Abstract
Abnormalities of brain white matter and oligodendroglia are among the most consistent findings in schizophrenia (Sz) research. Various gene expression microarray studies of postmortem Sz brains showed a downregulation of myelin transcripts, while imaging and microscopy studies demonstrated decreases in prefrontal cortical (PFC) white matter volume and oligodendroglia density. Currently, the extent to which reduced oligodendrocyte markers contribute to pathophysiological domains of Sz is unknown.
We exposed adolescent rats to cuprizone (CPZ), a copper chelator known to cause demyelination in mice, and examined expression of oligodendrocyte mRNA transcripts and PFC-mediated behavior. Rats on the CPZ diet showed decreased expression of mRNA transcripts encoding oligodendroglial proteins within the medial PFC, but not in the hippocampus or the striatum. These rats also displayed a specific deficit in the ability to shift between perceptual dimensions in the attentional set-shifting task, a PFC-mediated behavioral paradigm modeled after the Wisconsin Card Sorting Test (WCST). The inability to shift strategies corresponds to the deficits exhibited by Sz patients in the WCST. The results demonstrate that a reduction in oligodendrocyte markers is associated with impaired PFC-mediated behaviors. Thus, CPZ exposure of rats can serve as a model to examine the contribution of oligodendrocyte perturbation to cognitive deficits observed in Sz.
Keywords: oligodendrocytes, attentional set-shifting task, prefrontal cortex, hippocampus, schizophrenia, adolescence
Introduction
One of the most consistent observations in schizophrenia (Sz) research is an abnormality of white matter and oligodendroglia in the prefrontal cortex (PFC) and other brain areas (Davis et al., 2003; Karoutzou et al., 2008). Decreases in oligodendrocyte density were found in post-mortem studies (Uranova et al., 2001; Uranova et al., 2007; Vostrikov et al., 2007), a down-regulation of myelin genes was demonstrated in molecular studies (Hakak et al., 2001; Haroutunian et al., 2007; Mimmack et al., 2002), and brain imaging studies in living patients showed decreases in PFC white matter volume (Breier et al., 1992; Buchanan et al., 1998; Sanfilipo et al., 2000; Sigmundsson et al., 2001). White matter abnormalities have been observed at illness onset and in drug-free patients, suggesting that these changes are not due to medication effects (Bagary et al., 2003; Hakak et al., 2001).
Although symptoms associated with Sz are wide-ranging, recent emphasis has turned to cognitive deficits. This emphasis is driven by studies showing that cognitive function is one of the most critical determinants of quality of life for patients (Green et al., 2000). Many of the cognitive deficits are mapped to the PFC (Barch, 2005; Boeker et al., 2006), and Sz patients perform poorly on tasks that depend on PFC function, such as the Wisconsin Card Sorting Test (WCST), (Franke et al., 1992). To date, the extent to which reduced oligodendrocyte markers contribute to particular pathophysiological aspects of Sz and related disorders is unknown, although it is known that white matter is important for cognitive function (Dwork et al., 2007; Fields, 2008). Moreover, disorders associated with demyelination, such as metachromatic leukodystrophy or multiple sclerosis (MS) are frequently accompanied by psychosis (Feinstein, 2007; Hyde et al., 1992).
A number of rodent models were developed that induced PFC deficits through brain lesions, drug exposure and genetic preparations such as transgenic mice (for an in-depth review see Carpenter and Koenig, 2008). Many of these models cause severe pathologies beyond what is observed and expected in Sz. A specific model of the effect of oligodendroglial abnormalities on PFC function has not been examined to date.
Cuprizone (CPZ; biscyclohexanone oxalyldihydrazone) is a copper (Cu) chelator (Messori et al., 2007) which is used in mice to model demyelination disorders (Matsushima and Morell, 2001). Cu is an important catalytic and structural cofactor in a wide array of enzymatic processes with a narrow range of optimal concentration (Kim et al., 2008). Cu levels outside this range are highly neurotoxic (de Bie et al., 2007; Harrison et al., 2000; Menkes et al., 1962; Prohaska and Smith, 1982; Puig and Thiele, 2002). In mice, oral CPZ administration increases Cu and Zn concentrations in the brain (Zatta et al., 2005) and decreases the expression of myelin-specific genes (Morell et al., 1998; Pasquini et al., 2007; Seiwa et al., 2007). However, it is still not clear if CPZ-toxicity results from increased or decreased availability of Cu. Moreover, the exact reasons for the specific toxicity to myelin are still under investigation.
CPZ impairs spatial working memory in mice, which can be reversed by the antipsychotic drug quetiapine (Xiao et al., 2008). Although CPZ works well as a demyelinating agent in mice and as a model of MS, it is not used as a demyelinating agent in rats as it does not cause severe oligodendrocyte toxicity in this species (Adamo et al., 2006; Love, 1988; Matsushima and Morell, 2001; Purves et al., 1991).
The present experiments were designed to examine if CPZ exposure during adolescence reduces myelin transcripts in the rat comparable to what is observed in Sz, and the potential effect this has on PFC-mediated performance. We were interested in inducing a mild lesion of oligodendrocytes in the forebrain without inducing MS-like pathology.
Sprague-Dawley rats were exposed to a CPZ diet and gene expression patterns were examined in the PFC, hippocampus and striatum. The behavior of CPZ-treated rats was investigated in the Attentional Set Shifting Task (ASST), a modified version of the WCST which reveals impairments in Sz (Franke et al., 1992; Haut et al., 1996). One phase of the ASST, the extra-dimensional shift, is impaired by bilateral lesions of the medial frontal cortex and could thus reveal a decline in PFC function (Birrell and Brown, 2000; McAlonan and Brown, 2003).
The results demonstrate that CPZ exposure of the rat leads to a downregulation of myelin transcripts in the PFC and to decreased performance in the extra-dimensional shift of the ASST, while hippocampus and striatum are unaffected. This model might be useful to examine the contribution of oligodendrocytes to the cognitive deficits in various neuro-psychiatric disorders.
Materials and Methods
Subjects
Male Sprague Dawley rats (Charles River Laboratories, Wilmington, MA), n=12 for gene expression microarrays and Q-PCR on P43, n=11 for Q-PCR on P54, n=11 for locomotor activity, and n=24 for ASST, were housed in groups of 2–4. Beginning on P29 and throughout all experiments, rats were fed CPZ (0.2%) diet or control diet (custom manufactured by Harlan Teklad, Madison, WI). Rats examined in the ASST were maintained on a restricted diet of 16–25 g of food per rat per day, starting three days before behavioral testing (P44) until the end of testing (P57), with water available ad libitum (Figure 1A). Animals were weighed tri-weekly, and chow was adjusted to maintain weight during the food restriction phase (Figure 1B). All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and Vanderbilt University’s Institutional Animal Care and Use Committee guidelines.
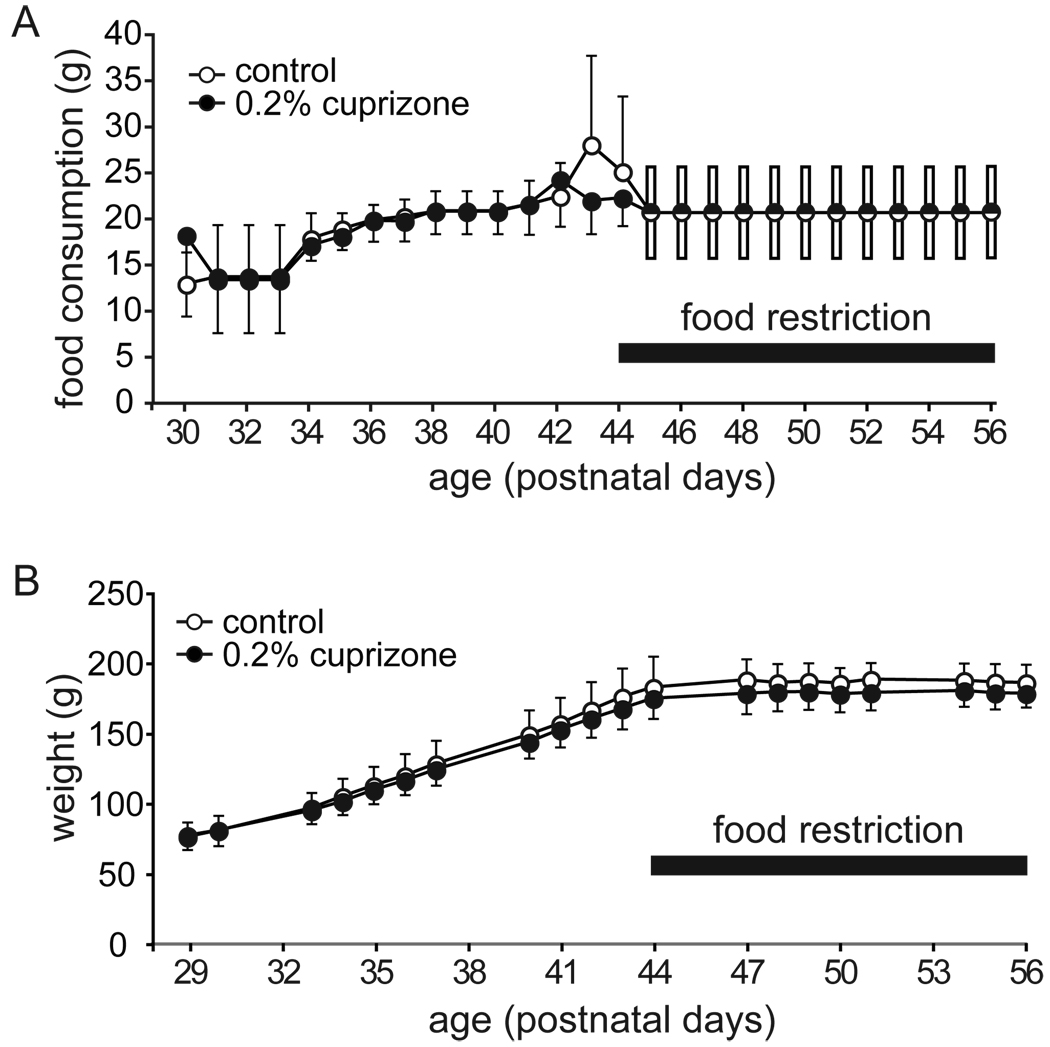
Figure 1.
CPZ intake and weight gain of ASST rats. A. Average food intake between P30 and P56. Chow was provided ad libitum until P44. Starting at P45, food restriction was implemented to increase motivation for food-digging in the ASST. During food restriction, enough food was provided to sustain weight. B. Representative growth curve for CPZ and control rats. Until P44, the growth curve was in line with the growth curve for male Sprague Dawley rats provided by Charles River Laboratories. All data in average ± SD of control (open circles; n=10) and CPZ-fed rats (black circles; n=8).
Gene expression microarrays and Q-PCR
One group of animals was killed on P43 (day 15 of CPZ administration), and a second group on P54. The brains were extracted and snap-frozen at –30°C in 2-methylbutane, and stored at –80°C. Medial PFC [cingulate cortex 1, prelimbic cortex, and infralimbic cortex; from bregma anterioposterior (AP), +4.2 to +2.2] (Paxinos and Watson, 1986), hippocampus [from bregma AP −0.8 to −4.3] and striatum [caudate putamen; from bregma AP +1.2 to −0.8] (Paxinos and Watson, 1986) were dissected on a freezing microtome using micropunches. Care was taken to avoid inclusion of the corpus callosum in the punches.
Gene array experiments
Samples from individual rats (P43) were hybridized to individual arrays. RNA was extracted from 10–15 mg of tissue using the PureLink kit (Invitrogen, Carlsbad, CA). Two µg of total RNA was used for complementary DNA (cDNA) synthesis with the SuperScript double-stranded cDNA synthesis kit (Invitrogen Corp., Carlsbad, CA) and an oligo (dT) primer with T7 promoter sequence. In vitro transcription and biotinylation were carried out with the Gene Chip Expression 3′ Amplification kit for IVT (Affymetrix, Santa Clara, CA). Biotinylated RNA was hybridized to the RAE230 2.0 array (Affymetrix), and washing and staining were performed according to company protocol.
Quality control criteria
Tissue preparation and RNA extractions were performed in a single batch by the same investigator. All quality control criteria defined by Affymetrix were met by the samples, and no significant differences between experimental groups were observed. For the PFC, noise was 1.38 ± 0.1, % present call 63.4 ± 1.6, and 3’/5’ GAPDH 1.21 ± 0.04 (average ± SD); for the hippocampus, noise was 1.16 ± 0.1, % present call 63.0 ± 2.1, and 3’/5’ GAPDH 1.15 ± 0.04 (average ± SD). Quality control criteria were furthermore examined in dChip (Li and Wong, 2001) and in RMA (Bolstad et al., 2003). Percent array outlier in dChip was 0.54 ± 0.1 for PFC, and 0.19±0.21 for hippocampus. One PFC sample with 3% array outliers, and one hippocampus sample with 24% array outliers were excluded from the analysis.
Q-PCR
cDNA was synthesized from 0.2 – 2 ug of total RNA from medial PFC, hippocampus and striatum (P43 and P54) using the iScript cDNA synthesis kit. A primer set for each gene was designed with the Primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi), for amplicons of 100–250 base pairs. Melt curve analysis and polyacrylamide gel electrophoresis were used to confirm the specificity of each primer pair. The iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) was used with an Opticon 2 real-time PCR detection system (Bio-Rad). Sample volume was 20 µl, with 4 µl of 1:10 diluted cDNA samples and 0.3 µM primers. The PCR cycling conditions were initially 95°C for 5 min followed by 39 cycles of 94°C for 10 s, 57°C for 15 s, and 72°C for 20 s. Data were collected between 72 and 84°C depending on amplicon melt temperature. A melt curve analysis was performed at the end of each Q-PCR experiment. Standard curves were generated for each primer pair in every experiment by diluting cDNA from a vehicle sample to a final concentration of 1.00, 0.2, 0.04, and 0.008. Blanks were assessed with each dilution curve to control for cross-contamination. Duplicates were used for dilution curves, blanks, and samples. Reported values were normalized to the average of three internal standards: beta-actin (Actb; GeneID: 81822), alpha 1A tubulin (Tuba1a; GeneID: 64158), and general transcription factor IIB (Gtf2b; GeneID: 81673) none of which were regulated by gene-array or showed group differences in Q-PCR analysis. Genes of interest and primer pairs are listed in supplemental table S1.
Locomotor activity and Attentional Set-Shifting Task
For all behavioral experiments, investigators were blinded for treatment group. Three separate cohorts of animals were tested.
Locomotor Activity
Rats were tested for one hour on P43 and P57 (day 15 and 29 of exposure to CPZ; prior to and after ASST and food restriction). Activity was recorded in 43.2 × 43.2 × 50 cm chambers with photodetector-LED detection in sound-attenuated chambers (Med Associates, St. Albans, VT). All animals were assessed for locomotor activity between 1100–1400h. Chambers were cleaned with 70% ethanol between animals.
Attentional Set-Shifting Task (ASST)
The ASST was employed to assess PFC function as described previously, with modifications (Black et al., 2006). Rats were first trained to discriminate texture and then shifted to odor discrimination. The testing apparatus was a semitranslucent plastic bin (63.5 × 41.9 × 23.2 cm) that was divided into two equal-sized compartments (31.5 × 41.9 cm), separated by a nontransparent divider. One compartment contained two terracotta pots adjacent to each other. The bottoms of all pots were filled with paraffin approximately 12 mm high, and covered by a layer of Frosted Cheerios (General Mills, Minneapolis, MN) held in place by a mesh screen which made the Cheerios inaccessible to the rat. This was done to match potential ‘Cheerio odor’ in all pots. Each pot contained distinct digging medium, and the reinforced pot contained an accessible buried food reward (half, stale, Frosted Cheerio). During scent trials, the pot and media were rubbed with a perfume oil (The Body Shop, Wake Forest, NC). Pairs of media were composed of identical material differing in shape or size to ensure that the smell of the material was not used for discrimination. Each phase of the ASST had unique exemplars (Table 1). During the task, a rat was placed in the empty compartment and the divider was raised to allow access to both pots.
Table 1.
Phases of the ASST, stimuli used, and age at which testing of rats was performed.
| Phase (day) | Dimension | Combination of Stimuli | Age | ||
|---|---|---|---|---|---|
| Relevant | Irrelevant | Positive | Negative | ||
| Introduction to Exemplars | Medium | -- | Eppendorf tube lids | Eppendorf tubes (w/o lids) | P54 |
| Simple discrimination (SD) | Medium | -- | Small chopped plastic tubing | Large chopped plastic tubing | P55 |
| Compound Discrimination (CD) | Medium | Odor | Small chopped plastic tubing + Tea Rose | Large chopped plastic tubing + Tea Rose | P55 |
| Small chopped plastic tubing + White Musk | Large chopped plastic tubing + White Musk | ||||
| Compound Discrimination Reversal (CD-Rev) | Medium | Odor | Large chopped plastic tubing + Tea Rose | Small chopped plastic tubing + Tea Rose | P55 |
| Large chopped plastic tubing + White Musk | Small chopped plastic tubing + White Musk | ||||
| Intradimensional Shift (IDS) | Medium | Odor | Lg Glass Beads + Mandarin | Sm Glass Beads + Mandarin | P56 |
| Lg Glass Beads + Patchouli | Sm Glass Beads + Patchouli | ||||
| Intradimensional Shift Reversal (IDS-Rev) | Medium | Odor | Sm Glass Beads + Mandarin | Lg Glass Beads + Mandarin | P56 |
| Sm Glass Beads + Patchouli | Lg Glass Beads + Patchouli | ||||
| Extradimensional Shift (EDS) | Odor | Medium | Vanilla + Lg Foil Balls | Jasmine + Lg Foil Balls | P56 |
| Vanilla + Sm Foil Balls | Jasmine + Sm Foil Balls | ||||
| Extradimensional Shift Reversal (EDS-Rev) | Odor | Medium | Jasmine + Lg Foil Balls | Vanilla + Lg Foil Balls | P56 |
| Jasmine + Sm Foil Balls | Vanilla + Sm Foil Balls | ||||
Beginning on P47 (day 19 of CPZ administration), after three days of food restriction, rats were habituated to chambers and trained to dig for five consecutive days (trial max/day = 30 min). For dig training, a single unscented pot filled with polypropylene pellets was continuously re-baited until the rat approached and dug in less than 10 seconds after the divider was raised. A "dig" was defined as any distinct displacement of the digging medium with either the paw or the nose. Thus, a rat could investigate the pot or the medium in the pot by sniffing or touching the medium with its whiskers, before executing what was scored as a dig response.
On P54, in the Introduction to Exemplars, rats were trained to dig in Eppendorf tube lids for the food reward and to ignore Eppendorf tube bottoms which were not rewarded. On P55, rats performed a Simple Discrimination (SD; Table 1). The first four trials of a session were defined as exploratory trials, where the rat was permitted to dig in both pots, regardless of whether the first choice was correct. During subsequent trials, once a "dig" was executed in one pot, the other pot was removed to prevent digging in both pots within one trial. The criterion for each phase was six consecutive correct trials.
After reaching criterion in SD, rats were challenged with a Compound Discrimination (CD) task by introducing the second perceptual dimension, odor. The same medium as in SD was reinforced, with the intent to have the rats ignore the ‘odor’ dimension. After completion of the CD, a stimulus reversal (CD-Rev) was introduced, in which the previously non-enforced medium carried the reward (Table 1).
The Intra-dimensional Shift (IDS) started on P56. During this phase of the task, novel stimuli were presented, with medium still as the relevant perceptual dimension. After the IDS, rats performed a stimulus reversal (IDS-Rev), followed by an Extra-dimensional Shift (EDS). In the EDS, a novel set of stimuli was presented, but the previously irrelevant perceptual dimension, odor, became the relevant dimension. The final discrimination was an odor stimulus reversal (EDS-Rev). Of the 24 rats, trained in three different groups, six did not learn to dig (control n=2, CPZ n=4) and could not be used for the task. This difference was not significant for treatment group (Fisher’s Exact Test p=0.640).
Data analysis
For gene array experiments, a number of programs were used for data analysis: GeneChip Operating Software (GCOS) (Affymetrix) was used for scanning and to obtain quality control data. RMAExpress (Bolstad et al., 2003; Irizarry et al., 2003) was used for quantile normalization and background correction to compute expression levels for all probe sets. DNAChip Analyzer (Li and Wong, 2001) was used for clustering and David 2.0 (Dennis et al., 2003) was used to group regulated genes into functional annotations. In addition we used GenMAPP (Salomonis et al., 2007) to built 1473 brain-specific gene groups (MAPPs) by categorizing all curated probe sets from the rat RAE 230 2.0 array, and the MAPPfinder program to analyze regulation of groups. We used the permuted p-value (calculated with non-parametric statistic based on 2000 permutations of the data) as the indicator of significant regulation. Expression values were log2-transformed for statistical analyses.
T-tests and Wilcoxon Rank-Sum tests were performed for Q-PCR, for the individual phases of the ASST and for locomotor measures. Analysis of covariance was furthermore carried out for locomotor measurements. Repeated measures ANOVA was carried out for weight and food intake studies.
Results
Body Weights and Food Consumption
Between P29 and P44 no difference between treatment groups was observed in weight gain [F(1,16) = 1.0, p=0.324]. During behavioral testing between P47 and P56, chow was adjusted for weight maintenance without weight gain [F(1,16) = 2.0, p=0.179], (Figure 1).
Effects of CPZ exposure during adolescence on gene expression in the medial PFC, hippocampus and striatum
Gene expression microarrays
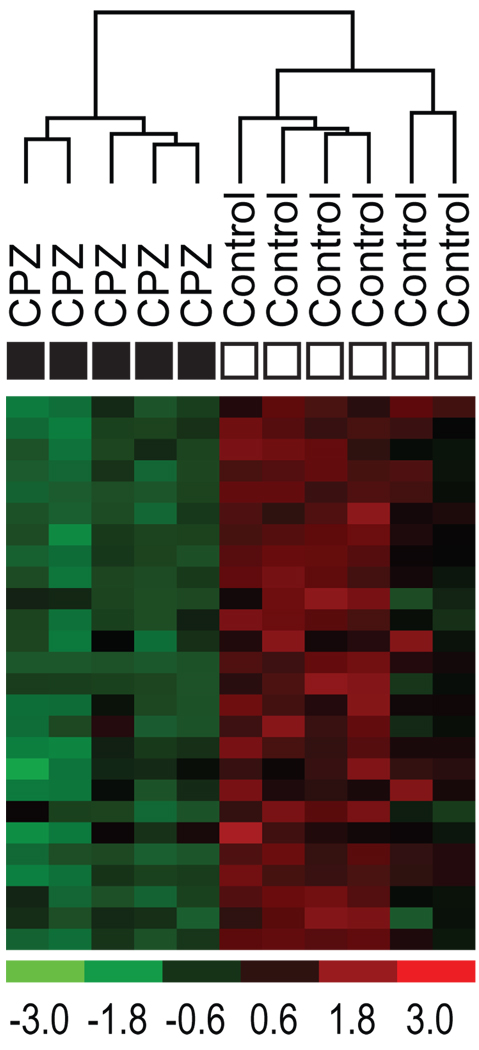
We chose gene expression microarrays for the initial molecular examination since they reveal broad, biological themes (Konradi, 2005). The RAE 230 2.0 Array contains over 31,000 probe sets. Of those, only probe sets with at least 40% ‘Present’ call in either group (control, CPZ), at least 1.2 fold difference in expression levels, and a p-value below 0.05 were used in the analysis. For the PFC, 337 probe sets matched these criteria, of which 149 were downregulated and 188 were upregulated. Using the NIH David Tool to examine the downregulated genes, one annotation cluster was above the minimum criteria. This annotation cluster had a high enrichment score (5.6) with the terms within that cluster related to myelination (Table 2; Figure 3B). Using our own 1473 brain-specific MAPPs and the MAPPfinder program (Salomonis et al., 2007), only 3 MAPPs reached significance, ‘oligodendrocyte markers’, ‘myelin’, and ‘glia’. Table 3 lists the data for all regulated probesets of myelin markers, and these probesets clustered control and CPZ genes into distinct groups (Eisen et al., 1998), (Figure 2). False discovery rate of the group of myelin genes, established with 2000 permutations, was 0.0%.
Table 2.
Significant annotation clusters as calculated with NIH-David (39, 60). An enrichment score of ‘2’ was used as cutoff. David provides batch annotation and gene term enrichment analysis to highlight the most relevant terms associated with a given gene list. The Functional Annotation Chart reduces the redundancy of annotations with related/identical terms by grouping similar annotations together. Although DAVID uses over 40 annotation categories, all significant enrichment scores came from the Gene Ontology (GO) database. The P-value in DAVID is a modified Fisher Exact P-Value. ‘Benjamini’ refers to the Benjamini-Hochberg procedure to control for the false discovery rate in multiple comparisons (61).
| Annotation clusters for upregulated genes | ||||
|---|---|---|---|---|
| Annotation Cluster 1 | Enrichment Score: 5.1 | Count | ||
| Database | Term of the annotation cluster | # of genes involved in term | P_Value | Benjamini |
| GO-Biological Process | organ development | 31 | 6.60E-08 | 3.30E-04 |
| GO-Biological Process | developmental process | 47 | 4.70E-07 | 1.20E-03 |
| GO-Biological Process | system development | 35 | 5.00E-07 | 8.40E-04 |
| GO-Biological Process | anatomical structure development | 37 | 2.10E-06 | 2.10E-03 |
| GO-Biological Process | cellular developmental process | 31 | 8.70E-06 | 6.20E-03 |
| GO-Biological Process | cell differentiation | 31 | 8.70E-06 | 6.20E-03 |
| GO-Biological Process | multicellular organismal development | 36 | 1.10E-05 | 7.20E-03 |
| GO-Biological Process | nervous system development | 18 | 1.80E-04 | 7.30E-02 |
| GO-Biological Process | multicellular organismal process | 47 | 1.50E-02 | 7.90E-01 |
| Annotation Cluster 2 | Enrichment Score: 3.5 | Count | P_Value | Benjamini |
| Database | Term of the annotation cluster | # of genes involved in term | ||
| GO-Biological Process | response to stress | 26 | 8.40E-07 | 1.10E-03 |
| GO-Biological Process | response to external stimulus | 19 | 1.70E-05 | 9.40E-03 |
| GO-Biological Process | response to wounding | 14 | 1.60E-04 | 7.10E-02 |
| GO-Biological Process | inflammatory response | 8 | 1.40E-02 | 7.70E-01 |
| GO-Biological Process | response to stimulus | 35 | 1.10E-01 | 9.90E-01 |
| Annotation clusters for downregulated genes | ||||
| Annotation Cluster 1 | Enrichment Score: 5.6 | Count | ||
| Database | Term of the annotation cluster | # of genes involved in term | P_Value | Benjamini |
| GO-Biological Process | ensheathment of neurons | 11 | 1.50E-13 | 3.80E-10 |
| GO-Biological Process | axon ensheathment | 11 | 1.50E-13 | 3.80E-10 |
| GO-Biological Process | regulation of action potential | 11 | 8.20E-13 | 1.40E-09 |
| GO-Biological Process | myelination | 10 | 3.90E-12 | 4.90E-09 |
| GO-Biological Process | nervous system development | 24 | 6.40E-11 | 6.40E-08 |
| GO-Biological Process | system development | 29 | 2.80E-07 | 2.40E-04 |
| GO-Biological Process | transmission of nerve impulse | 15 | 3.70E-07 | 2.70E-04 |
| GO-Biological Process | anatomical structure development | 31 | 5.50E-07 | 3.50E-04 |
| GO-Biological Process | regulation of biological quality | 19 | 2.40E-06 | 1.40E-03 |
| GO-Biological Process | multicellular organismal development | 29 | 1.10E-05 | 5.60E-03 |
| GO-Biological Process | cell-cell signaling | 16 | 1.20E-05 | 5.40E-03 |
| GO-Biological Process | developmental process | 34 | 2.60E-05 | 1.10E-02 |
| GO-Biological Process | neurogenesis | 9 | 2.10E-03 | 5.30E-01 |
| GO-Biological Process | cellular developmental process | 19 | 5.40E-03 | 8.00E-01 |
| GO-Biological Process | cell differentiation | 19 | 5.40E-03 | 8.00E-01 |
| GO-Biological Process | cell development | 13 | 4.90E-02 | 1.00E+00 |
| GO-Biological Process | multicellular organismal process | 33 | 6.70E-02 | 1.00E+00 |
| GO-Biological Process | system process | 20 | 7.40E-02 | 1.00E+00 |
| GO-Biological Process | neurological system process | 17 | 1.40E-01 | 1.00E+00 |
| GO-Biological Process | cell communication | 32 | 1.60E-01 | 1.00E+00 |
Figure 3.
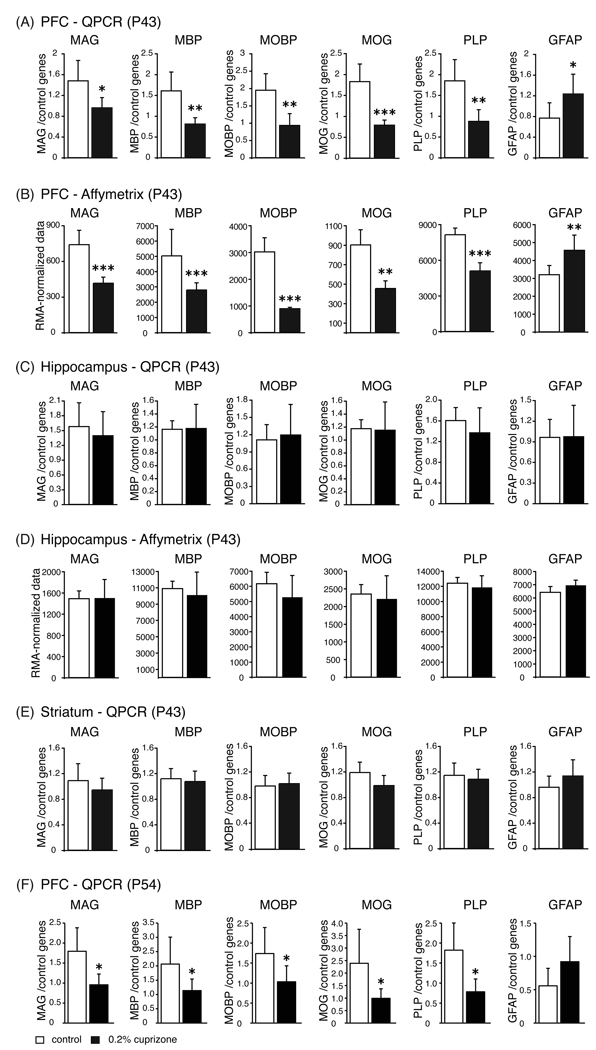
Real time quantitative PCR results after 15 days of exposure to 0.2% CPZ in PFC (A), hippocampus (C) and striatum (E), and gene array results in PFC (B) and hippocampus (D) of control rats (open bars), and rats fed with CPZ chow (black bars). (F) Real time quantitative PCR results after 26 days of exposure to 0.2% CPZ in the PFC. For Q-PCR, all transcript levels were normalized to an average of 3 loading controls, β-actin (Actb), alpha-tubulin (Tuba1a) and general transcription factor IIB (Gtf2b). Mean normalized expression values of six independent samples per group + SD are plotted. *p<=0.05, **p<=0.01, ***p<=0.001. MAG, myelin-associated glycoprotein; MBP, myelin basic protein; MOBP, myelin-associated oligodendrocytic basic protein; MOG, myelin oligodendrocyte glycoprotein; PLP, proteolipid protein; GFAP, glial fibrillary acidic protein.
Table 3.
Myelin markers found to be significantly regulated in CPZ treated rats. Fold-change was calculated with RMA. P-value was calculated with log2 transformed data. Affymetrix probe set IDs and Entrez gene ID numbers are provided. Transcripts shown in Figure 3 are denoted with ‘x’.
| Myelin markers (abbreviations) | fold change | P value (log2) | P call % of control | P call % of cuprizone | Entrez Gene ID | Affymetrix probe set ID | Verified with Q-PCR |
|---|---|---|---|---|---|---|---|
| myelin-associated glycoprotein (Mag) | −1.8 | 0.0002 | 100 | 100 | 29409 | 1368861_a_at | x |
| myelin-associated oligodendrocytic basic protein (Mobp) | −2.1 | 0.0000 | 100 | 100 | 25037 | 1368263_a_at | x |
| myelin-associated oligodendrocytic basic protein (Mobp) | −3.5 | 0.0025 | 100 | 100 | 25037 | 1370500_a_at | |
| myelin-associated oligodendrocytic basic protein (Mobp) | −3.4 | 0.0000 | 100 | 100 | 25037 | 1370434_a_at | |
| myelin basic protein (Mbp) | −1.8 | 0.0254 | 100 | 100 | 24547 | 1387341_a_at | x |
| myelin basic protein (Mbp) | −1.4 | 0.0002 | 100 | 100 | 24547 | 1368810_a_at | |
| myelin oligodendrocyte glycoprotein (Mog) | −2.0 | 0.0002 | 100 | 100 | 24558 | 1398257_at | x |
| myelin oligodendrocyte glycoprotein (Mog) | −1.3 | 0.0114 | 100 | 60 | 24558 | 1377352_at | |
| myelin and lymphocyte protein (Mal) | −3.8 | 0.0000 | 100 | 100 | 25263 | 1387040_at | |
| oligodendrocyte transcription factor 1 (Olig1) | −1.3 | 0.0005 | 100 | 100 | 60394 | 1387200_at | |
| plasma membrane proteolipid (Pllp) | −1.3 | 0.0138 | 100 | 100 | 64364 | 1386943_at | |
| proteolipid protein (Plp) | −1.6 | 0.0006 | 100 | 100 | 24943 | 1387112_at | x |
| claudin 11 (Cldn11) | −1.9 | 0.0305 | 100 | 100 | 84588 | 1369609_at | |
| claudin 11 (Cldn11) | −1.5 | 0.0029 | 100 | 100 | 84588 | 1376711_at | |
| CD9 antigen (Cd9) | −1.3 | 0.0119 | 100 | 100 | 24936 | 1371499_at | |
| v-erb-b2 erythroblastic leukemia viral oncogene homolog (Erbb3) | −1.6 | 0.0037 | 100 | 100 | 13867 | 1377821_at | |
| gelsolin (Gsn) | −1.4 | 0.0007 | 100 | 100 | 296654 | 1371414_at | |
| kallikrein 6 (Klk6) | −2.8 | 0.0000 | 100 | 40 | 29245 | 1368384_at | |
| response gene to complement 32 (Rgc32) | −1.3 | 0.0001 | 100 | 100 | 117183 | 1368080_at | |
| developmentally regulated protein TPO1 | −1.2 | 0.0479 | 100 | 100 | 170907 | 1386979_at | |
| transferrin | −1.6 | 0.0027 | 100 | 100 | 24825 | 1370228_at | |
| tetraspanin 2 (Tspan2) | −1.5 | 0.0002 | 100 | 100 | 64521 | 1368104_at | |
| tetraspanin 2 (Tspan2) | −1.4 | 0.0008 | 100 | 100 | 64521 | 1368105_at | |
| UDP galactosyltransferase 8 (Ugt8) | −2.3 | 0.0001 | 100 | 100 | 50555 | 1368858_at | |
| aspartoacylase (Aspa) | −1.4 | 0.0000 | 100 | 100 | 79251 | 1368563_at | |
| Astrocyte Marker | |||||||
| glial fibrillary acidic protein (Gfap) | 1.4 | 0.0128 | 100 | 100 | 24387 | 1368353_at | x |
Figure 2.
Hierarchical clustering of PFC samples with probesets of myelin transcripts. Twenty-six probesets of myelin transcripts (Table 3) were used for hierarchical clustering of all samples used for gene array analysis. Samples clustered according to treatment group, demonstrating the specific effect of CPZ on the expression of oligodendrocyte markers.
Upregulated genes in the PFC fell into two annotation clusters, related to the somewhat generic Gene Ontology terms of ‘stress’ and ‘development’. ‘Development’ encompasses terms like ‘organ development’, ‘developmental process’ and ‘system development’, and is among the most generic parent terms in the Gene Ontology database. ‘Developmental process’ contains over 20,000 gene products, ‘organ development’ almost 7,700. The genes in these categories did not seem to have a more specific function common to them. The genes in the second annotation cluster were related to stress response and included genes involved in inflammation (note that this group does not survive Benjamini-Hochberg correction) as well as the astrocyte marker glial fibrillary acidic protein (GFAP). In the MAPPfinder program, none of the brain-specific gene groups survived p-value permutation. Supplementary table S2 lists all regulated genes.
For the hippocampus, 194 probe sets matched the criteria, of which 115 were upregulated and 79 were downregulated. Neither set of genes was affiliated with any specific annotation in the DAVID database after Benjamini-Hochberg correction (Benjamini and Hochberg, 1995). Of the oligodendrocyte specific probe sets, only one, response gene to complement 32 (Rgc32), was downregulated (-23%; p=0.012); (Figure 3D). This finding was not significant for the myelin group (Fisher’s exact test). GFAP was unchanged.
Nuclear transcripts coding for mitochondrial proteins were not changed in either brain area.
Q-PCR
Six genes were chosen for verification with Q-PCR (Figure 3). The five chosen oligodendrocyte markers were significantly downregulated in the medial PFC after 15 days of 0.2% CPZ treatment (MAG, t10=-2.9, p<= 0.016; MBP, t10=-4.1, p <= 0.002; MOBP, t10=-4.2, p <= 0.002, MOG, t10 = −5.8, p <= 0.0002; PLP, t10 = −4.10, p <= 0.002) whereas the astroglial marker GFAP was significantly upregulated (t10 = 2.4, p <= 0.040), (Figure 3A; see Table 3 for abbreviations and Entrez Gene IDs). Comparable results were obtained with the gene arrays (Figure 3B; MAG, t9= −5.6, p <= 0.0003; MBP, t9= −6.4, p <= 0.0001; MOBP, t9= −8.9, p <= 0.0001; MOG, t9 = −5.8, p <= 0.003; PLP, t9 = −8.1, p <= 0.0001, GFAP, t9 = 3.3, p <= 0.009). A gene array analysis of the hippocampus (Figure 3D), or Q-PCR analyses of hippocampal or striatal mRNA, showed no changes (Figure 3C, Figure 3E). In the PFC, similar fold- reductions were observed after 26 days (P54) of exposure to 0.2% CPZ (Figure 3F; MAG, t9= −3.2, p <= 0.01; MBP, t9 = −2.2, p <= 0.05; MOBP, t9 = −2.2, p <= 0.05; MOG, t9 = −2.4, p <= 0.04; PLP, t9 = −3.1, p <= 0.01, GFAP, p = ns). No changes were seen in these transcripts in a Q-PCR analysis of the hippocampus or striatum at P54 (data not shown).
Effects of adolescent CPZ exposure on locomotor behavior
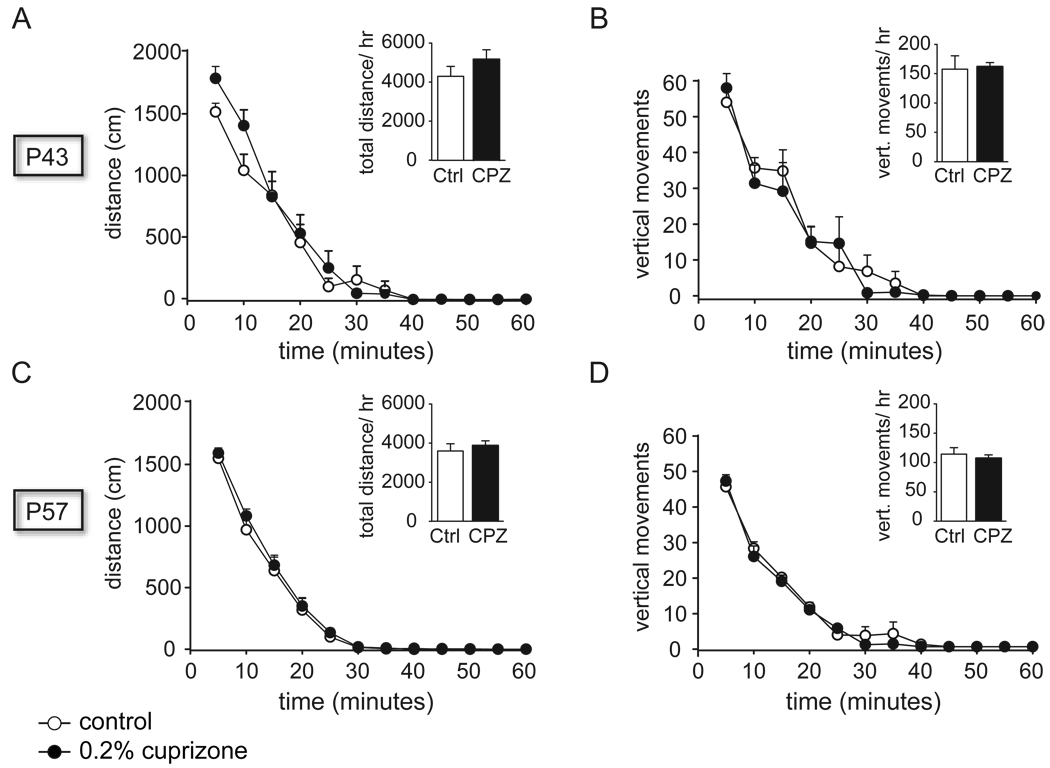
Animals on the 0.2% CPZ diet and control animals were examined for locomotor activity in open field chambers on P43 (day 15 of CPZ exposure) and P57 (day 29 of CPZ exposure). No effect of treatment was observed between the groups on distance traveled or vertical movements measured in 5 minute intervals (ANOVA: distance traveled; vertical rearing; ANCOVA: treatment*time; Figure 4). An effect of ‘time’ was observed over the one hour period within each treatment group (P43: distance traveled F(11,120)=79.9, p<=0.0001; vertical rearing F(11,120)=54.6, p<=0.0001; P57: distance traveled F(11,120)=222.3, p<=0.0001; vertical rearing F(11,120)=187.6, p<=0.0001).
Figure 4.
Locomotor activity in the open field. Open field distance traveled was measured in 5 minute bins on P43, after 2 weeks of CPZ exposure, (A), and on P57, after 4 weeks of CPZ exposure and completion of the ASST, (C). Vertical movements were charted in 5 minute bins on P43, (B), and on P57, (D). All inserts show data accumulated over one hour. All data in average + SEM of control (open bars; n=6) and CPZ-fed rats (black bars; n=5).
Effects of adolescent CPZ exposure on PFC-mediated behavior
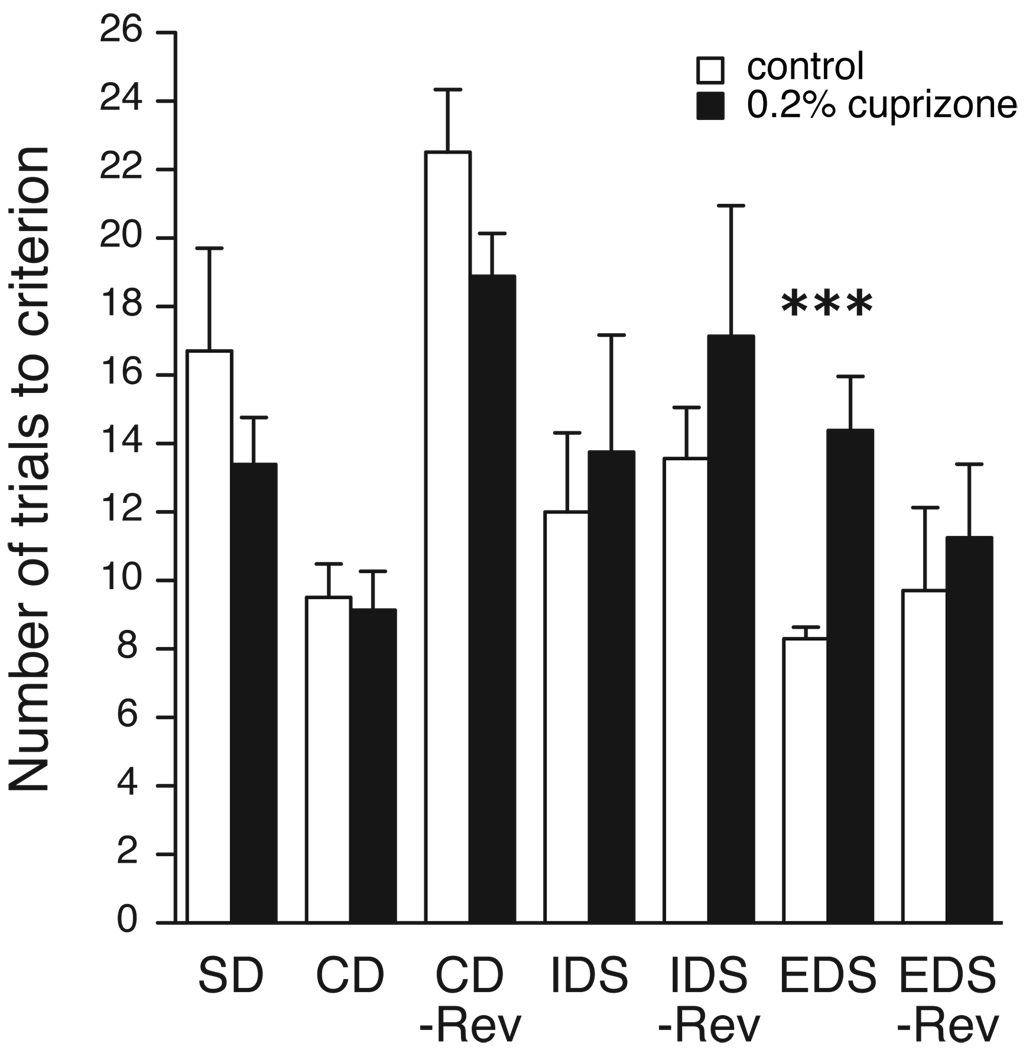
Animals underwent behavioral testing using the ASST between P44 (16 days on CPZ) and P56 (28 days on CPZ). Only one phase of the task, the EDS, was significantly different (t16 = −4.2, p = 0.0007; z=3.0, p=0.003), whereas none of the other phases reached significance (Figure 5). Since bilateral lesions of the medial frontal cortex result in impairment in shifting of attentional sets the data indicate a specific impairment of the medial PFC (Birrell and Brown, 2000).
Figure 5.
Trials to reach criterion performance for all phases of the ASST. Values represent the mean + SEM of control (open bars, n=10) and CPZ-intake rats (black bars, n=8). ***p<=0.001. See Table 1 for abbreviations.
Discussion
The PFC in humans is important for a large range of functions, including working memory, action planning, response inhibition, decision-making, reward processing, and social behavior (Miller and Cohen, 2001; Ridderinkhof et al., 2004). Many of these functions are impaired in people suffering from Sz (Barch, 2005). Because a number of Sz studies have demonstrated decreased levels of oligodendrocyte mRNA transcripts and abnormalities of white matter in the PFC, we were interested if a reduction of myelin transcripts in the rat can cause behavioral deficits of the PFC similar to the ones observed in Sz. While such a study cannot address if the lower expression of oligodendrocyte markers in Sz is a neurodevelopmental or neurodegenerative event, it can establish a correlation between disturbances in myelin and specific Sz-related behavioral symptoms. Adolescence was chosen for treatment-onset with the Cu chelator CPZ, since Sz is frequently first diagnosed during this time (Mueser and McGurk, 2004).
CPZ affected oligodendrocyte transcripts in the PFC but not the striatum or the hippocampus with similar abnormalities at both the beginning, and toward the end of the ASST. The lack of effect of CPZ in the striatum and the hippocampus is surprising and can be explained by the low levels of CPZ used in the current study and the general resilience of rats to CPZ toxicity (Love, 1988; Matsushima and Morell, 2001; Purves et al., 1991). The data demonstrate that the PFC has a higher sensitivity to CPZ than the other two brain areas, which could be caused by a higher accumulation of CPZ in the PFC or a narrower tolerance for abnormal Cu levels.
The decrease in oligodendrocyte transcripts was accompanied by an increased difficulty to shift attention from one perceptual dimension to another in the EDS phase of the ASST. This is comparable to the deficit Sz patients show in the WCST (Berman et al., 1986; Franke et al., 1992; Haut et al., 1996). Attentional set-shifting has been linked to PFC function and is impaired by damage to the PFC in monkeys, rats and humans (Birrell and Brown, 2000; Dias et al., 1996a, b, 1997). The specificity for PFC involvement was demonstrated by the fact that the EDS was the only phase of the ASST that was affected by CPZ, since bilateral lesions of the medial frontal cortex should result in impairment in shifting of attentional sets, but not in impairments of initial acquisition or reversal learning (Birrell and Brown, 2000).
The observed upregulation of the astrocyte marker GFAP in the PFC is an indicator of astrocytosis. Astrocytosis was described in murine studies of the superior cerebellar peduncle after prolonged, high-dose CPZ administration (Ludwin, 1978). During demyelination, insulinlike growth factor-I (IGF-I) mRNA is induced in astrocytes while the IGF-I receptor is transiently expressed in oligodendrocytes, leading to the suggestion that astrocytosis might be beneficial for oligodendrocyte regeneration (Komoly et al., 1992). Indeed, astrocytes and the IGF-I system play an important role in the recovery from demyelination (Mason et al., 2003; Mason et al., 2000).
The upregulated group of genes in the PFC fell into generic Gene Ontology database categories related to ‘development’ and ‘stress’. The genes in these categories did not seem to have a specific function or location common to them, and more specific child terms of the generic parent terms such as ‘inflammatory response’ did not survive Benjamini-Hochberg correction. Thus, while it is quite plausible that stress-related genes are upregulated, these groups of genes would need further exploration.
Transcripts for mitochondrial genes were normal in the gene array analysis of PFC and the hippocampus, an important control since mitochondrial enlargement was described under high CPZ administration (Love, 1988), and CPZ is used to study megamitochondria in the mouse liver (Flatmark et al., 1980; Petronilli and Zoratti, 1990).
Given the history of CPZ as a model for MS in mice, we were particularly concerned about motor abnormalities. This concern was further fueled by a report that weanling Wistar rats exposed to CPZ at doses of 1% and 2% exhibited generalized weakness and wasting (Love, 1988). However, we did not observe motor impairments, either by direct observation, or by locomotor activity assessment. Likely explanations include the lower concentrations of CPZ used in our study and the different rat strain. Another concern in the present study involved sensory and sensory-motor deficits, since these modalities are important covariates in the ASST. Because CPZ rats performed comparable to control rats in six out of the seven phases of the ASST, and only showed a deficit in the medial PFC-mediated EDS, an impairment in tactile or odorant systems by CPZ is unlikely.
Despite the correlation between myelin abnormalities and behavior in the PFC, we cannot exclude that other brain areas are playing a contributory role in the behavioral effect. In mice, CPZ treatment affects the corpus callosum, the hippocampus, the cortex and the cerebellum (Blakemore, 1972, 1973; Hiremath et al., 1998; Remington et al., 2007; Skripuletz et al., 2008; Stidworthy et al., 2003), all of which can participate in memory processing (Vertes, 2006). While we did not see any effects on oligodendrocyte gene expression in the hippocampus and striatum, further clarification of the potential contribution of other brain areas to PFC function in the ASST might be useful in the future. Given that reduced levels of oligodendrocyte transcripts are observed in a number of different brain areas in Sz, this would not diminish the usefulness of the CPZ model (Haroutunian et al., 2007; Sokolov, 2007; Stewart and Davis, 2004).
Current treatments for Sz fall short on addressing impairments in cognitive function. One major challenge is to develop translational preclinical models that are subtle in their pathology and deal with cognitive deficits. Such models are needed to screen for drugs with potential benefits in the treatment of Sz. Our results provide such a translational model of PFC white matter deficits combined with cognitive-behavioral abnormalities, which could be useful for novel strategies of drug discovery.
Supplementary Material
Primer Sequences for Q-PCR products.
All probe sets of the PFC with at least 40% ‘Present’ call in either group (control, CPZ), at least 1.2 fold difference in expression levels, and a p-value below 0.05 are shown. Downregulated transcripts are shown in alphabetical order, followed by upregulated transcripts in alphabetical order. Myelin markers are denoted with ‘x’.
Acknowledgement
The authors thank Karoly Mirnics, MD, for helpful discussions. The project described was supported by Award Number MH74000 from the NIMH (CK) and K12GM068543 from NIGMS (NH; PI Dr. Roger Chalkley). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutes or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure/Conflict of Interest: The authors have no conflict of interest
References
- Adamo AM, Paez PM, Escobar Cabrera OE, Wolfson M, Franco PG, Pasquini JM, Soto EF. Remyelination after cuprizone-induced demyelination in the rat is stimulated by apotransferrin. Exp Neurol. 2006;198:519–529. doi: 10.1016/j.expneurol.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Bagary MS, Symms MR, Barker GJ, Mutsatsa SH, Joyce EM, Ron MA. Gray and white matter brain abnormalities in first-episode schizophrenia inferred from magnetization transfer imaging. Arch Gen Psychiatry. 2003;60:779–788. doi: 10.1001/archpsyc.60.8.779. [DOI] [PubMed] [Google Scholar]
- Barch DM. The cognitive neuroscience of schizophrenia. Annu Rev Clin Psychol. 2005;1:321–353. doi: 10.1146/annurev.clinpsy.1.102803.143959. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Berman KF, Zec RF, Weinberger DR. Physiologic dysfunction of dorsolateral prefrontal cortex in schizophrenia II. Role of neuroleptic treatment, attention, and mental effort. Arch Gen Psychiatry. 1986;43:126–135. doi: 10.1001/archpsyc.1986.01800020032005. [DOI] [PubMed] [Google Scholar]
- Birrell JM, Brown VJ. Medial frontal cortex mediates perceptual attentional set shifting in the rat. J Neurosci. 2000;20:4320–4324. doi: 10.1523/JNEUROSCI.20-11-04320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black YD, Maclaren FR, Naydenov AV, Carlezon WA, Jr, Baxter MG, Konradi C. Altered attention and prefrontal cortex gene expression in rats after binge-like exposure to cocaine during adolescence. J Neurosci. 2006;26:9656–9665. doi: 10.1523/JNEUROSCI.2391-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore WF. Observations on oligodendrocyte degeneration, the resolution of status spongiosus and remyelination in cuprizone intoxication in mice. J Neurocytol. 1972;1:413–426. doi: 10.1007/BF01102943. [DOI] [PubMed] [Google Scholar]
- Blakemore WF. Remyelination of the superior cerebellar peduncle in the mouse following demyelination induced by feeding cuprizone. J Neurol Sci. 1973;20:73–83. doi: 10.1016/0022-510x(73)90119-6. [DOI] [PubMed] [Google Scholar]
- Boeker H, Kleiser M, Lehman D, Jaenke L, Bogerts B, Northoff G. Executive dysfunction, self, and ego pathology in schizophrenia: an exploratory study of neuropsychology and personality. Compr Psychiatry. 2006;47:7–19. doi: 10.1016/j.comppsych.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Breier A, Buchanan RW, Elkashef A, Munson RC, Kirkpatrick B, Gellad F. Brain morphology and schizophrenia A magnetic resonance imaging study of limbic, prefrontal cortex, and caudate structures. Arch Gen Psychiatry. 1992;49:921–926. doi: 10.1001/archpsyc.1992.01820120009003. [DOI] [PubMed] [Google Scholar]
- Buchanan RW, Vladar K, Barta PE, Pearlson GD. Structural evaluation of the prefrontal cortex in schizophrenia. Am J Psychiatry. 1998;155:1049–1055. doi: 10.1176/ajp.155.8.1049. [DOI] [PubMed] [Google Scholar]
- Carpenter WT, Koenig JI. The Evolution of Drug Development in Schizophrenia: Past Issues and Future Opportunities. Neuropsychopharmacol. 2008;33:2061–2079. doi: 10.1038/sj.npp.1301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelinrelated dysfunction. Arch Gen Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- de Bie P, Muller P, Wijmenga C, Klomp LW. Molecular pathogenesis of Wilson and Menkes disease: correlation of mutations with molecular defects and disease phenotypes. J Med Genet. 2007;44:673–688. doi: 10.1136/jmg.2007.052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dias R, Robbins TW, Roberts AC. Dissociation in prefrontal cortex of affective and attentional shifts. Nature. 1996a;380:69–72. doi: 10.1038/380069a0. [DOI] [PubMed] [Google Scholar]
- Dias R, Robbins TW, Roberts AC. Primate analogue of the Wisconsin Card Sorting Test: effects of excitotoxic lesions of the prefrontal cortex in the marmoset. Behav Neurosci. 1996b;110:872–886. doi: 10.1037//0735-7044.110.5.872. [DOI] [PubMed] [Google Scholar]
- Dias R, Robbins TW, Roberts AC. Dissociable forms of inhibitory control within prefrontal cortex with an analog of the Wisconsin Card Sort Test: restriction to novel situations and independence from "on-line" processing. J Neurosci. 1997;17:9285–9297. doi: 10.1523/JNEUROSCI.17-23-09285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwork AJ, Mancevski B, Rosoklija G. White matter and cognitive function in schizophrenia. Int J Neuropsychopharmacol. 2007;10:513–536. doi: 10.1017/S1461145707007638. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein A. Neuropsychiatric syndromes associated with multiple sclerosis. J Neurol. 2007;254(Suppl 2):II73–II76. doi: 10.1007/s00415-007-2017-2. [DOI] [PubMed] [Google Scholar]
- Fields RD. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatmark T, Kryvi H, Tangeras A. Induction of megamitochondria by cuprizone(biscyclohexanone oxaldihydrazone) Evidence for an inhibition of the mitochondrial division process. Eur J Cell Biol. 1980;23:141–148. [PubMed] [Google Scholar]
- Franke P, Maier W, Hain C, Klingler T. Wisconsin Card Sorting Test: an indicator of vulnerability to schizophrenia? Schizophr Res. 1992;6:243–249. doi: 10.1016/0920-9964(92)90007-r. [DOI] [PubMed] [Google Scholar]
- Green MF, Kern RS, Braff DL, Mintz J. Neurocognitive deficits and functional outcome in schizophrenia: are we measuring the "right stuff"? Schizophr Bull. 2000;26:119–136. doi: 10.1093/oxfordjournals.schbul.a033430. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroutunian V, Katsel P, Dracheva S, Stewart DG, Davis KL. Variations in oligodendrocyte-related gene expression across multiple cortical regions: implications for the pathophysiology of schizophrenia. Int J Neuropsychopharmacol. 2007;10:565–573. doi: 10.1017/S1461145706007310. [DOI] [PubMed] [Google Scholar]
- Harrison MD, Jones CE, Solioz M, Dameron CT. Intracellular copper routing: the role of copper chaperones. Trends Biochem Sci. 2000;25:29–32. doi: 10.1016/s0968-0004(99)01492-9. [DOI] [PubMed] [Google Scholar]
- Haut MW, Cahill J, Cutlip WD, Stevenson JM, Makela EH, Bloomfield SM. On the nature of Wisconsin Card Sorting Test performance in schizophrenia. Psychiatry Res. 1996;65:15–22. doi: 10.1016/0165-1781(96)02940-x. [DOI] [PubMed] [Google Scholar]
- Hiremath MM, Saito Y, Knapp GW, Ting JP, Suzuki K, Matsushima GK. Microglial/macrophage accumulation during cuprizone-induced demyelination in C57BL/6 mice. J Neuroimmunol. 1998;92:38–49. doi: 10.1016/s0165-5728(98)00168-4. [DOI] [PubMed] [Google Scholar]
- Hosack D, Dennis G, Sherman B, Lane H, Lempicki R. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:P4. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde TM, Ziegler JC, Weinberger DR. Psychiatric disturbances in metachromatic leukodystrophy Insights into the neurobiology of psychosis. Arch Neurol. 1992;49:401–406. doi: 10.1001/archneur.1992.00530280095028. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Karoutzou G, Emrich HM, Dietrich DE. The myelin-pathogenesis puzzle in schizophrenia: a literature review. Mol Psychiatry. 2008;13:245–260. doi: 10.1038/sj.mp.4002096. [DOI] [PubMed] [Google Scholar]
- Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- Komoly S, Hudson LD, Webster HD, Bondy CA. Insulin-like growth factor I gene expression is induced in astrocytes during experimental demyelination. Proc Natl Acad Sci U S A. 1992;89:1894–1898. doi: 10.1073/pnas.89.5.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C. Gene expression microarray studies in polygenic psychiatric disorders: applications and data analysis. Brain Res Brain Res Rev. 2005;50:142–155. doi: 10.1016/j.brainresrev.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S. Cuprizone neurotoxicity in the rat: morphologic observations. J Neurol Sci. 1988;84:223–237. doi: 10.1016/0022-510x(88)90127-x. [DOI] [PubMed] [Google Scholar]
- Ludwin SK. Central nervous system demyelination and remyelination in the mouse: an ultrastructural study of cuprizone toxicity. Lab Invest. 1978;39:597–612. [PubMed] [Google Scholar]
- Mason JL, Xuan S, Dragatsis I, Efstratiadis A, Goldman JE. Insulin-like growth factor (IGF) signaling through type 1 IGF receptor plays an important role in remyelination. J Neurosci. 2003;23:7710–7718. doi: 10.1523/JNEUROSCI.23-20-07710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Ye P, Suzuki K, D’Ercole AJ, Matsushima GK. Insulin-like growth factor-1 inhibits mature oligodendrocyte apoptosis during primary demyelination. J Neurosci. 2000;20:5703–5708. doi: 10.1523/JNEUROSCI.20-15-05703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima GK, Morell P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001;11:107–116. doi: 10.1111/j.1750-3639.2001.tb00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlonan K, Brown VJ. Orbital prefrontal cortex mediates reversal learning and not attentional set shifting in the rat. Behav Brain Res. 2003;146:97–103. doi: 10.1016/j.bbr.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Menkes JH, Alter M, Steigleder GK, Weakley DR, Sung JH. A sex-linked recessive disorder with retardation of growth, peculiar hair, and focal cerebral and cerebellar degeneration. Pediatrics. 1962;29:764–779. [PubMed] [Google Scholar]
- Messori L, Casini A, Gabbiani C, Sorace L, Muniz-Miranda M, Zatta P. Unravelling the chemical nature of copper cuprizone. Dalton Trans. 2007:2112–2114. doi: 10.1039/b701896g. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, McKenna PJ, Jones PB, Arai H, Starkey M, Emson PC, Bahn S. Gene expression analysis in schizophrenia: reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc Natl Acad Sci USA. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell P, Barrett CV, Mason JL, Toews AD, Hostettler JD, Knapp GW, Matsushima GK. Gene expression in brain during cuprizone-induced demyelination and remyelination. Mol Cell Neurosci. 1998;12:220–227. doi: 10.1006/mcne.1998.0715. [DOI] [PubMed] [Google Scholar]
- Mueser KT, McGurk SR. Schizophrenia. Lancet. 2004;363:2063–2072. doi: 10.1016/S0140-6736(04)16458-1. [DOI] [PubMed] [Google Scholar]
- Pasquini LA, Calatayud CA, Bertone Una AL, Millet V, Pasquini JM, Soto EF. The neurotoxic effect of cuprizone on oligodendrocytes depends on the presence of proinflammatory cytokines secreted by microglia. Neurochem Res. 2007;32:279–292. doi: 10.1007/s11064-006-9165-0. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic, San Diego. 1986 doi: 10.1016/0165-0270(80)90021-7. [DOI] [PubMed] [Google Scholar]
- Petronilli V, Zoratti M. A characterization of cuprizone-induced giant mouse liver mitochondria. J Bioenerg Biomembr. 1990;22:663–677. doi: 10.1007/BF00809070. [DOI] [PubMed] [Google Scholar]
- Prohaska JR, Smith TL. Effect of dietary or genetic copper deficiency on brain catecholamines, trace metals and enzymes in mice and rats. J Nutr. 1982;112:1706–1717. doi: 10.1093/jn/112.9.1706. [DOI] [PubMed] [Google Scholar]
- Puig S, Thiele DJ. Molecular mechanisms of copper uptake and distribution. Curr Opin Chem Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- Purves DC, Garrod IJ, Dayan AD. A comparison of spongiosis induced in the brain by hexachlorophene, cuprizone and triethyl tin in the Sprague-Dawley rat. Hum Exp Toxicol. 1991;10:439–444. doi: 10.1177/096032719101000613. [DOI] [PubMed] [Google Scholar]
- Remington LT, Babcock AA, Zehntner SP, Owens T. Microglial recruitment, activation, and proliferation in response to primary demyelination. Am J Pathol. 2007;170:1713–1724. doi: 10.2353/ajpath.2007.060783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderinkhof KR, van den Wildenberg WP, Segalowitz SJ, Carter CS. Neurocognitive mechanisms of cognitive control: the role of prefrontal cortex in action selection, response inhibition, performance monitoring, and reward-based learning. Brain Cogn. 2004;56:129–140. doi: 10.1016/j.bandc.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilipo M, Lafargue T, Rusinek H, Arena L, Loneragan C, Lautin A, Feiner D, Rotrosen J, Wolkin A. Volumetric measure of the frontal and temporal lobe regions in schizophrenia: relationship to negative symptoms. Arch Gen Psychiatry. 2000;57:471–480. doi: 10.1001/archpsyc.57.5.471. [DOI] [PubMed] [Google Scholar]
- Seiwa C, Yamamoto M, Tanaka K, Fukutake M, Ueki T, Takeda S, Sakai R, Ishige A, Watanabe K, Akita M, Yagi T, Tanaka K, Asou H. Restoration of FcRgamma/Fyn signaling repairs central nervous system demyelination. J Neurosci Res. 2007;85:954–966. doi: 10.1002/jnr.21196. [DOI] [PubMed] [Google Scholar]
- Sigmundsson T, Suckling J, Maier M, Williams S, Bullmore E, Greenwood K, Fukuda R, Ron M, Toone B. Structural abnormalities in frontal, temporal, and limbic regions and interconnecting white matter tracts in schizophrenic patients with prominent negative symptoms. Am J Psychiatry. 2001;158:234–243. doi: 10.1176/appi.ajp.158.2.234. [DOI] [PubMed] [Google Scholar]
- Skripuletz T, Lindner M, Kotsiari A, Garde N, Fokuhl J, Linsmeier F, Trebst C, Stangel M. Cortical demyelination is prominent in the murine cuprizone model and is strain-dependent. Am J Pathol. 2008;172:1053–1061. doi: 10.2353/ajpath.2008.070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? Int J Neuropsychopharmacol. 2007;10:547–555. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- Stewart DG, Davis KL. Possible contributions of myelin and oligodendrocyte dysfunction to schizophrenia. Int Rev Neurobiol. 2004;59:381–424. doi: 10.1016/S0074-7742(04)59015-3. [DOI] [PubMed] [Google Scholar]
- Stidworthy MF, Genoud S, Suter U, Mantei N, Franklin RJ. Quantifying the early stages of remyelination following cuprizone-induced demyelination. Brain Pathol. 2003;13:329–339. doi: 10.1111/j.1750-3639.2003.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uranova N, Orlovskaya D, Vikhreva O, Zimina I, Kolomeets N, Vostrikov V, Rachmanova V. Electron microscopy of oligodendroglia in severe mental illness. Brain Res Bull. 2001;55:597–610. doi: 10.1016/s0361-9230(01)00528-7. [DOI] [PubMed] [Google Scholar]
- Uranova NA, Vostrikov VM, Vikhreva OV, Zimina IS, Kolomeets NS, Orlovskaya DD. The role of oligodendrocyte pathology in schizophrenia. Int J Neuropsychopharmacol. 2007;10:537–545. doi: 10.1017/S1461145707007626. [DOI] [PubMed] [Google Scholar]
- Vertes RP. Interactions among the medial prefrontal cortex, hippocampus and midline thalamus in emotional and cognitive processing in the rat. Neuroscience. 2006;142:1–20. doi: 10.1016/j.neuroscience.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Vostrikov VM, Uranova NA, Orlovskaya DD. Deficit of perineuronal oligodendrocytes in the prefrontal cortex in schizophrenia and mood disorders. Schizophr Res. 2007;94:273–280. doi: 10.1016/j.schres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Xiao L, Xu H, Zhang Y, Wei Z, He J, Jiang W, Li X, Dyck LE, Devon RM, Deng Y, Li XM. Quetiapine facilitates oligodendrocyte development and prevents mice from myelin breakdown and behavioral changes. Mol Psychiatry. 2008;13:697–708. doi: 10.1038/sj.mp.4002064. [DOI] [PubMed] [Google Scholar]
- Zatta P, Raso M, Zambenedetti P, Wittkowski W, Messori L, Piccioli F, Mauri PL, Beltramini M. Copper and zinc dismetabolism in the mouse brain upon chronic cuprizone treatment. Cell Mol Life Sci. 2005;62:1502–1513. doi: 10.1007/s00018-005-5073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer Sequences for Q-PCR products.
All probe sets of the PFC with at least 40% ‘Present’ call in either group (control, CPZ), at least 1.2 fold difference in expression levels, and a p-value below 0.05 are shown. Downregulated transcripts are shown in alphabetical order, followed by upregulated transcripts in alphabetical order. Myelin markers are denoted with ‘x’.