Abstract
Methionine sulfoxide (MetO) is a common posttranslational modification to proteins occurring in vivo. These modifications are prevalent when reactive oxygen species levels are increased. To enable the detection of MetO in pure and extracted proteins from various sources, we have developed novel antibodies that can recognize MetO-proteins. These antibodies are polyclonal antibodies raised against an oxidized methionine-rich zein protein (MetO-DZS18) that are shown to recognize methionine oxidation in pure proteins and mouse and yeast extracts. Furthermore, mouse serum albumin and immunoglobulin (IgG) were shown to accumulate MetO as function of age especially in serums of methionine sulfoxide reductase A knockout mice. Interestingly, high levels of methionine-oxidized IgG in serums of subjects diagnosed with Alzheimer’s disease were detected by western blot analysis using these antibodies. It is suggested that anti-MetO-DZS18 antibodies can be applied in the identification of proteins that undergo methionine oxidation under oxidative stress, aging, or disease state conditions.
Keywords: Oxidative stress, post-translation modification, aging, neurodegenerative diseases, Alzheimer’s disease, prion protein, serum proteins
Introduction
Oxidative damage to proteins is considered to be one of the major causes of aging and age-related diseases, and thus mechanisms have evolved to prevent or reverse these modifications. Methionine (Met) oxidation is one of the most common posttranslational modifications to proteins mediated by reactive oxygen species (ROS) that may alter protein structure and function. During conditions of oxidative stress, there is an accumulation of proteins containing methionine sulfoxide (MetO) [1, 2]. Insufficient reversal of protein-MetO to Met by the methionine sulfoxide reductase system (Msr family consisting of MsrA and MsrB) could cause certain proteins to loose their function, aggregate, and be toxic to the cell [1–4]. This function of the Msr system is supported by the fact that Met oxidation can denature proteins and convert the hydrophobic properties of Met into hydrophilic properties; thereby causing structural alterations to selected proteins that may affect their function and may be reversed by the Msr system in vivo [5]. Thus far, Met residue oxidation has been shown to affect the activities of many proteins, mostly in vitro [3]. Examples include the potassium channel of the brain (as demonstrated in an ex vivo system) [6], an isoform of the inhibitory protein κB [7, 8], and calmodulin [9]. The general role of MsrA as a protein-function regulator and as antioxidant has been demonstrated in various tissues. For example, a recent report has emphasized the importance of MsrA in the reversal of calcium/calmodulin-dependent protein kinase II oxidation [10]. These findings suggest a key role for MsrA in controlling MetO reduction in proteins as well as the expression of genes involved in cellular antioxidant defense mechanisms. Identifying MetO-harboring proteins will greatly enhance the knowledge about processes leading to cellular malfunction associated with protein damage; thereby providing important information for the development of novel therapeutics against oxidative stress-associated diseases.
The endosperm of maize (Zea mays) seeds contains several classes of alcohol-soluble storage proteins called zeins, which together make up nearly 50% of the total seed protein content. Some zeins proteins are rich in the sulfur amino acids Met and cysteine (Cys). Therefore, these proteins could be very useful in designing a high rich-MetO protein. Zeins can be separated into four major classes (as determined by their sequence and apparent molecular mass following gel electrophoresis): α (19 and 22 kDa), β (15 kDa), γ (16 and 27 kDa) and δ (10 kDa) zeins. In this study, we have used a derivative of the gene encoding an 18 kDa Met-rich zein that is closely related to the 10 kDa zein protein [11]. The zeins proteins, 10 kDa (DZS10) and the 18 kDa (DZS18), share 80% percent similarity and 66% identity while the DZS18 contains 25% of Met residues compared with 23% of DZS10 [11]. The high percentage of Met residues in DZS18 (while having no Cys residues) makes it the most Met-rich protein among cereals and legumes [11]. Both δ-zein genes are coordinately and temporally up regulated during endosperm development. It is suggested that their corresponding proteins may be a good source for free Met supply needed during the plant’s later stages of development. Consequently, the zein proteins can serve as Met–rich substrate for oxidation resulting in MetO-rich proteins.
In the current study, we apply novel antibodies that can identify MetO in proteins (using MetO-zein as an antigen; patent pending) to probe for the presence of MetO in various pure proteins and in biological extracts. The newly developed antibodies will facilitate the determination and identification of proteins with increased levels of MetO from the aging process, conditions of oxidative stress, and disease state. Accordingly, we have used cellular protein extracts from organisms lacking the MsrA protein (msrA null mutant yeast and MsrA−/− mice) and their wild type controls, oxidized and scrapie prion protein, blood serum from young and aged mouse strains, and blood serum of healthy and patients diagnosed with Alzheimer’s disease (AD).
Materials and Methods
Overexpression and purification of the recombinant DZS18 protein
The DZS18 open reading frame was amplified by PCR reaction using 5′ forward primer harboring BamHI restriction site and 3′ reverse complementry primer harboring Hind III restriction site. The PCR reaction mixture contained the 5′-forward DNA primer starting at the 5′ end of the DZS18 open reading frame (66 bp downstream of the first ATG codon); the 3′-reverse complement DNA primer starting at the untranslated region forwarding towards the DZS18 stop codon, and cDNA for Zea mays (BioChain, Hayward, CA). The resulting PCR product was digested with BamHI and HindIII enzymes and subcloned into a pQE30 vector (Qiagen) at the complementary restriction sites. This created an open reading frame for a fusion protein between the 6xHis tag and the zein N-terminus, starting at the DZS18-DNA sequence corresponding to the 22nd amino acid of the protein. The pQE30 6xHis tag was inserted in frame with the N-terminus of the DZS18 and the construct was transformed and expressed in E. coli. Following overexpression of the fusion protein in bacteria, the recombinant protein was extracted from the bacterial cells and purified on an affinity nickel coulmn, according to the manufacture procedure (BD Biosciences). Then, the protein was oxidized with 0.3% hydrogen peroxide for 2 hours at 37°C. The oxidized protein (MetO-DZS18) was dialyzed against phosphate-buffered saline (PBS) at pH 7.4 to remove remaining oxidant and subjected to amino acid analysis, as previously described [2]. The protein was found to be fully oxidized in its Met moiety (by amino acid analysis [2]; data not shown) and consequently injected into a rabbit over several time intervals to produce anti-MetO-DZS18 antibodies.
Purified proteins serving as substrates for the anti-MetO-DZS18 antibodies
Glutamine synthetase and glyceraldehyde 3-phosphate dehydrogenase. Purified recombinant glutamine synthetase (GS; a gift from Mrs. Berlett, NHLBI) and glyceraldehyde 3-phosphate dehydrogenase (GAPD purchased from Sigma) were incubated in the presence and absence of 0.3% of H2O2 for 2 hours at 37°C. Thereafter, equal amounts of theses proteins were subjected to SDS-gel electrophoresis followed by western blot analysis using the anti-MetO-DZS18 antibodies.
Prion protein
Full length polypeptide chains corresponding to the hamster prion (PrP) sequence, lacking the N- and C-terminal sequences were expressed from their pET11a construct and folded to their α-form as previously described [12]. The enriched recombinant PrP protein, rSHaPrP (Residues 23–230), was then oxidized in the presence and absence of 50 mM H2O2 for 30 minutes at room temperature. For the purified Proteinase K resistant PrPSc samples, membranes from scrapie infected hamster brain homogenates were subjected to extraction by sarkosyl followed by ultracentrifugation, gradient enrichment, and PK digestion as previously described [13, 14]. Thereafter, equal amounts of theses proteins were subjected to SDS-gel electrophoresis followed by western blot analysis using either the anti-MetO-DZS18 antibodies, or α-PrP mAb IPCV1 (monoclonal antibodies against PrP) [15], or α-PrP mAb IPC2 (monoclonal antibodies which recognizes PrP with intact disulfide bond) [15].
Detection of MetO-containing proteins in tissue/cell extracts
Mouse tissues
To further characterize the anti-MetO-DZS18 antibodies for detection of MetO-containing proteins in biological extracts, protein extracts from several mouse tissues were treated with and without 100 mM H2O2 for 2 hours at 37°C in the presence of 2% SDS (to inhibit peroxidases activities). Thereafter, equal amounts of protein extracts were subjected to SDS-gel electrophoresis followed by western blot analysis using the anti-MetO-DZS18 antibodies.
Yeast cells
To determine whether the anti-MetO-DZS18 antibodies are capable of detecting oxidized proteins in yeast, two yeast strains were grown in the presence or absence of H2O2. One strain was enriched in its MetO-reduction ability (OP, an overproducing strain of MsrA) and one strain was compromised in its MetO-reduction ability (MT, a null mutant strain of msrA). The yeast strains OP and MT were grown in the presence or absence of 1 mM H2O2 till their growth rate reached 150 klett units. Following their growth, the cells were harvested, extensively washed with PBS, and disrupted in the presence of PBS and proteases inhibitors cocktail (Roche). Equal amounts of protein extracts from each strain were subjected to an SDS-gel electrophoresis followed by western blot analysis using the anti-MetO-DZS18 antibodies. To compete for the binding of the antibodies to the targeted proteins, the antibodies were incubated in the presence of 100 mM free L-MetO in a duplicate experiment (in parallel, L-MetO was substituted with L-Met to serve as a control experiment to the L-MetO binding competition to the antibodies).
Blood-serum proteins
Aliquots of blood samples (100–200 μL) were isolated from wild type and MsrA−/− mouse strains at ages 6, 12, and 16 months. Following serum extraction, equal amounts of serum protein from each sample (measured by the Bradford reagent (Bio-Rad)) were subjected to SDS-gel electrophoresis and western blot analysis using the anti-MetO-DZS18 antibodies.
Similarly, human serums from healthy and Alzheimer’s patients donors were analyzed as well (5 individuals from each group with matching ages between 70–80 years old, provided by Dr. Jeff Burns clinic KUMC, University of Kansas). Additionally, to determine MetO content in mouse serum albumin the protein was purified by separating mouse serum in SDS-gel-electrophoresis and extracting the corresponding band, digesting it with trypsin, and subjecting the resulting peptides to mass spectrometry analysis. A parallel gel was ran and analyzed for the presence of MetO in the serum albumin by western blot analysis using the anti-MetO-DZS18 antibodies.
In-gel digestion for mass spectrometry analysis
The mouse serum albumin band was excised from the gel and washed with 0.2 M ammonium bicarbonate/50% acetylnitrile at 37°C for 45 minutes with gentle agitation. After discarding the washing solution, the gel was dried in a CentiVap Concentrator (Labconco) and and incubated with 150 μl of 0.2 M ammonium bicarbonate and 20 μl of 100 mM DTT solution at 60°C for 30 minutes. Then, 40 μl of 100 mM iodoacetamide was added and the sample was incubated at room temperature for 30 minutes. After discarding the solution the gel was washed with 200 μl of 0.2 M ammonium prior to the incubation with 20 μl of the digestion buffer (0.2 M ammonium bicarbonate/5 mM CaCl2) containing 0.3–0.5 μg trypsin at 37°C overnight. The resulting digested peptides were analyzed by mass spectrometry techniques for their amino acid sequences.
Results
Antibodies produced against Meto-DZS18 show specificity towards protein-MetO
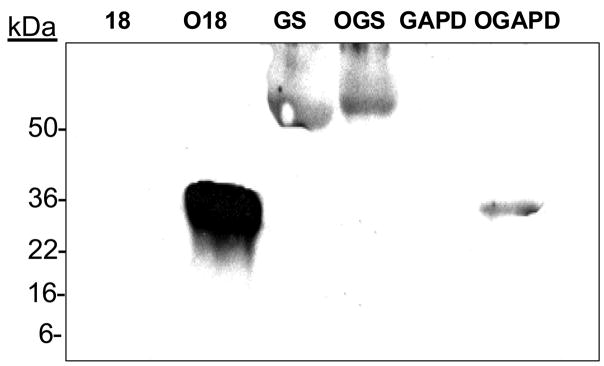
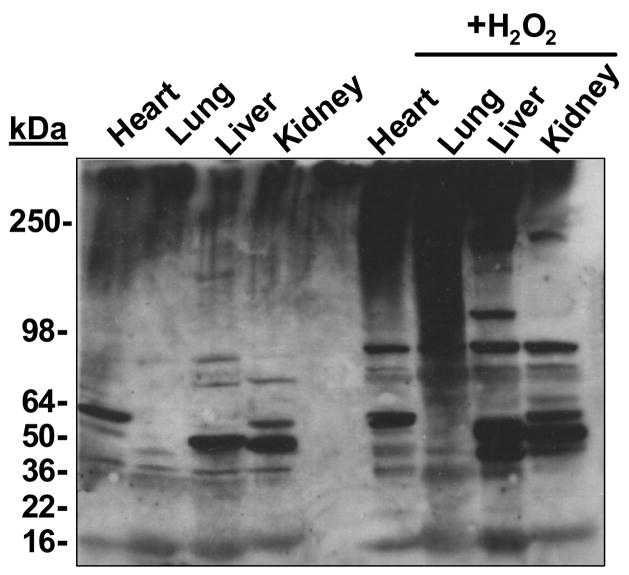
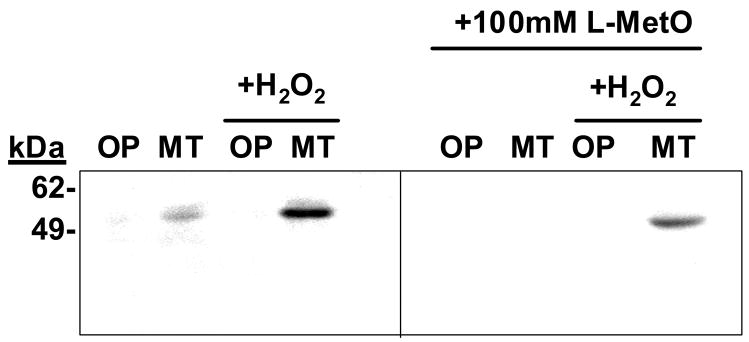
The antibodies showed specificity towrds the MetO-DZS18 protein and did not react with the non-oxidized form of the protein (DZS18). We have verified by using amino acid analysis for MetO detection [2] that indeed all Met residues in the MetO-DZS18 were oxidized while all other amino acid residues remained intact (data not shown). In addition, the antibodies reacted with several non-homologus proteins in their oxidized forms: glutamine synthetase (GS) and glyceraldehyde 3-phosphate dehydrogenase (GAPD) (Fig. 1), suggesting specific reactivity of the antibodies towards MetO residues (all the Met residues in the oxidized proteins were found to be oxidized, as judged by amino acid analysis; data not shown). The MetO-18 (Fig. 1, O18) migrates as a 36 kDa protein due to the mass contribution by the addition of 6xHis tag to its N-terminus and oxygen to all of its Met residues (this protein consists of 25% Met residues). The native GS shows reaction with antibodies (Fig. 1) as it is naturally oxidized under its recommended storage conditions at 4°C. However, the oxidized GS (OGS) reacted slightly stronger with the antibodies and migrated slower in the gel due its enhanced Met oxidation (Fig. 1). Moreover, the reaction intensity of the antibodies with the proteins seems to correlate with the percent of the Met residues in each tested protein (O18, 25%; OGS, 3.6%; and OGAPD, 3.0%), suggesting full Met oxidation in all proteins. To further characterize the anti-MetO-DZS18 antibodies for detection of MetO-containing proteins in biological extracts, protein extracts from several mouse tissues were treated with and without 100 mM H2O2 for 2 hours at 37°C in the presence of 2% SDS (to inhibit peroxidases activities). As expected, following western blot analysis using the anti-MetO-DZS18 antibodies, the reaction of the antibodies with existing and new protein bands was increased in all tissues that were treated with H2O2 in comparison to the non-treated proteins (Fig. 2). This result indicates that the anti-MetO-DZS18 antibodies are capable of detecting oxidized proteins in crude extracts due to MetO modification mediated by H2O2. The antigen MetO-DZS18 protein does not have any Cys nor other amino acids that are readily oxidized except Met. This protein did not show any actual amino acid oxidation except for Met (as determined by amino acid analysis (data not shown)); thereby making the MetO residues as the only possible binding target for the antibodies raised against MetO-DZS18. To determine whether the anti-MetO-DZS18 antibodies are capable of detecting oxidized proteins in vivo, the two yeast strains were exposed to H2O2. One strain was enriched in its MetO-reduction ability (OP, an overproducing strain of MsrA) and a one strain was compromised its MetO-reduction ability (MT, a null mutant strain of msrA). As expected, the MT strain that was more sensitive to Met oxidation produced a dominant protein band following reaction with the anti-MetO-DZS18 antibodies even without the H2O2 treatment. Moreover, the reaction with this protein band was enhanced following exposure of the cells to H2O2 (Fig. 3). Accordingly, the OP strain to that is resistant to Met oxidation showed no detectable reactions with the anti-MetO-DZS18 antibodies (Figure 3). The specificity of the antibodies to single MetO residues was supported by the fact that when free MetO was co-incubated with the anti-MetO-DZS18 antibodies (during the western blot analysis) the reactivity of the protein band with the antibodies was diminished (Fig. 3); suggesting a competition between the free amino acid and the protein MetO residues on the binding to the respected antibodies. The competition was not complete as the competitor is an amino acid rather than protein residue and the antibodies may have subclasses that can bind multiple MetO residues only. Moreover, an identical control experiment performed in the presence of free Met, instead of free MetO, did not show any change in the reaction of the antibodies with the targeted protein (relative to the reaction without any amino acid, data not shown). We believe that the additional presented data provide supportive evidence for the concept that indeed the anti-MetO-DZS18 antibodies are reacting specifically towards multiple and single MetO residues. It is important to note, that the average percent of total Met residues in a protein is only ~2%, while the percentage of the surface exposed Met is even lower as Met by its nature is a hydrophobic amino acid. Therefore, most of the Met residues are predicted to be buried within the molecular structure of a protein, thereby becoming less excessive to oxidation by most oxidants).
Figure 1. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in purified proteins.
Western blot analysis of purified recombinant glutamine synthetase (GS) and glyceraldehyde 3-phosphate dehydrogenase (GAPD) were incubated in the presence and absence of 0.3% of H2O2 for 2 hours at 37°C. Equal amounts of theses proteins were subjected to SDS-gel electrophoresis followed by western blot analysis using the anti-MetO-DZS18 antibodies. OGS, oxidized GS; OGAPD, oxidized GAPD; 18, DZS18; O18, oxidized DZS18. kDa, molecular mass indicators (in kDa). The presented data represents three independent experiments.
Figure 2. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in protein extracts of various mouse tissues.
Protein extracts from several mouse tissues were treated with and without 100 mM H2O2 for 2 hours at 37°C in the presence of 2% SDS (to inhibit peroxidases activities). Thereafter, equal amounts of protein extracts were subjected to SDS-gel electrophoresis followed by western blot analysis using the anti-MetO-DZS18 antibodies. kDa, molecular mass indicators (in kDa). The presented data represents three independent experiments.
Figure 3. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in yeast protein extracts.
Two yeast strains were grown in the presence or absence of H2O2. One strain was enriched in its MetO-reduction ability (OP, an overproducing strain of MsrA) and one strain was compromised in its MetO-reduction ability (MT, a null mutant strain of msrA). The yeast strains: OP (MsrA overproducing strain) and MT (null mutant msrA strain) were grown in the presence or absence of 1 mM H2O2 till their growth rate reached 150 klett units. Following their growth, the cells were harvested, extensively washed with PBS, and disrupted in the presence of PBS and proteases inhibitors cocktail (Roche). Equal amounts of protein extracts from each strain were subjected to SDS-gel electrophoresis followed by western blot analysis using the anti-MetO-DZS18 antibodies. To compete for the antibodies’ binding to the targeted proteins, the antibodies were incubated in the presence of 100 mM free L-MetO in a duplicate experiment. L-Met did not show an ability to compete on the antibodies’ binding (data not shown). kDa, molecular mass indicators (in kDa). The presented data represents three independent experiments.
Detection of MetO in prion protein
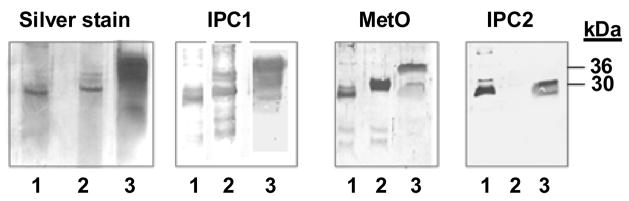
The prion type PrPSc is the modified form of the membranal prion protein (PrPC). PrPSc is considered to be a version of prion that is associated with transmissible spongiform encephalopathies. Stahl et al. have shown that either one or two Met residues (M206 and/or M213) were present in the form of MetO in PrPSc [16]. Additionally, PrPC has been assigned a role in the protection of cells from ROS [17, 18] and recombinant PrP (rPrP) was a target for oxidation [19, 20]. Recently, it has been demonstrated by Gabizon et al that brain PrPSc contains high levels of MetO, which are not detected in brain PrPC and its recombinant models [15]. Accordingly, we have examined the possibility of detecting MetO in rPrP, oxidized rPrP, and Proteinase K (PK) resistant hamster PrPSc by the anti-MetO-DZS18 antibodies. As shown in Fig. 4, the α-PrP mAb IPCV1 antibodies recognized all forms tested in the figure; α-PrP mAb IPC2 antibodies recognized PrP forms that were not oxidized at Met213 [15], and the anti-MetO-DZS18 antibodies recognized all forms of PrP tested, while exhibiting the strongest reaction against the oxidized form of rPrP. As seen in Fig. 4, while α-PrP mAb IPC2 antibodies reacted with non-oxidized rPrP and with the lower forms of hamster PrPSc, the anti-MetO-DZS18 antibodies recognized all the additional PrP forms that react with α-PrP mAb IPCV1 antibodies. The fact that the silver stain pattern of PrPSc is similar to the α-PrP mAb IPCV1 pattern of PrP recognition suggests the anti-MetO-DZS18 antibodies indeed reacted with the fully glycosylated PrPSc form not recognized by α-PrP mAb IPC2 antibodies (primarily the ~36 kDa band that is mainly presented in the PrPSc form). It is important to note that the gel-electrophoresis performed prior to the western blot using α-PrP mAb IPC2 antibodies was done in the absence of β-mercaptoethanol. This was due to the fact that α-PrP mAb IPC2 antibodies can recognize only PrP proteins with intact disulfide bridges [15]. However, oxidation of PrP causing MetO formation interferes with the protein recognition by the α-PrP mAb IPC2 antibodies (Fig. 4). Complementary to this observation, it is suggested that the presence of β-mercaptoethanol causes the MetO at position 213 to be more surface exposed to facilitating its recognition by the anti-MetO-DZS18 antibodies (Fig. 4). These results are consistent with the recent publication [15] indicating that several Met residues are oxidized in PrPSc. This suggests that the anti-MetO-DZS18 antibodies can be an important tool to study this modification in prion protein.
Figure 4. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in prion protein.
The following samples were subjected to SDS-gel-electrophoresis followed by silver staining and immunoblotting with several antibodies. Full length polypeptide chains corresponding to the hamster PrP sequence, lacking the N- and C-terminal sequences (rSHaPrP (23–230; denoted as rPrP) were expressed and folded to their α-form as described in Materials and Methods. The rPrP was then incubated in the presence and absence of 50 mM H2O2 for 30 min. For the purified PK resistant PrPSc samples, membranes from scrapie infected hamster brain homogenates were purified as described in Materials and Methods. Following SDS-gel electrophoresis, all samples (1: rPrP; 2: oxidized rPrP; 3: PK resistant hamster PrPSc) were subjected to either silver staining or immunoblotting with: α-PrP mAb IPCV1, which recognize all forms tested in the figure (IPC1); α-PrP mAb IPC2, which recognize PrP forms that are non-oxidized at Met 213 (IPC2), as well as the anti-MetO-DZS18 antibodies, which recognize MetO in proteins (MetO). kDa, molecular mass indicators (in kDa). The presented data represents three independent experiments.
Blood-serum proteins
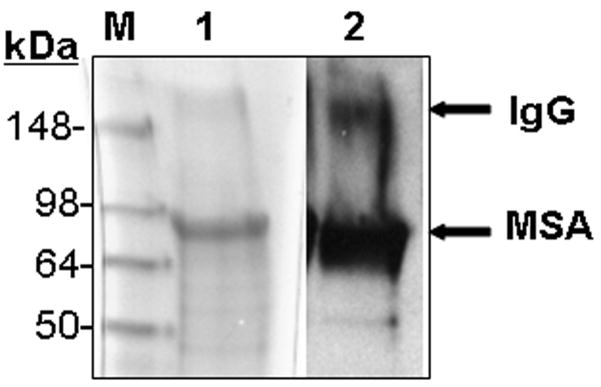
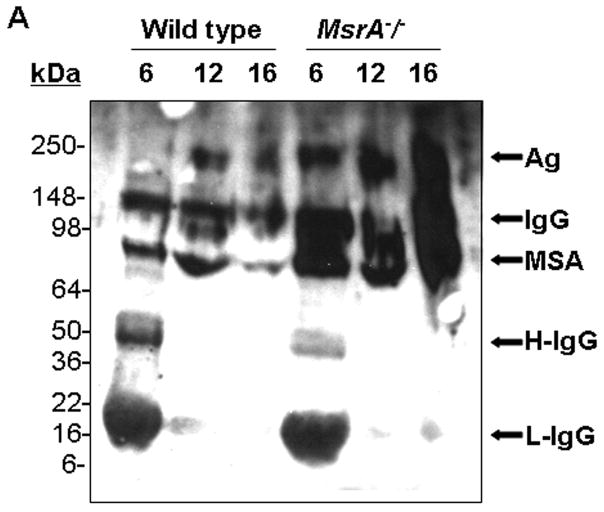
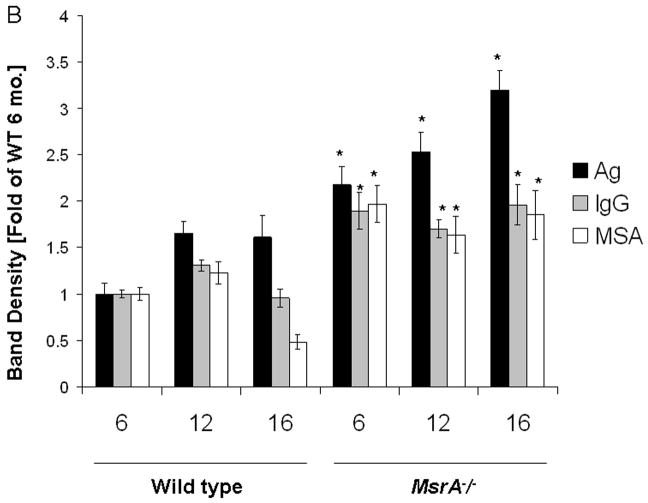
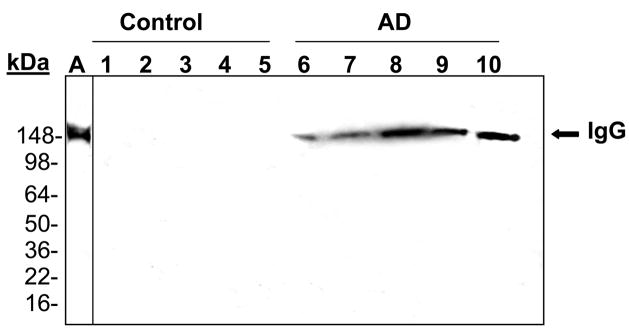
The major proteins in serum, mouse serum albumin (MSA) as well of intact IgG, were shown to react in western blot analysis with the anti-MetO-DZS18 antibodies (Fig. 5). The identification of the MSA was confirmed by mass spectrometry analysis using trypsin-digested peptides covering about 25% of the protein sequence. Among all peptides analyzed, Met572 was shown to be oxidized to MetO. Apparently, Met572 is conserved in all sequenced serum albumins and perhaps its structural location promotes its availability to oxidation. The identification of an intact IgG as the other target for Met oxidation (Fig. 5) was confirmed by probing a parallel western blot with primary rabbit anti-mouse IgG (data not shown). Further western blot analyses for Met oxidation in MSA and IgG were performed on serums from wild type and MsrA−/− mice taken at various ages. As shown in Fig. 6A, the oxidation of MSA and IgG in its intact form (~150 kDa) intensified in MsrA−/− compared to wild type serums. The SDS gel-electrophoresis was performed in the presence of β-mercapthoethanol that is expected to reduce the disulfide bonds between each IgG monomer as well as between the heavy (~50 kDa) and light (~25 kDa) chains of the protein. Accordingly, in serum of 6-month-old mice the IgG molecules were reduced to their monomeric components and shown to contain MetO by western blot analysis (Fig. 6A). In contrast, with older age, the disulfide bond reduction was inhibited and the relative oxidative levels of intact IgG (~150 kDa) and aggregated form of an unknown serum protein (~250 kDa suggested to be an aggregated form of IgG (Ag) were increased in both mouse strains as function of age (Fig. 6A). In comparison to wild type, the Met oxidation of the Ag form of IgG was mostly pronounced in MsrA−/− serums (Fig. 6A). Densitometry analysis for the intensities of the corresponding MSA, IgG, and AG bands have confirmed that the MetO levels of the selected proteins increases in MsrA−/− versus wild type serums (Fig. 6B). Moreover, the levels of the MetO-Ag were increased in an age-dependent manner in both strains, while showing the highest levels in the MsrA−/− strain (Fig. 6B). The latter observation may be due to the common nature of aggregated proteins of being more resistant to degradation, which fosters their accumulation with age. Similar to the observed effects of age and lack of MsrA on IgG MetO levels, intact forms of IgGs in serums of patients with AD were shown to contain MetO while none was observed in control serums, respectively (Fig. 7). It is noted that the limited number of human specimens analyzed for the presence of MetO in serum proteins requires further validation by larger group analyses.
Figure 5. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in mouse serum proteins.
Fifty microgram of mouse serum proteins were subjected to SDS-gel electrophoresis and either stained with coomassie blue (1), or probed with anti-MetO-DZS18 antibodies following western blot analysis (2). M, molecular mass markers (in kDa). IgG-Immunoglobulin G that was detected in a parallel gel by primary goat anti-mouse IgG antibodies (GenWay; data not shown). MSA, mouse serum albumin (detected by mass spectrometry analysis according to procedures described in Materials and Methods). The presented data represents three independent experiments.
Figure 6. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in mouse serum proteins in an age-dependent manner.
A. Western blot analysis on mouse serums as function of age (6, 12, and 16, represent age in months) using the anti-MetO-DZS18 antibodies. Ag, aggregated form of serum protein. IgG, native form of IgG. H-IgG, heavy chain of IgG. L-IgG, light chain of IgG. MSA, mouse serum albumin. kDa, molecular mass indicators (in kDa). B. Densitometry analysis for the intensities of the MSA, IgG, and Ag bands shown in panel A. The numbers 6, 12, 16 represent age in months. The (*) symbol represents significant statistical difference (P<0.05; t-test) between the age-matching bands of both mouse strains. Also, there were significant statistical differences (P<0.05; t-test) within the relative levels of the Ag bands in each mouse strain: between all age groups of MsrA−/− and between 6 and 12 or 16 months old of wild type mice.The shown data represent three independent experiments.
Figure 7. The anti-MetO-DZS18 antibodies detect methionine sulfoxide in IgG in serum proteins of Alzheimer’s disease human subjects.
Western blot analysis on human serums using the anti-MetO-DZS18 antibodies (1–5, samples from control human subjects; 6–10, samples of AD patients; all samples are aged matched (70–80 years old). A. A representative sample of human serum probed with primary rabbit anti-human IgG antibodies (GenWay). kDa, molecular mass indicators (in kDa). The presented data represents three independent experiments.
Discussion
In the current study the development and characterization of novel anti-MetO antibodies is described. These anti-MetO-DZS18 antibodies were able to identify oxidized forms of purified proteins (Fig. 1) as well as oxidized proteins in various tissue extracts, respectively (Fig. 2). Likewise, the reaction of the antibodies with a major detectable protein in yeast extract intensified when the yeast strain lacked MsrA and was exposed to H2O2 (Fig. 3). In contrast, the reaction of the antibodies with the same protein diminished completely in protein extracts of an MsrA-overexpressing yeast strain and was reduced in the presence of free L-MetO. Thus, the presented data (Figs. 1–3) support the specificity of the anti-MetO-DZS18 antibodies toward protein-MetO. Very recently, similar antibodies were developed but were not characterized for their ability to recognize MetO in various proteins [21]. Methionine oxidation in proteins that are associated with in neurodegenerative diseases is thought to play a role in their toxicity and structure. For example, α-synuclein in Parkinson’s disease, β-amyloid in Alzheimer’s disease and PrP in Creutzfeldt-Jakob disease (CJD) [4]. With respect to the latter protein, very limited literature is provided regarding the formation and consequence of MetO in PrP. One of the major problems in identifying MetO in PrPSc is that it becomes proteinase K resistant and aggregates into structures that interfere with commonly used analytical processes for MetO determination (like mass spectrometry and amino acid composition analyses). Accordingly, the ability of the anti-MetO-DZS18 antibodies to recognize MetO in both oxidized rPrP and PrPSc (Fig. 4) may serve as a tool to follow primary events that are causing the occurrence of high levels of oxidation and aggregation to PrPc, which in turn are prompting the appearance of PrPSc. In the search of a marker for Met oxidation in proteins in vivo, we have examined the possibility of monitoring the levels of MetO in serum proteins, like serum albumin and immunoglobulin (IgG) that are the major proteins in serum. Indeed, serum albumin (Figs. 5 and 6) and IgG were shown to be prone to Met oxidation, especially in an antioxidant-compromised mice (MsrA−/−; Fig. 6). The aggregated form of IgG (Ag) was also shown to exhibit age-dependent accumulations of MetO, with the highest levels observed in MsrA−/− mouse (Fig 6). It is suggested that with age or enhanced oxidative stress conditions (like in the case of MsrA−/− ), the levels of MetO in serum proteins are elevated and thus could serve as a marker to organism oxidative stress conditions and age (using the anti-MetO-DZS18 antibodies as a probe for MetO-proteins). For example, recently it was shown by mass spectrometry analyses that plasma albumin in patients treated with hemodialysis contained MetO [22]. Also, fully recombinant human monoclonal antibody was shown to contain MetO under chemical oxidation in vitro [23].
Neurodegenerative diseases are associated with increased conditions of oxidative stress leading to increase of protein oxidation. However, to best of our knowledge there is no reliable antibody that can show significant differences in serum protein oxidation between patients having AD and non-AD individuals. The use of the current novel antibodies showed that MetO in IgG is highly elevated in AD patients and thus may be applied in screening for the occurrence of AD following western blot analysis (Fig. 7). Further investigations with larger groups of AD patients and matched controls are under way to strengthen this observation. Nevertheless, in spite of the small number of tested samples, the striking relative high levels of MetO in serum IgG of individuals with AD suggest that the presence of MetO-IgG is associated with AD state and that although its levels are lower in human than in mouse serums they are still detectable by the anti-MetO-DZS18 antibodies. In summary, we present in this study novel anti-MetO antibodies that enable the identification of MetO-proteins, both as a mean to follow biological processes associated with oxidative stress and aging and as a screening tool for neurodegenerative diseases like AD.
Acknowledgments
This study was partially supported by the National Institute of Aging grant AG027363.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moskovitz J, Berlett BS, Poston JM, Stadtman ER. Proc Natl Acad Sci U S A. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER. Proc Natl Acad Sci U S A. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oien DB, Moskovitz J. Curr Top Dev Biol. 2008;80:93–133. doi: 10.1016/S0070-2153(07)80003-2. [DOI] [PubMed] [Google Scholar]
- 4.Moskovitz J. Biochim Biophys Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Chao CC, Ma YS, Stadtman ER. Proc Natl Acad Sci U S A 94. 1997:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciorba MA, Heinemann SH, Weissbach H, Brot N, Hoshi T. Proc Natl Acad Sci U S A. 1997;94:9932–9937. doi: 10.1073/pnas.94.18.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohri M, Reinach PS, Kanayama A, Shimizu M, Moskovitz J, Hisatsune T, Miyamoto Y. Invest Ophthalmol Vis Sci. 2002;43:3190–3195. [PubMed] [Google Scholar]
- 8.Midwinter RG, Cheah FC, Moskovitz J, Vissers MC, Winterbourn CC. Biochem J. 2006;396:71–78. doi: 10.1042/BJ20052026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Y, Chen B, Smallwood HS, Urbauer RJ, Markille LM, Galeva N, Williams TD, Squier TC. Biochemistry. 2006;45:14642–14654. doi: 10.1021/bi0612465. [DOI] [PubMed] [Google Scholar]
- 10.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swarup S, Timmermans MC, Chaudhuri S, Messing J. Plant J. 1995;8:359–368. doi: 10.1046/j.1365-313x.1995.08030359.x. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Iglesias R, Pajares MA, Ocal C, Espinosa JC, Oesch B, Gasset M. J Mol Biol. 2002;319:527–540. doi: 10.1016/S0022-2836(02)00341-8. [DOI] [PubMed] [Google Scholar]
- 13*.Prusiner SB, Hadlow WJ, Garfin DE, Cochran SP, Baringer JR, Race RE, Eklund CM. Biochemistry. 1978;17(23):4993–4999. doi: 10.1021/bi00616a021. LHM: This journal is received by the Medical Library, Ein Kerem *LHC: 1962- SHELF 4942. [DOI] [PubMed] [Google Scholar]
- 14.Caughey B, Raymond GJ, Priola SA, Kocisko DA, Race RE, Bessen RA, Lansbury PT, Jr, Chesebro B. Mol Biotechnol. 1999;13:45–55. doi: 10.1385/MB:13:1:45. [DOI] [PubMed] [Google Scholar]
- 15.Canello T, Engelstein R, Moshel O, Xanthopoulos K, Juanes ME, Langeveld J, Sklaviadis T, Gasset M, Gabizon R. Biochemistry. 2008;47:8866–8873. doi: 10.1021/bi800801f. [DOI] [PubMed] [Google Scholar]
- 16.Stahl N, Baldwin MA, Teplow DB, Hood L, Gibson BW, Burlingame AL, Prusiner SB. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 17.Brown DR. Folia Neuropathol. 2005;43:229–243. [PubMed] [Google Scholar]
- 18.Nadal RC, Abdelraheim SR, Brazier MW, Rigby SE, Brown DR, Viles JH. Free Radic Biol Med. 2007;42:79–89. doi: 10.1016/j.freeradbiomed.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 19.Redecke L, von Bergen M, Clos J, Konarev PV, Svergun DI, Fittschen UE, Broekaert JA, Bruns O, Georgieva D, Mandelkow E, Genov N, Betzel C. J Struct Biol. 2007;157:308–320. doi: 10.1016/j.jsb.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Requena JR, Dimitrova MN, Legname G, Teijeira S, Prusiner SB, Levine RL. Arch Biochem Biophys. 2004;432:188–195. doi: 10.1016/j.abb.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Le DT, Liang X, Fomenko DE, Raza AS, Chong CK, Carlson BA, Hatfield DL, Gladyshev VN. Biochemistry. 2008 doi: 10.1021/bi800422s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruschi M, Petretto A, Candiano G, Musante L, Movilli E, Santucci L, Urbani A, Gusmano R, Verrina E, Cancarini G, Scolari F, Ghiggeri GM. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;864:29–37. doi: 10.1016/j.jchromb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Chumsae C, Gaza-Bulseco G, Sun J, Liu H. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;850:285–294. doi: 10.1016/j.jchromb.2006.11.050. [DOI] [PubMed] [Google Scholar]