Abstract
Rearrangements of the MLL gene at chromosome 11q23 are uncommon in de novo myelodysplastic syndrome (MDS). Here we describe molecular findings in a patient with multilineage dysplasia and t(11;17)(q23;q25) who responded to decitabine therapy. Fluorescent in situ hybridization (FISH) demonstrated rearrangement of MLL, while RT-PCR analysis and sequencing identified the transcript fusion partner as SEPT9, a member of the septin family of GTPases. MLL-SEPT9 fusion appears to be rare, having been described to date in only 7 cases of AML and not, to our knowledge, in MDS. Analysis of MLL-septin family member fusion products such as the one seen here may provide further insights into the etiology of myeloid neoplasia, and MLL-SEPT9 fusion may be worth seeking in other cases of MLL rearrangements with a translocation partner on chromosome 17q.
Keywords: MLL, Myelodysplastic syndrome, Septin family proteins, translocations
Introduction
Clonally restricted cytogenetic abnormalities are important prognostic determinants in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). In MDS, about 50% of patients with de novo disease and more than 80% of those with secondary (i.e., therapy-related) disease harbor a karyotypic abnormality.(1) While most of these MDS-associated chromosomal rearrangements are large deletions or numerical alterations, recurrent reciprocal translocations also occur. In a few cases (e.g., t(8;21), t(15;17), and inv(16)), the translocation is considered to be AML-defining, regardless of the marrow blast proportion.
Recurrent translocations involving band q23 of chromosome 11 are common in AML — especially among patients previously exposed to a topoisomerase II inhibiting agent— but have proven to be quite rare in de novo MDS.(2, 3) Most 11q23 translocations can be shown to involve the MLL gene locus, although other genes localized to 11q23 may also be rearranged in some cases. Clinical outcomes with 11q23 rearrangements are heterogeneous, and likely depend in large part on the specific fusion partners involved.(4)
At present, more than 87 different MLL reciprocal translocations have been described, and for 51 of these the translocation partner gene has been characterized at the molecular level.(3) MLL and its translocation partners are almost always fused in-frame, and in most cases the fusion partner is suspected to have specific functional significance. Occasional patients with AML have t(11;17)(q23;q11-q25) translocations, and several potential fusion partners for MLL have been identified on chromosome 17q.(5) These AML-associated fusion partners include MLLT6 (formerly known as AF17; 17q21), LASP1 (17q12), ACACA (17q21), and SEPT9 (formerly known as MSF; 17q25).(3, 5)
We recently evaluated a woman who presented with de novo MDS and a t(11;17)(q23;q25) translocation. Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis demonstrated a MLL-SEPT9 fusion, a rare translocation in general that has, to our knowledge, not previously been associated with de novo MDS.
Case Report and Methods
The patient consented to molecular analysis of her sample and reporting of the results, and the study was approved by the Institutional Review Board of the Mayo Clinic.
A previously healthy 61-year-old woman presented to a physician for bilateral arm paresthesias causally related to a worn mattress, and she was incidentally found to have neutropenia. Complete blood count (CBC) showed a leukocyte count of 1.6 × 109/L without circulating blasts or any other earlier forms, absolute neutropenia (0.67 × 109/L), a low-normal platelet count and hemoglobin level, and an MCV of 98 fL. She had no history of infections, and previous CBCs had been normal. Serum chemistry values were unremarkable, and vitamin B12 and folate levels were normal. A bone marrow aspiration and biopsy showed decreased myeloid precursors with moderately left-shifted granulopoiesis, impaired hemoglobinization, increased marrow iron stores without ringed sideroblasts, normocellularity, and a blast count of 2%. Cytogenetic analysis showed 46,XX,t(11;17)(q23;q25) in 2 of 6 metaphases. The patient then presented to our institution, and a repeat bone marrow examination (3 weeks after the initial study) showed hypercellular marrow with extensive trilineage dysplasia and a blast count of 8%. A diagnosis of de novo MDS, refractory anemia with excess blasts, subtype I (RAEB-I), was given. The marrow karyotype was again abnormal, with 46,XX,t(11;17)(q23;q25) observed in 13 of 20 metaphases. JAK2 and FLT3 gene mutation analysis and T-cell receptor gene rearrangement studies were all negative.
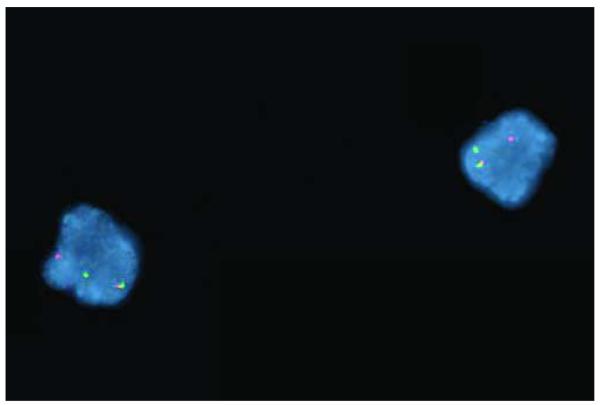
A clinically validated break-apart fluorescent in situ hybridization (FISH) probe set (Vysis, Inc., Downers Grove, Illinois) showed that 12.5% of 200 cells counted had abnormal separation of the MLL gene probes (Figure 1), consistent with the translocation observed by chromosome studies. Another FISH probe set demonstrated that RARA at 17q21 was intact.
Figure 1. Breakapart FISH analysis of interphase cells from a patient with de novo RAEB-I, demonstrating rearrangement of MLL.
Red probe corresponds to the 5′ end of MLL, whereas the green probe corresponds to the 3′ end of MLL. Intact MLL is associated with an overlapping red/green (i.e., yellow) fusion signal. In this case, each cell demonstrates separation of one set of the red and green probes, corresponding to a translocation involving MLL.
Unfractionated bone marrow total RNA was extracted using a resin-based kit (RNAEasy, Qiagen, Valencia, California). RNA quality was validated with an Agilent 2100 Bioanalyzer (Santa Clara, California), and cDNA generated using iScript cDNA synthesis kit (Biorad, Hercules, California). RT-PCR was performed using a series of oligonucleotide primer pairs designed to amplify candidate MLL fusion transcripts involving partner genes on chromosome 17q, as described.(5) RT-PCR amplification conditions are available on request.
Results and Discussion
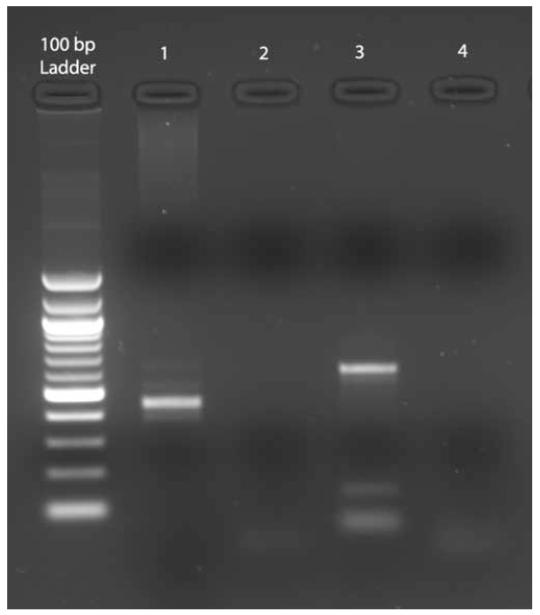
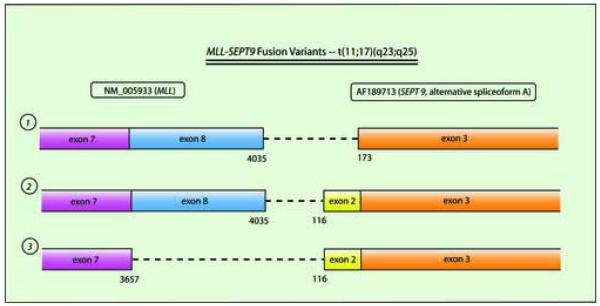
RT-PCR analysis demonstrated an abnormal fusion product corresponding to MLL and SEPT9 (Figure 2). Direct fluorescent dye chemistry-based sequencing showed fusion between exon 7 of MLL (including up to base pair 3657 of GenBank Accession NM_005933.2) and exon 2 of SEPT9 (beginning at base pair 116 of AF189713, the alternative splice variant corresponding to SEPT9 isoform ε) (Figure 3). Published nomenclature concerning exon numbering of MLL and SEPT9 is variable, so here we use that of Nilson et al(6) for MLL, and of McIlhatton et al(7) for SEPT9. The particular MLL and SEPT9 exon fusion in our case was identical to that detected in a solitary case of de novo AML M4 in an 8 year old boy (Table) (personal communication, Dr Sabine Strehl, Vienna, Austria, November 2006).(5) RT-PCR reactions for other potential MLL-chromosome 17q fusion partners were all negative. Our patient was treated with decitabine therapy (20 mg/m2/day × 5 days), and experienced normalization of the marrow blast count and cytogenetics after a single treatment cycle. She was then referred for unrelated-donor hematopoietic stem cell transplantation.
Figure 2. Agarose gel electrophoresis of RT-PCR products demonstrating MLL-SEPT9 fusion.
Lanes: 100 bp molecular weight ladder (New England Biolabs, Cambridge, Massachusetts); 1, MDS patient sample RT-PCR product, amplified using primer pair: 5′-CCTCTTGCTCCACCCATCAAAC-3′ (MLL exon 6, sense strand) and 5′-CGCCCAGGTCCTGGAATTT-3′ (SEPT9 exon 3, antisense); 2, wild-type control with same primer pairs; 3, patient PCR product using primer pair: MLL exon 6, sense, as above, and 5′-TTCTCCACCTGCTTGGACGAG-3′ (SEPT9 exon 3, antisense); 4, wild-type control with same primer pairs. Transcript variant 3 (Figure 3) is predicted to have a 608 bp product using the primer set in lane 3 and 448 bp product using the primer set in lane 1, which is what is observed. Product identity was verified by DNA sequencing.
Figure 3. Schematic of the 3 described MLL-SEPT9 fusions.
The particular fusion seen in this case is depicted at bottom. Exon 8 of MLL is 378 bp and exon 2 of SEPT9 is 57 bp; thus, all fusion transcripts remain in frame. Transcript variants are numbered (1, 2, and 3) and are referred to in the Table. AF189713 represents transcript encoding isoform ε of SEPT9.(7)
Table.
Characteristics of Patients With MLL-SEPT9 Fusion
| Case Number |
Age (y) and Sex |
Diagnosis |
MLL- SEPT9 Variant* |
Clinical Outcome |
Reference |
|---|---|---|---|---|---|
| 1 | 10 F | t-AML** | 1 | NR | (9) |
| 2 | 24 M | AML M4*** | 1 | Death without achieving remission |
(10) |
| 3 | 4 months F | De novo AML M5 | 1 | Alive >5 years with multiagent chemotherapy followed by cord blood transplant |
(10) |
| 4 | 64 M | De novo AML M4 | 1 | Septic death during relapse after brief chemotherapy- induced remission |
(11) |
| 5 | 8 M | De novo AML M4 | 3 | NR | (5) |
| 6 | 60 F | De novo AML M5 | 2 | NR | (5) |
| 7 | 50 F | De novo AML M2 | 1 | NR | (5) |
| 8 | 61 F | MDS (unclassifiable, then RAEB-I) |
3 | Clinical response to decitabine, referred for stem cell transplantation |
This report |
Abbreviations: M, male; F, female; y, years of age; AML, acute myeloid leukemia; t-AML, therapy-related AML; MDS, myelodysplastic syndrome; NR, not reported.
See Figure 1B.
Following therapy for Hodgkin disease.
Patient was previously treated for AML M2 with t(8;21) AML1-ETO fusion.
The function of SEPT9 in normal cell biology and tumorigenesis is unclear. SEPT9 is a member of the septin family of GTPases that have diverse cellular activity, including roles in cytokinesis, apoptosis, and vesicle trafficking.(5, 8) Inactivating germline mutations in SEPT9 have recently been associated with hereditary neuralgic amyotrophy, whereas abnormal expression or deletion of SEPT9 has been linked to ovarian and breast neoplasia. SEPT9 has many alternative spliceoforms, the functional significance of which is not clearly understood.(7) Several other septin family members (e.g., SEPT2 and SEPT6) have been involved in translocations with MLL, and it is possible that the mechanism of MLL-associated leukemogenesis for the various septin family members is similar.(3, 5)
In this case, an unusual MLL-SEPT9 fusion was associated with trilineage myelodysplasia and a rapidly increasing marrow blast proportion (from 2% to 8% in 3 weeks). It is possible that our case actually represented evolving AML that was incidentally discovered earlier than usual, due to the occurrence of an unrelated symptom that led to performance of a CBC, rather than being a more typical de novo MDS that would have exhibited stability for many months. However, the marrow dysplasia observed was extensive, and without knowledge of the cytogenetic findings, the case otherwise appeared to be a typical instance of RAEB-I. To our knowledge, there are no comparable reports of MLL-SEPT9 fusion in MDS, and a search of the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman, verified November 15, 2006) revealed only two reports of MDS cases with t(11;17)(q23;q25), neither of which were mapped at the molecular level. Knowledge of the specific fusion product may be useful in monitoring minimal residual disease in our patient post-transplantation by RT-PCR at a greater level of diagnostic sensitivity than is possible with FISH alone, and may be worth seeking in other cases of MLL rearrangement with a translocation partner on chromosome 17q.
Acknowledgments
This study was supported by National Cancer Institute K12 CA90628 to DPS and by a grant from the Robert A. Kyle Hematologic Malignancies Fund.
This study was funded by National Cancer Institute award K12 CA90628 to DPS and by a grant from the Robert A. Kyle Hematologic Malignancies Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no potential conflicts of interest to report.
REFERENCES
- 1.Sole F, Espinet B, Sanz GF, Cervera J, Calasanz MJ, Luno E, et al. Incidence, characterization and prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Grupo Cooperativo Espanol de Citogenetica Hematologica. Br J Haematol. 2000;108(2):346–56. doi: 10.1046/j.1365-2141.2000.01868.x. [DOI] [PubMed] [Google Scholar]
- 2.Pappa V, Young BD, Economopoulos T, Papageorgiou E, Panani A, Lilington D, et al. Absence of MLL gene rearrangement in de novo myelodysplastic syndromes (MDS) Ann Hematol. 2004;83(3):170–5. doi: 10.1007/s00277-003-0818-7. [DOI] [PubMed] [Google Scholar]
- 3.Meyer C, Schneider B, Jakob S, Strehl S, Attarbaschi A, Schnittger S, et al. The MLL recombinome of acute leukemias. Leukemia. 2006;20(5):777–84. doi: 10.1038/sj.leu.2404150. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Chessells JM, Camitta B, Baruchel A, Biondi A, Boyett JM, et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia. 2003;17(4):700–6. doi: 10.1038/sj.leu.2402883. [DOI] [PubMed] [Google Scholar]
- 5.Strehl S, Konig M, Meyer C, Schneider B, Harbott J, Jager U, et al. Molecular dissection of t(11;17) in acute myeloid leukemia reveals a variety of gene fusions with heterogeneous fusion transcripts and multiple splice variants. Genes Chromosomes Cancer. 2006;45(11):1041–9. doi: 10.1002/gcc.20372. [DOI] [PubMed] [Google Scholar]
- 6.Nilson I, Lochner K, Siegler G, Greil J, Beck JD, Fey GH, et al. Exon/intron structure of the human ALL-1 (MLL) gene involved in translocations to chromosomal region 11q23 and acute leukaemias. Br J Haematol. 1996;93(4):966–72. doi: 10.1046/j.1365-2141.1996.d01-1748.x. [DOI] [PubMed] [Google Scholar]
- 7.McIlhatton MA, Burrows JF, Donaghy PG, Chanduloy S, Johnston PG, Russell SE. Genomic organization, complex splicing pattern and expression of a human septin gene on chromosome 17q25.3. Oncogene. 2001;20(41):5930–9. doi: 10.1038/sj.onc.1204752. [DOI] [PubMed] [Google Scholar]
- 8.Macara IG, Baldarelli R, Field CM, Glotzer M, Hayashi Y, Hsu SC, et al. Mammalian septins nomenclature. Mol Biol Cell. 2002;13(12):4111–3. doi: 10.1091/mbc.E02-07-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osaka M, Rowley JD, Zeleznik-Le NJ. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25) Proc Natl Acad Sci U S A. 1999;96(11):6428–33. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taki T, Ohnishi H, Shinohara K, Sako M, Bessho F, Yanagisawa M, et al. AF17q25, a putative septin family gene, fuses the MLL gene in acute myeloid leukemia with t(11;17)(q23;q25) Cancer Res. 1999;59(17):4261–5. [PubMed] [Google Scholar]
- 11.Yamamoto K, Shibata F, Yamaguchi M, Miura O. Fusion of MLL and MSF in adult de novo acute myelomonocytic leukemia (M4) with t(11;17)(q23;q25) Int J Hematol. 2002;75(5):503–7. doi: 10.1007/BF02982114. [DOI] [PubMed] [Google Scholar]