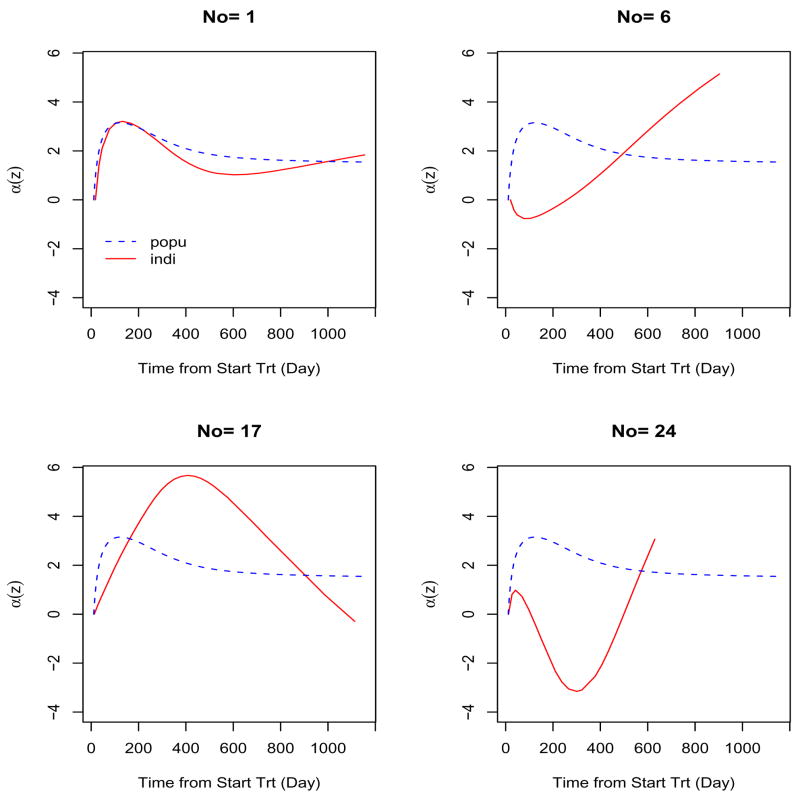
Recalling the viral load of each subject shown in Figure 1, we note that the between-subject variation can not be ignored and the individual curves, αi(z) = θ(z) + ci(z), may not follow the pattern of the population curve. We present the results for 4 individual subjects, and these results illustrate the principal advantage of the proposed model and developed methods in that the estimates can be obtained for both population and individuals. Because the patterns for the same individual under scenarios (iii) and (iv) are similar, we report in detail only the results for the case in which measurement errors in CD4+ cell counts are ignored. Figure 3 shows the individual estimates from 4 patients. For comparison, the corresponding population estimate is also plotted. The population and individual estimates are different not only in magnitude but also in patterns of change. The individual curve for subject 1 almost follows the population curve. The individual curves of the other 3 subjects are totally different from the population curve. Curves of subjects 6 and 24 decline sharply at first and then rebound later, to the end of treatment, but have different rebounding times. In contrast, the curve of subject 17 follows a convex shape. Given this large between-subject variation, the estimated trajectories of the individual curves become very important for individualizing treatment management and care for AIDS patients.
Figure 3.
Selected individual curve estimates (solid line), with the population curve estimate (dashed line) provided for comparison.
4 Discussion
In order to study the relation between binary virologic variables and immunologic variables, repeatedly measured indices of success of suppression of viral load (the virologic variable) and CD4+ cell counts (the immunologic variable) in AIDS clinical trials, we proposed a semiparamet-ric mixed-effects model, which can parsimoniously reflect both population and individual relationships between the two longitudinal variables. Similar models have been reported in the literature. However, most published studies focused only on the estimation of the population characteristic, although between-subject and within-subject variations have been incorporated. The method proposed in this article extends existing methods and allows us to estimate the population profile and individual diversities.
In step 1 we minimized the objective function with respect to α, β, bi and νi. Alternatively, we may specify initial values β(0) and , and maximize the objective function (a similar form to (2.2) but replacing β and bi with β(0) and ) with respect to α and νi. This alternative may increase efficiency (see Carroll et al., 1997 for a related discussion), but it increases computation efforts.
When considering measurement errors in CD4 cell counts, we used a mixed-effects based approach to calibrate the effect of measurement error. This calibration may also be achieved by using a SIMEX-based procedure such as Liang and Ren (2005). However, additional information is required to estimate the covariance matrix cov(X) in implementing the SIMEX procedure. This investigation also increases computation efforts.
In this article, we develop a method using local linear regression. There are a lot of alternative ways for local linear approximation in step 1, including higher degree local polynomial kernel methods and smoothing and regression splines. The details for these methods require a further investigation in our setting. We chose the local linear smoother because theoretical results can be derived and the estimators of nonparametric components do not suffer from boundary effects (Fan and Gijbels, 1996).
Model (2.1) may be extended to a generalized additive partially linear mixed-effect model, of the form
The study of this model is interesting and requires tedious efforts, but is beyond the scope of this paper.
Acknowledgments
The author appreciates the Editor, Associate Editor, and two referees for their valuable comments and suggestions. This research was partially supported by NIH/NIAID grants AI62247 and AI59773.
Appendix
A.1 Proof of Theorem 1
Let cn = (Nh)−1/2,
and let f(z) denote the density function of Zij. Denote further . (2.3) implies that β̂* maximizes
with respect to β*. By the concavity of the function ℓn(β*) and Taylor expansion of the function Q{g−1(·), Yij} with respect to β*, we obtain that
| (A.1) |
where and . Furthermore, let . Without loss of generality, assume that the means of random-effects terms b̂i − bi, ĉ(z0) − ci(z0), and are zeros. Then
| (A.2) |
where ζij is between and η̄ij. Note that . Using an argument similar to the proof of (A.9) of Carroll et al. (1997) and condition (i) yield that the second term in the (A.2) is of order o(h). We therefore have
In a similar way, we obtain that
Let
We write and deal with the first two moments of . Note that . We find for the first moment, with η̄ = XTβ + ATb + θ(z0) + θ(z0)(Z − z0) + c(z0) + c′(z0)(Z − z0),
| (A.3) |
The first term in (A.3) is of order O(h2). It follows that
| (A.4) |
In an analogous way we can show that
| (A.5) |
Combining (A.4) and (A.5), we obtain
| (A.6) |
Therefore, by (A.1),
| (A.7) |
By applying the convexity lemma (Pollard, 1991), we obtain that . Hence the asymptotic normality of β̂* will follow from that of , which we will establish below. By the definition of and using Taylor’s expansion we get for the first moment,
| (A.8) |
It follows from these statements and (A.6) that
where with sij = q1(ηij, yij)Kh(Zij − z0). We have
where
It is easy to see that
and
Furthermore, for j1 ≠ j2, by condition (h)
which, combined with condition (g), yields that
It follows that
Finally, using condition (f), it can be shown that Liapounov’s condition is satisfied and hence Theorem 1 holds, as claimed.
A.2 Proof of Theorem 2
First of all, we note that under the specified condition, in a proof similar to that for theorem 2 of Carroll et al. (1997), we can establish
| (A.9) |
Let ζ̂ = N1/2(β̂ − β0), , and . Then, ζ̂ maximizes
| (A.10) |
with respect to ζ. By Taylor’s expansion, we have
| (A.11) |
where
with ζnij and between 0 and , independent of Yij, and with . It can be shown that
| (A.12) |
Using similar arguments as for obtaining (A.12), we get
By (A.9), the second term in the above expression can be expressed as
Tn1 can further be simplified as
| (A.13) |
Combining (A.10)–(A.13) we obtain that
where
By the convexity lemma (Pollard, 1991) we find that , from which it follows that
as claimed.
References
- Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. J Am Statist Assoc. 1993;88:9–25. [Google Scholar]
- Buonaccorsi JP, Demidenko E, Tosteson TD. Estimation in longitudinal random effects models with measurement error. Statist Sinica. 2000;10:885–904. [Google Scholar]
- Carroll RJ, Fan J, Gijbels I, Wand MP. Generalized partially single-index models. J Am Statist Assoc. 1997;92:477–489. [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA. Nonlinear Measurement Error Models. New York: Chapman and Hall; 1995. [Google Scholar]
- Davidian M, Giltinan D. Nonlinear Models for Repeated Measurement Data. New York: Chapman and Hall; 1995. [Google Scholar]
- Eubank RL. Nonparametric Regression and Spline Smoothing. New York: Marcel Dekker; 1999. [Google Scholar]
- Fan J, Gijbels I. Local Polynomial Modeling and Its Applications. London: Chapman and Hall; 1996. [Google Scholar]
- Fuller WA. Measurement Error Models. New York: John Wiley; 1987. [Google Scholar]
- Higgins KM, Davidian M, Giltinan DM. A two-step approach to measurement error in time-dependent covariates in nonlinear mixed-effects models, with application to IGF-I pharmacokinetics. J Am Statist Assoc. 1997;92:436–448. [Google Scholar]
- Ke CL, Wang YD. Semiparametric nonlinear mixed-effects models and their applications (with discussions) J Am Statist Assoc. 2001;96:1272–1298. [Google Scholar]
- Laird NM, Ware JH. Random effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Liang H, Ren HB. Generalized partially linear measurement error models. J Comp Graph Statist. 2005;14:237–250. [Google Scholar]
- Liang H, Wu HL, Carroll RJ. The relationship between virologic and immunologic responses in AIDS clinical research using mixed-effect varying-coefficient semiparamet-ric models with measurement error. Biostatistics. 2003;4:297–312. doi: 10.1093/biostatistics/4.2.297. [DOI] [PubMed] [Google Scholar]
- Lin XH, Carroll RJ. Semiparametric regression for clustered data using generalized estimating equations. J Am Statist Assoc. 2001;96:1045–1056. [Google Scholar]
- Lin XH, Zhang DW. Inference in generalized additive mixed models by using smoothing splines. J R Statist Soc B. 1999;61:381–400. [Google Scholar]
- Pinheiro JC, Bates DM. Mixed-Effects Models in S and S-PLUS. New York: Springer; 2000. [Google Scholar]
- Pollard D. Asymptotics for least absolute deviation regression estimators. Economet Theory. 1991;7:186–199. [Google Scholar]
- Ruppert D. Empirical-bias bandwidths for local polynomial nonparametric regression and density estimation. J Am Statist Assoc. 1997;92:1049–1062. [Google Scholar]
- Rice JA, Wu CO. Nonparametric mixed effects models for unequally sampled noisy curve. Biometrics. 2001;57:253–259. doi: 10.1111/j.0006-341x.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- Schall R. Estimation in generalized linear models with random effects. Biometrika. 1991;78:717–727. [Google Scholar]
- Scott ZA, Chadwick EG, Gibson LL, et al. Infrequent detection of HIV-1-specific, but not cytomegalovirus-specific, CD8+T cell responses in young HIV-1-infected infants. J Immunology. 2001;167:7134–7140. doi: 10.4049/jimmunol.167.12.7134. [DOI] [PubMed] [Google Scholar]
- Severini TA, Staniswalis JG. Quasilikelihood estimation in semiparametric models. J Am Statist Assoc. 1994;89:501–511. [Google Scholar]
- Shi M, Weiss RE, Taylor JMG. An analysis of pediatric CD4+ counts for acquired immune deficiency syndrome using flexible random curves. Applied Statistics. 1996;45:151–163. [Google Scholar]
- Stefanski LA, Cook JR. Simulation-extrapolation: the measurement error jackknife. J Am Statist Assoc. 1995;90:1247–1256. [Google Scholar]
- Wang NS, Lin XH, Gutierrez RG, Carroll RJ. Bias analysis and SIMEX approach in generalized linear mixed measurement error models. J Am Statist Assoc. 1998;93:249–261. [Google Scholar]
- Wu HL, Kuritzkes DR, Clair MS, et al. Characterizing individual and population viral dynamics in HIV-1-infected patients with potent antiretroviral therapy: correlations with host-specific factors and virological endpoints. J Infectious Disease. 1999;179:799–897. [Google Scholar]
- Wu HL, Zhang JT. Semiparametric nonlinear mixed-effects models for longitudinal data application to an AIDS clinical study. Statist Med. 2002;21:3655–3675. doi: 10.1002/sim.1317. [DOI] [PubMed] [Google Scholar]
- Wu L. A joint model for nonlinear mixed-effects models with censoring and covariates measured with error, with application to AIDS studies. J Am Statist Assoc. 2002;97:955–964. [Google Scholar]
- Zeger SL, Karim MR. Generalized linear models with random effects: a gibbs sampling approach. J Am Statist Assoc. 1991;6:79–86. [Google Scholar]
- Zhang D. Generalized linear mixed models with varying coefficients for longitudinal data. Biometrics. 2004;60:8–15. doi: 10.1111/j.0006-341X.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- Zhang D, Lin X, Raz J, Sowers M. Semiparametric stochastic mixed models for longitudinal data. J Am Statist Assoc. 1998;93:710–719. [Google Scholar]