Abstract
Background:
Loss of growth inhibitory response to transforming growth factor-β (TGF-β) is a common feature of epithelial cancers. Recent studies have reported that genetic lesions and overexpression of oncoproteins in TGF-β/Smads signalling cascade contribute to the TGF-β resistance. Here, we showed that the overexpressed FOXG1 was involved in attenuating the anti-proliferative control of TGF-β/Smads signalling in ovarian cancer.
Methods:
FOXG1 and p21WAF1/CIP1 expressions were evaluated by real-time quantitative reverse-transcription polymerase chain reaction (RT–PCR), western blot and immunohistochemical analyses. The effect of FOXG1 on p21WAF1/CIP1 transcriptional activity was examined by luciferase reporter assays. Cell lines stably expressing or short hairpin RNA interference-mediated knockdown FOXG1 were established for studying the gain-or-loss functional effects of FOXG1. XTT cell proliferation assay was used to measure cell growth of ovarian cancer cells.
Results:
Quantitative RT–PCR and western blot analyses showed that FOXG1 was upregulated and inversely associated with the expression levels of p21WAF1/CIP1 in ovarian cancer. The overexpression of FOXG1 was significantly correlated with high-grade ovarian cancer (P=0.025). Immunohistochemical analysis on ovarian cancer tissue array was further evidenced that FOXG1 was highly expressed and significantly correlated with high-grade ovarian cancer (P=0.048). Functionally, enforced expression of FOXG1 selectively blocked the TGF-β-induced p21WAF1/CIP1 expressions and increased cell proliferation in ovarian cancer cells. Conversely, FOXG1 knockdown resulted in a 20–26% decrease in cell proliferation together with 16–33% increase in p21WAF1/CIP1 expression. Notably, FOXG1 was able to inhibit the p21WAF1/CIP1 promoter activity in a p53-independent manner by transient reporter assays.
Conclusion
Our results suggest that FOXG1 acts as an oncoprotein inhibiting TGF-β-mediated anti-proliferative responses in ovarian cancer cells through suppressing p21WAF1/CIP1 transcription.
Keywords: FOXG1, p21WAF1/CIP1, TGF-β, ovarian cancer
The transforming growth factor-β (TGF-β) family consists of multifunctional cytokines that control a wide variety of biological activities including cell proliferation, differentiation, apoptosis, cell adhesion, extracellular matrix formation, immune cell regulation and embryonic development through activating TGF-β receptors and Smad transducer proteins (Hu et al, 1998; Massague, 1998). The complicated role of TGF-β in mediating these cellular activities depends on cell types, growth environment, concentration of ligands and the presence of other growth factors (Hu et al, 1998; Massague, 1998; Savage-Dunn, 2005). Inhibition of cell proliferation is one of the biological effects of TGF-β on normal epithelial cells, suggesting that TGF-β acts as a tumour suppressor (Moses et al, 1987). Loss of autocrine activity and/or responsiveness to TGF-β is frequently found in human cancers during tumour progression. Mutations, deletions or methylation of members of TGF-β signalling pathway leading to TGF-β resistance in human cancers have been extensively reported. For examples, decreased expressions or mutations in TGF-β R1, TGF-β RII and Smad4 have been frequently detected in a number of human cancers. (Eisma et al, 1996; Kim et al, 1996; Ko et al, 1998; Korchynskyi et al, 1999; Matsushita et al, 1999; Venkatasubbarao et al, 2000; Maurice et al, 2001; Paik et al, 2003; Sakaguchi et al, 2005; Perttu et al, 2006; Zhong et al, 2006).
On the other hand, elevated expression of proto-oncogenes or oncogenes may also cause TGF-β resistance in tumour cells. For example, upregulation of the c-Ski and SnoN represses the growth inhibitory function of the Smad proteins (He et al, 2003); increased expression of the oncoprotein HER2/Neu activates Smad7 transcription (Dowdy et al, 2003), and overexpression of BCL6 disrupts the Smad-p300 interaction and represses the transcriptional activity of Smad4 (Wang et al, 2008). Moreover, increased expression of the EWS/Fli1 oncogene, the Tax oncoprotein from HTLV-1, the E1A oncoprotein of DNA virus and the E7 oncoprotein of HPV resulted in reduced TGF-β responsiveness (Datta and Bagchi, 1994; Hahm et al, 1999; Mori et al, 2001; Lee et al, 2002). Given the important tumour suppressor functions of the TGF-β pathway, it is expected that other oncoproteins could promote tumourigenesis by counteracting this pathway.
FOXG1, also known as brain factor-1, is a member of Forkhead box family of transcription factors (Arden, 2004; Obendorf et al, 2007). FOXG1 contains a highly conserved DNA-binding domain, which binds to specific DNA sequences and regulates gene expression (Yao et al, 2001). Recent studies have shown that FOXG1 acts as a negative regulator of TGF-β signalling pathway by specifically binding to the Smad MH2 domain and associates with Smad -1, -2, -3 and -4 (Dou et al, 2000; Seoane et al, 2004). This association blocks the binding of Smad proteins to DNA and results in the inhibition of TGF-β signalling (Rodriguez et al, 2001). In addition, FOXG1 has been shown to inhibit expression of the cyclin-dependent kinase (CDK) inhibitor p21WAF1/CIP1, which is normally transcriptionally activated by TGF-β signalling, in glioblastoma and the neuroepithelium (Pardali et al, 2000; Seoane et al, 2004). However, the functions of FOXG1 in other human cancers remain unclear.
In this study, we reported that the FOXG1 was overexpressed in ovarian cancer. The overexpressed FOXG1 was significantly correlated with high-grade ovarian cancer. More importantly, we showed that the increased expression of FOXG1 significantly suppressed the expression of p21WAF1/CIP1 and increased cell proliferation of ovarian cancer cells. Altogether, these findings suggest that FOXG1 was associated with attenuating the TGF-β anti-proliferative response in ovarian cancer.
Materials and methods
Clinical samples and cell lines
Sixty-seven ovarian cancer tissues and 49 normal ovarian tissues were obtained from the Department of Obstetric and Gynaecology at Queen Mary hospital. The histological subtypes and disease stages of the ovarian tumours were classified according to the International Federation of Gynaecology and Obstetrics (FIGO) criteria. All the clinical specimens used in this study were approved by the local institutional ethics committee (Institutional Review Board number: UW05-143 T1806). Four immortalised human ovarian surface epithelial (HOSE) cells were used in this study: HOSE 6-3, HOSE 10-2, HOSE 11-12, HOSE 17-1 and HOSE 11-24 (from Prof George Tsao, the University of Hong Kong). Ovarian cancer cell lines OV2008, C13*, A2780s, A2780cp (gift from Prof Benjamin Tsang, University of Ottawa), OVCAR3, SKOV3, OV420, OV429 and OV433 (American Type Culture Collection, Rockville, MD, USA) were used in this study. All were grown at 37°C in 5% CO2 in minimum essential medium or Dulbecco's modified Eagle medium supplemented with 10% foetal bovine serum.
Plasmids and cell transfection
The pCMV2-Flag-FOXG1-expressing plasmid (gift from Dr Stefano Stifani from McGill University, Montreal, Quebec, Canada) was used for ectopic expression of Flag-tagged FOXG1. The short hairpin RNA interference (shRNAi) targeting FOXG1 (target sequence: TCTGTCCCTCAACAAGTGC) was ligated into pTER vector (gift from Dr Marc van de Wetering, Centre for Biochemical Genetics, the Netherlands) to generate pTER-shFOXG1 plasmid. A human mutant p21WAF1/CIP1 promoter luciferase construct (pWWP) containing a truncated p21WAF1/CIP1 promoter with deleted p53-binding sites (gift from Dr Mark Feitelson, Mercer Laboratory, Thomas Jefferson University, Philadelphia, PA, USA) was used for luciferase reporter assay. LipofectAMINE 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) and Fugene6 Transfection Reagent (Roche Biosciences, Indianapolis, IN, USA) were used for cell transfection according to the manufacturer's instructions. The pcDNA3 and pTER empty vectors were used as mock transfection, respectively, in enforced expression and knockdown assays, whereas pRL-SV40 (Promega, Madison, WI, USA) was served as an internal control in luciferase reporter assay. Stably overexpressed Flag-tagged FOXG1 or FOXG1 knockdown clones were established by drug selection using G418 at 400 μg ml−1 for 2 weeks or puromycin at 2 μg ml−1 for 1 week, respectively. Positive clones were randomly chosen for cell number expansion and verified by western blot analysis.
Quantitative and semi-quantitative reverse-transcription polymerase chain reaction
Total RNA from each cell line was prepared by TRIzol reagent (Invitrogen). First strand cDNA was synthesised by random hexamers and Taqman reverse transcription reagent kit (Applied Biosystems, Foster City, CA, USA). For real-time quantitative reverse-transcription polymerase chain reaction (RT–PCR) (Q-PCR), the amount of FOXG1 and p21WAF1/CIP1 genes were quantified by TaqMan Gene Expression Assays and in an ABI 7700 system (Applied Biosystems) using the FOXG1 and p21WAF1/CIP1 primers and probe from Applied Biosystems (FOXG1, assay ID: Hs00702391_s1; p21WAF1/CIP1, assay ID: Hs00355782; GAPDH, assay ID: Hs99999905_m1). Each sample was performed in triplicate and normalised with human GAPDH (assay ID: Hs99999905_m1; Applied Biosystem). For semi-quantitative RT–PCR, the p21WAF1/CIP1 mRNA level was evaluated by a pair of primers (p21WAF1/CIP1-Sense 5′-ACCATGTGGACCTGTCACTGTCTT-3′ and p21WAF1/CIP1-Antisense 5′-AGAAGATGTAGAGCGGGCCTTTGA-3′) with the following conditions for 30–35 cycles: denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 30 sec. The relative amount of p21WAF1/CIP1 was normalised using GAPDH mRNA with the following primers (GAPDH-Sense: 5′-ACGCATTTGGTCGTATTGGG-3′ and GAPDH-Antisense: 5′-TGATTTTGGAGGGATCTCGC-3′) and amplification at 25 cycles: denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 1 min.
Immunohistochemical and western blot analyses
Immunohistochemical staining for FOXG1 was performed on an ovarian cancer tissue array (OVC961) (Pantomics Inc, San Francisco, CA, USA). The section was immunostained with primary rabbit polyclonal anti-FOXG1 antibody (Abcam Inc, Cambridge, MA, USA) in 1 : 20 dilution. For negative controls, the primary antibody was replaced with Tris-buffered saline. The intensity of staining was scored as 0 (negative), 1+ (faint), 2+ (moderate), 3+ (strong) and 4+ (marked).
For western blot analysis, samples containing equal amounts of protein were separated by SDS–PAGE and electroblotted onto Hybond-P membranes (Amersham Pharmacia Biotech, Cleveland, OH, USA). Blots were blotted with 5% skimmed milk and analysed by immunoblotting with antibodies specific for anti-FOXG1 (Abcam), anti-Flag, anti-β-actin and anti-phosphoserine (Sigma Chemical Co, St Louis, MO, USA), anti-p21WAF1/CIP1, anti-Smad3 and anti-phospho-Smad3 (Cell Signalling Technology, Darvers, MA, USA) and anti-Histone H1 (Santa Cruz Biotechnology Inc, Santa Cruz, CA, USA). Blots were then incubated with goat anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Amersham) and visualised by enhanced chemiluminescence (ECL) (Amersham).
Subcellular fractionation and phosphorylation studies
The cellular localisation of endogenous FOXG1 or Flag-tagged FOXG1 in ovarian cancer cells was examined by western blot analysis on subcellular extracts prepared from NE-PER nuclear and cytoplasmic extraction reagents according to the manufacturer's protocol (Pierce Biotechnology, Rockford, IL, USA). The serine phosphorylation status of FOXG1 in ovarian cancer cells was analysed according to Regad et al (2007). Briefly, ovarian cancer cells were collected in NET lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl and 5 mM EDTA) with 1% NP40, pH 8.0, 0.1 mM PMSF and 1 mM Complete TM protease inhibitor cocktail (Roche). FOXG1 or Flag-tagged FOXG1 was immunopreciptated from 500 ng whole cell lysates by 0.5 μg of anti-FOXG1 (Santa Cruz) by incubation with Protein A/G Plus-Agarose beads (Santa Cruz) at 4°C. Immunoprecipitates on the beads were washed four times with NET lysis buffer, followed by elution using SDS sample buffer (Cell Signalling) and denature by boiling before electrophoresis.
Luciferase reporter assay
Cells were seeded in 24-well plates and transiently transfected with various amounts of pCMV2-Flag-FOXG1 vector with pWWP-luciferase reporter construct. Cells were incubated with the transfection reagents for at least 24 h and were then replaced with fresh medium containing 100 pM TGF-β. After 5 h of TGF-β incubation, cells were lysed for luciferase activity analysis using the Dual-Luciferase Reporter Assay System (Promega). The transfection efficiency was normalised with Renilla luciferase activity. All experiments were repeated three times.
Cell viability analysis
Cell viability was measured by Cell Proliferation kit II (XTT) for 5 days according to the manufacturer's instructions (Roche). The experiment was performed in triplicate for each time point. Human TGF-β1 was purchased as lyophilised samples from R&D systems (Minneapolis, MN, USA). Sterile 4 mM HCl (Merck, Darmstadt, Germany) containing 0.1% (v/v) bovine serum albumin (Pierce) was added to prepare a stock solution of 10 μg ml−1.
Statistical analysis
Student's t test (for parametric data) and the Mann–Whitney test (for non-parametric data) were used. Correlation of gene expression was analysed using Spearman's rho non-parametric statistics. Statistical analyses on clinicopathological correlation were performed using the SPSS version 13.0 software (SPSS, Chicago, IL, USA). A P-value was considered significant when <0.05.
Results
Overexpression of FOXG1 in ovarian cancer
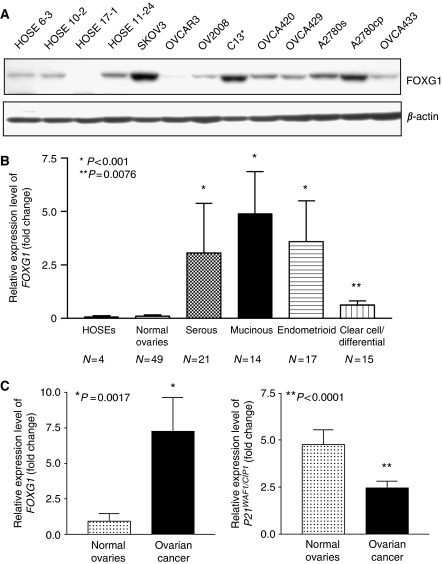
Our preliminary data using cDNA microarray analysis has shown that the expression of FOXG1 in ovarian cancer cell lines was 2.2-folds higher than that in HOSE cells (Liu et al, unpublished data). In this study, we found that four out of nine ovarian cancer cell lines (SKOV3, C13*, A2780s and A2780cp) expressed relatively higher levels of FOXG1 as compared with four HOSEs cell lines by western blotting (Figure 1A). To further confirm the upregulation of FOXG1 in ovarian cancer, we evaluated the expression status of FOXG1 in ovarian cancer tissues (n=67) and normal ovarian tissues (n=49) by Q-PCR analysis. On the basis of comparative CT method using GAPDH as the endogenous control, very low expression level of FOXG1 was detected in normal ovaries and HOSEs (Figure 1B). Conversely, FOXG1 expression was significantly higher in serous, mucinous, endometrioid (P<0.001) and clear cell/undifferentiated (P=0.0076) subtypes of ovarian tumours when compared with the normal ovaries (Figure 1B).
Figure 1.
FOXG1 is underexpressed and is inversely correlated with p21WAF1/CIP1 in normal ovaries and ovarian cancer tissues. (A) Western blot analysis showed the expression of FOXG1 in ovarian cancer cell lines and immortalised normal ovarian epithelial cell lines (HOSEs). (B) The relative expression levels of FOXG1 was evaluated by quantitative RT–PCR on HOSEs, normal ovaries and four histological subtypes of ovarian cancers (*P<0.001; **P=0.0076). N is the number of subtype cases. (C) Quantitative RT–PCR showed the expression level of FOXG1 and p21WAF1/CIP1 in normal ovaries and ovarian cancer tissues. The values were obtained by mean±s.e.m. (*P=0.001; **P<0.0001).
As earlier studies have shown that FOXG1 is able to block the transcription of p21WAF1/CIP1 by counteracting TGF-β-induced signalling pathway and hence promoting cell proliferation (Seoane et al, 2004; Adesina et al, 2007a), it would be interesting to examine the gene expression of p21WAF1/CIP1 in normal ovaries and ovarian cancer tissues. Using Q-PCR analysis, we found that p21WAF1/CIP1 showed higher expression levels in normal ovarian tissues when compared with ovarian tumour (P<0.0001) (Figure 1C). In addition, the expression status of p21WAF1/CIP1 was inversely associated with FOXG1 (Figure 1C). However, no statistical significant negative correlation was found for FOXG1 and p21WAF1/CIP1 expressions using non-parametric Spearman rho test (r=−0.05, P>0.05).
Clinicopathological correlation of FOXG1 and ovarian cancer
On clinicopathological correlation, we found that the FOXG1 overexpression (>1.5-fold) in ovarian cancer was significantly correlated with high-grade tumour (P=0.025) (Table 1). However, there was no association with any histological subtypes of ovarian cancers (serous, mucinous, endometrioid and clear cell), tumour stage, recurrence and age (Table 1), as well as the patient's survival (data not shown). This indicates that the overexpression of FOXG1 does not link to any specific subtypes, tumour stage and patient's survival but is involved in high-grade tumours of ovarian cancer.
Table 1. Clinicopathological correlation of FOXG1 expression in ovarian cancer patients.
|
FOXG1 expression (fold)
|
||||
|---|---|---|---|---|
| Characteristics | Total | ⩽1.5 | >1.5 | P |
| All cases | 67 | 33 (49.25%) | 34 (50.75%) | |
| Age (y) | ||||
| <55 | 39 | 16 (41.03%) | 23 (58.97%) | |
| >55 | 28 | 17 (60.71%) | 11 (39.29%) | 0.112 |
| Stage | ||||
| Early | 21 | 12 (57.14%) | 9 (42.86%) | |
| Late | 43 | 21 (48.84%) | 22 (51.16%) | 0.532 |
| Grade | ||||
| 1 and 2 | 20 | 14 (70.00%) | 6 (30.00%) | |
| 3 | 29 | 11 (37.93%) | 18 (62.07%) | 0.027* |
| Histological subtypes | ||||
| Serous+/−papillary | 21 | 13 (61.90%) | 8 (38.10%) | |
| Others | 46 | 20 (43.48%) | 26 (56.52%) | 0.162 |
| Mucinous | 14 | 5 (35.71%) | 9 (64.29%) | |
| Others | 53 | 28 (52.83%) | 25 (47.17%) | 0.255 |
| Endometrioid | 17 | 7 (41.18%) | 10 (58.82%) | |
| Others | 50 | 26 (52.00%) | 24 (48.00%) | 0.441 |
| Clear cells/undifferentiated | 15 | 8 (53.33%) | 7 (46.67%) | |
| Others | 52 | 25 (48.08%) | 27 (51.92%) | 0.720 |
| Recurrence | ||||
| + | 31 | 15 (48.39%) | 16 (51.61%) | |
| − | 31 | 16 (51.61%) | 15 (48.39%) | 0.799 |
Immunohistochemical analysis of FOXG1 expression in ovarian cancer tissue array
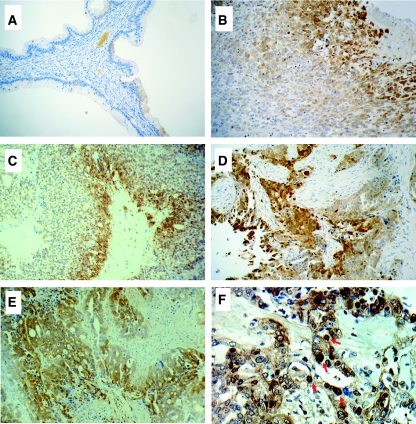
Expression of FOXG1 was also assessed by immunohistochemical staining in ovarian cancer tissue array (OVC961) (Pantomics Inc), which has 31 cases of epithelial ovarian cancer with different histological subtypes (serous, mucinous, endometrioid, clear cell/undifferentiated) and 6 cases of normal and benign tumour tissues. FOXG1 protein expression was evaluated as 0 (negative), 1+ (faint), 2+ (moderate), 3+ (strong) and 4+ (marked). FOXG1 staining was observed in 80.6% (25 out of 31) of ovarian carcinomas examined whereas positive staining was rarely observed in the epithelial cells of normal ovarian and benign tumour tissues (Figure 2A) (Table 2). Positive staining of FOXG1 was observed in 100.0% (9 out of 9) serous, 66.7% (4 out of 6) mucinous, 84.6% (11 out of 13) endometrioid and 33.3% (1 out of 3) clear cell/undifferentiated ovarian cancer (Table 2) (Figure 2B–E). Of 29 ovarian cancer cases with tumour grade and stage scores, high intensity (>3+) of FOXG1 was significantly correlated with high-grade tumour (P=0.048) but no association was found for tumour stage (data not shown). This finding was consistent with the quantitative RT–PCR data. Besides, FOXG1 expression was mainly localised in the cytoplasm, but nuclear localisation of FOXG1 was also detected (Figure 2F), suggesting that FOXG1 exhibits nucleocytoplasmic shuttling and has different functions in cells.
Figure 2.
Immunohistochemical analyses of FOXG1 expression in ovarian cancer tissues. FOXG1 staining in (A) benign, (B) clear cell/undifferentiated, (C) mucinous, (D) serous and (E) endometrioid ovarian cancers. (F) Cytoplasmic and nuclear localisation of FOXG1. Arrow, nuclear localisation of FOXG1. Magnification: × 200 (A, B, C, D and E), and × 400 (F).
Table 2. Immunohistochemical scores of FOXG1 on tissue array of ovarian cancer.
|
Expression level (%)
|
||||
|---|---|---|---|---|
| Histological types | Number of cases | Negative | 1+/2+ | 3+/4+ |
| Normal/benign tumour | 6 | 5 (58.3) | 1 (16.7) | 0 |
| Clear cell/undifferentiated | 3 | 2 (66.7) | 0 | 1 (33.4) |
| Serous | 9 | 0 | 2 (22.2) | 7 (77.8) |
| Mucinous | 6 | 2 (33.3) | 3 (50.0) | 1 (16.7) |
| Endometrioid | 13 | 2 (15.4) | 2 (15.4) | 9 (69.4) |
The effect of TGF-β on ovarian cancer cells
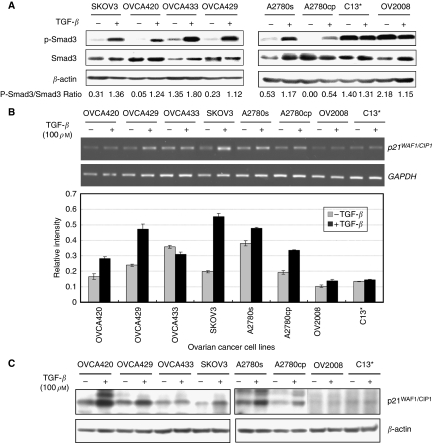
To examine whether the overexpressed FOXG1 exerts inhibitory role on TGF-β-induced p21WAF1/CIP1 expression similar to earlier reports (Seoane et al, 2004; Adesina et al, 2007a), several ovarian cancer cell models were used. Genetic and/or epigenetic lesions usually deregulate TGF-β-mediated cell growth inhibitory effects in various human cancers, including ovarian cancer (Sakaguchi et al, 2005). To exclude ovarian cancer cell lines with malfunctioned TGF-β signalling, we tested the TGF-β/Smad signalling component, Smad3 and the p21WAF1/CIP1 gene induction under TGF-β treatment for the selected cell lines. To test the effect of TGF-β on ovarian cancer cell lines, cells were cultured in the presence or absence of 100 pM TGF-β in serum-free medium for 5 h and were then collected for immunoblotting. As Smad3 is one of the key effectors of TGF-β signalling, the levels of Smad3 and phosphorylated Smad3, which represent the activated form of Smad3, were examined.
On TGF-β treatment, activation of Smad3 was observed in OVCA420, OVCA429, OVCA433, SKOV3, A2780s and A2780cp cells, but activation of Smad3 was not observed in OV2008 and C13* after TGF-β treatment (Figure 3A). Moreover, by semi-quantitative RT–PCR and western blot analyses, induction of p21WAF1/CIP1 expression was observed in OVCA420, OVCA429, SKOV3, A2780s and A2780cp (Figure 3B and C). As these five cell lines also showed Smad3 activation on TGF-β treatment, we believe that these cell lines are still responsive to the inhibitory effect of TGF-β signalling. However, OVCA433, which showed Smad3 activation in the presence of TGF-β, failed to show p21WAF1/CIP1 induction after TGF-β addition. It is suspected that the TGF-β signalling pathway downstream of Smad3 has been disrupted in OVCA433. Taken together, five of the ovarian cancer cell lines studied (OVCA420, OVCA429, SKOV3, A2780s and A2780cp) acquire a functional TGF-β signalling pathway and are able to drive the transcriptional activation of p21WAF1/CIP1 on TGF-β treatment.
Figure 3.
Effects of TGF-β on ovarian cancer cell lines. (A) Activation of Smad3 on TGF-β treatment. Ovarian cancer cell lines were cultured in the presence (+) or absence (−) of 100 pM TGF-β in serum-free medium for 5 h and were then collected for immunoblotting using antibodies specific to Smad3 and phosphorylated form of Smad3. Loading was compared with the internal control using antibody against β-actin. Semi-quantitative RT–PCR (B) and western blot (C) analyses showed the induction of p21WAF1/CIP1 expression by TGF-β. For semi-quiantitative RT–PCR analysis, the relative p21WAF1/CIP1 mRNA level normalised by the level of GAPDH was then plotted and presented as a bar chart.
FOXG1 inhibits p21WAF1/CIP1 promoter activity on TGF-β treatment
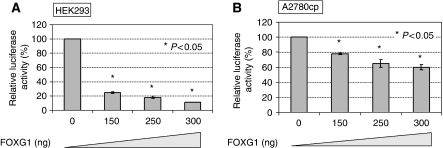
To determine whether FOXG1 regulates p21WAF1/CIP1 expression at the transcriptional level, luciferase reporter assay was conducted. As it has been well known that p21WAF1/CIP1 is a downstream target of p53, truncated p21WAF1/CIP1 promoter without p53-binding site (pWWP) was used to ensure that the change of p21WAF1/CIP1 promoter activity was independent of p53 regulation. On transfection of FOXG1 at increasing amounts (0, 150, 250 and 300 ng) and treatment of TGF-β (100 pM), the relative luciferase activity of p21WAF1/CIP was reduced from 100% to 25%, 19% and 11%, respectively, in HEK293T cells (P<0.05), and from 100% to 79%, 65% and 60%, respectively, in A2780cp cells (P<0.05) (Figure 4A and B). These data suggest that FOXG1 regulates the expression of p21WAF1/CIP1 at the transcriptional level in HEK293T as well as A2780cp ovarian cancer cells.
Figure 4.
FOXG1 inhibits p21WAF1/CIP1 promoter activity on TGF-β treatment. (A) HEK293T, (B) A2780cp cells were transfected with fixed amount of the reporter plasmids pWWP and pRL-SV40 and varied amounts of pCMV2-Flag-FOXG1 and empty vector pCMV2. 100 pM TGF-β was added 24 h after transfection and was incubated for another 5 h before measuring the luciferase activity. The promoter activity of empty vector pCMV2 transfection was set to be 100% and the promoter activity with FOXG1 plasmids addition was expressed as a percentage relative to empty vector control.
Enforced expression of FOXG1 inhibits p21WAF1/CIP1 induction and increases cell proliferation on TGF-β treatment
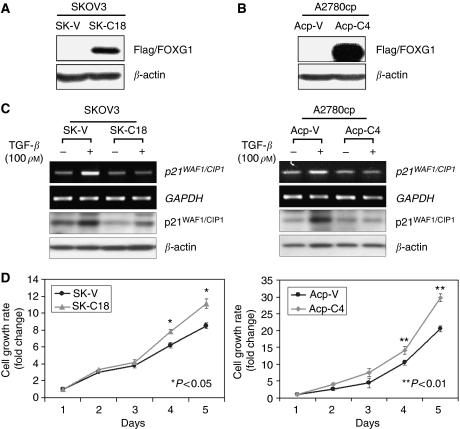
To investigate the inhibitory function of FOXG1 on TGF-β-induced p21WAF1/CIP1 expression and cell proliferation, we first generated Flag-tagged FOXG1 stable-expressing clones from two ovarian cancer cell lines, SKOV3 and A2780cp (Figure 5A and B). On TGF-β treatment, a 2.6-fold and 1.5-fold induction of p21WAF1/CIP1 mRNA and protein levels were observed in SKOV3 and A2780cp vector controls, respectively (Figure 5C). However, there was no change in p21WAF1/CIP1 levels on TGF-β treatment for two stable clones expressing Flag-tagged FOXG1 of both cell lines (SK-C18 and Acp-C4) (Figure 5C). This suggests that the overexpressed FOXG1 suppresses TGF-β-induced p21WAF1/CIP1 expression. We next examined the effect of enforced FOXG1 expression on cell growth using in vitro proliferation assay (XTT assay). In the presence of 10 pg TGF-β1 in cell culture medium, enforced expression of FOXG1 in both stable clones exhibited a higher cell proliferation rate (38% for SK-C18, P<0.05; and 48% for Acp-C4, P<0.01) as compared with their vector control cell lines (Figure 5D). These results suggest that FOXG1 promotes the cell proliferation of ovarian cancer cells, which is consistent with the role of FOXG1 in the inhibition of anti-proliferative effect of TGF-β reported in earlier studies.
Figure 5.
Enforced expression of FOXG1 blocks the induction of p21WAF1/CIP1 and promotes cell proliferation on TGF-β treatment. (A, B) Western blot analysis showed the expression of Flag-tagged FOXG1 in two stable clones, SK-C18 and Acp-C4, of SKOV3 and A2780cp ovarian cancer cell lines, respectively. SK-V and Acp-V are their corresponding vector controls. (C) The vector controls and Flag-tagged FOXG1 stable clones of SKOV3 (left) and A2780cp (right) were cultured in the presence (+) or absence (−) of 100 pM TGF-β for 24 h. The p21WAF1/CIP1 mRNA and protein levels were analysed by semi-quantitative RT–PCR and western blot analyses, respectively. (D) The vector controls and Flag-tagged FOXG1 stable clones of SKOV3 (left) and A2780cp (right) were cultured in media containing 10 pM TGF-β for 5 days. XTT assay showed lower cell proliferation rate in FOXG1-overexpressed clones (38% for SK-C18, P<0.05; and 48% for Acp-C4, P<0.01) as compared with their vector controls.
Depletion of FOXG1 sensitises TGF-β-mediated p21WAF1/CIP1 induction and cell growth inhibition
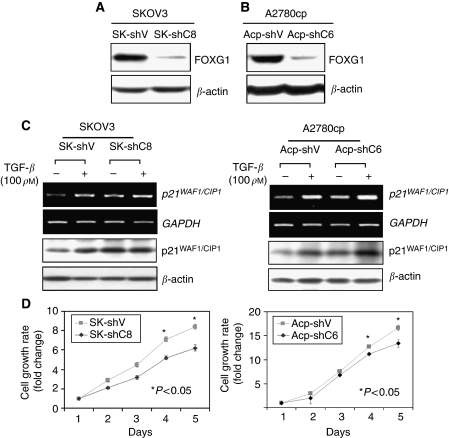
To further confirm the inhibitory effect of FOXG1 on TGF-β signalling in ovarian cancer cells, we used the vector-based RNAi technique and successfully knockdown >75% endogenous FOXG1 in an FOXG1 overexpressing cell lines, SKOV3 and A2780cp (Figure 6A and B). On TGF-β treatment, additional increase of p21WAF1/CIP1 expressions was observed in the FOXG1-depleted clones of SKOV3 (16% increase in SK-shC8) and A2780cp (33% increase in Acp-shC6) cells as compared with empty vector controls (Figure 6B). XTT assay also demonstrated that depletion of FOXG1 could reduce 26% (SK-shC8) and 20% (Acp-shC6) cell proliferation rate (P<0.05) in the presence of 10 pg TGF-β in SKOV3 and A2780cp cells, respectively (Figure 6C). These data further support that FOXG1 counteracts TGF-β-mediated cell growth arrest through regulation of p21WAF1/CIP1 induction.
Figure 6.
Depletion of FOXG1 sensitises TGF-β mediated of p21WAF1/CIP1 induction and cell growth inhibition. (A, B) Western blot analysis showed the reduction of endogenous FOXG1 by FOXG1 shRNAi plasmid, pTER-shFOXG1, SKOV3 and A2780cp ovarian cancer cell lines, respectively. SK-shV and Acp-shV are their corresponding vector controls. (C) The vector controls and FOXG1 stable knockdown clones of SKOV3 (SK-shV and SK-shC8) (left), and A2780cp (Acp-shV and Acp-shC6) (right) were cultured in the presence (+) or absence (−) of 100 pM TGF-β for 24 h. The p21WAF1/CIP1 mRNA and protein levels were analysed by semi-quantitative RT–PCR and western blot analyses, respectively. (D) FOXG1 knockdown clones of SKOV3 (SK-shV and SK-shC8) (left), and A2780cp (Acp-shV and Acp-shC6), cultured in the medium supplemented with 10 pM TGF-β for 5 days. XTT assay shown higher cell proliferation rate in FOXG1-depleted clones (26% for SK-shC8 and 20% for Acp-shC6, P<0.05) as compared with their vector controls.
Inhibition of p21WAF1/CIP1 induction because of increased nuclear localisation of FOXG1
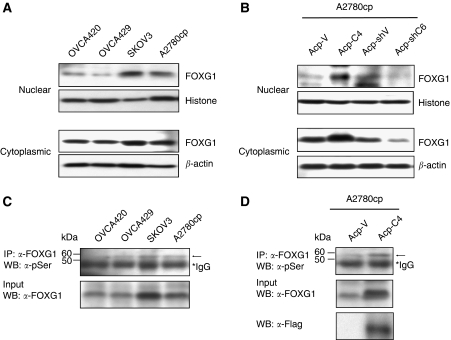
It has been shown that FOXG1 is able to block p21WAF1/CIP1 induction by interacting with FoxO–Smad complexes in nucleus (Seoane et al, 2004). To examine whether the inhibition of p21WAF1/CIP1 induction in overexpressed FOXG1 ovarian cancer cells was due to the increased accumulation of nuclear FOXG1, we conducted western blot analysis on subcellular extracts from ovarian cancer cell lines. Of four ovarian cancer cell lines, a relatively higher level of nuclear FOXG1 was observed in SKOV3 and A2780cp cells, which expressed relatively higher levels of FOXG1 as compared with OVCA420 and OVCA429 cells (Figures 1A and 7A). We also found that a significant increase of nuclear Flag-tagged FOXG1 was observed in Flag-tagged FOXG1 stably expressing clone (Acp-C4) (Figure 7B). Conversely, a reduction in nuclear FOXG1 was found in FOXG1 knockdown clone (Acp-shC6) of A2780cp cells (Figure 7B). A recent finding has shown that the elevation of phosphorylation at Ser19 of FOXG1 promotes nuclear import (Regad et al, 2007). Thus, we attempted to evaluate the serine phsophorylation levels of FOXG1 according to the similar protocol of Regad et al (2007). By immunopreciptation and western blot assays, an increase level of serine phosphorylation of FOXG1 was detected in SKOV3 and A2780cp cells, and in Flag-tagged FOXG1 stably expressing clone (Acp-C4) (Figure 7C and D). These results show that the inhibition of p21WAF1/CIP1 induction in FOXG1-overexpressed ovarian cancer cells is due to the increased serine phosphorylation and nuclear localisation of FOXG1.
Figure 7.
Subcellular localisation and phosphorylation of FOXG1. Western blot analysis showed the expression levels of cytoplasmic and nuclear FOXG1 in ovarian cancer cell lines (OVCA420, OVCA429, SKOV3 and A2780cp) (A), and the expression of Flag-tagged FOXG1 and FOXG1 in Flag-tagged FOXG1 stable clone (Acp-C4) and FOXG1 knockdown clone (Acp-shC6) of A2780cp (B). Histone H1 and β-actin were used as internal controls of nuclear and cytoplasmic extracts, respectively. (C) Western blot analysis showed higher expression levels of phosphoserine FOXG1 (arrow) in SKOV3 and A2780cp cell lines. (D) An increased phosphoserine FOXG1 was detected in Flag-tagged FOXG1 stable clone (Acp-C4) as compared with vector control (Acp-C4) of A2780cp.
Discussion
Loss of responsiveness to the growth inhibitory effect of TGF-β is a substantial mechanism in cancer development (Moses et al, 1987). Apart from genetic and/or epigenetic lesions in TGF-β receptors or TGF-β/Smad transducers that can contribute to TGF-β resistance in human cancers (Arai et al, 1998; Ko et al, 1998; Matsushita et al, 1999; Venkatasubbarao et al, 2000; Maurice et al, 2001; Fukushima et al, 2003; Sakaguchi et al, 2005; Zhong et al, 2006), emerging data have also suggested that the expressions of viral proto-oncogenes and oncogenes are capable of inducing TGF-β resistance through blocking the functions of Smads (Datta and Bagchi, 1994; Hahm et al, 1999; Mori et al, 2001; Lee et al, 2002; Dowdy et al, 2003; He et al, 2003; Wang et al, 2008). In this study, we found that FOXG1 was overexpressed in ovarian cancer cell lines and tissue samples. Using ovarian cancer cell models, we showed that overexpressed FOXG1 could suppress the TGF-β/Smad pathway-induced p21WAF1/CIP1 expression and enhanced cell proliferation in ovarian cancer cells. These data suggest that FOXG1 is another oncogene participating in TGF-β resistance through suppressing p21WAF1/CIP1 expression mediated by TGF/Smad signalling in ovarian cancer cells.
FOXG1 is highly expressed in the telencephalon and has a crucial role in both proliferation and differentiation of neocortical progenitors during brain development (Hanashima et al, 2002; Muzio and Mallamaci, 2005). Indeed, v-Qin and c-Qin, the orthologs of FOXG1, have been reported to induce oncogenic transformation of chicken embryo fibroblasts, suggesting FOXG1 may function as an oncogene in human cancer (Chang et al, 1996; Li et al, 1997). This is further evidenced by the current reports that overexpression of FOXG1 is associated with the development of human medulloblastoma and glioblastoma through abolishing TGF-β-mediated growth inhibition by suppressing the transcription activation of p21WAF1/CIP1 (Seoane et al, 2004; Adesina et al, 2007b). However, these reports just indicate the important roles of FOXG1 in the development of brain cells and neural tumourigenesis. The roles and the oncogenic potential of FOXG1 are rarely reported in other human epithelial-derived cancers. In this study, we showed that FOXG1 exercises an oncogenic function in attenuating the anti-proliferative control of TGF-β through negative regulation of p21WAF1/CIP1 expression in ovarian cancer cells. This suggests that the oncogenic role of FOXG1 contributing to TGF-β resistance is not only restricted in human medulloblastoma and glioblastoma but also extends to other human epithelial-derived cancers.
TGF-β exerts cell growth inhibitory effects through induction of the CDK inhibitors p15Ink4b and p21WAF1/CIP1 expressions through Smad-mediated transcriptional activities (Voss et al, 1999; Feng et al, 2000). These inhibitors are able to block cyclin and CDKs from phosphorylating the retinoblastoma protein (Rb), as well as preventing the progression of the cell cycle (Reynisdottir et al, 1995; Massague et al, 2000). FOXG1 has been shown to inhibit Smad-mediated induction of p21WAF1/CIP1 expression through association with FoxO–Smad complexes in the nucleus (Rodriguez et al, 2001; Seoane et al, 2004; Adesina et al, 2007b). To investigate the cell growth inhibitory mechanism of FOXG1 on ovarian cancer cells, we evaluated the p21WAF1/CIP1 expression by quantitative RT–PCR, western blot and immunohistochemical analyses. Although we observed that there was an inverse relationship between the expressions of FOXG1 and p21WAF1/CIP1, no significant statistical correlation was found. However, this is not surprising because p21WAF1/CIP1 is usually downregulated in human cancers and is regulated by p53-dependent and -independent pathways (O'Reilly, 2005). Therefore, to exclude the effect of p53 on p21WAF1/CIP1 expression, we used p21WAF1/CIP1 promoter (truncated without p53-binding site) luciferase reporter assay to study the effect of FOXG1 on p21WAF1/CIP1 in HEK293T and A2780cp cell models. Consistent with the findings from FOXG1 on human medulloblastoma and glioblastoma (Rodriguez et al, 2001; Seoane et al, 2004; Adesina et al, 2007b), FOXG1 could reduce TGF-β-mediated p21WAF1/CIP1 expression in a dose-dependent manner. We also showed that the overexpression or RNAi-mediated depletion of FOXG1 could alter the expression of p21WAF1/CIP1 in ovarian cancer cells. This suggests that FOXG1 is able to inhibit TGF-β-mediated p21WAF1/CIP1 induction and supports its growth promoting function in ovarian cancer cells.
Recent studies have shown that the phosphorylation of Ser 19 at the N-terminus of FOXG1 promotes nuclear imports of FOXG1 (Regad et al, 2007). The nuclear FOXG1, in turn, blocks TGF-β-mediated p21WAF1/CIP1 induction (Seoane et al, 2004). To investigate whether the inhibitory effect on p21WAF1/CIP1 induction is due to this mechanism, we evaluated subcellular localisation and serine phosphorylation status of FOXG1. Our data showed that higher levels of serine phosphorylation of FOXG1 were consistent with the accumulation of nuclear FOXG1 in FOXG1-overexpressed ovarian cancer cell lines (SKOV3 and A2780cp) and Flag-tagged FOXG1 enforced-expressing cells. However, no change of FOXG1 nuclear localisation and serine phosphorylation levels were observed in ovarian cancer cells under TGF-β treatment (see Supplementary Figure). This indicates that the increased serine phosphorylation and nuclear localisation of FOXG1 in FOXG1-overexpressed ovarian cancer cells attribute the inhibition of TGF-β-mediated p21WAF1/CIP1 induction.
Finally, apart from inhibition on cell proliferation, TGF-β also induces differentiation and apoptosis in many normal epithelial cells (Elliott and Blobe, 2005). Loss of TGF-β responsiveness has been found in high grade with poor differentiated human cancers (Tang et al, 2003; Elliott and Blobe, 2005). Intriguingly, the overexpression of FOXG1 was also significantly correlated with a higher grade of ovarian cancer and in agreement with the above findings. These findings suggest that overexpressed FOXG1 suppresses TGF-β responsiveness in ovarian cancer.
Acknowledgments
We thank Prof Benjamin Tsang from the Department of Obstetrics and Gynaecology, University of Ottawa, Canada for providing four ovarian cancer cell lines, OV2008, C13*, A2780s, A2780cp; Professor George Tsao from the Department of Anatomy, the University of Hong Kong for providing four HOSEs cell lines; Dr Stefano Stifani from McGill University, Montreal, Quebec, Canada for providing the pCMV2-Flag-FOXG1-expressing plasmid; Dr Mark Feitelson from Mercer Laboratory, Thomas Jefferson University, Philadelphia, PA, USA for providing the human mutant p21WAF1/CIP1 promoter luciferase construct (pWWP). This study was supported by HKU Seed Funding Programme for Basic Research (200711159085) and Wong Check She Charitable Foundation.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Adesina AM, Nguyen Y, Guanaratne P, Pulliam J, Lopez-Terrada D, Margolin J, Finegold M (2007a) FOXG1 is overexpressed in hepatoblastoma. Hum Pathol 38: 400–409 [DOI] [PubMed] [Google Scholar]
- Adesina AM, Nguyen Y, Mehta V, Takei H, Stangeby P, Crabtree S, Chintagumpala M, Gumerlock MK (2007b) FOXG1 dysregulation is a frequent event in medulloblastoma. J Neurooncol 85: 111–122 [DOI] [PubMed] [Google Scholar]
- Arai T, Akiyama Y, Okabe S, Ando M, Endo M, Yuasa Y (1998) Genomic structure of the human Smad3 gene and its infrequent alterations in colorectal cancers. Cancer Lett 122: 157–163 [DOI] [PubMed] [Google Scholar]
- Arden KC (2004) FoxO: linking new signaling pathways. Mol Cell 14: 416–418 [DOI] [PubMed] [Google Scholar]
- Chang HW, Li J, Vogt PK (1996) Domains of the qin protein required for oncogenic transformation. Oncogene 13: 441–444 [PubMed] [Google Scholar]
- Datta PK, Bagchi S (1994) Repression of transforming growth factor beta 1 promoter by the adenovirus oncogene E1A. Identification of a unique GC-rich sequence as a target for E1A repression. J Biol Chem 269: 25392–25399 [PubMed] [Google Scholar]
- Dou C, Lee J, Liu B, Liu F, Massague J, Xuan S, Lai E (2000) BF-1 interferes with transforming growth factor beta signaling by associating with Smad partners. Mol Cell Biol 20: 6201–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SC, Mariani A, Janknecht R (2003) HER2/Neu- and TAK1-mediated up-regulation of the transforming growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol Chem 278: 44377–44384 [DOI] [PubMed] [Google Scholar]
- Eisma RJ, Spiro JD, von Biberstein SE, Lindquist R, Kreutzer DL (1996) Decreased expression of transforming growth factor beta receptors on head and neck squamous cell carcinoma tumor cells. Am J Surg 172: 641–645 [DOI] [PubMed] [Google Scholar]
- Elliott RL, Blobe GC (2005) Role of transforming growth factor Beta in human cancer. J Clin Oncol 23: 2078–2093 [DOI] [PubMed] [Google Scholar]
- Feng XH, Lin X, Derynck R (2000) Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J 19: 5178–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Mashiko M, Takita K, Otake T, Endo Y, Sekikawa K, Takenoshita S (2003) Mutational analysis of TGF-beta type II receptor, Smad2, Smad3, Smad4, Smad6 and Smad7 genes in colorectal cancer. J Exp Clin Cancer Res 22: 315–320 [PubMed] [Google Scholar]
- Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG, Sorensen P, Thiele C, Kim S (1999) Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet 23: 222–227 [DOI] [PubMed] [Google Scholar]
- Hanashima C, Shen L, Li SC, Lai E (2002) Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci 22: 6526–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tegen SB, Krawitz AR, Martin GS, Luo K (2003) The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J Biol Chem 278: 30540–30547 [DOI] [PubMed] [Google Scholar]
- Hu PP, Datto MB, Wang XF (1998) Molecular mechanisms of transforming growth factor-beta signaling. Endocr Rev 19: 349–363 [DOI] [PubMed] [Google Scholar]
- Kim IY, Ahn HJ, Zelner DJ, Shaw JW, Lang S, Kato M, Oefelein MG, Miyazono K, Nemeth JA, Kozlowski JM, Lee C (1996) Loss of expression of transforming growth factor beta type I and type II receptors correlates with tumor grade in human prostate cancer tissues. Clin Cancer Res 2: 1255–1261 [PubMed] [Google Scholar]
- Ko Y, Banerji SS, Liu Y, Li W, Liang J, Soule HD, Pauley RJ, Willson JK, Zborowska E, Brattain MG (1998) Expression of transforming growth factor-beta receptor type II and tumorigenicity in human breast adenocarcinoma MCF-7 cells. J Cell Physiol 176: 424–434 [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, Landstrom M, Stoika R, Funa K, Heldin CH, ten Dijke P, Souchelnytskyi S (1999) Expression of Smad proteins in human colorectal cancer. Int J Cancer 82: 197–202 [DOI] [PubMed] [Google Scholar]
- Lee DK, Kim BC, Kim IY, Cho EA, Satterwhite DJ, Kim SJ (2002) The human papilloma virus E7 oncoprotein inhibits transforming growth factor-beta signaling by blocking binding of the Smad complex to its target sequence. J Biol Chem 277: 38557–38564 [DOI] [PubMed] [Google Scholar]
- Li J, Thurm H, Chang HW, Iacovoni JS, Vogt PK (1997) Oncogenic transformation induced by the Qin protein is correlated with transcriptional repression. Proc Natl Acad Sci USA 94: 10885–10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J (1998) TGF-beta signal transduction. Annu Rev Biochem 67: 753–791 [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS (2000) TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103: 295–309 [DOI] [PubMed] [Google Scholar]
- Matsushita M, Matsuzaki K, Date M, Watanabe T, Shibano K, Nakagawa T, Yanagitani S, Amoh Y, Takemoto H, Ogata N, Yamamoto C, Kubota Y, Seki T, Inokuchi H, Nishizawa M, Takada H, Sawamura T, Okamura A, Inoue K (1999) Down-regulation of TGF-beta receptors in human colorectal cancer: implications for cancer development. Br J Cancer 80: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice D, Pierreux CE, Howell M, Wilentz RE, Owen MJ, Hill CS (2001) Loss of Smad4 function in pancreatic tumors: C-terminal truncation leads to decreased stability. J Biol Chem 276: 43175–43181 [DOI] [PubMed] [Google Scholar]
- Mori N, Morishita M, Tsukazaki T, Giam CZ, Kumatori A, Tanaka Y, Yamamoto N (2001) Human T-cell leukemia virus type I oncoprotein Tax represses Smad-dependent transforming growth factor beta signaling through interaction with CREB-binding protein/p300. Blood 97: 2137–2144 [DOI] [PubMed] [Google Scholar]
- Moses HL, Coffey Jr RJ, Leof EB, Lyons RM, Keski-Oja J (1987) Transforming growth factor beta regulation of cell proliferation. J Cell Physiol (Suppl 5): 1–7 [DOI] [PubMed]
- Muzio L, Mallamaci A (2005) Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci 25: 4435–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly MA (2005) Redox activation of p21Cip1/WAF1/Sdi1: a multifunctional regulator of cell survival and death. Antioxid Redox Signal 7: 108–118 [DOI] [PubMed] [Google Scholar]
- Obendorf M, Meyer R, Henning K, Mitev YA, Schroder J, Patchev VK, Wolf SS (2007) FoxG1, a member of the forkhead family, is a corepressor of the androgen receptor. J Steroid Biochem Mol Biol 104: 195–207 [DOI] [PubMed] [Google Scholar]
- Paik SY, Park YN, Kim H, Park C (2003) Expression of transforming growth factor-beta1 and transforming growth factor-beta receptors in hepatocellular carcinoma and dysplastic nodules. Mod Pathol 16: 86–96 [DOI] [PubMed] [Google Scholar]
- Pardali K, Kurisaki A, Moren A, ten Dijke P, Kardassis D, Moustakas A (2000) Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J Biol Chem 275: 29244–29256 [DOI] [PubMed] [Google Scholar]
- Perttu MC, Martikainen PM, Huhtala HS, Blauer M, Tammela TL, Tuohimaa PJ, Syvala H (2006) Altered levels of Smad2 and Smad4 are associated with human prostate carcinogenesis. Prostate Cancer Prostatic Dis 9: 185–189 [DOI] [PubMed] [Google Scholar]
- Regad T, Roth M, Bredenkamp N, Illing N, Papalopulu N (2007) The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat Cell Biol 9: 531–540 [DOI] [PubMed] [Google Scholar]
- Reynisdottir I, Polyak K, Iavarone A, Massague J (1995) Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev 9: 1831–1845 [DOI] [PubMed] [Google Scholar]
- Rodriguez C, Huang LJ, Son JK, McKee A, Xiao Z, Lodish HF (2001) Functional cloning of the proto-oncogene brain factor-1 (BF-1) as a Smad-binding antagonist of transforming growth factor-beta signaling. J Biol Chem 276: 30224–30230 [DOI] [PubMed] [Google Scholar]
- Sakaguchi J, Kyo S, Kanaya T, Maida Y, Hashimoto M, Nakamura M, Yamada K, Inoue M (2005) Aberrant expression and mutations of TGF-beta receptor type II gene in endometrial cancer. Gynecol Oncol 98: 427–433 [DOI] [PubMed] [Google Scholar]
- Savage-Dunn C (2005) TGF-beta signaling. Wormbook 1–12 [DOI] [PMC free article] [PubMed]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117: 211–223 [DOI] [PubMed] [Google Scholar]
- Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM (2003) TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest 112: 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubbarao K, Ahmed MM, Mohiuddin M, Swiderski C, Lee E, Gower Jr WR, Salhab KF, McGrath P, Strodel W, Freeman JW (2000) Differential expression of transforming growth factor beta receptors in human pancreatic adenocarcinoma. Anticancer Res 20: 43–51 [PubMed] [Google Scholar]
- Voss M, Wolff B, Savitskaia N, Ungefroren H, Deppert W, Schmiegel W, Kalthoff H, Naumann M (1999) TGFbeta-induced growth inhibition involves cell cycle inhibitor p21 and pRb independent from p15 expression. Int J Oncol 14: 93–101 [DOI] [PubMed] [Google Scholar]
- Wang D, Long J, Dai F, Liang M, Feng XH, Lin X (2008) BCL6 represses Smad signaling in transforming growth factor-beta resistance. Cancer Res 68: 783–789 [DOI] [PubMed] [Google Scholar]
- Yao J, Lai E, Stifani S (2001) The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol Cell Biol 21: 1962–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D, Morikawa A, Guo L, Colpaert C, Xiong L, Nassar A, Chen C, Lamb N, Dong JT, Zhou W (2006) Homozygous deletion of SMAD4 in breast cancer cell lines and invasive ductal carcinomas. Cancer Biol Ther 5: 601–607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.