Abstract
Dengue virus (DV) and West Nile virus (WNV) have become a global concern due to their widespread distribution and their ability to cause a variety of human diseases. Antiviral immune defenses involve natural killer (NK) cells. In the present study, we investigated the interaction between NK cells and these two flaviviruses. We show that the NK-activating receptor NKp44 is involved in virally-mediated NK activation through direct interaction with the flavivirus envelope protein. Recombinant NKp44 directly binds to purified DV and WNV envelope proteins and specifically to domain III of WNV envelope protein (EIII); it also binds to WNV virus-like particles (VLPs). These WNV-VLPs and WNV-EIII directly bind NK cells expressing high levels of NKp44. Functionally, interaction of NK cells with infective and inactivated WNV results in NKp44-mediated NK de-granulation. Finally, WNV infection of cells results in increased binding of recombinant NKp44 that is specifically inhibited by anti-WNV serum. WNV-infected target cells induce IFNγ secretion and augmented lysis by NKp44-expressing primary NK cells that are blocked by anti-NKp44 antibodies. Our findings show that triggering of NK cells by flavivirus is mediated by interaction of NKp44 with the flavivirus envelope protein.
Keywords: Human, Natural Killer Cells, Viral, Cell Surface Molecules, Cell Activation
Introduction
The Flavivirus genus contains a large group of arthropod-borne viruses responsible for many human diseases worldwide. Two important members of this genus are the dengue virus (DV) and West Nile virus (WNV). DV causes dengue fever (DF) and dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS), the most prevalent mosquito-borne viral diseases in humans with an estimated 2.5 billion people who live in areas at risk for epidemic transmission (1). WNV is the causative agent of West Nile encephalitis in humans. The virus was first detected in the Western Hemisphere in 1999 during an outbreak of encephalitis in New York City and has since spread across the continent (2). DV and WNV virions, like those of other flaviviruses, contain two viral surface proteins named envelope (E) and prM/M, a capsid protein, and the positive-stranded genomic RNA (3). The E protein mediates virus attachment by interacting with cellular receptor molecules. During DV and WNV life cycles, immature non-infectious particles are formed in the lumen of the endoplasmic reticulum. Subsequently, maturation of these particles occurs in the trans-Golgi network, in which they undergo a pH-induced irreversible conformational change involving the prM and E proteins followed by cleavage of prM by furin.
Innate, humoral and cellular responses are all involved in the protection against flavivirus infection (4). Natural killer (NK) cells are an important component of the innate immune response, as they play a crucial role in the host first line of defense against several viral infections and other pathogens (5). NK cell activity is a balance between signals delivered by their inhibitory and activating receptors. Engagement of inhibitory receptors by major histocompatibility complex (MHC) class I molecules expressed on normal cells down regulates NK cells’ effector function (6, 7). We have recently shown that expression of dengue non-structural proteins induces MHC class I up-regulation and as a result inhibits NK activity (8). NK major activating receptors are NKG2D and the natural cytotoxicity receptors (NCRs) NKp30, NKp46, and NKp44 (9, 10). NKp44 is expressed only on activated NK cells (11) and was reported to be involved in triggering NK cells’ cytolytic activity against both tumor and virus-infected cells (6, 12). We have shown that NKp44 is involved in the functional recognition of H1-, H3- and H5-subtype of the influenza virus (IV) and the hemagglutinin-neuraminidase (HN) of the Sendai virus (13–15). Binding of NKp44 to a putative ligand on the surface of mycobacteria was recently reported (16), yet the functional relevance of this binding was not fully addressed. The role of NK cells in flavivrus infection is not clear. While flavivirus-induced MHC class I enhancement/aggregation inhibits NK cell activity (8, 17), NK cells have been shown to be activated early in humans infected with dengue virus (18, 19). Moreover, Kurane et al. reported that human NK cells are cytotoxic against DV-infected cells via direct cytolysis and antibody-dependent cell-mediated cytotoxicity (20).
In this study, we explored the interaction between NK cells and two members of the flavivirus genus, DV and WNV. We show that WNV and DV E proteins bind directly to NKp44 and that WNV virus-like particles (VLPs) and WNV-infected cells induce NKp44-mediated NK cell activation.
Materials and Methods
Cells and viruses
Cell lines used in this work were as follows: The 293T line was derived from SV40 large T antigen-transfected HEK293 cells. The Vero cell line was derived from an African green monkey (ATCC no.CCL-81, www.atcc.org). CHOK1 – a Chinese hamster ovarian line (ATCC no. CCL-61). NK-92 (ATCC no. CRL-2407) is a human natural killer lymphoma. The NK-92 cell line, transduced by retrovirus to express high levels of wild-type NKp44 (designated as NK92-44), has been characterized in detail elsewhere (21). The Drosophila melanogaster Schneider 2 (S2) cell line was purchased from Invitrogen. S2 cells were incubated at 27°C.
The Kimron Veterinary Institute (KVI) strain of WNV was isolated from goose brain in Israel in 1998 and has over 99% homology to the WNV strain isolated in New York in 1999. The goose brain-isolated KVI WNV was kindly provided by Dr. Itzchak Samina from the state laboratory for vaccine control, Kimron Veterinary Institute, Beit Dagan, Israel. In all of the experiments performed in this study, the WNV-KVI was used after two passages in Vero cells. Cells infected by WNV and Vero or 293T cells were plated one day prior to infection. Next, cells were infected with KVI WNV (MOI ~ 1–5) in plain DMEM medium for one hour, after which 5 ml of DMEM supplemented with 5% fetal calf serum was added. Duration of infection is indicated in the experiment legends.
Antibodies and fusion-Ig proteins
The following antibodies were used in this study: The 4G2 monoclonal antibody that recognizes the flavivirus E-protein (ATCC no. HB-112), mouse anti-WNV serum (22), anti-WNV hyperimmune mouse ascites fluids (HMAF) (23), the pan-HLA-I-specific mAb W6/32, anti-NKp44 mAb (R&D SYSTEMS, MAB22491), anti-NKp30 mAb (R&D SYSTEMS, MAB18491), anti-His tag (His-probe, Santa Cruz Biotechnology, sc-8036) and biotin-conjugated mouse anti-human CD107a/LAMP-1 mAb (Southern Biotech, 9835-08). Anti-ILT2 HP-F1 mAb was kindly provided by Dr. Miguel Lopez-Botet (Servicio de Inmunologia, Hospital Universitario de la Princesa, Madrid, Spain) (24). Generation of mouse polyclonal anti-NKp44 or anti-NKp46D2 was previously described (14, 25). AO4BO8 is a phage-display Ab that recognizes heparan sulfate; MPB49V is a phage-display Ab that does not recognize heparan sulfate and was employed as negative control for AO4BO8. Production and characterization of these Abs was described in previous work (26). To generate NKp44-Ig, the sequence encoding the extracellular portion of NKp44 (accession number NM_004828, residues 1–169, www.ncbi.nlm.nih.gov/nuccore) was amplified by PCR from cDNA isolated from human NK clones. The corresponding PCR fragment, containing kozak sequence and leader sequence of CD5, was cloned into a pcDNA3.1-Ig vector encoding the CH2+CH3 regions of human IgG1. The sequences for truncated fusion proteins, NKp44D-Ig (residues 1–111) and NKp44LP-Ig (residues 109–169) were amplified by PCR from the NKp44-Ig-encoding plasmid and the corresponding PCR fragments, containing the kozak sequence and leader sequence of CD5, were back-cloned into pcDNA 3.1-Ig vector. Sequencing of the constructs revealed that all cDNAs were in frame with the human Fc DNA. CHO cells were transfected with these expression vectors and G418-selected clones were screened for highest protein production. Re-cloned high producer clones were grown in BIO-CHO-1 medium (Biological Industries, Kibbutz Beit Haemek, Israel) and supernatants were collected daily and purified on protein-G columns using FPLC. SDS-PAGE analysis revealed that all Ig fusion proteins were approximately 95% pure and of the proper molecular mass. The generation of NKp30-Ig and NKp46D2-Ig in CHO cells (27, 28) and the generation of LIR1-Ig, NKG2D-Ig and KIR2DS4-Ig produced in HEK 293T cells (25, 28) was previously described (NKG2D construct was a kind gift from Dr. Adelheid Cerwenka, German Cancer Center, Heidelberg, Germany). Human IgG1 (hIgG1 kappa, PHP010) was purchased from Serotec, Oxford, UK, and Human IgG was purchased from Sigma (I4506).
Construction and expression of recombinant soluble DV sE and WNV sE proteins
The genomic RNAs of clinical isolates FGA/89 of dengue virus-1 (DV1; Genbank access 226687, www.ncbi.nlm.nih.gov/Genbank) and 63632 of dengue virus-4 (DV4; Burma, 1976) were extracted from purified virions grown on mosquito AP61 cells and reverse transcribed using Titan One-Step RT-PCR kit (Roche Molecular Biochemicals) using specific PCR primers. The coding sequences for prM followed by the E ectodomain (E-1 to E-395) were digested with Bgl II and Not I and then inserted into the unique Bgl II and Not I sites of the pMT/Bip/V5-HisA plasmid (Invitrogen) to generate pMT/BiP/DV1.sE and pMT/BiP/DV4.sE. The WN coding sequence for the internal E translocation signal (prM-151 to prM 166) followed by the E sequence deleted from both transmembrane domains (E-1 to E-441) has been previously described (29). By directed mutagenesis, WN E sequence was inserted into the Bgl II and Not I sites of the shuttle vector to generate pMT/BiP/WN.sE. The flaviviral sequences were placed in-frame with a BiP sequence that directs recombinant protein to the secretory pathway. In the expression vector, the viral E sequences are followed at their C-terminus by the V5 epitope and six histidines for affinity purification using nickel chelate affinity chromatography. Drosophila S2 cells (Invitrogen) were transfected by the recombinant plasmid pMT/BiP/DV1.sE, pMT/BiP/DV4.sE or pMT/BiP/WN.sE using the Calcium Phosphate Transfection Kit (Invitrogen). Stably transfected cells were selected by adding 25 μg/ml blasticidin over several weeks. Cultured S2 cells expressing viral proteins were adapted in serum-free growth medium containing 10 μg/ml blasticidin. CuSO4 was added to a final concentration of 500 μM to induce synthesis and secretion of recombinant soluble E protein (sE). Accumulation of secreted sE proteins in the culture medium was maximal 10 days after addition of CuSO4. The cell culture supernatants were passed on 0.2 μM filters. Recombinant sE proteins were purified from cell culture supernatant on equilibrated chelating column chromatography (HiTrap Chelating HP, Amersham). The column was washed several times with washing buffer (0.5 M NaCl, 50 mM sodium phosphate buffer, [pH 8.0]) and bound sEs were eluted with increasing concentration of imidazole. Fractions containing DV1-sE, DV4-sE or WN-sE were pooled and dialyzed in PBS. Flaviviral sE proteins were assessed by Coomassie blue staining on SDS-PAGE and immunoblot analysis with 4G2 mAb or anti-WNV HMAF.
Construction and production of the H6-PhoA and H6-WNV-EIII-PhoA hybrids
The plasmids pVP5 (30) and pLIP5GN-H6 (31) have been previously described. pVP5 carries a partial cDNA clone of the West-Niles virus (strain IS98-ST1; GenBank accession N° AT481864, www.ncbi.nlm.nih.gov/Genbank), coding for domain 3 (EIII) of its envelope protein. Plasmid pLIP5GN-H6 carries the lacIq gene, and a mutant allele of the phoA gene from E. coli under control of the promoter ptac. This allele codes for an alkaline phosphatase (PhoA) with improved catalytic properties. pLIP5GN-H6 also carries six codons of histidine (H6) and a multiple cloning site (MCS) region between codons 27 (CCT) and 28 (GTT) of phoA.
Plasmid pLIP5GN-H6 does not express the PhoA enzyme because the MCS region introduces a frameshift. We constructed a derivative of pLIP5GN-H6 that expressed PhoA by deleting the dinucleotide TT that is located immediately downstream of the MCS region, at codon 28 (GTT) of the wild type phoA gene, using oligonucleotide site-directed mutagenesis. We named the resulting control plasmid pEBLØ it coded for a hybrid H6-PhoA between a hexahistidine and PhoA. The H6-WNV-EIII-phoA hybrid gene, coding for a hybrid protein between a hexahistidine, the EIII domain of WNV and PhoA, was constructed as follows. Plasmid pLIP5GN-H6 was digested with the restriction enzyme SalI and SmaI, within the MCS region. The WNV-EIII gene was amplified by PCR from plasmid pVP5. The primer that hybridized at the 5′-end of the WNV-EIII gene, brought in a SalI site and the primer that hybridized at the 3′-end, a ScaI site. The PCR product was digested with SalI and ScaI and then recombined by ligation between the SalI and SmaI sites of pLIP5GN-H6. We named the recombinant plasmid pEBL15. The sequences of pEBLØ and pEBL15 were checked with oligonucleotide primers that hybridized outside of the cloning region.
The H6-PhoA and H6-WNV-EIII-PhoA hybrids were expressed from plasmids pEBLØ and pEBL15 in the E. coli strain XL1-Blue after induction of the ptac promoter with 0.5 mM IPTG. The proteins were purified from periplasmic extracts of the producing strain by nickel ion chromatography (NiNTA resin, Qiagen) as previously described (30). Purification fractions were analyzed by SDS-PAGE (12% acrylamide, NuPage Novex system, Invitrogen) in reducing conditions. The purest fractions were pooled and dialysed against PBS. The final protein preparations were homogeneous at > 95 %. They were snap-frozen and kept at −80°C. The extinction coefficient ε280nm of each hybrid was calculated from its amino acid sequences with the subroutine Pepstats of the software suite EMBOSS (32). The specific activity for each hybrid for the hydrolysis of para-Nitro Phenyl Phosphate (pNPP) was assayed and found consistent with the published values (31). These phosphatase activities showed that both hybrids were in a dimeric state as the wild type PhoA.
Generation of WNV VLPs
To generate a Tet-Off cell line that allows tetracycline-inducible expression of the prM and E genes from WN virus strain IS-98-ST1, the MEF/3T3.Tet-Off cells (Clontech) were transfected with pTRE2hyg/prME.WN#4 constructed by cloning the cDNA coding for WN prM and E into the pTRE2Hyg plasmid DNA. Transfected cells were subjected to selection with 100 μg/ml Geneticin and 100 μg/ml Hygromycin B in the presence of 1 μg/ml doxycycline (Tet) and resistant cell clones were selected as previously described (33). Cell clones were then cultured in the presence of 1 μg/ml tetracycline (Tet). A number of cell clones expressing the WN prM and E cassette were capable of Virus-Like Particle (VLP) production in absence of Tet. The most efficient cell clone was designated MEF/3T3.Tet-Off/prME.WN#h2 and used in this study. To produce VLPs, the medium of MEF/3T3.Tet-Off/prME.WN#h2 cells was replaced with fresh medium without Tet. Three days after removal of Tet, the VLPs present in the supernatant of induced MEF/3T3.Tet-Off/prME.WN#h2 cells were precipitated with 7% (wt:vol) polyethylene glycol (PEG) 6,000 (Fluka) at 4°C for 4 hrs. After centrifugation, the pellet was resuspended in TNE buffer (20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 2 mM EDTA) and centrifuged on a discontinuous sucrose gradient composed of 60% (wt:vol) and 20% (wt:vol) sucrose at 39,000 rpm at 4°C for 2 hrs. The visible band at the interface was harvested and diluted in TNE buffer. The VLPs were further purified on a continuous 11–52% (wt:vol) sucrose gradient at 35,000 rpm at 4°C for 18 hrs. Fractions were collected from the top and WN antigens were detected by indirect ELISA with anti-WNV HMAF. Fractions containing VLPs were pooled and stored at −80°C. The total production of WN VLPs reached equivalent 8.7 log FFU/ml.
ELISA
ELISA binding assay was carried out to demonstrate direct binding of E-protein and NKp44-Ig. Plates were coated overnight at 4°C with a titration quantity of DV-sE DV4-sE and WNV-sE protein as indicated (diluted with PBS, final volume 50 μl/well). Blocking buffer (PBS supplemented with 3% dry skim milk, 200 μl/well) was applied for 1.5 hrs, after which plates were washed with PBS with 0.05% Tween 20 (PBST) and incubated with 5 μg/ml of fusion-Ig or hIgG in diluted blocking solution (0.6% skim milk 0.05% Tween 20) for 1.5 hrs at 37°C (75 μl/well). Following washing with PBST, 75 μl/well of biotin-conjugated goat anti-human IgG, Fcγ fragment-specific (Jackson, 109-066-098) was added for 1 hr at 1:2000 dilution in diluted blocking solution. Following washing with PBST, 75 μl/well of Streptavidin-HRP (Jackson, 016-030-084) diluted 1:1000 in blocking solution was added for 30 min. Following washing with PBST, 75 μl/well of TMB was added and color was allowed to develop at room temperature in the dark. Optical density was read at 650 nm (Thermo Electron Corporation Multiskan Spectum). The binding of NKp44 to WNV VLPs was performed by a similar ELISA procedure except that wells were coated with PBS only or with WNV VLPs diluted with PBS overnight at 4°C. In some experiments the WNV VLPs were pre-treated at a low pH by incubating them in Tris-Maleate pH 6.2 for 15 min at 37°C, followed by NaOH neutralization to pH~7.5. The binding of NKp44 to WNV-EIII was assayed using two complementary protocols. In the first protocol wells were coated with WNV-H6-EIII-PhoA dimer or mock-H6-PhoA dimer (4 μg/ml) and detection was performed with different fusion-Igs and anti-human IgG as described above for WNV-sE. The second protocol involved coating with different fusion-Igs (4 μg/ml), blocking with skim milk, and detection with WNV-H6-EIII-PhoA dimer or mock-H6-PhoA dimer (4 μg/ml) followed by pNPP substrate for AP (optical density was read at 405 nm).
Flow cytometry
Cells were incubated with various fusion-Igs (40 μg/ml) for 1.5–2 hrs at 4°C, washed and stained with APC-conjugated F(ab′)2 goat anti-human-IgG (Jackson, 109-136-098). Staining and washing buffer consisted of 0.5% (w/v) BSA and 0.05% sodium azide in PBS. When staining with 4G2 mAb, W6/32 mAb or polyclonal mouse serum antibodies, cells were incubated with the indicated antibodies for 45 min at 4°C, washed and stained with APC-conjugated F(ab′)2 goat anti-mouse IgG (Jackson, 115-136-068). In these experiments, staining and washing buffer consisted of 2% FCS and 0.05% sodium azide in PBS. Propidium iodide (PI) was added prior to reading for exclusion of dead cells. Flow cytometry was performed using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA) and fluorescence data was acquired using logarithmic amplification. Data files were acquired and analyzed using BD CELLQuest™ 3.3 software. Staining protocol for heparan sulfate has been previously described (26). Some experiments were performed using FACS Canto II and results were analyzed using FlowJo software.
Isolation and culture of primary NK cells
NK cells were isolated from the peripheral blood of healthy donors using the human NK cell isolation kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). NK purity was assayed using B-D MultTEST antibodies cocktail (anti-CD3/CD16+56/CD45/CD19). NK cells were more then 90% CD3− and CD56+. In order to obtain long-term activated polyclonal NK cells, purified NK cells were cultured on irradiated feeder cells and maintained in Cellgro SCGM serum-free medium (CellGenix, Freiburg, Germany) supplemented with 10% heat inactivated human plasma obtained from healthy donors, 1 mM sodium pyruvate, 2 mM L-glutamine, MEM non-essential amino acids, 1% penicillin/streptomycin, 10 mM HEPES (all supplements from Gibco) and 300 IU/ml of human IL-2 (Biological Industries).
CD107a degranulation assay for NK activity
NK degranulation assay was previously described (27, 34); briefly: NK cells (pre-incubated with final concentration of 20 U/ml human rIL-2) were incubated with WNV, anti-NKp30 (0.04 μg/ml) or anti-NKp44 (0.01 μg/ml) for 4 hrs at 37°C, the medium included 0.3 μl/well CD107a-biotin (Southern Biotech, 9835-08). After incubation cells were washed and incubated for 30 min on ice with 0.4 μl/well CD107a-biotin. CD107a expression was detected with streptavidin-conjugated APC (Jackson, 016-130-084) using a FACS Canto flow cytometer. In some experiments the WNV was inactivated prior to the experiment as follows: WNV was irradiated for 10 min with a UV lamp, which resulted in a total of 1200 W/m2. WNV inactivation was verified by infecting Vero cells with treated and untreated WNV. UV-treated WNV infected Vero cells significantly less than untreated WNV, as detected by flow cytometry.
Cytotoxicity assay and IFNγ assay
Cytotoxicity assays: 24 hrs after WNV infection, target cells were radioactively labeled for 12 hrs. Then, the cytotoxic activity of NK92 and NK92-44 cell lines and IL-2 activated primary human NK cells was assessed against labeled non-infected and WNV infected cells in a 5-h 35S-release assay, as previously described (35). To prevent class I HLA binding the inhibitory LIR1/ILT2 receptor, NK cells (NK92 and NK92-44) were pre-incubated with anti-ILT2 HP-F1 mAb for 30 min; alternatively, target cells were incubated with W6/32 to block ligands for NK inhibitory receptors. For additional blocking with antibodies, NK cells were pre-incubated on ice for 30 min with polyclonal mouse serum diluted 1:80 before being combined with target cells. In all experiments shown, the spontaneous release was less than 25% of maximal release. IFNγ assays: human IFNγ concentrations in the supernatants of NK cells co-incubated with target cells for 24 hrs were assayed by standard ELISA according to the manufacturer’s instructions (BioLegend, San Diego, CA).
Results
Direct binding of NKp44-Ig to flavivirus E glycoproteins
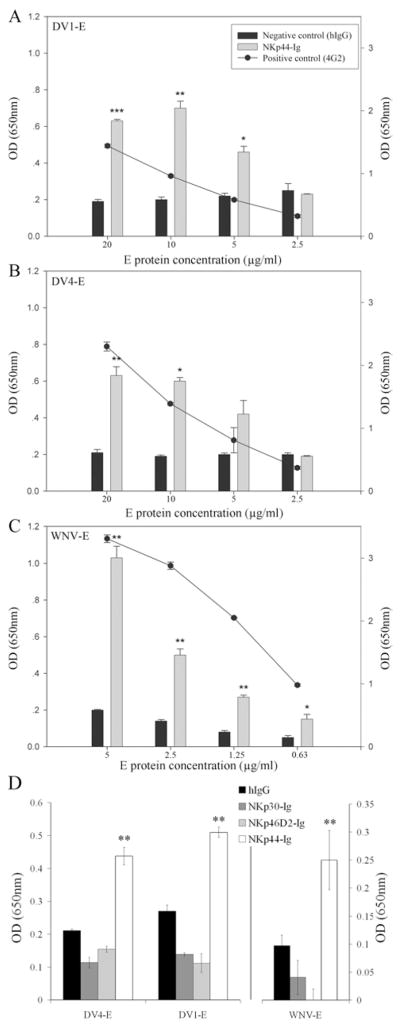
Several clinical studies have reported that human NK cells are activated following DV infection (18, 19, 36). Since NKp44 receptor surface expression is induced in activated NK cells, we tested whether NKp44 interacts with the E protein from DV of distinct serotypes 1 and 4. To begin, we studied the interaction between recombinant NKp44 and the soluble secreted form of E (sE) protein produced in invertebrate S2 cells. Given that flavivirus E glycoproteins share high structural similarity (37), we also assessed the binding of purified WNV-sE protein to NKp44. ELISA assays were performed to compare the binding of NKp44-Ig and hIgG to purified DV1-sE, DV4-sE or WNV-sE. Human IgG served as a negative control since the fusion-Igs contain the Fc portion of hIgG1, and for positive control we employed the flavivirus E protein-specific antibody 4G2. ELISA wells were coated with titrated quantities of sE proteins and the binding of 4G2, hIgG and NKp44-Ig was measured. NKp44-Ig, but not the control hIgG, bound to the sE protein from DV1, DV4 and WNV (Fig. 1). The binding of NKp44-Ig and 4G2 antibody correlated with the quantity of coated sE protein. The other two NCRs (NKp30 and NKp46) did not interact with the sE from DV and WNV, compared to hIgG (Fig. 1D).
Figure 1. NKp44-Ig binds flavivirus E protein specifically.
ELISA plates were coated with titrated concentrations of purified E protein from DV1 (A), DV4 (B) and WNV (C), followed by incubation with 5 μg/ml NKp44-Ig and hIgG1. Bar graphs indicate the binding of NKp44-Ig (gray) and hIgG1 (black) to the three E proteins (left y-axis). The curves in A, B and C show the binding of anti-flavivirus E protein 4G2 antibody to the three E proteins (right y-axis). (D) ELISA plates were coated with 5 μg/ml DV1-sE and DV4-sE and 1.25 μg/ml WNV-sE, followed by incubation with 5 μg/ml fusion-Ig and hIgG1. Bar graphs indicate the binding of fusion-Igs and hIgG1 to the three E proteins. Bars ± SD. One representative experiment of 3 performed is shown. *** P-value < 0.001, ** P-value < 0.01, * P-value < 0.05.
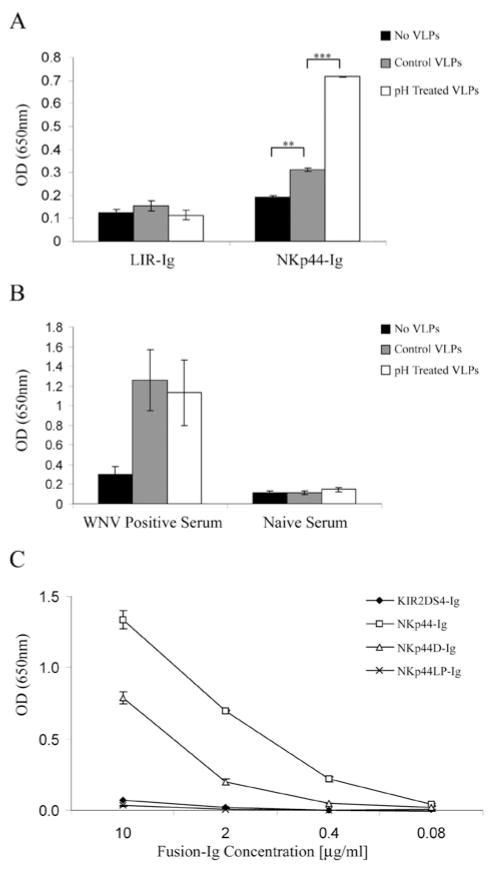
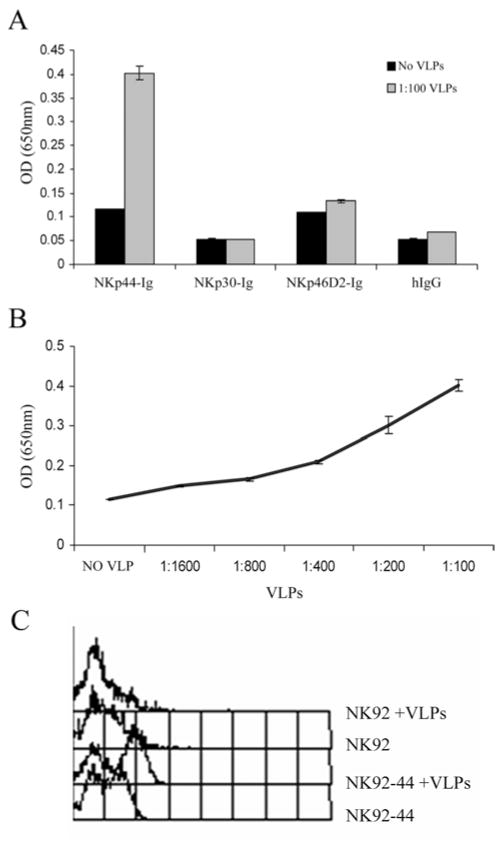
Since NKp44 manifested a strong binding to WNV-sE protein, we further explored this interaction by testing the binding of NKp44-Ig to WNV-VLPs, which are recombinant virus-like particles containing only the E and prM/M proteins and the lipid membrane. Flavivirus-VLPs were previously used as a reliable model for studying the structure and the membrane fusion properties of flavivirus. Furthermore, flavivirus-VLPs were also shown to induce protective immunity (reviewed in (37)). Therefore, we set out to check whether NKp44 also recognizes the flavivirus-VLPs; secreted WNV-VLPs were recovered from supernatant of an induced mouse fibroblastic MEF/3T3 cells expressing WNV prM and E glycoproteins. ELISA wells were coated with highly purified WNV-VLPs diluted 1:100 and binding of NKp44-Ig, NKp30-Ig, NKp46D2-Ig and hIgG1 was assayed. NKp44-Ig, but not the other Ig-proteins, bound the WNV-VLPs compared to their background binding to uncoated wells (Fig. 2A). Moreover, the binding of NKp44-Ig correlated with the quantity of coated WNV-VLPs (Fig. 2B). To assess the binding of WNV-VLPs to membrane-associated NKp44, we employed NKp44-transfected NK-92 cells that express high levels of transfected wild-type NKp44 (designated as NK92-44). NK92 and NK92-44 cells were incubated on ice in buffer only or with WNV-VLPs followed by mouse anti-WNV serum. WNV-VLPs bound only to NK92-44 cells as analyzed by flow cytometry, but not to NK92 cells that express none to little membrane-associated NKp44 (15) (Fig. 2C).
Figure 2. Direct binding of NKp44-Ig to WNV VLPs.
(A) ELISA wells were coated with WNV VLPs diluted 1:100 with PBS or with PBS alone (no VLPs), followed by incubation with 4 μg/ml of the indicated fusion-Igs and hIgG1. Bound fusion-Igs were detected by HRP-conjugated anti-human IgG. (B) Titration of WNV VLPs. ELISA wells were coated with diluted WNV VLPs, followed by incubation with 4 μg/ml NKp44-Ig. Results are one representative experiment of 3 performed. (C) Binding of WNV VLPs to NK92 and NK92-44 cells; cells were incubated on ice for two hours in buffer only or with 2μl WNV VLPs, followed by mouse anti-WNV serum. Then, cells were washed and incubated with biotin-conjugated donkey anti-mouse followed by streptavidin-conjugated APC. Binding results were analyzed by flow cytometry (FACSCalibur).
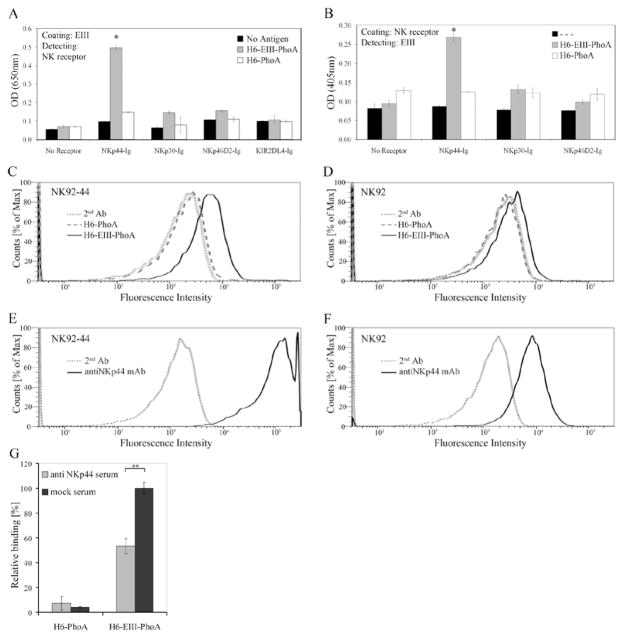
To further explore the interaction of NKp44 with the E protein, we studied whether NKp44 can bind to domain III of the WNV-E protein. Dimers of WNV-EIII fused to alkaline phosphatase (PhoA) and His tag (H6) were produced in bacteria. We first assayed the binding of NKp44-Ig to H6-EIII-PhoA and to mock H6-PhoA using two ELISA-based approaches (Fig. 3A, B). We coated the wells with either H6-EIII-PhoA or H6-PhoA and detected with the different NK-receptor Igs (Fig. 3A). We also coated the wells with the different NK-receptor Igs and detected with either H6-EIII-PhoA or H6-PhoA using substrate to PhoA AP (Fig. 3B). In both approaches significant interaction was observed only for NKp44 and H6-EIII-PhoA as compared to NKp44 and mock H6-PhoA. Other NK receptors including NKp46 and NKp30 did not manifest notable interaction with H6-EIII-PhoA as compared to mock. The observation that NKp44 binds to EIII explains the inability of mAb4G2 to block this interaction (data not shown) as 4G2 binds at the interface between the EI and EII domains (38). To assess the interaction of NK-expressed NKp44 with WNV-EIII we stained NK92 and NK92-44 cells with either H6-EIII-PhoA or mock H6-PhoA complexes. WNV-EIII-including complexes specifically stained NK cells as compared to mock complexes (Fig. 3C, D) and this staining correlated with NKp44 expression on the NK cell surface (Fig. 3E, F). Anti-NKp44 serum significantly blocked the binding of H6-EIII-PhoA to wells coated with NKp44 (Fig. 3B, G). To summarize, these findings demonstrate direct interaction between NKp44 and DV1-, DV4- and WNV-envelope proteins.
Figure 3. WNV-EIII binds to NKp44.
NKp44-Ig interacts with WNV-EIII. (A) ELISA wells were coated with either H6-EIII-PhoA or H6-PhoA, and NK receptor-fusion-Igs were applied. Bound fusion-Igs were detected by HRP-conjugated anti-human IgG. (B) ELISA plates were coated with fusion-Igs, and either H6-EIII-PhoA or H6-PhoA were applied. Bound PhoA activity was detected by pNPP substrate. Bars ± SD. * P-value < 0.001. (C–D) H6-EIII-PhoA or H6-PhoA were incubated with anti-His mAb at 2:1 molar ratio for 1 h on ice to produce complexes that were used to stain NK92-44 (C) and NK92 (D) cells. Cell-bound complexes were detected with APC-labeled goat anti-mouse IgG. (E-F) NKp44 expression on NK92-44 (E) and NK92 (F) was assessed with anti-NKp44 mAb. Binding results were analyzed by flow cytometry (FACS Canto II). Results are representative of 3 performed. (G) Binding of H6-EIII-PhoA or H6-PhoA to NKp44-Ig-coated wells in the presence of anti-NKp44 or mock serum (1:200 dilution). Bound PhoA activity was detected by pNPP substrate. Results are summary of 2 experiments, normalized according to H6-EIII-PhoA binding in the presence of mock serum. Bars ± SD. ** P-value < 0.005.
Binding of NKp44-Ig to WNV-infected cells is inhibited by anti-WNV serum
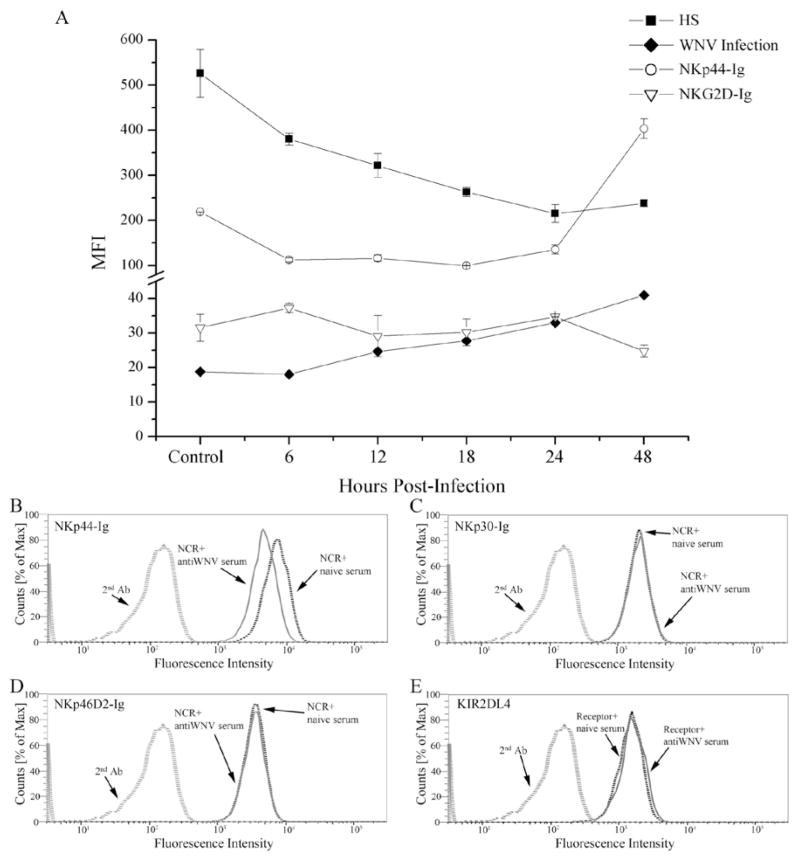
Next, the binding of NKp44-Ig and NKG2D-Ig to WNV infected and non-infected Vero cells was assayed at different time points post-infection by flow cytometry. Surprisingly, the binding of NKp44-Ig to Vero cells following WNV infection was already reduced to half (MFI values: 219 to 112) at 6 hrs post-infection and continued to decrease at 12 and 18 hrs post-infection (MFI values: 116 and 99, respectively). However, at 24 hrs post-infection, NKp44-Ig showed enhanced binding to WNV-infected Vero cells, which reached a maximal binding at 48 hrs post-infection (in MFI values: 404). Compared to Vero non-infected cells, the binding of NKp44-Ig to WNV-infected cells at 48 hrs post-infection was enhanced by almost two-fold. However, NKG2D-Ig binding to WNV-Vero infected cells and non-infected cells was comparable at all time points (Fig. 4A).
Figure 4. NKp44-Ig binding to WNV-infected cells is inhibited by anti-WNV serum.
(A) Kinetics of NKp44 binding to, membrane-associated heparin sulfate (HS) and WNV-E expression by WNV-infected Vero cells. Vero cells were infected with the WNV KVI strain at about 4 MOI. Cells were harvested at indicated hours post-infection and stained with WNV serum (WNV infection), anti-HS antibody AO4B08, NKp44-Ig and NKG2D-Ig. Positive binding was detected by flow cytometry (using a FACSCalibur flow cytometer) and presented as geometric mean fluorescence intensity (MFI). Results are the average of 2 samples assayed in the same experiment that are representative of 3 performed. Bars ± SD. (B–E) Binding of NK receptor-Igs (NKp44-Ig, NKp30-Ig, NKp46D2-Ig and KIR2DL4-Ig) to WNV-infected Vero in the presence of anti-WNV and naive mouse sera. Thirty hours after WNV infection, Vero cells were first incubated with the mouse sera (1:60 dilution, 30 minutes), then fusion-Igs were added followed by APC-labeled goat anti-hIgG as described in methods. Binding results were detected by FACS Canto II and are presented as overlays of staining histogram. Results are representative of 2 performed.
NKp44 is involved in the recognition of both cellular and viral ligands (12). Therefore, the overall binding of NKp44-Ig to WNV-infected Vero cells could be affected by the expression of cellular and viral ligands. We previously showed that membrane-associated heparan sulfate epitopes are co-ligands of NKp44, involved in its target cell recognition (34). We thus tested whether the unexpected reduced staining of WNV-infected Vero cells by NKp44-Ig early post-infection is associated with down-regulation of cell surface heparan sulfate. Indeed, staining of WNV-infected Vero cells with anti-heparan sulfate antibody AO4B08 resulted in a similar reduction, compared to staining with NKp44-Ig. Yet, while at 48 hrs post-infection staining with NKp44-Ig was markedly enhanced, only a slight enhancement in heparan sulfate staining at 48 hrs was observed compared to staining at 24 hrs; the expression of heparan sulfate was still low compared to non-infected Vero cells (Fig. 4A). Therefore, NKp44-Ig enhanced binding at 48 hrs post-infection is most likely the result of the increased expression of cell surface WNV-E protein as detected by staining of WNV-infected Vero cells with mouse anti-WNV serum (Fig. 4A), or with anti-E protein specific antibody 4G2 (data not shown). To prove this assumption we took WNV-infected Vero cells 30 hrs following infection and incubated the cells with either mouse anti-WNV or naive serum. Next, we stained them with NKp44-Ig and other NK receptor-Igs including NKp46 and NKp30 (Fig. 4B–E). Vero cells express cellular ligands to all of these receptors, thus all tested NK-receptor-Igs positively stained the cells. However, only NKp44-Ig staining of WNV-infected Vero cells was inhibited in the presence of anti-WNV serum as compared to naive serum (Fig. 4B–E). Therefore, the enhanced NKp44-Ig staining of WNV-infected cells observed at the second day following infection is due to the presence of WNV-E protein on the membrane of the infected cells.
Functional recognition of WNV by NK cells is mediated via NKp44
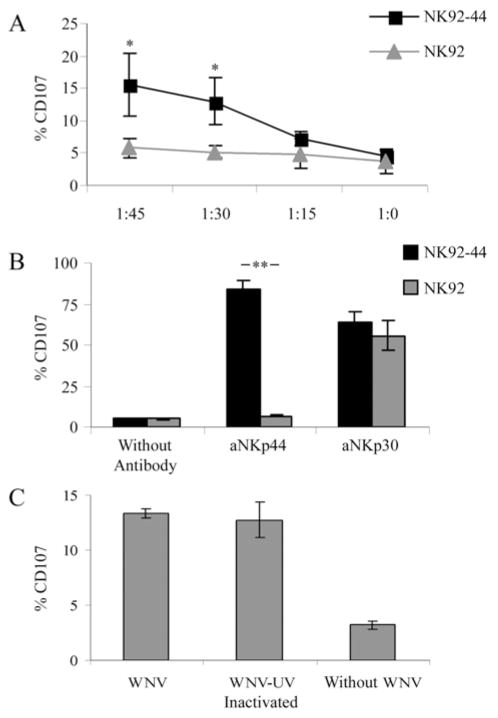
To test whether the interaction between NKp44 and WNV sE protein might lead to NK cell activation, we co-cultured NK92 and NK92-44 with the WNV-KVI strain, which was used throughout this study. To assess the activation we stained the cells for CD107a cell surface expression, which is a marker for NK cell degranulation and functional activity (39). NK cells and WNV were co-cultured for four hrs at different NK:WNV ratios, calculated according to the multiplicity of infection (MOI) value, and the percentage of CD107a positive (CD107a+) cells was analyzed. There was no significant increase in the percentage of CD107a+ NK-92 cells even when co-cultured with high doses of WNV. Yet, the percentage of CD107a+ NK92-44 cells was significantly enhanced when co-cultured with WNV and correlated with the increasing quantity of the virus (*P < 0.05; Fig. 5A). When stimulated by plastic-bound anti-NKp44, the percentage of CD107a+ cells in the NK92-44, but not in the NK92, cell line was significantly increased (** P < 0.01; Fig. 5B). Yet, no substantial difference was observed between NK92 and NK92-44 cell lines in the percentage of CD107a+ cells when triggered by anti-NKp30 (Fig. 5B). These results imply a similar activation potential between NK92 and NK92-44 cells when stimulated by a triggering receptor expressed on both cells, while triggering the cells via NKp44 activated only the NK92-44 cells and not NK92. Thus, expression of functional wild type NKp44 by NK cells directly correlated with their activation by WNV.
Figure 5. WNV activates NK cells via the NKp44 receptor.
Comparative analysis of CD107a expression on NK92 and NK92-44 cells co-incubated with WNV KVI strain (A) or with anti-NKp44 or anti-NKp30 antibodies (B). (A) NK92 and NK92-44 cells were co-incubated with WNV for four hours at the indicated NK:WNV ratio calculated as multiplicity of infection (MOI). Cells were then washed and stained for CD107a cell surface expression. Results shown are the average of 3 experiments. (B) NK92 and NK92-44 cells were incubated for four hrs in uncoated wells or wells pre-coated with anti-NKp44 (aNKp44; 0.01 μg/ml) or anti-NKp30 (aNKp30; 0.04 μg/ml) antibodies before staining for CD107a expression. One representative experiment of three performed is shown. (C) CD107a cell surface expression on NK92-44 cells after four hrs exposure to infective and UV-inactivated WNV at 1:45 NK:WNV ratio. Graph includes data from two different experiments. Bars ± SD. ** P-value < 0.01, * P-value < 0.05.
NK cells have been shown to be directly infected in vitro by HIV and HSV (40). To assess whether the activation of NK92-44 cells by WNV involved their direct infection, we tested whether UV-irradiated virus could induce degranulation of NK92-44 cells. UV treatment of WNV significantly reduced the virus’ ability to infect Vero cells, compared to the non-treated WNV (data not shown). As shown in Figure 5C, comparable percentages of NK92-44 CD107a+ cells were induced by UV-irradiated and non-irradiated WNV (p = 0.726, Tukey post hoc test that followed a one way ANOVA, p < 0.001). Therefore, the NKp44-mediated activation of NK92-44 cells by WNV did not involve NK cell infection and only required the exposure of NK92-44 cells to the virus.
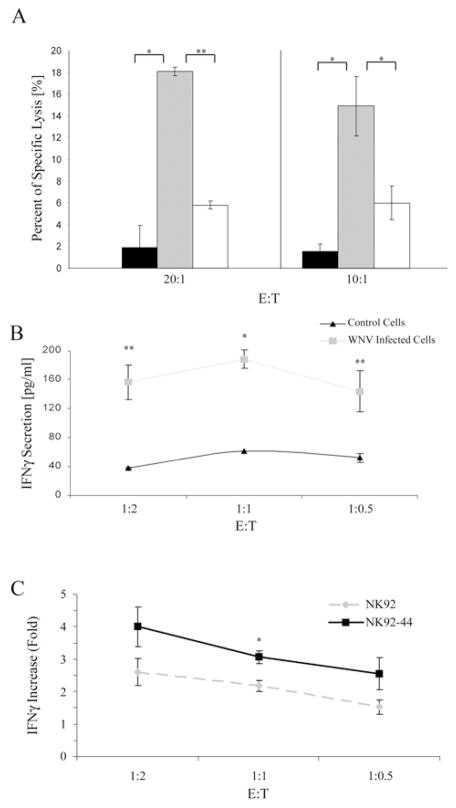
To investigate whether the enhanced binding of NKp44 to WNV-infected Vero cells would increase their susceptibility to NK cell lysis, we compared the killing of non-infected and WNV-infected Vero cells by NK92-44 cells. It is known that the cytolytic activity of NK cells is inhibited by the expression of MHC class I molecules on the surface of target cells (41). In addition, flaviviruses are known to up-regulate the expression of MHC class I molecules on the cell surface of host cells (17). Indeed, we observed MHC class I up-regulation in Vero cells induced by WNV infection, as detected by staining with W6/32 antibody (data not shown). Thus, we first blocked the LIR-1 receptor, which is the only inhibitory receptor expressed on NK92-44 cells, by pre-incubation with anti-LIR-1 HP-F1 mAb. This blocking was necessary to demonstrate NKp44-mediated killing of WNV-infected Vero cells. Second, we used WNV-infected target cells 36 hrs post infection (including the labeling time) to ensure sufficient expression of WNV-E on the infected cell membrane (Fig. 4A). The results in Figure 6A show that NK92-44-killing of WNV-infected Vero is augmented compared to non-infected cells, and that this enhanced killing is significantly inhibited by anti-NKp44 antibodies, but not by control serum. However, NK92 cytolytic activity against WNV-infected and non-infected Vero cells was comparable (data not shown). These results indicate that NKp44 is directly involved in the killing of WNV infected Vero cells.
Figure 6. NKp44 is involved in the activation of NK cell lines by WNV-infected cells.
(A) Vero cells were infected with the WNV KVI strain. After 24 hrs WNV infected cells or Vero uninfected cells were radioactively labeled for 12 hrs, washed and used as target cells in a standard 5-h 35S-release assay. NK92-44 effector cells were pre-incubated with control mouse serum or anti-NKp44 mouse serum diluted 1:80 before being added to target cells. Uninfected Vero (black histogram); WNV infected Vero in the presence of control serum (gray histogram); WNV infected Vero in the presence of anti-NKp44 serum (white histogram); each time point constitutes the mean of the percent specific lysis of four separate wells. Results are a summary of two different experiments. Bars ± SD. ** P-value < 0.01, * P-value < 0.0002 (B, C) 293T cells were infected with WNV KVI strain. After 24 hrs cells were plated in a 96-well plate and co-incubated with effector NK92-44 or NK-92 cells at indicated E:T ratios. After 24 hrs 100 μl of the culture medium was assayed for the presence of human IFNγ (pg/ml) by standard ELISA. (B) IFNγ secretion by NK92-44 cells co-incubated with WNV infected 293T cells and non-infected 293T cells (control cells). (C) IFNγ secretion ratio between NK cells co-incubated with WNV infected 293T cells and NK cells co-incubated with control cells. Results are shown for both NK92 and NK92-44. One representative experiment of 2 performed is shown. Bars ±SD. In panels A, B and C, NK cells were pre-incubated with anti-LIR-1 antibodies. ** P-value < 0.05, * P-value < 0.005
The secretion of IFN-γ by NK cells is crucial for their immune-mediated response to pathogen infection (5). Therefore, we further analyzed the influence of WNV infection on NKp44-mediated IFN-γ secretion by NK cells. 293T cells were infected with WNV or left uninfected. After 24 hrs cells were harvested and co-incubated for 24 hrs with NK92 and NK92-44 cells and IFN-γ secretion into the culture medium was assayed. Again, in order to demonstrate the NKp44-mediated activation of WNV-infected 293T cells, we blocked the LIR-1 receptor on NK92 and NK92-44 cells by pre-incubation with anti-LIR-1 HP-F1 mAb. Low levels of IFN-γ secretion by NK92-44 cells were detected when co-incubated with non-infected 293T cells. However, co-incubation of NK92-44 cells with WNV-infected 293T cells resulted in elevated levels of IFN-γ secretion in different E:T ratios (Fig. 6B). As shown in Figure 6C, induction of IFN-γ secretion by NK92 cells was also observed when co-incubated with WNV-infected 293T cells compared to non-infected 293T cells. Thus, NK receptor(s), in addition to NKp44, might be involved in WNV-mediated activation of NK cells. Yet, the magnitude of increase in IFN-γ secretion by NK92-44 resulting from WNV infection was significantly greater compared to the magnitude of increase in IFN-γ secretion by NK92.
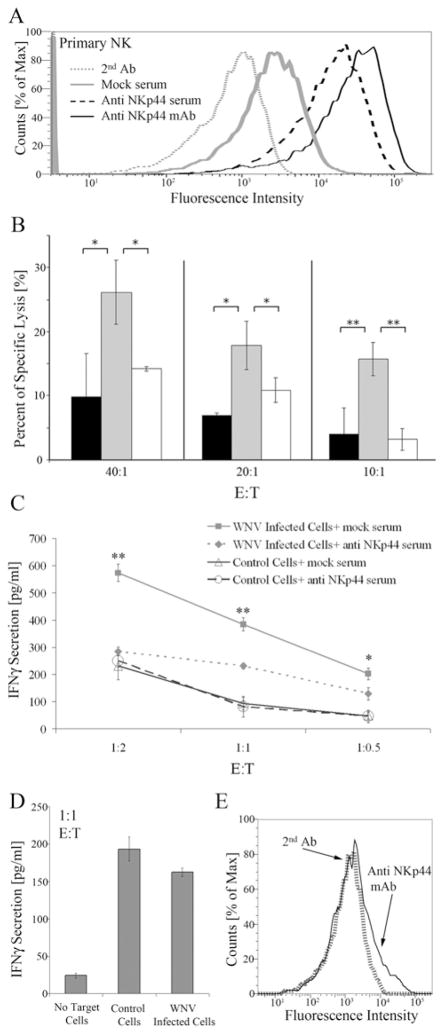
Finally, we studied the involvement of NKp44 in the functional activation of primary human NK cells incubated with WNV-infected cells. NK cells purified from human PBMC and activated with IL-2 for 3 days express NKp44 on their cell membrane (Fig. 7A). We thus studied their capacity to lyse 36 hrs-WNV-infected Vero cells that express WNV-E on their cell membrane (Fig. 4A). As observed for NK92-44 cells: (i) NKp44-expressing primary NK cells lysed WNV-infected Vero cells significantly better compared to non-infected cells; and (ii) application of anti NKp44 sera significantly suppressed the binding as compared to mock sera (Fig. 7B). We also studied IFNγ secretion by IL-2-activated, NKp44-expressing primary NK cells following incubation with WNV-infected and control non-infected cells. Supernatants of NK cells incubated with control cells had moderate levels of IFNγ that were not affected by the addition of soluble anti-NKp44 sera as compared to mock sera (Fig. 7C). In contrast, supernatants of NK cells incubated with WNV-infected Vero cells had significantly higher levels of IFNγ that were markedly reduced when soluble anti-NKp44 serum was present as compared to mock serum (Fig. 7C). IL-2-activated primary human NK cells that did not enhance NKp44 expression were further incubated with WNV-infected and non-infected target cells; in conformity with the above results, IFNγ induction was similar for infected and non-infected target cells (Fig. 7D, E).
Figure 7. NKp44 is involved in the activation of primary NK cells by WNV-infected cells.
(A) IL-2 activated primary human NK cells were stained with anti NKp44 mAb, mouse anti-NKp44 serum and mock serum followed by APC-goat anti mouse IgG. Binding results were detected by FACS Canto II and are presented as overlays of staining histogram. (B–D) Vero cells were infected with the WNV KVI strain. After 24 hrs Vero uninfected cells or WNV-infected cells were harvested and used as target cells. IL-2 activated primary human NK cells were pre-incubated with control mouse serum or anti-NKp44 mouse serum diluted 1:80 before being added to target cells. (B) 24 hrs following infection, target cells were radioactively labeled for 12 hrs, washed and incubated with W6/32 to block MHC-I ligands for NK inhibitory receptors. Then, target cells’ lysis by NK cells was assayed in a standard 5-h 35S-release assay. Uninfected Vero (black histogram); WNV-infected Vero in the presence of control serum (gray histogram); WNV infected Vero in the presence of anti-NKp44 serum (white histogram); each time point constitutes the mean of the percent specific lysis of four separate wells. Bars ± SD. ** P-value < 0.01, * P-value < 0.05. Primary human NK cells were activated with IL-2 for 3 days (C) and for 24 hrs (D). IFNγ levels in the culture supernatants of target cells and activated NK cells were measured after co-incubation for 24 h. ** P-value < 0.00001, * P-value < 0.001. (E) NKp44 expression by the primary human NK cells used for the experiment described in 7D. One representative experiment of 2 performed is shown.
Taken together, the above results clearly suggest that NKp44 binds to WNV infected cells and that augmented binding correlates with WNV-E protein expression on the surface of infected cells. Moreover, using anti-NKp44 serum we show that NKp44 is directly involved in NK cell activation induced by WNV infection.
Partial characterization of NKp44-interaction with WNV and DV sE protein
During flavivirus maturation, irreversible conformational change induced by low pH conditions occurs in the trans-Golgi network. This structural change involves the prM and E proteins. VLPs can consist of both pH-mature and-immature particles (37). Therefore, we tested whether low-pH treatment of WNV-VLPs would affect the interaction with NKp44. This treatment significantly enhanced the binding of NKp44 to WNV-VLPs; yet it did not affect the background binding of the LIR1-Ig negative control (Fig. 8A), nor did it enhance the binding of anti-WNV sera (Fig. 8B). Therefore, NKp44 interacts efficiently with the mature form of the E protein, as expected from its binding to the mature soluble form of WNV-sE and DV-sE proteins (Fig. 1).
Figure 8. Characterization of NKp44-E protein interaction.
(A, B) ELISA wells were coated with PBS (no VLPs), WNV VLPs diluted with PBS (Control VLPs) or WNV VLPs pretreated in low pH=6 buffer (pH treated VLPs). Wells were then incubated with 4 μg/ml fusion-Igs (A) or with polyclonal mouse anti-WNV positive or naïve serum (B). Positive binding was detected by HRP-conjugated secondary antibody. Graph includes data from two (A) or three (B) different experiments. Bars ± SD. (C) ELISA wells were coated with purified E protein from DV4 followed by incubation with titrated concentrations of indicated fusion-Igs. One representative experiment of 2 performed is shown. Bars ± SD. *** P-value < 0.001, ** P-value < 0.005
We showed that NKp44 and NKp46 proteins recognize the HA of IV and that binding of NKp44 and NKp46 to the HA is required for lysis of IV-infected cells expressing HA (14, 15, 35). The recognition of the HA required the sialylation of oligosaccharides of NKp44 and NKp46 (14, 15, 35). We further showed that this recognition was restricted to one of the three oligosaccharides found on the membrane-proximal domain and the linker peptide of NKp46 (no glycosylation sites are present in the membrane-distal domain) (13). Unlike NKp46, all the glycosylation sites predicated for NKp44 (13 O-sites and 1 N-site) are located in the linker peptide, while the extracellular domain has none (42, 43). Similar to HA of IV, flavivirus-E protein is a viral fusion protein that facilitates the fusion of the viral membrane with the target cell membrane (44). Therefore, we studied whether NKp44 binds the flavivirus-E protein via the linker peptide region (NKp44LP) and/or the extracellular domain (NKp44D). We compared the binding of titrated concentrations of NKp44-Ig, NKp44D-Ig, NKp44LP-Ig and KIR2DS4-Ig to DV4-sE protein in ELISA assay. As described above, positive binding of NKp44-Ig to DV4-sE was observed and correlated with the concentration of NKp44-Ig (Fig. 8C). A similar pattern of binding was also observed for NKp44D-Ig, although binding intensity was lower compared to NKp44-Ig. However, only background recognition, similar to that of KIR2DS4-Ig, was observed for NKp44LP-Ig. Yet, when testing the binding of NKp44-Ig, NKp44D-Ig and NKp44LP-Ig to IV-infected and uninfected 1106 melanoma cells, we observed enhanced binding of NKp44-Ig and NKp44LP-Ig, but not of NKp44D-Ig, to IV-infected cells (unpublished data).
These findings demonstrate that the binding region of DV-sE protein is located within the extracellular domain of NKp44 and not in the linker peptide region of the receptor. In addition, they suggest that the binding of NKp44 to DV-sE is not sialic-acid associated, since the extracellular domain has no predicted glycosylations sites.
Discussion
Natural killer (NK) cells constitute a key frontline defense against a range of viruses and bacteria. Different studies have demonstrated the importance of NK cells in controlling human viral infection, in particular viruses that belongs to the herpes virus genus, such as human cytomegalovirus, Epstein–Barr virus and herpes simplex virus (reviewed in (5, 45)). Yet, the involvement of NK cells in the immune response to flaviviruses is poorly appreciated. In the current study, we tested the interaction between the NK-activating receptor NKp44 and DV and WNV-E protein. We showed (i) direct protein-protein interaction between recombinant NKp44-Ig and soluble S2-produced DV-E and WNV-E proteins (Fig. 1), (ii) direct protein-particle interaction between recombinant NKp44 and WNV-VLPs expressing the E protein (Fig. 2), (iii) direct binding of WNV-VLPs to NK cells expressing high levels of wild type NKp44 (Fig. 2), (iv) direct binding of NKp44-Ig and soluble WNV-EIII domain (blocked by anti-NKp44 serum) as well as binding of WNV-EIII to NKp44-expressing NK cells (Fig. 3), (v) enhanced binding of NKp44-Ig to WNV-infected cells that correlated with the expression of WNV-E protein on the surface of infected cells and is blocked by anti-WNV serum (Fig. 4), (vi) that the interaction between West Nile virions or WNV-infected cells and NKp44 expressed by either NK cell line or primary NK cells resulted in NK activation (Figs. 5–7) and (vii) that the extracellular domain of NKp44 mediated the binding of NKp44 to mature WNV-E protein (Fig. 8). Taken together, these findings suggest that flavivirus E protein activates NK cells via NKp44.
Both soluble oligomeric (sE, EIII) and multimeric (VLP) forms of WNV-E glycoprotein reproduced the interaction with NKp44, and low pH-matured VLPs enhanced the binding of VLPs to NKp44 (Figs. 1–3 and 8). This reproduction of binding results suggests that the ectodomain of the E and particularly the EIII domain is necessary and sufficient for the binding to NKp44. It is likely that the N-oligosaccharide profile of the E-glycoprotein does not play a critical role in the binding to NKp44 since WNV-sE brings a single trimannosyl core on each monomer, whereas VLPs-expressed E brings mammalian-like complexed antennary N-glycans. Consistent with this conclusion, DV1-sE brings two trimannosyl cores on each monomer, whereas DV4-sE brings a single one; yet both manifested similar interaction with NKp44 (Fig. 1). The binding of NKp44 to bacterially-produced WNV-EIII (Fig. 3) further substantiate the observation that the glycan profile of the E protein is not involved in the interaction with NKp44.
The other side of the coin is whether the glycosylation of the NKp44 glycoprotein is involved in the interaction of NKp44 and the E protein. We previously reported that the recognition of HA of IV required the sialylation of the NKp44 and NKp46 receptors and that this recognition mediated the lysis of virus-infected cells by NK (14, 35). The linker peptide region of NKp44 contains 13 predicted O-glycosylation sites and a single putative N-glycosylation site, while the extracellular domain of NKp44 does not contain any glycosylations (42, 43)). These multiple glycosylation sites probably express the sialic acid that is required for the recognition of IV-infected cells by the linker peptide. Consistent with this, the linker peptide region, but not the extracellular domain of NKp44, mediated binding to IV-infected cells (unpublished data). However, the extracellular domain of NKp44-mediated binding to DV-sE protein as opposed to the linker peptide region (Fig. 8). Yet, the binding of the NKp44D was reduced compared to the whole NKp44 extracellular domain. The linker peptide sequence is typical of hinge-like sequences that allow an extended open conformation (42). Furthermore, the dense glycosylation of the linker peptide could confer structural rigidity to NKp44 (46, 47). Therefore, we assume that the glycosylated linker peptide contributes to the conformation or flexibility of the NKp44 and this results in a better apparent binding. The finding that the binding region of DV-sE protein is located within the extracellular domain of NKp44 suggests that the flavivirus E protein recognition by NKp44 is not sialic-acid associated. It also explains why NKp46 did not bind to DV or WNV-sE protein, to WNV-EIII or to WNV VLPs (Figs 1–3), and implies a different pattern recognition from the sialic acid-mediated binding of NKp44 and NKp46 to viral hemagglutinins.
Although the capability of NK cells to directly kill virally infected cells and to produce inflammatory cytokines can potentially limit infection by flaviviruses, only a few studies have been published that described activation of NK cells following flavivirus infection. In humans, early activation of NK cells in DV-infected patients was reported to be associated with mild dengue disease (18) and activation of NK cells was also observed in children with dengue hemorrhagic fever (19). A study of human liver paraffin embedded blocks from patients who died with yellow fever has found increased numbers of NK cells, mainly in yellow fever damaged area of the liver (48). Our results suggest that NKp44-expressing activated NK cells are triggered through NKp44-E protein engagement following interaction with the flavivirus or flavivirus-infected cells. It is likely that a complex balance exists between flavivirus-induced triggering of NK cells and subversion of NK killing by flavivirus-induced enhanced MHC class I expression on infected cells. Consistent with this idea, it was necessary in our study to block the LIR-1 receptor on NK cells in order to show the functional relevance of NKp44 binding to WNV-infected cells (Fig. 4). Another possible mechanism evolved by flaviviruses to evade NK cell response is to reduce heparan sulfate cell surface expression following virus infection of the cell, as was detected in Vero cells following WNV-infection (Fig. 4A). Heparan sulfate has been shown to be a possible co-ligand of all the members of the NCRs family: NKp30, NKp46 and NKp44 (25, 34). Thus, down-regulation of heparan sulfate cell surface expression would subsequently result in a reduced recognition of the infected cell by all of the NCRs, as was observed for NKp44 (Fig. 4A), and therefore for reduced susceptibility to NK cell lysis. Yet, following E protein up-regulation, binding of NKp44 to WNV-infected cells was enhanced and subsequently lysis of WNV-infected cells. Therefore, the NKp44-mediated outcome of the interaction between NK cell and flavivirus-infected cell is affected by the expression of the cellular and viral ligands on the cell surface. Of course, it is possible that the reduced expression of heparan sulfate following WNV infection is a by-product of the virus entry into the cells. Interestingly, WNV-E protein was reported to specifically block the dsRNA-induced cytokine production by murine macrophages in a TLR3-independent manner (49). These findings suggest an additional mechanism by which WNV impairs innate immunity response.
In mice, NK cell activity was transiently increased following flavivirus infection (50). Yet, antibody depletion of NK cells in mice did not alter morbidity or mortality after WNV infection (51). So far, no homolog of NKp44 has been identified in mice, and NKp46 that does have a mouse homologue did not interact with the E-protein (Figs. 1–3). In accordance with our results, the lack of murine NKp44 homologue could account for the different reports on NK contribution to flavivirus protection in mice and humans.
As two mosquito-borne flaviviruses classed into two different serocomplexes, WNV is the leading cause of meningitis-encephalitis, whereas DV is the leading cause of viral hemorrhagic fever. Moreover, DV1 and DV4 are two very distinct serotypes. Therefore, our results suggest that NKp44-E glycoprotein interaction might be common for flavivirus and NK cells regardless the origin of the virus and the E protein (grown in mammalian or invertebrate cells) and the virus tropism in human (neurotropism versus viscerotropism). This is in accordance with previous studies providing evidence that flaviviruses have evolved common mechanisms to evade NK triggering through MHC-I enhancement (8, 17). This may reflect selective pressure towards viruses that are capable of replicating rapidly and reaching the high blood titers required for transmission to its arthropod vector (8, 17). Our current study further stresses the need of flaviviruses to evade NK through MHC-I enhancement, as the E protein is found to interact with the NK triggering receptor, NKp44.
Acknowledgments
Support: This work was supported by grant from the European Commission (INCO-DEV, contract DENFRAME N 51711).
Abbreviations used in this paper
- DV
dengue virus
- DV1
dengue virus-1
- DV4
dengue virus-4
- E protein
envelope protein
- HA
hemagglutinin
- IV
influenza virus
- NKp44D
NKp44 extracellular domain
- NKp44LP
NKp44 linker peptide
- sE
soluble envelope
- VLPs
virus-like particles
- WNV
West Nile virus
Footnotes
Conflict of interest: the authors declare no conflict of interest or financial interests.
References
- 1.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 2.Hayes EB, Komar N, Nasci RS, Montgomery SP, O’Leary DR, Campbell GL. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stiasny K, Heinz FX. Flavivirus membrane fusion. J Gen Virol. 2006;87:2755–2766. doi: 10.1099/vir.0.82210-0. [DOI] [PubMed] [Google Scholar]
- 4.Diamond MS, Shrestha B, Mehlhop E, Sitati E, Engle M. Innate and adaptive immune responses determine protection against disseminated infection by West Nile encephalitis virus. Viral Immunol. 2003;16:259–278. doi: 10.1089/088282403322396082. [DOI] [PubMed] [Google Scholar]
- 5.Lodoen MB, Lanier LL. Natural killer cells as an initial defense against pathogens. Curr Opin Immunol. 2006;18:391–398. doi: 10.1016/j.coi.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 7.Kirwan SE, Burshtyn DN. Regulation of natural killer cell activity. Curr Opin Immunol. 2007;19:46–54. doi: 10.1016/j.coi.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Hershkovitz O, Zilka A, Bar-Ilan A, Abutbul S, Davidson A, Mazzon M, Kummerer BM, Monsoengo A, Jacobs M, Porgador A. Dengue virus replicon expressing the nonstructural proteins suffices to enhance membrane expression of HLA class I and inhibit lysis by human NK cells. J Virol. 2008;82:7666–7676. doi: 10.1128/JVI.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porgador A. Natural cytotoxicity receptors: pattern recognition and involvement of carbohydrates. ScientificWorldJournal. 2005;5:151–154. doi: 10.1100/tsw.2005.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. Embo J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnon TI, Markel G, Mandelboim O. Tumor and viral recognition by natural killer cells receptors. Semin Cancer Biol. 2006;16:348–358. doi: 10.1016/j.semcancer.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Arnon TI, Achdout H, Lieberman N, Gazit R, Gonen-Gross T, Katz G, Bar-Ilan A, Bloushtain N, Lev M, Joseph A, Kedar E, Porgador A, Mandelboim O. The mechanisms controlling the recognition of tumor and virus infected cells by NKp46. Blood. 2004;103:664–672. doi: 10.1182/blood-2003-05-1716. [DOI] [PubMed] [Google Scholar]
- 14.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 15.Ho JW, Hershkovitz O, Peiris M, Zilka A, Bar-Ilan A, Nal B, Chu K, Kudelko M, Kam YW, Achdout H, Mandelboim M, Altmeyer R, Mandelboim O, Bruzzone R, Porgador A. H5-type influenza virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKp44. J Virol. 2008;82:2028–2032. doi: 10.1128/JVI.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esin S, Batoni G, Counoupas C, Stringaro A, Brancatisano FL, Colone M, Maisetta G, Florio W, Arancia G, Campa M. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surfaces of mycobacteria and other bacteria. Infect Immun. 2008;76:1719–1727. doi: 10.1128/IAI.00870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobigs M, Mullbacher A, Regner M. MHC class I up-regulation by flaviviruses: Immune interaction with unknown advantage to host or pathogen. Immunol Cell Biol. 2003;81:217–223. doi: 10.1046/j.1440-1711.2003.01161.x. [DOI] [PubMed] [Google Scholar]
- 18.Azeredo EL, De Oliveira-Pinto LM, Zagne SM, Cerqueira DI, Nogueira RM, Kubelka CF. NK cells, displaying early activation, cytotoxicity and adhesion molecules, are associated with mild dengue disease. Clin Exp Immunol. 2006;143:345–356. doi: 10.1111/j.1365-2249.2006.02996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green S, Pichyangkul S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Nisalak A, Kurane I, Rothman AL, Ennis FA. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis. 1999;180:1429–1435. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 20.Kurane I, Hebblewaite D, Brandt WE, Ennis FA. Lysis of dengue virus-infected cells by natural cell-mediated cytotoxicity and antibody-dependent cell-mediated cytotoxicity. J Virol. 1984;52:223–230. doi: 10.1128/jvi.52.1.223-230.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell KS, Yusa S, Kikuchi-Maki A, Catina TL. NKp44 triggers NK cell activation through DAP12 association that is not influenced by a putative cytoplasmic inhibitory sequence. J Immunol. 2004;172:899–906. doi: 10.4049/jimmunol.172.2.899. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Nathan D, Lustig S, Tam G, Robinzon S, Segal S, Rager-Zisman B. Prophylactic and therapeutic efficacy of human intravenous immunoglobulin in treating West Nile virus infection in mice. J Infect Dis. 2003;188:5–12. doi: 10.1086/376870. [DOI] [PubMed] [Google Scholar]
- 23.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloushtain N, Qimron U, Bar-Ilan A, Hershkovitz O, Gazit R, Fima E, Korc M, Vlodavsky I, Bovin NV, Porgador A. Membrane-Associated Heparan Sulfate Proteoglycans Are Involved in the Recognition of Cellular Targets by NKp30 and NKp46. J Immunol. 2004;173:2392–2401. doi: 10.4049/jimmunol.173.4.2392. [DOI] [PubMed] [Google Scholar]
- 26.Dennissen MA, Jenniskens GJ, Pieffers M, Versteeg EM, Petitou M, Veerkamp JH, van Kuppevelt TH. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- 27.Hershkovitz O, Jarahian M, Zilka A, Bar-Ilan A, Landau G, Jivov S, Tekoah Y, Glicklis R, Gallagher JT, Hoffmann SC, Zer H, Mandelboim O, Watzl C, Momburg F, Porgador A. Altered glycosylation of recombinant NKp30 hampers binding to heparan sulfate: a lesson for the use of recombinant immunoreceptors as an immunological tool. Glycobiology. 2008;18:28–41. doi: 10.1093/glycob/cwm125. [DOI] [PubMed] [Google Scholar]
- 28.Zilka A, Landau G, Hershkovitz O, Bloushtain N, Bar-Ilan A, Benchetrit F, Fima E, van Kuppevelt TH, Gallagher JT, Elgavish S, Porgador A. Characterization of the heparin/heparan sulfate binding site of the natural cytotoxicity receptor NKp46. Biochemistry. 2005;44:14477–14485. doi: 10.1021/bi051241s. [DOI] [PubMed] [Google Scholar]
- 29.Desprès P, Combredet C, Frenkiel MP, Lorin C, Brahic M, Tangy F. Live measles vaccine expressing the secreted form of the West Nile virus envelope glycoprotein protects against West Nile virus encephalitis. J Infect Dis. 2005;191:207–214. doi: 10.1086/426824. [DOI] [PubMed] [Google Scholar]
- 30.Lisova O, Hardy F, Petit V, Bedouelle H. Mapping to completeness and transplantation of a group-specific, discontinuous, neutralizing epitope in the envelope protein of dengue virus. J Gen Virol. 2007;88:2387–2397. doi: 10.1099/vir.0.83028-0. [DOI] [PubMed] [Google Scholar]
- 31.Boulain JC, Ducancel F. Expression of recombinant alkaline phosphatase conjugates in Escherichia coli. Methods Mol Biol. 2004;267:101–112. doi: 10.1385/1-59259-774-2:101. [DOI] [PubMed] [Google Scholar]
- 32.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 33.Kajaste-Rudnitski A, Mashimo T, Frenkiel MP, Guénet JL, Lucas M, Desprès P. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J Biol Chem. 2006;281:4624–4637. doi: 10.1074/jbc.M508649200. [DOI] [PubMed] [Google Scholar]
- 34.Hershkovitz O, Jivov S, Bloushtain N, Zilka A, Landau G, Bar-Ilan A, Lichtenstein RG, Campbell KS, Kuppevelt TH, Porgador A. Characterization of the Recognition of Tumor Cells by the Natural Cytotoxicity Receptor, NKp44. Biochemistry. 2007;46:7426–7436. doi: 10.1021/bi7000455. [DOI] [PubMed] [Google Scholar]
- 35.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 36.Chau TN, Quyen NT, Thuy TT, Tuan NM, Hoang DM, Dung NT, Lien LB, Quy NT, Hieu NT, Hieu LT, Hien TT, Hung NT, Farrar J, Simmons CP. Dengue in Vietnamese Infants--Results of Infection-Enhancement Assays Correlate with Age-Related Disease Epidemiology, and Cellular Immune Responses Correlate with Disease Severity. J Infect Dis. 2008;198:516–524. doi: 10.1086/590117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 38.Serafin IL, Aaskov JG. Identification of epitopes on the envelope (E) protein of dengue 2 and dengue 3 viruses using monoclonal antibodies. Arch Virol. 2001;146:2469–2479. doi: 10.1007/s007050170017. [DOI] [PubMed] [Google Scholar]
- 39.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Orange JS, Fassett MS, Koopman LA, Boyson JE, Strominger JL. Viral evasion of natural killer cells. Nat Immunol. 2002;3:1006–1012. doi: 10.1038/ni1102-1006. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-Botet M, Llano M, Navarro F, Bellon T. NK cell recognition of non-classical HLA class I molecules. Semin Immunol. 2000;12:109–119. doi: 10.1006/smim.2000.0213. [DOI] [PubMed] [Google Scholar]
- 42.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, Moretta L, Moretta A, Biassoni R. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantoni C, Ponassi M, Biassoni R, Conte R, Spallarossa A, Moretta A, Moretta L, Bolognesi M, Bordo D. The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure (Camb) 2003;11:725–734. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 44.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scalzo AA. Successful control of viruses by NK cells--a balance of opposing forces? Trends Microbiol. 2002;10:470–474. doi: 10.1016/s0966-842x(02)02441-1. [DOI] [PubMed] [Google Scholar]
- 46.Chaturvedi P, Singh AP, Batra SK. Structure, evolution, and biology of the MUC4 mucin. FASEB J. 2008;22:966–981. doi: 10.1096/fj.07-9673rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuchroo VK, Umetsu DT, DeKruyff RH, Freeman GJ. The TIM gene family: emerging roles in immunity and disease. Nat Rev Immunol. 2003;3:454–462. doi: 10.1038/nri1111. [DOI] [PubMed] [Google Scholar]
- 48.Quaresma JA, V, Barros L, Pagliari C, Fernandes ER, Guedes F, Takakura CF, Andrade HF, Jr, Vasconcelos PF, Duarte MI. Revisiting the liver in human yellow fever: virus-induced apoptosis in hepatocytes associated with TGF-beta, TNF-alpha and NK cells activity. Virology. 2006;345:22–30. doi: 10.1016/j.virol.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 49.Arjona A, Ledizet M, Anthony K, Bonafe N, Modis Y, Town T, Fikrig E. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J Immunol. 2007;179:8403–8409. doi: 10.4049/jimmunol.179.12.8403. [DOI] [PubMed] [Google Scholar]
- 50.Vargin VV, Semenov BF. Changes of natural killer cell activity in different mouse lines by acute and asymptomatic flavivirus infections. Acta Virol. 1986;30:303–308. [PubMed] [Google Scholar]
- 51.Shrestha B, Samuel MA, Diamond MS. CD8+ T cells require perforin to clear West Nile virus from infected neurons. J Virol. 2006;80:119–129. doi: 10.1128/JVI.80.1.119-129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]