Abstract
The polyketide natural product (+)-discodermolide is a potent microtubule stabilizer that has generated considerable interest in its synthetic, medicinal and biological chemistry. It progressed to early clinical oncology trials where it showed some efficacy in terms some disease stabilization but also some indications of causing pneumotoxicity. Remarkably, there are no reports of its metabolism. Here, we examined its fate in mixed human liver microsomes. Due to limited availability of the agent, we chose a nanoflow liquid chromatography-electrospray ionization-mass spectrometry analytical approach employing quadrupolar ion trap and quadrupole-quadrupole-time of flight instruments for these studies. (+)-Discodermolide was rapidly converted to eight metabolites, with the left side lactone (net oxidation) and the right side diene (epoxidation followed by hydrolysis, along with an oxygen insertion product) being the most metabolically labile sites. Other sites of metabolism were the allylic and pendant methyl moieties in the C12-C14 region of the molecule. The results provide information on the metabolic soft spots of the molecule and can be used in further medicinal chemistry efforts to optimize discodermolide analogs.
(+)-Discodermolide is a polypropionate natural product originally isolated from the marine sponge Discodermia dissoluta.1 It was first introduced as an immunosuppressive agent,2–4 but then found to be a potent microtubule stabilizer5–7 that binds with remarkable affinity to the taxoid site on β-tubulin in microtubules.7,8 Unlike paclitaxel, the prototypical microtubule-stabilizing anticancer drug with clinical utility, (+)-discodermolide is not substrate of the ABCB1 transporter (P-glycoprotein pump).7 (+)-Discodermolide retains antiproliferative potency against β-tubulin mutant cell lines that are resistant to taxanes and epothilones.7,9 Furthermore, (+)-discodermolide and paclitaxel can act synergistically, and the combination significantly increases activity as well as decreases toxicity both in cell lines and xenograft-bearing mice.10 Its biological potency and novel structure made it a promising candidate for clinical development as an anticancer drug. After an astonishing full synthesis of 60 g of the agent,11–15 (+)-discodermolide progressed into early phase clinical oncology trials, but work on the agent was abruptly stopped when some indications of pneumotoxicity (3 of 32 patients) appeared.16
Discodermolide can be thought of as a C24:4 fatty acid C1-C5 δ-lactone with three of the double bonds in a Z geometric configuration and the fourth at the terminus of the carbon chain. This backbone bears eight pendant methyl groups, four hydroxy groups and an oxygen at C19 that is esterified with carbamic acid. The compound is therefore rife with sites potentially susceptible to not only hydrolysis (lactone, carbamate) but also to cytochrome P450-mediated oxidation (double bonds, allylic carbons). Although human pharmacokinetics of the parent drug have been reported,14 no reports of discodermolide’s metabolism have appeared. We therefore examined its fate in mixed human liver microsomes. Due to the fact that we had only very limited quantities of (+)-discodermolide for such studies, we chose to do the analyses using nanoLC-electrospray ionization-MS methods, and herein present the results of these qualitative studies.
Results and Discussion
MSn Spectra of (+)-Discodermolide
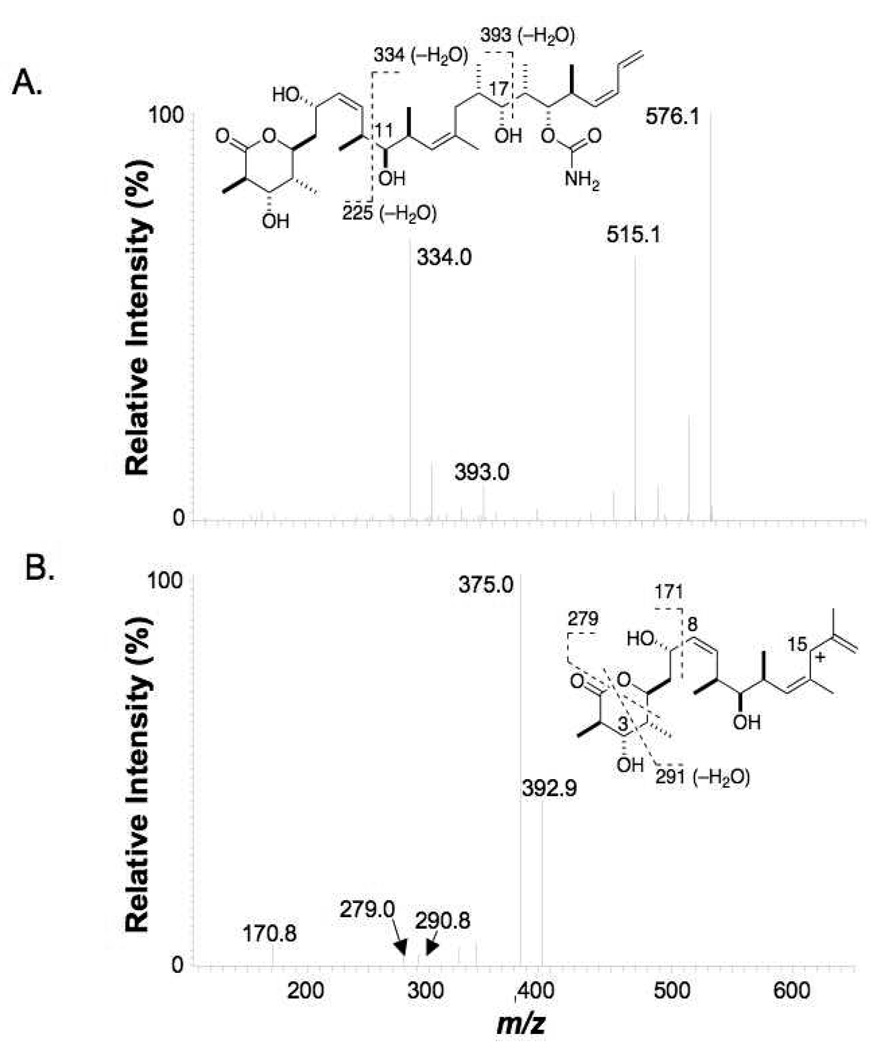
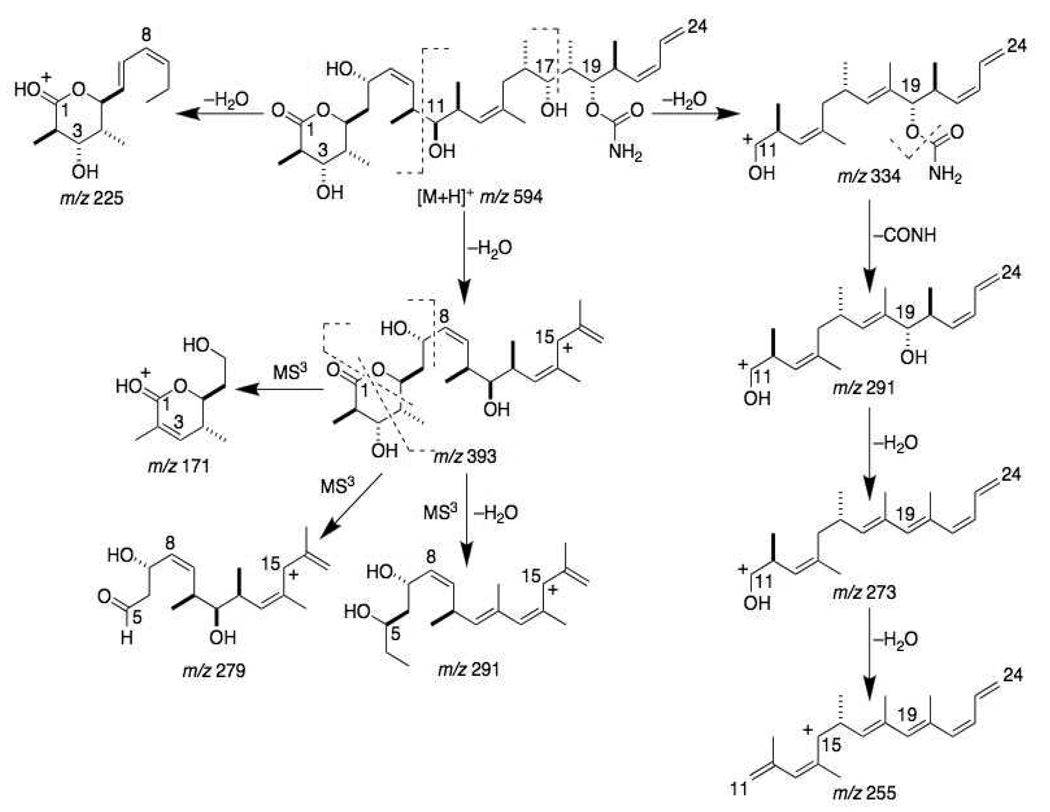
Typical fragmentation patterns of the [M+H]+ ion of discodermolide (m/z 594) in both QIT and QqTOF MS/MS spectra showed cleavage at: 1) the C-C bond adjacent to the 11-hydroxy group (C10-C11) to give the fragment ions m/z 352, m/z 334 (−H2O) and m/z 225 (−H2O); and 2) the C17-C18 bond adjacent to the 17-hydroxy group to give the fragment ions m/z 411 and m/z 393 (−H2O) (Figure 1A). Other major ions observed were at m/z 576, 558, 534, 515, 498 and 352. The m/z 393 ion could be fragmented further in MS3 experiments with the QIT instrument by cleavage of the C7-C8 bond near the 7-hydroxy group to form the m/z 171 ion (Figure 1B). The lactone ring of the m/z 393 ion was opened in MS3 experiments by cleavage of the C1-O bond and the C4-C5 bond to form the m/z 279 ion, and by cleavage of the C1-O bond and the C3-C4 bond followed by loss of a water molecule to form the m/z 291 ion. Because of the presence of a single nitrogen atom in the molecule (in the carbamate moiety on the right side of the drug), the MS/MS and in the MS3 fragment ions discussed above were important in the identification of the structures of metabolites via application of the nitrogen rule. The proposed fragmentation pattern important for metabolite structure identification is shown in Figure 2.
Figure 1.
A) MS/MS and B) MS3 (m/z 594→m/z 393→) spectra of (+)-discodermolide (obtained on the QIT instrument).
Figure 2.
MS/MS and MS3 fragmentation pathways of the [M+H]+ ion of discodermolide.
Microsomal Metabolism
Because of the limited amount of (+)-discodermolide we had (all experiments discussed and shown here were achieved with < 0.5 mg of the agent), we chose a drug concentration of 10 µM as this was considered to be the highest likely serum concentration that could be achieved with the drug. No evidence of chemical alteration was found by nanoLC-ESI-MS analysis when (+)-discodermolide was incubated at 37 °C with microsomes in the absence of NADPH or with heat-inactivated microsomes + NADPH (data not shown). When NADPH was present and intact microsomes were used, we found the human liver microsomal metabolism of (+)-discodermolide to be very rapid, with appreciable levels of eight metabolites appearing within 5 min and near-complete consumption of parent drug by 1 h (data not shown, as we deemed that we had insufficient drug to do a detailed kinetics analysis with the needed number of replicate experiments). We therefore chose 15 min incubations for the analyses presented here, as at that time point some parent drug remained and all eight detectable metabolites were apparent. Note that these same eight metabolites appeared in every repetition of the microsomal incubations.
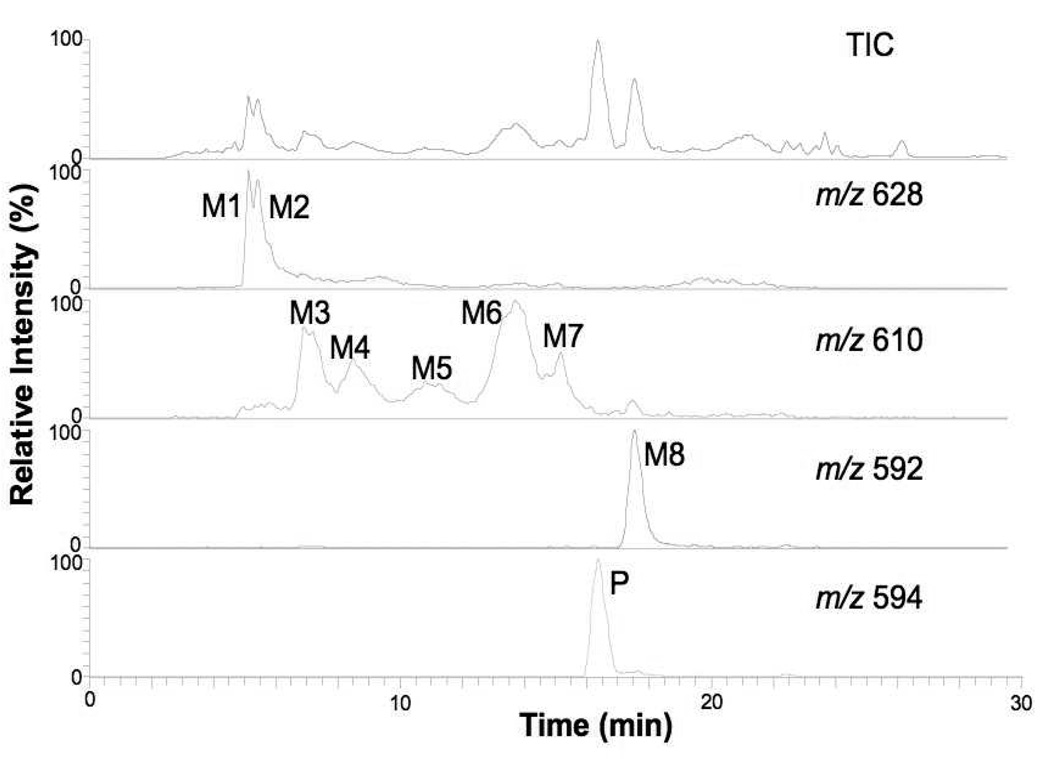
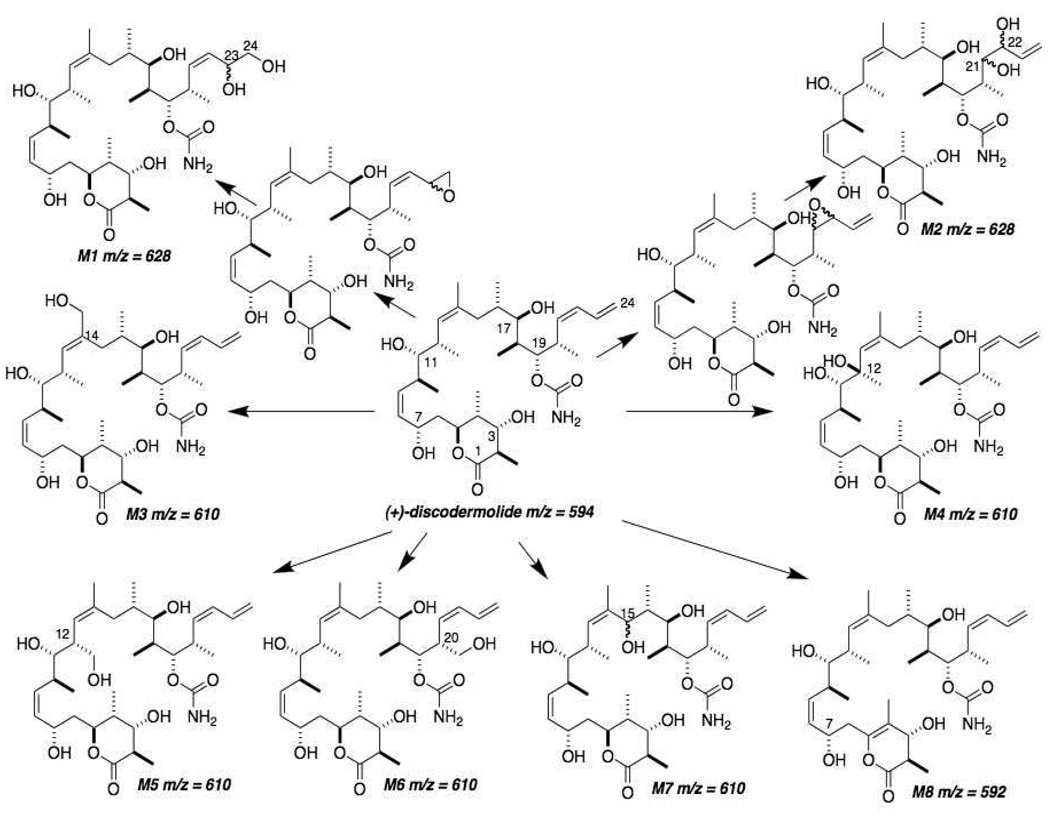
Figure 3 shows the total ion and extracted ion chromatograms from nanoLC-ESI-MS experiments of the parent drug (P) and its human liver metabolites after 15 min of incubation with microsomes. A replete search for mass shifts known to occur in microsomal systems, as well for products of hydrolysis of the parent as well as their oxygenated and/or oxidized products was made with the mass spectral data obtained. A total of eight metabolites (“M” with numerals denoting their respective order of C18 HPLC elution) were confidently detected. Their apparent (based on ion current) order of abundance was M8>M1~M2>M6>M3>M4~M7>M5. Metabolite M8 was clearly an oxidation product due to its [M+H]+ signal being −2 m/z from that of the parent drug. Metabolites M3–M7 arose from the metabolic addition of one oxygen atom (+16 m/z from the parent), and metabolites M1–M2 were from the addition of an oxygen atom and one molecule of water (+34 m/z).
Figure 3.
Positive ion LC-MS total ion chromatogram and extracted ion chromatograms of (+)-discodermolide and its metabolites after 15 min incubation at 10 µM with pooled human liver microsomes.
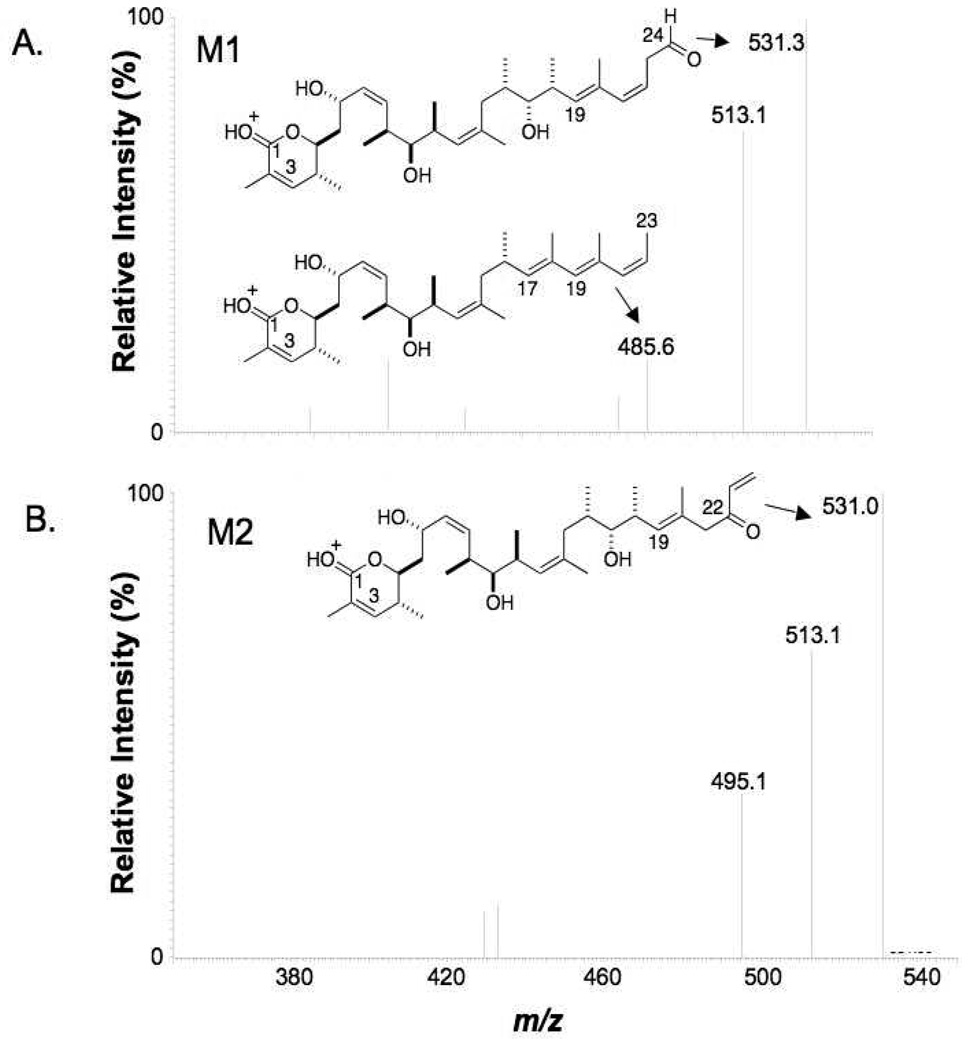
Metabolites M1 and M2
The most polar, first-eluting metabolites, M1 and M2, gave the same molecular ion [M+H]+ at m/z 628. The accurate masses detected by QqTOF experiments for these metabolites were, respectively, m/z 628.4010 and m/z 628.3999. Both corresponded to products of monooxygenation followed by hydrolysis, most likely catalyzed by epoxide hydrolase in the microsomal system, with an elemental composition of C33H57NO10+H (theoretical exact mass of 628.4061). Both M1 and M2 gave MS/MS fragmentation patterns similar to the parent drug (Table 1). MS/MS cleavage occurred at the C10-C11 and C17-C18 bonds. High abundance fragment ions were detected at m/z 368 and m/z 393, which suggested that the microsomal oxygenation occurred on the right side of the molecule. The proposed fragmentation pattern is shown in Figure 4. Metabolites M1 and M2 also showed a fragment ion at m/z 549, which corresponds to loss of two H2O and one CONH from the molecular ion (m/z 549 = m/z 628–36–43) (Table 1). The MS3 spectrum obtained from the m/z 549 ion of M1 gave a most informative fragment ion at m/z 485, which corresponds to a loss of C=O and two water molecules (m/z 485 = m/z 549–28–36) (Figure 4A). The presence of a C=O group in the m/z 549 ion strongly suggested that oxidation occurred at the C23-C24 terminal double bond of the right side of the molecule, such that M1 could be assigned as 23,24-dihydroxydiscodermolide (see Figure 9 for structures of the metabolites). The MS3 spectrum of the M2 m/z 549 ion did not show the same diagnostic fragment (Figure 4B), meaning the probable epoxidation + hydrolysis to give a vicinal diol occurred at the C21-C22 double bond, i.e., M2 was assigned as 21,22-dihydroxydiscodermolide.
Table 1.
Precursor and Fragment Ions in NanoLC-ESI-MS and -MS/MS Analyses of (+)-Discodermolide and Its Human Liver Microsomal Metabolites.
| Analyte | [M+H]+ ion, m/z |
Fragment ions, m/z |
|---|---|---|
| Discodermolide | 594 | 393, 334 |
| M1 | 628 | 610, 592, 567, 549, 531, 513, 411, 393, 385, 368, 225 |
| M2 | 628 | 610, 592, 567, 549, 531, 513, 411, 393, 385, 368, 225 |
| M3 | 610 | 592, 574, 549, 531, 513, 495, 477, 409, 391, 368, 350, 337, 307, 289, 271, 225 |
| M4 | 610 | 592, 574, 549, 531, 513, 495, 477, 409, 391, 368, 350, 337, 322, 289, 261 |
| M5 | 610 | 592, 574, 549, 531, 513, 495, 409, 391, 350, 337, 307, 289 |
| M6 | 610 | 592, 574, 549, 531, 513, 495, 393, 350, 337, 307, 289 |
| M7 | 610 | 592, 574, 549, 531, 513, 409, 350, 307, 289 |
| M8 | 592 | 391, 334 |
Figure 4.
MS3 (m/z 628→m/z 549→) product ion spectra of M1 and M2 (QIT instrument). The loss of two water molecules from the ion at m/z 549 of M1 produced a formaldehyde moiety in the right side and further neutral loss of CO from the formaldehyde group introduced the fragment ion at m/z 485. For M2, the loss of water could not form such a formaldehyde group.
Figure 9.
Proposed biotransformation pathways of (+)-discodermolide by pooled human liver microsomes. m/z values are for each analyte’s [M+H]+ ion. M1 and M2 can be rationalized by initial epoxidation of the double bond on the terminal diene system followed by epoxide hydrolysis. M3, M4, M5, M6 and M7 were formed by hydrogen abstraction followed by oxygen rebound. In metabolites M5 and M6, it is possible that delocalization of the unpaired electron of the initial allyl radical generated by cytochrome P450 could cause double bond migration, as some of the mass spectrometric data suggested. Dehydrogenation of the lactone ring, either directly or as a result of oxygen insertion followed by loss of water, formed the metabolite M8.
Metabolites M3–M7
Metabolites M3–M7 eluted after the dihydroxylation products M1–M2 but before the parent drug P. These metabolites all gave the same [M+H]+ ion at m/z 610. The observed accurate masses of M3, M4, M5, M6 and M7 were m/z 610.3917, m/z 610.3926, m/z 610.3912, m/z 610.3919 and m/z 610.3922, respectively. These m/z values corresponded to monooxygenation products with an elemental composition of C33H55NO9+H (theoretical exact mass of 610.3955).
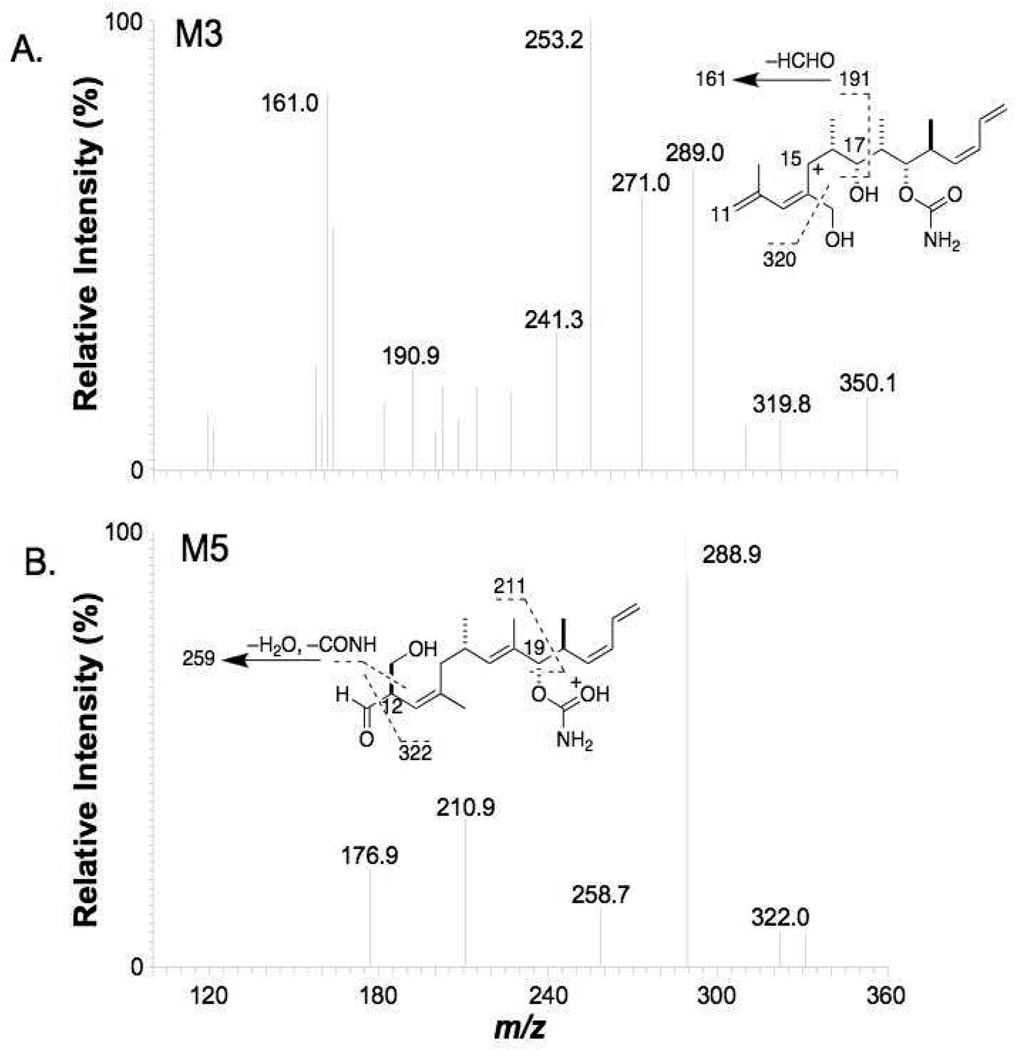
The abundant M3 fragment ions of m/z 391 and m/z 350 (Table 1) indicated that oxidation occurred in the middle section of discodermolide. The most informative fragment ions detected were m/z 320, m/z 241 and m/z 161 in the MS3 spectrum of the m/z 350 precursor ion, corresponding to a loss of formaldehyde (HCHO) from m/z 350, m/z 271 and m/z 191, respectively (Figure 5A). The ion at m/z 271 was produced by a loss of CONH and two water molecules from the m/z 350 precursor. The ion at m/z 191 was produced by the cleavage of C19-C20 bond followed by loss of CONH and two water molecules from the m/z 350 precursor. This indicated the presence of a CH2OH group and that oxidation occurred on the pendant methyl groups near the C13-C14 double bond to produce, suggesting M3 to be 14-(1’-hydroxy)discodermolide.
Figure 5.
MS3 (m/z 610→m/z 350→) product ion spectra of A) M3 and B) M5 (QIT instrument).
The abundant fragment ions of m/z 391 and m/z 350 found in the MS/MS data from M4 also indicated that oxidation to produce this metabolite occurred in the middle portion of discodermolide. The MS/MS product ion spectrum of M4 also showed unusual, highly abundant fragment ions at m/z 322 and m/z 261 (Figure 6). The most probable structure for M4 is 12-hydroxydiscodermolide. Based on this structure, the fragment ion at m/z 322 can be explained by cleavage of the C11-C12 bond and loss of water, followed by loss of water and CONH to give the fragment ion at m/z 261.
Figure 6.
MS/MS spectrum of M4 (QqTOF instrument).
The product ion spectrum of M5 was dominated by the fragment ions at m/z 391 and m/z 350 (Table 1), suggesting again that oxidation took place in the middle of discodermolide. A unique fragment ion at m/z 322 was detected in the MS3 spectrum from the m/z 350 ion (Figure 5B), which corresponded to the loss of a CO moiety. This implied the presence of a formaldehyde group in the m/z 350 ion. Another interesting feature in the MS3 spectrum of the M5 m/z 350 ion was the fragment ion at m/z 259, which corresponded to the loss of CONH, one water molecule and HCHO. This indicated the presence of a CH2OH group and suggested the oxidation occurred on the terminal methyl group of the m/z 350 ion. Since the C14 methyl group is the only methyl group near the double bond in the middle of discodermolide and oxidation at this position was already assigned to M3, it was considered highly possible that the hydroxy group of M5 was on the pendant C12 methyl group. The C13-C14 double bond also appeared to have migrated to C12-C13. Based on this structure, the fragment ion at m/z 322 can be explained by the loss of CO from a formaldehyde group on the ion at m/z 350. We rationalized the formation of the formaldehyde group in the ion at m/z 350 after the cleavage of the C10-C11 bond because the lack of a hydrogen on C12 negated the possibility of loss of water after cleavage of C10-C11. Since it is most likely that the double bond migration occurred in the mass spectrometer (and not as a result of microsomal metabolism), we assigned M5 to be 12-(1’-hydroxy)discodermolide.
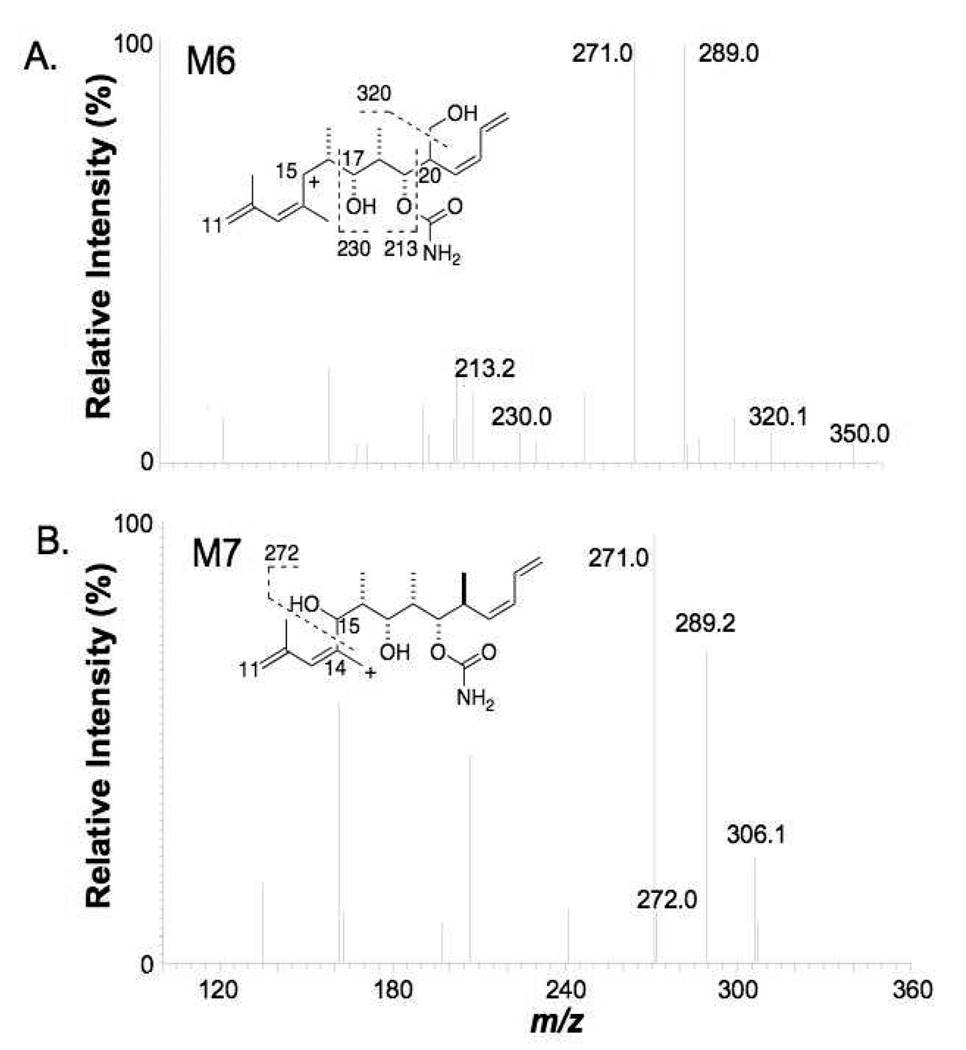
The product ion spectrum of M6 showed fragment ions at m/z 393 and m/z 350 (Table 1), suggesting that oxidation took place on the right side of discodermolide. Further fragmentation of the ion at m/z 350 (Figure 7A) gave the most informative fragment ion at m/z 320, which indicated loss of formaldehyde. The loss of this HCHO group strongly suggested that the oxidation occurred on the on a pendant methyl group of the right side of discodermolide. As there is only one pendant methyl group on the right side of discodermolide, the most likely structure that was consistent with the formation of the ion at m/z 320 was considered to be 20-(1’-hydroxy)discodermolide. Oxygenation at the methyl group at C20 could lead to double bond migration, which would generate an additional terminal methyl group at C24, so we cannot therefore exclude the possibility that the hydroxy group of M6 was situated on the terminal methyl group on C24 after double bond migration (see Figure 7A and Figure 9).
Figure 7.
MS3 (m/z 610→m/z 350→) spectra of A) M6 and B) M7 (QIT instrument).
The MS/MS spectra of M7 showed fragment ions at m/z 391 and m/z 350 (Table 1), indicating that hydroxylation occurred in the middle portion of discodermolide. Only one position, C15, remained after the assignments discussed above. Further fragmentation of the m/z 350 ion (Figure 8) produced a telling ion at m/z 272. The fact that this ion was an even number implied it contained a nitrogen atom. We envision that the fragment ion at m/z 272 was generated by the cleavage of the C14-C15 bond near the added hydroxy group. Therefore, the most probable structure of M7 is the 15-hydroxydiscodermolide. This assigned structure was consistent with all of the abundant fragment ions in the MS/MS and MS3 spectra from M7.
Figure 8.
MS3 (m/z 592→391→) spectrum of M8 (QIT instrument).
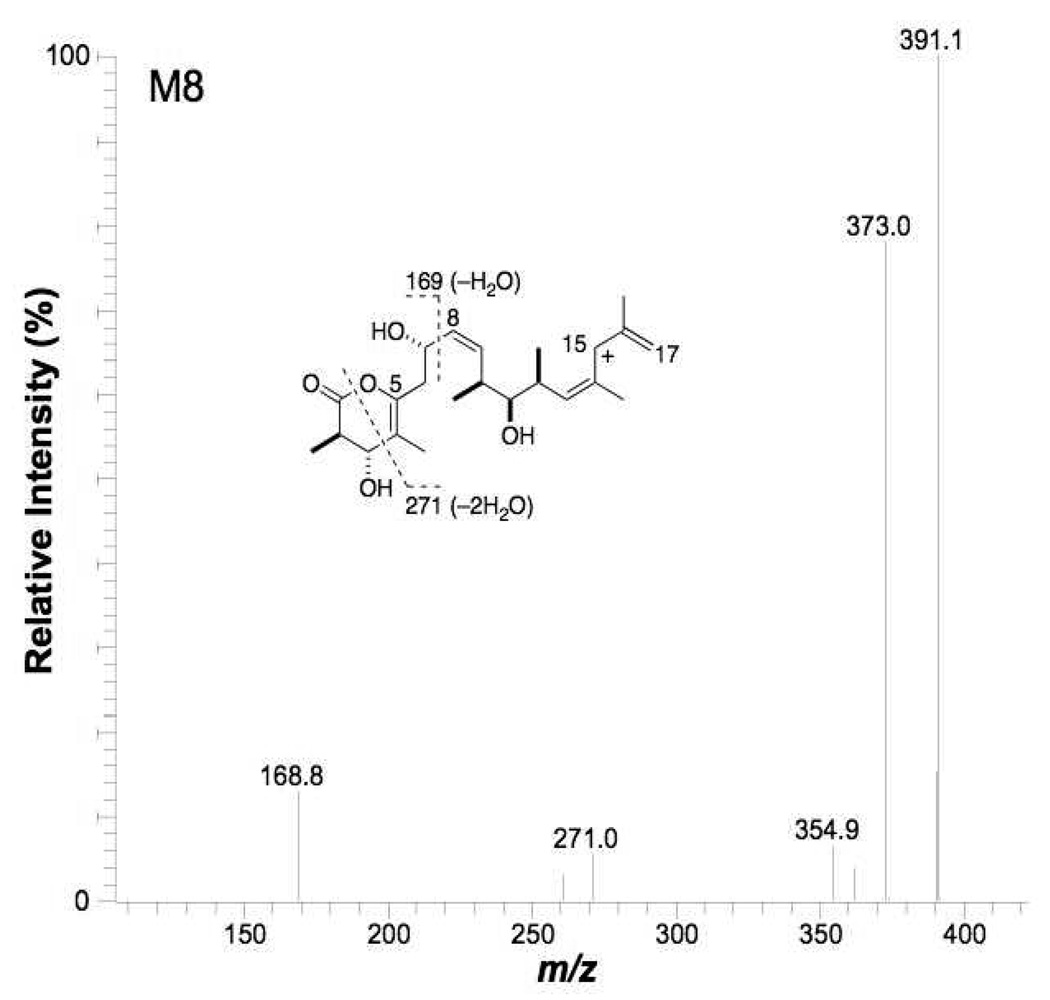
Metabolite M8
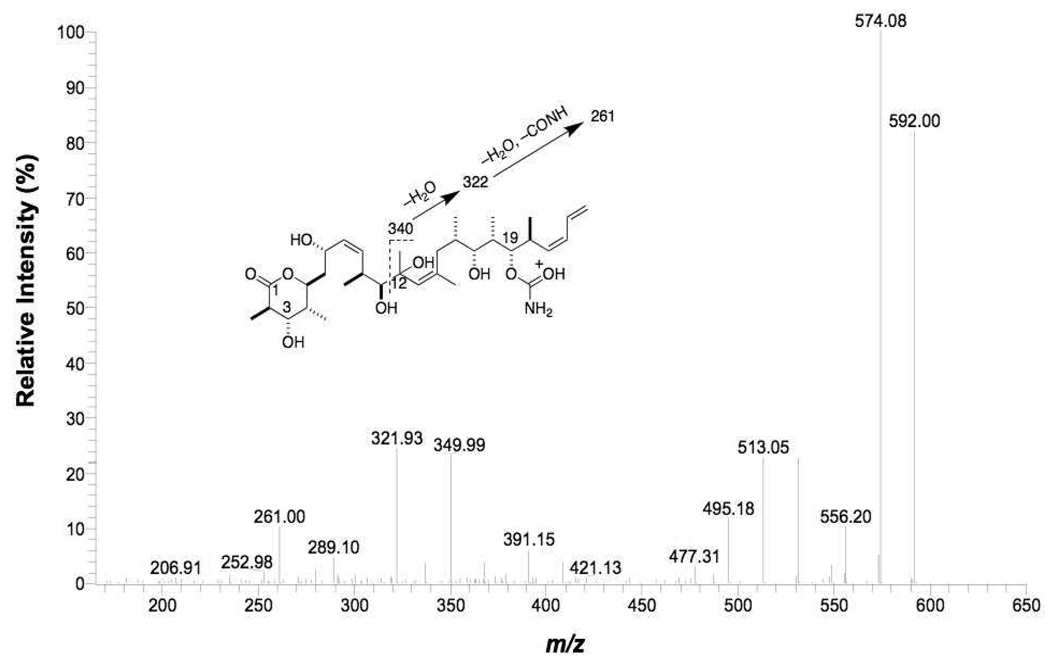
High resolution MS determination of the protonated molecular ion of M8 gave a value of m/z 592.3848, which corresponded to a net loss of two hydrogen atoms (theoretical exact mass of C33H53NO8+H of 592.3849) from discodermolide. This biotransformation may be result of direct dehydrogenation, or monooxygenation followed by loss of a water molecule. In addition, the retention time of M8 was longer than that of the parent compound P, indicating that polarity of M8 was lower. The MS/MS product ion spectrum of M8 showed abundant ions at m/z 391 and m/z 334 (Table 1), which suggested that right side and middle portion were unchanged, and therefore the net dehydrogenation had taken place in the left side of the molecule. In contrast to the parent drug, cleavage of the C4-C5 bond was not observed in the MS3 spectrum of m/z 391 from M8 (Figure 8); however, the cleavage of C1-O and C3-C4 was detected in this MS3 spectrum. The cleavage of C1-O and C3-C4 followed by loss of water formed the ion at m/z 271, indicating that the dehydrogenation occurred at the C4-C5 position of the lactone ring. Loss of two water molecules was relatively facile from the m/z 391 ion after cleavage of the C1-O and C3-C4 bonds (in the MS3 spectrum of discodermolide, only one hydroxy group was eliminated by loss of water after the same cleavage). This suggested the presence of a double bond at C4-C5 such that loss of an additional water molecule from the C7-hydroxy group would form a six double bond conjugation system into which the positive charge of the ion at m/z 271could be delocalized. Another important diagnostic fragment was the high abundance ion at m/z 169 (compared with the fragment at m/z 171 from the discodermolide product ion of m/z 393 in its MS3 spectrum). We propose that the ion at m/z 169 originated from homolytic cleavage of the C7-C8 bond followed by loss of water, and that this ion was stabilized through the resulting highly conjugated system. These water losses discussed above argued against the metabolite being the result of oxidation of an alcohol moiety (at, e.g., C7) to a ketone. Based on these considerations, we identified M8 as C4,C5-didehydrodiscodermolide.
The LC-MS/MS analyses provided some useful information about the fragmentation patterns of the parent drug and its metabolites, such as the ions at m/z 350 and m/z 393. These ions were not, however, sufficient for identification of the structures of metabolites. We therefore further employed MS3 experiments in a quadrupolar (3D) ion trap to tentatively identify the structures of the human liver microsomal metabolites of discodermolide. Of course, fully confident identification of the structures would require NMR analyses for both positional as well as stereochemical assignment, which was far beyond our reach as we had at our disposal only minute quantities of the parent drug, as well as independent syntheses, which would represent quite the substantial undertaking. Regardless, the MS/MS and MSn experiments gave great insight into the regions of the molecule that were modified by the microsomal system.
Human liver microsomes reproducibly converted discodermolide into at least eight metabolites as summarized in Figure 9. These metabolites were either dehydrogenation or oxygenation products. Most likely, cytochrome P450-mediated epoxidation of the diene moiety of the right side of discodermolide was followed by hydrolysis to give the high abundance vicinal diols M1 and M2. The predominance of this metabolic pathway is not surprising given the steric availability for oxygenation at this site. The solution and crystal structure of (+)-discodermolide are such that the shape of the chemical can be described as a tilde or helical, a result of the inflexible, curved nature of the middle portion of the molecule that must avoid A1,3 and A1,2 strain and syn-pentane interactions due to the substitution pattern of, and that of regions flanking, the C13-C14 trisubstituted Z double bond.1,17,18 The diene group of discodermolide is thus relatively exposed and therefore accessible to the attack by cytochromes P450. This is also true for the left side δ-lactone and the region near the apex of the curved middle, C10-C12. Interestingly, the C19-carbamoyl ester is also relatively sterically available, but we did not see any indication of metabolism or hydrolysis at this site. These findings might be significant in terms of compound optimization. An intact C21-C24 terminal diene system does not seem to be required for the biological activity of discodermolide, although a linear hydrophobic moiety of appropriate length does seem to be required.19,20 Replacing the terminal diene group with a saturated or blocked carbon system would increase metabolic stability of the compound without sacrificing the biological activity. The limited nature of the medicinal chemistry exercises so far reported around the pendant C20 methyl group, the site of metabolism in M6, give few clues as to what modifications would be tolerated there.
An interesting observation was the detection of the possible double bond migrations of C13-C14 and C21-C22 seen in the mass spectra from metabolites M5 and M6. Although assignment of these migrations in the metabolites themselves are at best tentative, if they do occur they might be caused by the delocalization of the unpaired electrons of radicals, generated by hydrogen atom abstraction by cytochrome P450, into the pi system of other carbons within the respective allylic systems.
The most abundant metabolite detected was M8, which we have assigned as the C4-C5 didehydro δ-lactone form of discodermolide. Elegant medicinal chemistry has shown the left side lactone of discodermolide can be altered considerably while retaining biological activity.21,22 It is therefore likely that M8, if formed in vivo, retains much or all of the biological activity of (+)-discodermolide.
Although discodermolide has been examined for many years as a potential anticancer drug, there have been no reports of its metabolism. Our goal here was to determine its metabolically labile sites and extent of metabolism, features that will have an impact on clinical and further medicinal chemistry development. In the oncology clinical trials of discodermolide, pneumotoxicity was noted in ~10% of pancreatic cancer patients after dose escalation, plus a rapid clearance rate from plasma was observed (16). Our experiments suggest that the half-life of discodermolide in human liver microsomes is short, and that the drug is highly susceptible to oxidative biotransformation. The metabolically labile “soft spots,” perhaps responsible for high clearance, were identified; it is unknown at this point if it is the parent drug or the metabolites that are responsible for the pneumotoxicity.
Experimental Section
Chemicals
Discodermolide, prepared by full synthesis, was kindly provided by Novartis Pharma AG. HPLC grade CH3CN and H2O were purchased from Burdick & Jackson (Muskegon, MI) and Fisher Scientific (Pittsburgh, PA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Human Liver Microsomes
Pooled human liver microsomes were purchased from BD Gentest (Wobum, MA). The cytochrome P450 content was 0.33 nmol/mg of protein.
Microsomal Incubations
A typical incubation mixture (0.5 mL) contained 0.2 mg/mL of microsomal protein, 10 µM discodermolide and 1 mM NADPH in 100 mM phosphate buffer (pH 7.4). The incubation was carried out for 15 min at 37 °C and stopped by addition of 0.5 mL of cold (chilled at −20 °C) CH3CN to precipitate proteins. Samples were centrifuged and the supernatant was evaporated in a SpeedVac with no heating. Controls were incubations without microsomal protein or without NADPH.
HPLC
Reversed phase HPLC separations were carried out using a prepacked PicoFrit Biobasic C18 column (5 µm, 300 Åpore, 75 µm × 10 cm, New Objectives, Inc.). The column was eluted with 0.1% formic acid in H2O (solvent A) and 0.1% formic acid in CH3CN (solvent B). The initial condition was 23% solvent B. The gradient was as follows: isocratic at 23% B for 5 min, 23% to 70% B over 10 min, 70% B over 10 min, 70% to 95% B over 1 min, then isocratic at 95% B for 4 min. The flow rate was 160 nL/min. The injection volume was 1 µL.
LC-MS/MS and –MS3 Analyses
A Thermo Finnigan LCQ Deca XP Plus quadrupolar (3D) ion trap (QIT) mass spectrometer equipped with a nanospray source and a Surveyor ProteomeX HPLC was used for MSn (n=1–3) analysis of discodermolide metabolites. Spray voltage was set at 1.43kV, capillary temperature was 180 °C and capillary voltage was 40V. The normalized collision energy was 35%. Accurate mass analysis was performed with a QSTAR Elite QqTOF system (Applied Biosystems/MDS Sciex, Foster City, CA) equipped with a NanoSpray II source and a Dionex Ultimate 3000 HPLC. The electrospray voltage was 1.9kV and the ion source heat temperature was 100 °C. The QqTOF was tuned to 13,000 resolution (half maximum). The [M+H]+ ion of reserpine (m/z 609.2812) was used as the autocalibration signal. The obtained mass accuracy of metabolites was < 10 ppm error.
Acknowledgment
Supported by grants from the NIH (CA078039) and DoD TATRC (W81XWH-05-2-0066). The authors thank Drs. Ken Bair and Fred Kinder at Novartis for the gift of (+)-discodermolide.
References and Notes
- 1.Gunasekera SP, Gunasekera M, Longley RE, Schulte GK. J. Org. Chem. 1990;55:4912–4915. [erratum J. Org. Chem. 1991, 56, 1346.] [Google Scholar]
- 2.Longley RE, Caddigan D, Harmody D, Gunasekera M, Gunasekera SP. Transplantation. 1991;52:650–656. doi: 10.1097/00007890-199110000-00014. [erratum Transplantation 1993, 55, 236] [DOI] [PubMed] [Google Scholar]
- 3.Longley RE, Caddigan D, Harmody D, Gunasekera M, Gunasekera SP. Transplantation. 1991;52:656–651. doi: 10.1097/00007890-199110000-00015. [DOI] [PubMed] [Google Scholar]
- 4.Longley RE, Gunasekera SP, Faherty D, McLane J, Dumont F. Ann. N.Y. Acad. Sci. 1993;696:94–107. doi: 10.1111/j.1749-6632.1993.tb17145.x. [DOI] [PubMed] [Google Scholar]
- 5.Ter Haar E, Kowalski RJ, Hamel E, Lin CM, Longley RE, Gunasekera SP, Rosenkranz HS, Day BW. Biochemistry. 1996;35:243–250. doi: 10.1021/bi9515127. [DOI] [PubMed] [Google Scholar]
- 6.Hung DT, Chen J, Schreiber SL. Chem. Biol. 1996;3:287–293. doi: 10.1016/s1074-5521(96)90108-8. [DOI] [PubMed] [Google Scholar]
- 7.Kowalski RJ, Giannakakou P, Gunasekera SP, Longley RE, Day BW, Hamel E. Mol. Pharmacol. 1997;52:613–622. [PubMed] [Google Scholar]
- 8.Buey RM, Isabel Barasoain I, Jackson E, Meyer A, Giannakakou P, Paterson I, Mooberry S, Andreu JM, Díaz JF. Chem. Biol. 2005;12:1269–1279. doi: 10.1016/j.chembiol.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 9.He L, Yang C-PH, Horwitz SB. Mol. Cancer Ther. 2001;1:3–10. [PubMed] [Google Scholar]
- 10.Huang GS, Lopez-Barcons L, Freeze BS, Smith AB, III, Goldberg GL, Horwitz SB, McDaid HM. Clin. Cancer Res. 2006;12:298–304. doi: 10.1158/1078-0432.CCR-05-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mickel SJ, Sedelmeier GH, Niederer D, Daeffler R, Osmani A, Schreiner K, Seeger-Weibel M, Brod B, Schaer K, Gamboni R, Chen S, Chen W, Jagoe CT, Kinder FR, Jr., Loo M, Prasad K, Repic O, Shieh W-C, Wang R-M, Waykole L, Xu DD, Xue S. Org. Proc. Res. Dev. 2004;8:92–100. [Google Scholar]
- 12.Mickel SJ, Sedelmeier GH, Niederer D, Schuerch F, Grimler D, Koch G, Daeffler R, Osmani A, Hirni A, Schaer K, Gamboni R, Bach A, Chaudhary A, Chen S, Chen W, Hu B, Jagoe CT, Kim H-Y, Kinder FR, Jr., Liu Y, Lu Y, McKenna J, Prashad M, Ramsey TM, Repic O, Rogers L, Shieh W-C, Wang R-M, Waykole L. Org. Proc. Res. Dev. 2004;8:101–106. [Google Scholar]
- 13.Mickel SJ, Sedelmeier GH, Niederer D, Schuerch F, Koch G, Kuesters E, Daeffler R, Osmani A, Seeger-Weibel M, Schmid E, Hirni A, Schaer K, Gamboni R, Bach A, Chen S, Chen W, Geng P, Jagoe CT, Kinder FR, Jr., Lee GT, McKenna J, Ramsey TM, Repic O, Rogers L, Shieh W-C, Wang R-M, Waykole L. Org. Proc. Res. Dev. 2004;8:107–112. [Google Scholar]
- 14.Mickel SJ, Sedelmeier GH, Niederer D, Schuerch F, Seger M, Schreiner K, Daeffler R, Osmani A, Bixel D, Loiseleur O, Cercus J, Stettler H, Schaer K, Gamboni R, Bach A, Chen G-P, Chen W, Geng P, Lee GT, Loeser E, McKenna J, Kinder FR, Jr., Konigsberger K, Prasad K, Ramsey TM, Reel N, Repic O, Rogers L, Shieh W-C, Wang R-M, Waykole L, Xue S, Florence G, Paterson I. Org. Proc. Res. Dev. 2004;8:113–121. [Google Scholar]
- 15.Mickel SJ, Niederer D, Daeffler R, Osmani A, Kuesters E, Schmid E, Schaer K, Gamboni R, Chen W, Loeser E, Kinder FR, Jr., Konigsberger K, Prasad K, Ramsey TM, Repic O, Wang R-M, Florence G, Lyothier I, Paterson I. Org. Proc. Res. Dev. 2004;8:122–130. [Google Scholar]
- 16.Mita A, Lockhart AC, Chen T-L, Bochinski K, Curtright J, Cooper W, Hammond L, Rothenberg M, Rowinsky E, Sharma S. J. Clin. Oncol. 2004;22(14S):2025. [Google Scholar]
- 17.Smith AB, III, LaMarche MJ, Falcone-Hindley M. Org. Lett. 2001;3:695–698. doi: 10.1021/ol006967p. [DOI] [PubMed] [Google Scholar]
- 18.Canales A, Matesanz R, Gardner NM, Andreu JM, Paterson I, Díaz JF, Jiménez-Barbero J. Chemistry. 2008;14:7557–7569. doi: 10.1002/chem.200800039. [DOI] [PubMed] [Google Scholar]
- 19.Gunasekera SP, Longley RE, Isbrucker RA. J. Nat. Prod. 2002;65:1830–1837. doi: 10.1021/np0203234. [DOI] [PubMed] [Google Scholar]
- 20.Shaw SJ. Mini-Rev. Med. Chem. 2008;8:276–284. doi: 10.2174/138955708783744137. [DOI] [PubMed] [Google Scholar]
- 21.Smith AB, III, Xian M. Org. Lett. 2005;23:5229–5232. doi: 10.1021/ol052070m. [DOI] [PubMed] [Google Scholar]
- 22.Shaw SJ, Sundermann KF, Burlingame MA, Myles DC, Freeze BS, Xian M, Brouard I, Smith AB., III J. Am. Chem. Soc. 2005;127:6532–6533. doi: 10.1021/ja051185i. [DOI] [PubMed] [Google Scholar]