Abstract
Parkinson’s disease has been long linked to environmental factors, such as transition metals and recently to α-synuclein, a presynaptic protein. Using tryptophan-containing peptides, we identified the minimal Cu(II)-binding sequence to be within the first four residues, MDV(F/W), anchored by the α-amino terminus. In addition, mutant peptide 1–10 (Lys→Arg) verified that neither Lys6 or Lys10 are necessary for Cu(II) binding. Interestingly, Trp4 excited-state decay kinetics measured for peptides and proteins reveal two quenching modes, possibly arising from two distinct Cu(II)-polypeptide structures.
Keywords: Parkinson’s disease, tryptophan fluorescence, amyloid
INTRODUCTION
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder, characterized by selective loss of dopaminergic neurons in the substantia nigra pars compacta.1–3 Often considered a protein conformational disorder, PD is distinguished histopathologically by an increase in intraneuronal inclusions of amyloid fibrils enriched in α-synuclein (α-syn), a small (~14 kD) presynaptic protein of ill-defined function.4, 5 Although unstructured in solution, it is a “protein chameleon”6 due to its ability to adopt both helical and β-sheet conformation in the membrane-associated and disease state, respectively. Thus, there has been considerable interest in studying α-syn conformations and dynamics.7–10 Remarkably, aggregation and accumulation of abnormally misfolded protein deposits are found in other neurodegenerative ailments such as Alzheimer’s disease (AD),11 amyotrophic lateral sclerosis,12–14 and the prion encephalopathies.15
Much research effort has been focused on the pathogenic mechanism of redox-active metals16, 17 and in particular, the copper binding properties of α-syn.18–21 Specific copper-protein chemistry also has been implicated in other amyloidogenic polypeptides such as the Alzheimer’s Aβ peptide,22–27 prion,28–32 and superoxide dismutase.12, 33 The relevance of transition metals in PD is bolstered by epidemiological studies correlating chronic environmental exposure to copper, iron, or manganese.34 Furthermore, Cu(II) has been shown to stimulate α-syn fibrillation under a wide range of concentrations in vitro.16, 35 Molecular triggers of α-syn aggregation such as environmental factors that promote oxidative stress and aberrant redox-active metal chemistry are therefore of great importance.
Numerous studies on the copper binding properties of α-syn have reported specific metal-protein interactions and have provided insights into the nature of metal coordination chemistry.18–21, 35 Using NMR spectroscopy, His50 was initially identified as a primary ligand, along with participation of residues near the vicinity of 3–9.35 Subsequent work however, suggests that His50 is not necessary for Cu(II) binding and likely is involved in an independent binding site.18, 19, 21 Additionally, this N-terminal site has been postulated to contain both N- and O-ligands involving the α-amino group of Met1 (NH2), deprotonated amides (N−), carboxylate sidechain of Asp2 (β-COO−), and water (H2O).19, 21, 36 Nevertheless, detailed EPR analyses propose that there are two N-terminal binding modes, one with and without His50 coordination20 consistent with previous models from potentiometric measurements on synthetic mutant peptides.37 In contrast, recent results from isothermal titration calorimetry and six synthetic 25 amino-acid fragments spanning the full length protein disputes the involvement of the N-terminus; only peptides corresponding to sequences 46–70 and 116–140 bound Cu(II) with measurable affinity.38 This observation is particularly perplexing since all available data point to the participation of the N-terminal region. Notably, potentiometric measurements showed that isolated residues 1-17 were capable of binding Cu(II).36 A clear consensus has yet to emerge regarding the exact nature of Cu(II) binding in α-syn, particularly on the role of His50 and whether it is a ligand in one or multiple sites.39
In our prior efforts, we have exploited a fluorescent amino acid, Trp, as a site-specific probe of Cu(II)-protein interaction. In particular, we found that Trp4 is the most responsive reporter of Cu(II), indicating a high-affinity N-terminal site with an apparent dissociation constant (Kd(app) ~ 100 nM, pH 7).19 We also demonstrated that the substitution of His50 with a Ser residue has little effect on the Cu(II)-affinity of the protein. Here, we sought to identify the minimal amino acid sequence necessary to preserve Cu(II) affinity by employing a series of synthetic Trp-containing α-syn peptides: α-syn1-10 (MDVWMKGLSL), α-syn1-6(MDVWMK), and α-syn1-4(MDVW). We examined the roles of specific amine ligands (NH2 (α-amino terminus) and ε-NH2 (Lys 6 and Lys10)). We also broadened the scope of our study to determine the effects of solution pH on Cu(II)-binding and Cu(II)-α-syn conformational heterogeneity.
EXPERIMENTAL SECTION
Materials
Copper(II) sulfate pentahydrate (99.999%), N-acetyl-tryptophanamide, 2-(N-morpholino)ethanesulfonic acid (MES, BioUltra, > 99.5%), and 3-(N-morpholino)propanesulfonic acid (MOPS, SigmaUltra, > 99.5%) were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. All peptides (95% purity) were purchased from Anaspec, Inc. (San Jose, CA) and used as received.
Protein Expression and Purification
The wild-type human α-syn expression plasmid (pRK172) was provided by M. Goedert (Medical Council Research Laboratory of Molecular Biology, Cambridge, U.K.).40 Recombinant α-syn was expressed and purified according to published procedures.19 Protein concentrations were determined using a molar extinction coefficient estimated on the basis of amino-acid content: ε280 nm = 10,810 M−1cm−1 (F4W and F4W/H50S). Because α-synuclein is unstructured in solution, it is extremely difficult to assess any direct mutational effects on the protein. However, since the protein is found to be associated with synaptic vesicles in vivo, we have demonstrated the F4W mutation does not perturb the protein’s affinity for membrane mimics (micelles and vesicles). The purity of all protein samples was assessed by SDS-PAGE on a Pharmacia Phastsystem (Amersham Biosciences) visualized by silver-staining methods. The protein molecular weights were confirmed by ESI-MS. Absorption spectra were measured on a CARY 300 Bio UV-Vis spectrophotometer (Varian). Buffer solutions were filtered (0.22 μm) to remove any particulate matter. All purified proteins were concentrated using Centriprep YM-3(MWCO 3kD) (Millipore), stored at −80 °C, and filtered through Microcon YM-100 (MWCO 100kD) (Millipore) spin filter units to remove any oligomeric material prior to experiments.
Copper(II) Titrations
Copper(II) concentrations were determined spectrophotometrically (ε833 nm = 11 M−1cm−1). Prior to experiments, all protein samples were exchanged into the appropriate buffer (20 mM MES (pH 5–6) or MOPS (pH 7) with 100 mM NaCl) using gel filtration chromatography (PD-10 column, GE Healthcare). Stock peptide solutions were gravimetrically prepared, dissolved in Milli-Q water, and then diluted with the appropriate buffer to achieve final working solutions (0.5–1 μM). All titrations were performed on deoxygenated samples (vide infra) to minimize deleterious metal-oxygen chemistry and photobleaching. Tryptophan was excited at 295 nm and fluorescence was monitored from 300 to 500 nm (Fluorolog-3 spectrofluorimeter, HORIBA Jobin Yvon Inc.). All experiments were conducted at 25 °C using a temperature-controlled cuvette holder. Integrated Trp emission intensity was determined (336–400 nm) from background subtracted spectrum and normalized to initial (no Cu(II)) fluorescence. A model fluorophore, N-acetyl-tryptophanamide was used to ensure that there was no deleterious photochemistry in the presence of Cu(II). A strong chelator, EDTA also was used to check for fluorescence reversibility in the proteins after Cu(II) titrations (typically >85% recovery).
pH Dependence of Copper(II) Binding
Protein samples (1 μM) were prepared in buffer (20 mM MOPS, 100 mM NaCl, pH 7) to a final volume of 20 mL, deoxygenated through repeated vacuum/Ar-refill cycles on a Schlenk line, and maintained continuously under an Ar atmosphere in a three-neck round bottom flask. One equivalent of Cu(II) was added (8.9 μL of 2.25 mM Cu(II) stock) and allowed to equilibrate for 15 min with gentle stirring; the pH of the solution was then sequentially adjusted with HCl (2 μL additions of 5.8 or 11.6 M HCl, pH 7 to 5) and monitored by an Orion 9802BN Micro pH electrode (Thermo Scientific). Aliquots (1.5 mL) were removed for fluorescence measurements. To ensure reversibility, we also reproduced the titration from pH 5 to 7 on the same sample (adjusting pH with 2 μL additions of 2.5 or 5 M NaOH). We did not account for any dilution factors since volume changes were insignificant (<0.11%).
Time-resolved Fluorescence Measurements
Tryptophan fluorescence decay kinetics at varying concentrations of Cu(II) were measured by using the fourth harmonic (293 nm) of a regeneratively amplified femtosecond Ti:sapphire (Clark-MXR) pumped optical parametric amplifier laser (Light Conversion) as an excitation source and a picosecond streak camera (Hamamatsu C5680) was used in the photon-counting mode for detection. Tryptophan emission was selected by the combination of an edge (REF-325) and a short-pass (UG-11) filter (CVI Laser) (325<λobs<400 nm). Protein and peptide samples (1 μM) were deoxygenated by repeated evacuation/Ar-fill cycles on a Schlenk line. All experiments were conducted at 25 °C using a temperature-controlled cuvette holder.
Data Analysis
Trp Fluorescence Decays
Measured Trp fluorescence kinetics were logarithmically compressed (100 points per time decade) and normalized (I(t = 0) = 1). We have fit the kinetics data using a MATLAB (The Math Works, Inc.) algorithm (LSQNONNEG; hereafter referred to as NNLS) that minimizes the sum of the squared deviations (χ2) between observed and calculated values of I(t), subject to a nonnegativity constraint on the probability distribution of rates constants, P(k). NNLS fitting produces the narrowest P(k) distributions.8 Average Trp excited state lifetimes (<τ>) were calculated by two methods: integrating over the normalized decay curves (∫I(t)/I(t =0)) or summing rate constants extracted from NNLS fits (Σk P(k)/k).
Copper(II) Binding Curves
Estimation of apparent dissociation constants (Kd(app)) were extracted as previously described for a simple two-state model:19
where P ≡ free α-syn, Cu ≡ free copper, PCu ≡ 1:1 complex of copper and α-syn. The total steady-state Trp fluorescence quenching data were fit according to the following equation:
where I(Cu) is the measured fluorescence intensity in the presence of Cu, I(Cu0) is the measured fluorescence intensity for the free protein, and α is relative fluorescence intensity of PCu. Data fitting were performed using IGOR Pro 6.01 (Wavemetrics).
pH Titration Curves
Estimation of apparent pKa values and the number of protons involved in the process, n, were extracted from fits to the Henderson-Hasselbalch equation:
where I is the measured fluorescence intensity in the presence Cu at a given pH, IpH7 is the relative fluorescence intensity at pH 7, and IpH5 is the relative fluorescence intensity at pH 5. Data fitting were performed using IGOR Pro 6.01 (Wavemetrics).
RESULTS AND DISCUSSION
Minimal Cu(II)-binding α-synuclein Sequence
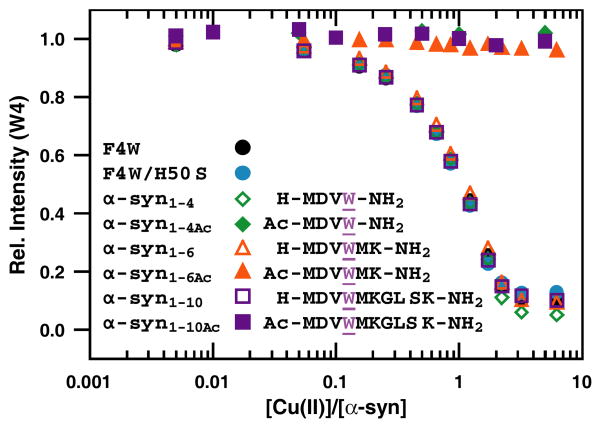
We sought to identify the minimal N-terminal sequence that is capable of retaining affinity for Cu(II) ions (as previously characterized in the full length F4W and F4W/H50S protein) by employing a series of synthetic α-syn peptides (α-syn1-10, α-syn1-6, α-syn1-4; Figure 1). All peptides contain a tryptophan substitution at position 4 (F→W) as a fluorescent probe and an amidated C-terminus to avoid the coordination of the C-terminal carboxylate. Upon addition of Cu(II) (5 nM–6 μM in 20 mM MOPS, 100 mM NaCl, pH 7, 25 °C), tryptophan fluorescence ([peptide] = 1.0 ± 0.2 μM) is dramatically quenched, attributable to direct Cu(II)-peptide interactions. Surprisingly, the removal of more than 130 residues has relatively little effect on metal-protein interactions; the titration curves for the three peptides and proteins are nearly indistinguishable. Indeed, our data strongly indicate that only the first four residues are essential to preserve Cu(II) affinity and we also note that α-syn1-4 may have a slightly increased affinity (≤5% more Trp quenching is observed at >2 eq. Cu(II)). These results clearly support our initial proposal that His50 is not required for N-terminal Cu(II) binding. Furthermore, our data indicate that even if His50 does interact with the copper center, it would contribute minimally to the stability of the metal-protein complex because the copper is chelated by the N-terminus and backbone amides.
Figure 1.
Comparison of Cu(II)-binding induced tryptophan fluorescence quenching of synthetic peptides and full length α-syn variants. Tryptophan fluorescence quenching was not observed when the α-amino termini of the peptides were acetylated ([peptide/protein] = 1.0 ± 0.2 μM in deoxygenated 20 mM MOPS, 100mM NaCl, pH 7.0 buffer). All C-termini in the peptides are amidated (denoted as −NH2).
Titration curves were measured at higher peptide concentrations (5–25 μM) to evaluate binding stoichiometry; nearly identical behaviors were obtained for all three model peptides reaching saturation ~1:1 protein:Cu complex (Figure S1). In addition, titrations performed under different solution pH conditions for α-syn1-10 are indistinguishable from that of the proteins (vide infra, Figure S2). Taken together, these data provide evidence that this N-terminal 1-4 peptide could serve as a minimal model to provide structural insights into the Cu(II)-α-syn complex.
Roles of Specific Amine Ligands
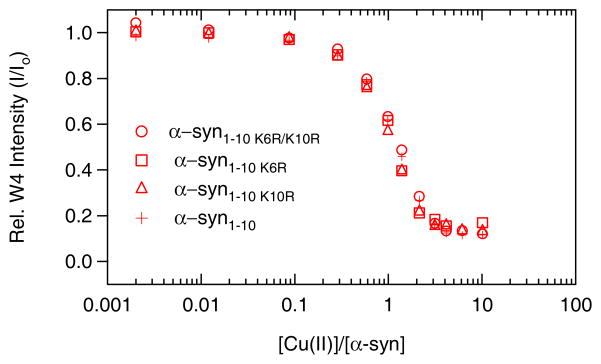
We further evaluated the roles of specific amine ligands: Lys at positions 6 and 10 as well as the α-amino terminus. Consistent with our hypothesis that the essential ligands for Cu(II) reside in the first four residues, binding curves for mutant peptides containing Arg substitutions (α-syn1-10K6R, α-syn1-10K10R, α-syn1-10K6RK10R) are nearly identical to α-syn1-10, ruling out the binding of Lys6 and/or Lys10 (Figure 2). To assess the participation of the α-amino moiety, the N-terminus was acetylated on all peptides. This modification completely disrupts Cu(II) coordination and assigns the α-amino group as the critical ligand (Figure 1). Our results provide direct evidence that for α-syn, a free N-terminal NH2 is required for Cu(II) binding. The finding that the α-amino terminus is crucial for Cu(II) binding in α-syn leads us to suggest a known motif in Cu(II)-polypeptide interactions: Cu(II) is anchored by the free amino-terminal nitrogen and chelates to adjacent backbone amides.41–43 Our data indicate that through post-translational modification of the N-terminus, copper-protein chemistry could be modulated in vivo. Recent studies have provided evidence for considerable α-amino terminal acetylation (to what extent remains unclear) in α-syn extracted from different cellular sources.44, 45 It is therefore difficult to assess the significance of N-terminal-Cu(II)-binding in α-syn function and its associated disease state.
Figure 2.
Trp4 fluorescence intensity of Lys substituted (Lys→Arg) α-synuclein peptides as a function of added Cu(II). [peptide] = 0.5 μM in deoxygenated 20 mM MOPS, 100 mM NaCl, pH 7 buffer.
pH Titrations of Cu(II)-F4W and Cu(II)-F4W/H50S α-synucleins
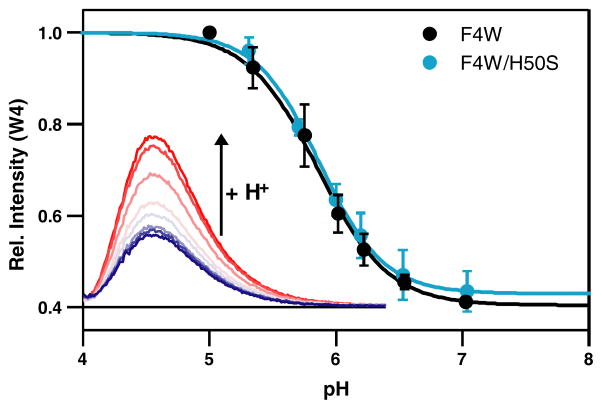
To gain further insights into Cu(II) binding to α-syn, we turned to the full length proteins and examined the effects of solution pH on Cu(II)-α-syn (Figure 3). At pH 7, Trp4 emission decreases upon the addition of one equivalent of Cu(II) (60% quenching at [α-syn] = [Cu(II)] = 1 μM in 20 mM MOPS, 100 mM NaCl, 25 °C) confirming direct metal interaction in both F4W and F4W/H50S proteins. Upon acidification, Trp4 fluorescence intensity of both proteins begins to recover with an apparent transition midpoint at pH 5.8 and at pH 5, nearly all of the bound Cu(II) ions are released, in accord with a model in which at least one of the ligands contain an ionizable group.
Figure 3.
Comparison of Trp4 fluorescence intensity of Cu(II)-F4W and Cu(II)-F4W/H50S α-syn ([α-syn] = [Cu(II)] = 1 μM) as a function of pH in deoxygenated 20 mM MOPS, 100 mM NaCl buffer. Error bars represent standard deviations. Fits are shown as solid lines. (Inset) Representative Trp fluorescence data (buffer background has been subtracted).
Notably, the titration curves are very similar for the two variants, completely reversible and independent of initial pH and thus revealing that His50 has negligible effects on the pH dependence of Cu(II) binding. Using the Henderson-Hasselbalch equation, we can fit the titration curves to obtain the apparent pKa (this is an apparent pKa because it involves coupled equilibria of deprotonation and metal-binding) and the number of protons, n, involved in the process (F4W: pKa = 5.8(2), n = 1.6(5); F4W/H50S: pKa = 5.9(1); n = 1.7(5)). Because our Trp4 fluorescence quenching is due to its close proximity to bound Cu(II), these values likely reflect two nearby ligand protonations. Indeed, pH titrations are fully reproduced for the α-syn1-4 peptide (data not shown). In particular, these numbers are consistent with deprotonation constants reported for amine and amide protons (pKa value ~4.4–6.0) for α-syn fragments.36, 37 Another possible candidate is the carboxylate side chain of Asp2.
To ensure that this observation is independent of Cu(II):protein stoichiometry, we also performed complete Cu(II) titrations (50 nM–1 mM Cu(II)) at selected pH solution conditions (pH 5–7) and obtained similar results for both proteins (Figure S2). As anticipated, the binding affinity is reduced substantially at lower pH (Kd(app) increases by two orders of magnitude from ~200 nM (pH 7) to 20 μM (pH 5), corresponding to competition between protonation and metal coordination (Table 1).
Cu(II)-α-synuclein Conformational Heterogeneity Revealed by Time-resolved Trp Fluorescence
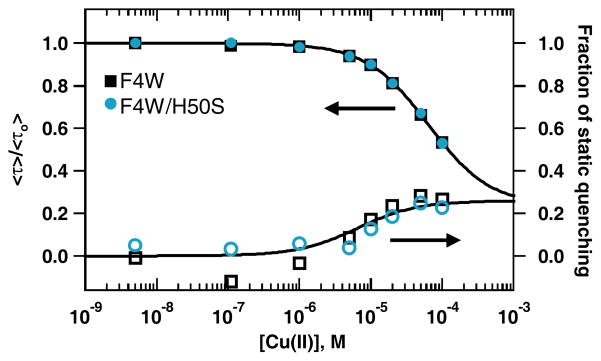
From our prior time-resolved fluorescence measurements at neutral pH, we observed the presence of two quenching modes reflecting Cu(II)-α-syn conformational heterogeneity.19 In order to investigate whether or not these quenching modes could represent different ionizable ligands, we examined the Trp excited-state decay kinetics under acidic solution conditions (20 mM MES, 100 mM NaCl, pH 5). We find the presence of both static- and dynamic-quenching modes is unaffected by solution pH: upon addition of Cu(II) ions, the average tryptophan lifetime shortens (<τ>no Cu ~2.5 ns; <τ>100 μM Cu ~1.1 ns) accounting only for ~80% reduction in the measured fluorescence intensity (Figure 4). Interestingly, differences emerge between titration curves extracted from the two quenching modes. For the static-quenching fraction, the midpoint transition is shifted to a lower Cu(II) concentration, revealing tighter Cu(II)-protein interactions (Kd(app)(static) ~7(3) μM; Kd(app)(dynamic) ~63(4) μM). This deviation between the two populations also is observed at pH 7; there is greater uncertainty associated with the static component at pH 7 so we cannot reliably determine its dissociation constant (Figure S3). Notably, the relative populations of dynamic vs. static quenching remain unperturbed by solution pH suggesting photophysical behavior of Trp4 is not sensitive to ligand protonation events.
Figure 4.
Average Trp4 fluorescence lifetimes (top) and fraction of static quenching (bottom) extracted from NNLS analyses of F4W(■) and F4W/H50S( ) α-synuclein variants ([protein] = 1 μM) as functions of added Cu(II) in deoxygenated 20 mM MES, 100 mM NaCl, pH 5 buffer. Fits to a single binding site model are shown as solid lines.
) α-synuclein variants ([protein] = 1 μM) as functions of added Cu(II) in deoxygenated 20 mM MES, 100 mM NaCl, pH 5 buffer. Fits to a single binding site model are shown as solid lines.
We attribute the Trp fluorescence quenching by Cu(II) to electron-transfer processes involving the metal ion and the singlet-excited state. The dual quenching modes could then arise from two distinct Cu(II)-protein structures, one in which Trp4 and Cu(II) are within static-quenching distance, and the other where Trp4 is sufficiently close to Cu(II) to make contact during the ~3-ns excited-singlet lifetime. Because of this relatively short lifetime, Trp must be in close proximity to the metal ion.10 The static-quenching population may be due to the formation of a nonluminescent ground state via outersphere association of Trp with the Cu(II)-protein complex, reminiscent of the Cu(II) site in the octarepeat domain of the prion peptide.46 Alternatively, the complex fluorescence quenching behavior may reflect the broad distribution of Trp4-Cu(II) distances in a structurally heterogeneous polypeptide. Nonetheless, our data reveal the presence of at least two N-terminal binding modes (conformations) that are not affected by the presence of His50. Tryptophan excited-state decay kinetics measured for all peptides at pH 7 retain comparable fractions of static and dynamic quenching as the proteins; it is clear that local polypeptide structure dictates Trp4 fluorescence properties (Figure S3).
CONCLUSIONS
Our data clearly demonstrate that Trp fluorescence is a sensitive molecular probe of Cu(II) and N-terminal α-syn interactions. Using site-directed mutagenesis and synthetic peptides, we show that His50 has little or no contribution to the formation of the high affinity Cu(II) coordination complex. Specifically, we identify the first four residues, MDV(F/W), as the essential binding site, anchored by the α-amino terminus. This peptide binds Cu(II) with a submicromolar dissociation constant comparable to the full length protein. These experiments do not completely preclude the possibility of Cu(II)-His50 interaction (the unstructured nature of α-syn allows for long range interactions), but rather highlight that it is not required for the N-terminal site that we have characterized. Nevertheless, elucidating the nature of Cu-α-synuclein interactions in the presence of other biomolecules, e.g. phospholipid bilayers, is necessary in defining the role of metal ions in PD, as well as other synucleinopathies.
Supplementary Material
Table I.
Apparent dissociation constants (Kd(app)) for Cu(II) binding to α-synuclein variants
| Kd(app) (μM)a | |||
|---|---|---|---|
| pHb | F4W | F4W/H50S | Syn1-10 |
| 5.0 | 17 ± 4 | 17 ± 5 | 19 ± 2 |
| 5.5 | 2.0 ± 0.1 | 3.4 ± 0.8 | 2.4 ± 0.1 |
| 5.8 | 0.8 ± 0.1 | 0.9 ± 0.2 | 0.7 ± 0.1 |
| 6.0 | 0.5 ± 0.2 | 0.9 ± 0.2 | 0.4 ± 0.1 |
| 7.0c | 0.2 ± 0.2 | 0.2 ± 0.2 | 0.2 ± 0.2 |
The Kd(app) values are obtained from fits of tryptophan fluorescence data shown for the full-length variants (Figure S2) using a two state binding model Syn-Cu ⇆ Syn + Cu.
All buffers were filtered through a 0.22 μm membrane. 20 mM MES and MOPS buffers containing 100 mM NaCl were used for pH 5 to 6 and pH 7, respectively.
Reliable Kd(app) values cannot be extracted because protein/peptide concentration used in these experiments (1 μM) are 10 fold greater than Kd(app).
Acknowledgments
Supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute. We thank Hank Fales (Laboratory of Applied Mass Spectrometry, NHLBI) for technical assistance with ESI-MS, Candace Pfefferkorn for purifying the F4W protein, and Heather Lucas for helpful discussions.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dawson TM, Dawson VL. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 2.Cookson MR. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 3.Tofaris GK, Spillantini MG. Cell Mol Life Sci. 2007;64:2194–2201. doi: 10.1007/s00018-007-7217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forno LS. J Neuropathol Exp Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Beal MF. Nat Rev Neurosci. 2001;2:325–332. doi: 10.1038/35072550. [DOI] [PubMed] [Google Scholar]
- 6.Uversky VN. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 7.Eliezer D, Kutluay E, Bussell R, Browne G. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 8.Lee JC, Langen R, Hummel PA, Gray HB, Winkler JR. Proc Natl Acad Sci U S A. 2004;101:16466–16471. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. J Am Chem Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 10.Lee JC, Lai BT, Kozak JJ, Gray HB, Winkler JR. J Phys Chem B. 2007;111:2107–2112. doi: 10.1021/jp068604y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lansbury PT. Acc Chem Res. 1996;29:317–321. [Google Scholar]
- 12.Valentine JS, Hart PJ. Proc Natl Acad Sci U S A. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Xu GL, Borchelt DR. J Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brannstrom T, Gredal O, Nilsson P, Marklund SL. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 15.Chien P, Weissman JS, DePace AH. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN, Li J, Fink AL. J Biol Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 17.Yamin G, Glaser CB, Uversky VN, Fink AL. J Biol Chem. 2003;278:27630–27635. doi: 10.1074/jbc.M303302200. [DOI] [PubMed] [Google Scholar]
- 18.Sung YH, Rospigliosi C, Eliezer D. BBA-Proteins Proteomics. 2006;1764:5–12. doi: 10.1016/j.bbapap.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Lee JC, Gray HB, Winkler JR. J Am Chem Soc. 2008;130:6898–6899. doi: 10.1021/ja711415b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drew SC, Leong SL, Pham CLL, Tew DJ, Masters CL, Miles LA, Cappai R, Barnham KJ. J Am Chem Soc. 2008;130:7766–7773. doi: 10.1021/ja800708x. [DOI] [PubMed] [Google Scholar]
- 21.Binolfi A, Lamberto GR, Duran R, Quintanar L, Bertoncini CW, Souza JM, Cervenansky C, Zweckstetter M, Griesinger C, Fernandez CO. J Am Chem Soc. 2008;130:11801–11812. doi: 10.1021/ja803494v. [DOI] [PubMed] [Google Scholar]
- 22.Barnham KJ, Bush AI. Curr Opin Chem Biol. 2008;12:222–228. doi: 10.1016/j.cbpa.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Karr JW, Szalai VA. Biochemistry. 2008;47:5006–5016. doi: 10.1021/bi702423h. [DOI] [PubMed] [Google Scholar]
- 24.Karr JW, Kaupp LJ, Szalai VA. J Am Chem Soc. 2004;126:13534–13538. doi: 10.1021/ja0488028. [DOI] [PubMed] [Google Scholar]
- 25.Shearer J, Szalai VA. J Am Chem Soc. 2008;130:17826–17835. doi: 10.1021/ja805940m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarell CJ, Syme CD, Rigby SEJ, Viles JH. Biochemistry. 2009;48:4388–4402. doi: 10.1021/bi900254n. [DOI] [PubMed] [Google Scholar]
- 27.Hatcher LQ, Hong L, Bush WD, Carducci T, Simon JD. J Phys Chem B. 2008;112:8160–8164. doi: 10.1021/jp710806s. [DOI] [PubMed] [Google Scholar]
- 28.Jackson GS, Murray I, Hosszu LLP, Gibbs N, Waltho JP, Clarke AR, Collinge J. Proc Natl Acad Sci U S A. 2001;98:8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millhauser GL. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shearer J, Soh P. Inorg Chem. 2007;46:710–719. doi: 10.1021/ic061236s. [DOI] [PubMed] [Google Scholar]
- 31.Shearer J, Soh P, Lentz S. J Inorg Biochem. 2008;102:2103–2113. doi: 10.1016/j.jinorgbio.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klewpatinond M, Davies P, Bowen S, Brown DR, Viles JH. J Biol Chem. 2008;283:1870–1881. doi: 10.1074/jbc.M708472200. [DOI] [PubMed] [Google Scholar]
- 33.Gaggelli E, Kozlowski H, Valensin D, Valensin G. Chem Rev. 2006;106:1995–2044. doi: 10.1021/cr040410w. [DOI] [PubMed] [Google Scholar]
- 34.Gorell JM, Johnson CC, Rybicki BA, Peterson EL, Kortsha GX, Brown GG, Richardson RJ. Neurotoxicology. 1999;20:239–247. [PubMed] [Google Scholar]
- 35.Rasia RM, Bertoncini CW, Marsh D, Hoyer W, Cherny D, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO. Proc Natl Acad Sci U S A. 2005;102:4294–4299. doi: 10.1073/pnas.0407881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalik-Jankowska T, Rajewska A, Wisniewska K, Grzonka Z, Jezierska J. J Inorg Biochem. 2005;99:2282–2291. doi: 10.1016/j.jinorgbio.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Kowalik-Jankowska T, Rajewska A, Jankowska E, Grzonka Z. Dalton Trans. 2006:5068–5076. doi: 10.1039/b610619f. [DOI] [PubMed] [Google Scholar]
- 38.Brown DR. Biochem Biophys Res Commun. 2009;380:377–381. doi: 10.1016/j.bbrc.2009.01.103. [DOI] [PubMed] [Google Scholar]
- 39.Brown DR. Dalton Trans. 2009:4069–4076. doi: 10.1039/b822135a. [DOI] [PubMed] [Google Scholar]
- 40.Jakes R, Spillantini MG, Goedert M. FEBS Lett. 1994;345:27–32. doi: 10.1016/0014-5793(94)00395-5. [DOI] [PubMed] [Google Scholar]
- 41.Sigel H, Martin RB. Chem Rev. 1982;82:385–426. [Google Scholar]
- 42.Rozga M, Sokolowska M, Protas AM, Bal W. J Biol Inorg Chem. 2007;12:913–918. doi: 10.1007/s00775-007-0244-8. [DOI] [PubMed] [Google Scholar]
- 43.Harford C, Sarkar B. Acc Chem Res. 1997;30:123–130. [Google Scholar]
- 44.Li W, West N, Colla E, Pletnikova O, Troncoso JC, Marsh L, Dawson TM, Jäkälä P, Hartmann T, Price DL, Lee MK. Proc Natl Acad Sci USA. 2005;102:2162–2167. doi: 10.1073/pnas.0406976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang JP, Kling K, Lee M, Diep L, Keim PS, Shen XF, Chataway T, Schlossmacher MG, Seubert P, Schenk D, Sinha S, Gai WP, Chilcote TJ. J Biol Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 46.Burns C, Aronoff-Spencer E, Dunham C, Lario P, Avdievich N, Antholine W, Olmstead M, Vrielink A, Gerfen G, Peisach J, Scott W, Millhauser G. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.