Abstract
Csk and Src protein tyrosine kinases are structurally homologous, but use opposite regulatory strategies. The isolated catalytic domain of Csk is intrinsically inactive and is activated by interactions with the regulatory SH3 and SH2 domains, while the isolated catalytic domain of Src is intrinsically active and is suppressed by interactions with the regulatory SH3 and SH2 domains. The structural basis for why one isolated catalytic domain is intrinsically active while the other is inactive is not clear. In this current study, we identify the structural elements in the N-terminal lobe of the catalytic domain that render the Src catalytic domain active. These structural elements include the α-helix C region, a β-turn between the β-4 and β-5 strands, and an Arg residue at the beginning of the catalytic domain. These three motifs interact with each other to activate the Src catalytic domain, but the equivalent motifs in Csk directly interact with the regulatory domains that are important for Csk activation. The Src motifs can be grafted to the Csk catalytic domain to obtain an active Csk catalytic domain. These results, together with available Src and Csk tertiary structures, reveal an important structural switch that determines the kinase activity of a catalytic domain and dictates the regulatory strategy of a kinase.
Keywords: Csk, Src, domain-domain interaction, kinase regulation, chimeric kinases
Protein tyrosine kinases (PTKs)1 are a large family of enzymes that catalyze the same chemical reaction: phosphorylation of proteins on Tyr residues. Yet each PTK plays a specific role in the signaling network of a mammalian cell. A key to the functional specificity of each kinase is its ability to respond to different regulatory signals. Even though the regulatory mechanisms in the kinome are highly variable, they can be classified into two general categories. Some kinases contain intrinsically inactive catalytic domains, such as Csk (1,2) and Fps (3), and the regulation of such kinases is based on the activation of the catalytic activity. Other kinases, such as Src (6) and Abl (7–8), contain intrinsically active catalytic domains (4, 5) and their regulation is often based on inactivation of the catalytic activity. Recent studies reveal that the SH2 domain of Abl may also stimulate the kinase activity through interaction with the N-terminal lobe of the catalytic domain (3). In all these scenarios, the intrinsic activity of the catalytic domain is a central feature that dictates the regulatory strategy of a kinase.
Src and Csk are two protein tyrosine kinases that typify these opposite regulatory strategies, even though they have a similar structural organization of SH3-SH2-catalytic domains. The catalytic domain of Src alone is highly active (4, 5), and the activity is suppressed by domain-domain interactions when a C-terminal Tyr (Tyr527) is phosphorylated (9, 10). This inactive form is the predominant and basal form in most cells, and can be activated by several mechanisms (11). For example, binding of the SH2 (12, 13) or the SH3 domain (14, 15) to external ligands, autophosphorylation in the activation loop (on Tyr416) (16), or binding to an activated α subunit of some G-proteins (17) can activate Src. In contrast, the catalytic domain of Csk alone displays minimal kinase activity (1). Extensive interactions between the catalytic domain and the regulatory SH3 and SH2 domains activate the catalytic activity (1, 2, 18–20). Furthermore, the Csk SH2 domain can bind to the Csk-binding protein, which further activates Csk activity (18, 21, 22). Thus the domain-domain interactions in Csk also serve as a basic platform for Csk regulation, albeit in a way generally opposite to that in Src. The crystal structures of Csk and Src family kinases reflect the two regulatory strategies. In full-length Csk structure (23), the SH3 and SH2 domains are located on top of the catalytic domain, and form direct interactions with residues from the N-terminal lobe of the catalytic domain. The catalytic domain alone adopts a conformation typical for an inactivated kinase (24). Several crystal structures of inactive Src and other Src family kinases (10, 25, 26) and one activated full-length Src (27) are available, providing clues to how domain-domain interactions regulate Src kinase activity.
A key question regarding kinase regulation that is yet to be answered is why some catalytic domains are intrinsically active while others are intrinsically inactive. The catalytic domain of a kinase contains two lobes, a smaller N-terminal lobe (N-lobe: about 80 aa) and a larger C-terminal lobe (C-lobe: about 180 aa). The cleft between the two lobes contains the ATP-binding site and harbors many catalytically essential residues (28). The C-lobe contains the motifs for recognizing protein substrates (29–32). The N-lobe contains most of the residues in the ATP-binding pocket and many of the catalytically essential residues, and is considered more conserved among PTKs. However, two recent studies suggest that the N-lobe may contain structural determinants that dictate the kinase activity of a catalytic domain. Using molecular dynamics free energy calculations, Banavali and Roux (33) show that the N-terminal end of the Hck catalytic domain contains a conformational switch implicated in long-range allosteric regulation. A recent study (34) of Csk and Src chimeric kinases indicates that the N-lobe dictates the kinase activity. Both studies implicate the N-lobe of the catalytic domain as the key determinant for the regulatory strategy of a kinase.
In the current study, we performed mutagenic and grafting experiments to identify the structural motifs that dictate the opposing regulatory mechanisms in Csk and Src family kinases. The results demonstrate that the N-lobe of Src contains regulatory structural elements that enable the catalytic domain to be intrinsically active, and such structural motifs can be grafted to the Csk catalytic domain to result in an active Csk catalytic domain. These results provide insights into the structural divergence in the N-lobes that dictates the regulatory strategy of kinases.
Results
Strategy for dissecting the regulatory architecture of Csk and Src
A central question in understanding the different regulatory mechanisms of Src and Csk is why the Src catalytic domain is active while the Csk catalytic domain is inactive. A previous study (27) reveals that the N-lobe dictates the activity level of a catalytic domain. While the catalytic domain of Csk has minimal kinase activity, a chimeric kinase composed of the Src N-lobe and the Csk C-lobe has kinase activity comparable to that of the catalytic domain of Src. These observations indicate that the Src N-lobe contains structural elements that render a catalytic domain active.
Figure 1A shows a comparison between the amino acid sequences of the N-lobes of Csk and Src. The N-lobe of a protein tyrosine kinase catalytic domain contains five β-strands, one α-helix and several loops and turns connecting them. Shown in Figure 1B is the secondary structure of Src N-lobe. To identify the activating structural motifs in the Src N-lobe, we employed the following strategy. The inactive catalytic domain of Csk (C0) was used as the basal form into which various portions of the Src N-lobe, individually or in combination, were grafted to replace the Csk counterparts (Figure 2). Analysis of kinase activity of these chimeric kinases helps locate the motifs of Src that would activate the Csk catalytic domain. When appropriate, the identified motifs will be investigated further by more specific mutations on the background of Src or Csk catalytic domains. By comparing and contrasting the activities of various mutants, the role of specific motifs in regulating the kinase activity can be deciphered. The ultimate goal is to identify the minimal structural elements from Src that can activate the Csk catalytic domain and to understand the role of these motifs in the regulation of Src and Csk.
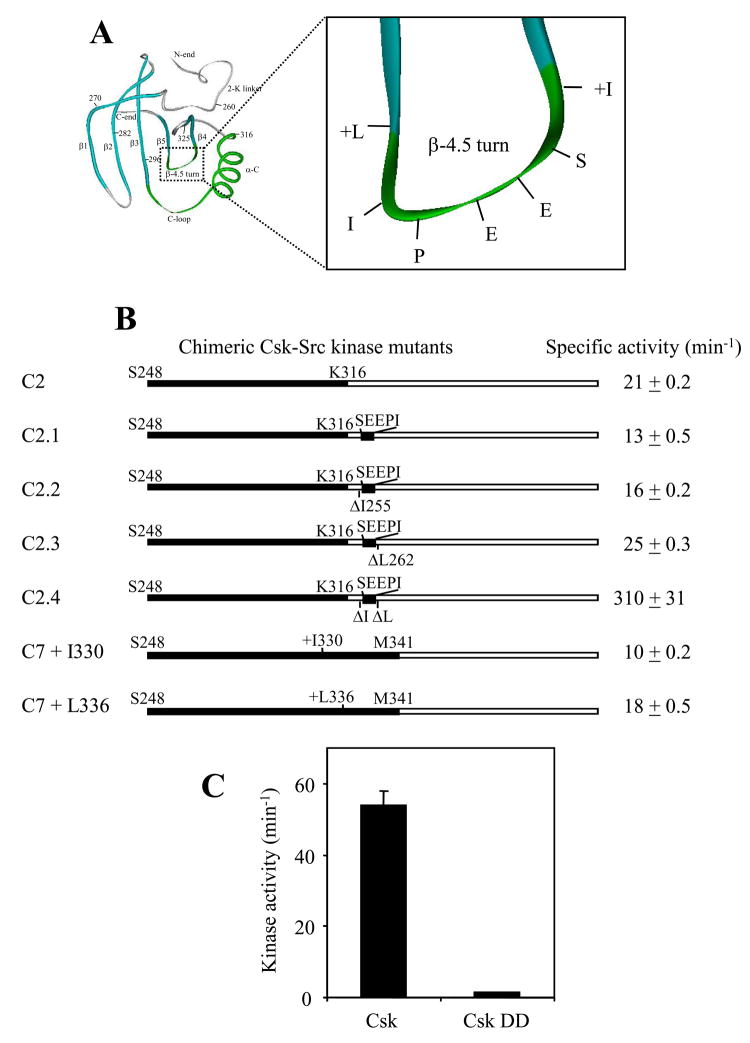
Figure 1. Primary and secondary structures of the kinase N-lobe.
A. Alignment of the amino acid sequences of the N-lobes of the catalytic domains of Csk and Src. The Csk residue numbering and the Src residue numbering are indicated above and below the sequences, respectively. The secondary structures of the Src N-lobe are indicated at the bottom of the illustration. The three motifs of particular interest to this study are colored green. B. A ribbon structure of the N-lobe of Src catalytic domain with all relevant secondary structures indicated. The residue R264 is also shown.
Figure 2. Chimeric walking to identify important regions of the Src N-lobe for its activating function.
A. The ribbon structure of the Src N-lobe. The joining residues of various constructs in B are indicated by the residue numbers. B. Chimeric constructs of Csk and Src catalytic domains. The black bars represent the portions of the Src catalytic domain and the white bars represent the portions of the Csk catalytic domain. The residues numbers indicate the starting and ending residues of the Src portions that replace the counterparts of Csk. The kinase activities with standard errors refer to the specific activity (min−1) under standard assay conditions as defined in Materials and Methods. The reported values are calculated from at least three independent determinations. For reference, full-length Src and Csk have kcat values of 260 ± 10 min−1 and 150 ± 27 min−1, respectively, with ATP as the variable substrate at 1 mg ml−1 polyE4Y (34).
Walking chimeras suggest motifs in the N-lobe that are crucial for regulation
The chimeric constructs of Csk catalytic domain containing various grafts of Src are illustrated in Figure 2. Replacing various fragments of the Csk catalytic domain with the Src counterparts resulted in recovery of different levels of kinase activity, and analysis of these mutants suggests three regions of Src that are potentially important for activating a catalytic domain. Region 1 includes S248-E270 (based on Src numbering). A comparison between C6 and C7 in Figure 2 suggests that this region is crucial for the function of the Src N-lobe. Region 1 includes part of the linker between the SH2 domain and the catalytic domain (S248-A259) and the beginning portion of the catalytic domain that contains the highly conserved Trp residue (W260-E270). The amino acid sequences in this region differ significantly between Csk and Src (Figure 1). Region 2 is the fragment of T297-K316, which results in an increase of activity from 0.03 min−1 (C1) to 21 min−1 (C2). This region includes the α-helix C and a loop N-terminal to the α-helix C (C-loop). The α-helix C is a crucial motif for kinase regulation, and its conformation directly correlates to the activity of a catalytic domain (34). Region 3 is the fragment of Y326-M341, which causes an increase in kinase activity from 20 to 170 min−1 (C3 versus C7). Region 3 contains parts of the β4 and β5 strands, and the turn between them (β-4.5 turn). The identification of these regions may not be exhaustive and other portions of the N-lobe could also contribute to the activation of a catalytic domain. However, this analysis provides clues to the potentially important motifs for further analysis.
Region 3: β-4.5 turn is a major motif that differentiates Src and Csk regulation
There are several structural differences between Csk and Src in region 3. One difference is the β-4.5 turn that has the sequence of SEEPI in Src and EEKGG in Csk. Both the β-4 and β-5 strands are also longer by one residue in Csk than in Src. In addition, two residues in Csk, L252G, are replaced by Y326A in Src. To investigate the role of the β-4.5 turn region, these structural features of Src were individually added into C2 to determine what structural features are important for the kinase activation (Figures 3A and 3B). The Csk β-4.5 turn in C2 (EEKGG) was first replaced by the counterpart of Src (SEEPI), however, this mutation resulted in a slight decrease in the kinase activity (C2.1). This suggests that either the β-turn sequence is not important or the correct sequence in the β-4.5 turn needs to be positioned correctly for it to stimulate the kinase activity. We then deleted the residues I255 and L263 individually and in combination from C2.1 to test if shorter β-4 andβ-5 strands are important for the activation. Deletion of I255 resulted in a recovery of activity from 13 min−1 to 16 min−1, while deletion of L262 resulted in an increase to 25 min−1. When both residues were deleted, the mutant displayed an activity of 310 min−1. These results indicate that the shorter β4 and β5 strands are crucial for a functional Src N-lobe.
Figure 3. Dissection of region 3.
A. Illustration of the region and residue studied in region 3, i.e., the β-4.5 turn region. B. Kinase mutants based on C2 and C7 from Figure 2 were generated to determine what structural features of the region 3 are important for the activating function of the Src-N-lobe. A “+” in front of residue indicates an insertion and a “Δ” indicates a deletion. Otherwise the indicated mutation is a substitution of the Src motif for the Csk counterpart. C. Kinase activity of full-length Csk and a Csk mutant that contains a double deletion of I255 and L262 (Csk DD).
To confirm that the shorter β4 and β5 are indeed important for the activity of Src, two mutants based on C7 were generated, inserting a single residue, Ile after V329, (C7+I330) or Leu after I335 (C7+L336) (Figure 3B). If the shorter β4 and β5 strands are important for the activating function of the Src N-lobe, then insertions to lengthen the strands should inactivate Src N-lobe. Indeed, each insertion decreased the activity of C7 over 90%, indicating that the longer β4 or β5 strands severely impair the function of the N-lobe of Src.
The crucial role of the shorter β4 and β5 strands for the Src N-lobe raises the question why Csk has the longer β4 and β5 strands. We tested if the longer β4 and β5 are essential for the activation of full-length Csk (Figure 3C). Double deletion of I255 and L262 in full length Csk resulted in a decrease in kinase activity from 54 to 1.3 min−1, confirming that the longer β4 and β5 strands are essential for the activation of full-length Csk. In the full-length Csk structure, the β-4.5 turn directly interacts with an α-helix from the SH2 domain, and the shorter β4 and β5 would likely disable this interaction, abolishing the activation of Csk by the SH2 domain. Indeed mutation of the residue from the SH2 domain α-helix, E154A, in full length Csk also abolishes Csk activity (18). These results collectively indicate that the interaction between β-4.5 turn and the SH2 domain in Csk is important for the activation of Csk. Together, the above analysis indicates that the β-4.5 turn is a crucial structure for the regulation of both Src and Csk, but plays distinct roles in these two enzymes.
An intriguing observation from the previous section was that C2.4 was even more active than C7 (Figure 3B). This was surprising because the less active C7 contains the whole Src N-lobe, while C2.4 is similar to C7 with four N-lobe residues replaced by the Csk counterparts. The four residues in C7, E320, K321, Y326 and A327, are replaced by Ser, Asn, Leu and Gly, respectively in C2.4. E320 and K321 are located in the loop connecting the α-helix C and β-4, while Y326 and A327 are located in the middle of β-4. This suggests that some of these four Src residues might suppress the kinase activity in Src. To test this hypothesis, two sets of mutants were constructed (Table 1). First these four residues in C7 were individually mutated into the Csk counterparts. Three of these mutations activated C7, indicating that these residues indeed suppress Src activity. As an independent test for this conclusion, the four equivalent Csk residues in C2.1 were mutated to the Src residues (Table 1). In all cases the activity significantly decreased, indicating that the Csk residues at these four positions were positive for the kinase activity. These results indicate that even though the N-lobe of Src activates a catalytic domain, it also contains inhibitory residues. Further research is needed to ascertain the suppressive mechanism implicated by these observations.
Table 1. The function of several residues in region three on the kinase activity of C7 and C2.1.
On the left, several indicated residues in C7 were individually mutated from the Src residue to their Csk counterparts. On the right, the equivalent Csk residues were individually mutated to the Src counterparts.
| Src to Csk mutation | Kinase activity (min−1) | Csk to Src mutation | Kinase activity (min−1) |
|---|---|---|---|
| C7 | 170 ± 3 | C2.1 | 13 ± 0.5 |
| C7.E320S | 228 ± 3.3 | C2.1.S246E | 2.1 ± 0.2 |
| C7.K321N | 210 ± 1.4 | C2.1.N247K | 4.5 ± 0.1 |
| C7.Y326L | 346 ± 3.2 | C2.1.L252Y | 1 ± 0.6 |
| C7.A327G | 177 ± 1.2 | C2.1.G253A | 0.4 ± 0.005 |
Region 2: the role of the α-helix C
Region 2 includes the α-helix C and a loop N-terminal to the α-helix C (C-loop). In numerous tertiary structures of various protein kinases, including PTKs and protein Ser/Thr kinases, the conformation of the α-helix C closely correlates to the activity of a kinase (35). When the α-helix C moves closer to the active site, a universally conserved Glu in α-helix C forms a salt bridge with a universally conserved Lys in the active site. This salt bridge is a ubiquitous conformational signature of an active kinase. Thus it is not surprising that the α-helix C region plays an important role in the activation of the Src catalytic domain. The crucial question is what structural features in the α-helix C region contribute to the regulatory differences in Csk and Src.
Two structural features in this region differ in Src and Csk. First, the C-loop contains five residues in Src (P209-S303) and four residues in Csk (N226-T229), and the two C-loops share no sequence homology (Figures 1 and 4A). In addition, the first two residues, in the α-helix C are also different in Src (P304 and E305) and Csk (A230 and Q231). In the tertiary structure of Src, this loop interacts with the β-4.5 turn. In contrast, the Csk C-loop interacts with an α-helix from the SH2 domain. To test the importance of C-loop in Src, the five residues in C-loop and the first two residues in the α-helix C in C7 as a block were mutated to the corresponding Csk residues (PGTMSPE to NDATAQ, the mutant is named C7-C-loop) (Figure 4A). The activity decreased to approximately 20 min−1 from 170 min−1 (Figure 4B). This suggests that these residues may play a significant regulatory role, but are not structurally or catalytically essential. The activity of C7-C-loop is similar to the C2 mutant that differs from C7 in the β-4.5 turn, while C7 contains a Src β-4.5 turn, C2 contains a Csk β-4.5 turn. Considering that the C-loop directly interacts with the β-4.5 turn, and mutating either motif has similar effect on the kinase activity, it appears that that the interaction between C-loop and β-4.5 turn activates the kinase. The C-loop of Csk has been previously tested by mutagenesis (18). Mutations of K225, D227 and T229 in the Csk C-loop to Ala cause severe losses of activity consistent with removing the regulatory SH2 domains, indicating that the interaction of this region with the SH2 domain is especially important for the function of Csk. Taken together, these results suggest that the C-loops are important motifs for the regulation of both Src and Csk, but they play different roles in the regulation.
Figure 4. Effects of mutations in the α-helix C region in chimeric kinase C7 and full-length Csk.
A. Diagram showing the Src residues studied in this experiment. PGTMSPE were mutated in block to the Csk counterparts, NDATAQ, in C7 C-loop, while four Src residues (Q309, Q312, K315 and K316) on the α-helix C in C7 were mutated to Csk residues (A, S, T, and M, respectively) (C7 QQKK). B. Effects of the mutations on the kinase activity of C7. C. Unique residues in the α-helix C of full-length Csk were individually mutated to Ala, and the effects of these mutations were determined.
The second difference between Src and Csk in region 2 is the α-helix C. While the α-helix C is largely conserved between Csk and Src, the two α-helixes differ by four residues in addition to the two N-terminal residues. The Src α-helix C contains Q309, Q312, K315 and K316, while the Csk α-helix C contains A235, S238, T241 and Q242 at the corresponding positions (Figure 4A). Two of these Csk residues, T241 and Q242, form hydrogen bonds with the peptide backbone of the linker between the SH3 and the SH2 domains in full length Csk (23). These structural observations provide clues to the different roles of the α-helix C in Csk and Src. To test the functional importance of the sequence variation in the α-helix C, Q309, Q312, K315 and K316 in C7 were mutated into the equivalent residues in Csk (Ala, Ser, Thr and Gln, respectively, the mutant is referred to as C7 QQKK in Figure 4B). This mutant displays a kinase activity level of approximately 35 min−1, indicating that these residues are also important for the activation of Src activity. These results indicate that the Src-specific residues play important roles for the activation of Src. We next tested if the interactions of the α-helix C residues with the SH3-SH2 linker were important for the Csk activity. The mutations of T241 or Q242 to Ala decreased the activities to 1 and 7 min−1, respectively from the activity of more than 50 min−1 for the full-length Csk (Figure 4C). These results indicate that the interactions between the α-helix C and the SH2-SH3 linker are important for Csk activation. Taken together, this mutational analysis indicates that the α-helix C region in Csk and Src have acquired distinct functionalities suited for the specific regulation of each kinase.
Region 1: R264 is an important residue for Src activation by interacting with β-4.5 turn
We next examined the contribution by region 1. This region appeared to be essential for the ability of Src N-lobe to activate Csk based on our initial walking data (Figure 2). The C7 construct contains 12 residues N-terminal to the start (W260) of the catalytic domain. We first deleted these 12 residues (S248-A259), resulting in C8 (Figure 2). C8 displayed a kinase activity of 84 ± 12 min−1, about half of that of C7 (Figure 2). Ala scanning was performed on the region 1 residues (W260 through S266) in C8 that are different from those in Csk. The mutation of R264A had the most effect among these mutants in decreasing the kinase activity of the Src-Csk chimera (Figure 5B). An examination of the Src crystal structure indicates that R264 points to the β-4.5 turn (10, 27), and forms hydrogen bonds with the peptide backbone oxygens of E331 and P333, and a side chain oxygen of E331 (Figure 5C). Both E331 and P333 are located in the β-4.5 turn. Both the structural and mutagenic data support the notion that R264 is involved in activating Src by its interaction with the β-4.5 turn. Indeed, Abl, whose catalytic domain alone is active, also contains an Arg residue (R239) at the position equivalent to R264 of Src. R239 of Abl also forms hydrogen bonds with the backbone oxygens of two residues from the β-4.5 turn, E308 and P310. These residues are equivalent to E331 and P333 of Src. In contrast, the catalytic domains that are not active in the absence of the regulatory domains, Csk and Fps, do not contain an Arg residue at the equivalent position. These results and correlations further support the notion that the interaction between R264 and β-4.5 turn are important for an active catalytic domain.
Figure 5. The kinase activity of Ala scanning mutants in region 1.
A. The N-lobe of Src catalytic domain. The seven-residue fragment, W260 through S266 studied in this experiment is indicated. B. The kinase activity of Src-Csk chimeric kinase C8 and designated Ala scanning mutants based on C8 are shown. All the activities are expressed as a percentage of that of C8. C. Interaction between R264 and the β-4.5 turn by four hydrogen bonds.
Acquisition of the Src structural elements from the N-lobe activates the catalytic domain of Csk
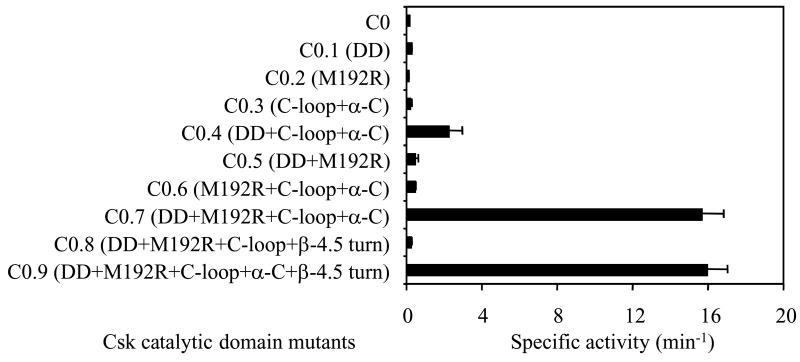
The above analysis identified three regulatory structural motifs that are crucial for the activating role of the Src N-lobe: the β-4.5 turn, the α-helix C region, and R264. To test if these elements are sufficient for the activation of a catalytic domain, we grafted various combinations of these motifs from Src to the Csk catalytic domain and analyzed what combination would be sufficient to activate the Csk catalytic domain. Grafting any one of these three structural elements individually or any combination of two did not activate the Csk catalytic domain (Figure 6). However, the combination of the double deletion of I255 and L262, M129R point mutation, and the transfer of the α-helix C region resulted in a significant activation of Csk catalytic domain (Figure 6, C0.9). It is interesting to note that the precise sequence of the β-4.5 turn appears insignificant for the activation of the Csk catalytic domain, as indicated by the comparable kinase activities of C0.7 and C0.9. These two constructs differ only in the β-4.5 turn, while C0.7 contains the original Csk β-4.5 turn, C0.9 contains the Src α-4.5 turn. A comparison between C0.8 and C0.9 indicates that the Src α-helix C is also necessary for the activation, even though the Src and Csk α-helix C’s differ by only four residues (Q, Q, K, and K in Src versus A, S, T, and Q in Csk).
Figure 6. Grafting of various Src N-lobe structural motifs to the catalytic domain of Csk.
Various Src motifs, and their combinations were grafted to the catalytic domain of Csk to determine if any motifs individually or in combination can activate the Csk catalytic domain. DD, double deletion of I255 and L262; M129R, M129 of catalytic domain mutated to R. Other motifs (C-loop, α-C, β-4.5 turn) are as indicated in Figure 1.
The above analysis of the kinase activity of various mutants has used polyE4Y as a substrate. PolyE4Y is a commercial random copolymer of Glu and Tyr in the ratio of 4:1, and contains a mixture of ill-defined phosphorylation sites. While phosphorylation of polyE4Y provides a convenient way to measure the kinase activity, its interaction with Csk does not represent the docking-based recognition between Csk and Src (29–32). Thus, we wanted to test if the artificially activated Csk catalytic domain could phosphorylate the physiological substrate of Csk, Src (a kinase defective version of Src, kdSrc, was used) (Table 2). C0.9 displayed a kcat of 136 min−1 and a Km of 30 μM. The Km value is somewhat higher than those of the full-length and the catalytic domain of Csk, but roughly comparable. The kcat is approximately 160-fold higher than that of the catalytic domain of Csk (C0 in Table 2), and comparable to that of full length Csk. These results demonstrate that not only the catalytic domain of Csk is activated by the grafted structural elements from Src, it also retains the ability to specifically phosphorylate its physiological substrate. This is consistent with the finding that the site of substrate recognition of Csk is located in the C-lobe of the catalytic domain (29–32).
Table 2. Catalytic parameters of full-length Csk (Csk), the catalytic domain of Csk (C0) and the catalytic domain of Csk with Src structural elements (C0.9).
The catalytic parameters of Csk and mutants using kdSrc as a variable substrate (2–10 μM) and ATP as a fixed substrate (0.2 mM) were determined as described in Materials and Methods. C0 refers to the catalytic domain of Csk, and C0.9 refers to the catalytic domain of Csk with all grafted Src elements (DD+M192R+C-loop+α-C +β-4.5 turn) as shown in Figure 6.
| Enzyme | kcat (min−1) | Km for kdSrc (μM) |
|---|---|---|
| Csk | 113 ± 23 | 10 ± 0.5 |
| C0 | 0.84 ± 0.3 | 8.3 ± 3 |
| C0.9 | 136 ± 14.8 | 30 ± 8.6 |
Discussion
Protein kinases, containing more than 500 members, represent the largest family of enzymes in the human proteome (36). All protein kinases catalyze a similar reaction, the phosphorylation of proteins on Tyr, Ser, or Thr residues. Yet each kinase plays a specific role in cellular signaling, due to its individual substrate specificity and the spectrum of regulatory signals each kinase responds to. Each kinase evolutionarily acquires unique structural elements that confer unique substrate specificity and regulation without compromising the general catalytic machinery. Identifying and understanding the functional implications of the structural motifs are important for understanding signaling by kinases. In this study, we focused on identifying unique structural elements that dictate the kinase activity levels of the Csk and Src catalytic domains, which dictate their regulatory strategies.
The catalytic activity of a kinase can be determined at three levels: substrate recognition, catalysis or regulation. Substrate recognition plays a major role in restricting the catalytic activity of Csk toward certain substrates. While Csk is efficient in phosphorylating Src family kinases, such as Src and Lck, it displays approximately 50,000 fold less activity toward small peptides mimicking the phosphorylation site of these protein substrates (37). Even toward optimized consensus peptide substrate for Csk, the Km is still more than 100 times higher than the protein substrates. Two reasons account for this deficiency in Csk activity toward peptide substrates. First, Csk recognition of Src family proteins relies on extensive remote docking interactions a peptides substrate cannot fulfill (29–32). Second, Csk has a shorter activation loop than other kinases, which disrupts the anchoring of peptide substrates (32). The combination of these two mechanisms restricts Csk substrates to those that can be positioned for phosphorylation through extensive docking interactions. PolyE4Y is an excellent substrate for Csk most likely for the same reason, even though the specific interactions between Csk and polyE4Y have not been identified. However, the lack of activity by the Csk catalytic domain is not likely solely due to a lack of substrate recognition, as the catalytic domain of Csk contains all the substrate-docking motifs and is fully capable of recognizing Src protein as a substrate (32). Furthermore, the Csk catalytic domain also contains all the catalytically important residues, thus the lack of activity by the Csk catalytic domain is likely due to a conformational rather than a primary structural defect. Based on these considerations, we hypothesized that the lack of kinase activity by the catalytic domain of Csk is either the result of missing regulatory mechanisms that maintain an active conformation, or inhibitory motifs that keep Csk catalytic domain at an inactive conformation.
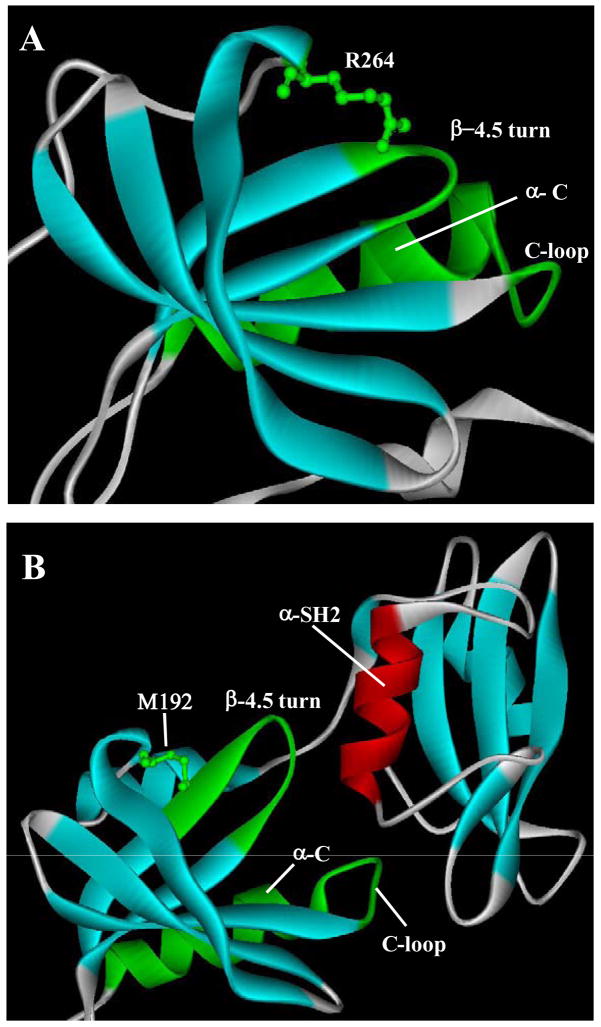
Previously we have identified the N-lobe of the catalytic domain to be responsible for controlling the kinase activity of Csk and Src (34). In this current study, we conducted an extensive dissection of the Src and Csk N-lobe to identify the responsible structural elements. Using a structure-guided grafting approach, we identified three important motifs in the N-lobe that differ structurally in Src and Csk and play distinct roles in their regulation: the α-helix C region, the β-4.5 turn and R264 in Src. These motifs and their interactions are highly divergent in Src and Csk. In the Src structure, the β-4.5 turn interacts with R264 on one side and the C-loop on the other (Figure 7A). Mutation to any of these elements would disable the activating function of the Src N-lobe. It is likely that the interactions among these motifs collectively dictate the conformation of the α-helix C and the active site residues to result in an active catalytic domain. In contrast, the equivalent motifs in Csk are different in primary structure from those of Src, and they do not have interactions similar to those in Src (Figure 7B). First, the β-4 and β-5 strands are longer in Csk, sending the β-4.5 turn further out. Furthermore, Csk contains a Met residue at the position equivalent to the R264 in Src, thus the interaction between the β-4.5 turn and the Arg residue is not present. Second, the combination of changes in the β-4.5 turn and the C-loop region also disables the interaction between the turn and the loop in Csk. Thus, the interactions that are important for the activating function of the Src N-lobe are not present in Csk, resulting in an inactive Csk catalytic domain. Indeed, the signature interactions and conformations of an active kinase, such as the conformation of the DFG motif and the salt bridge between Glu236 and Lys222 are not observed in the catalytic domain of Csk (24). The unique structures of the identified motifs in Csk also appear to set up the activating interactions with the regulatory SH2 domain, as if some of the activating functionalities in Src catalytic domain are transferred to the regulatory domain in Csk. First, the longer β-4 and β-5 strands send the β-4.5 turn further to directly interact with an α-helix in the SH2 domain (α-SH2 in Figure 7B). Second, the C-loop also directly interacts with the α-SH2. Third, the α-helix C in Csk has acquired residues that directly interact with the linker between the SH3 and the SH2 domains. Data presented in this and a previous study (18) demonstrates that each of these observed interactions is important for the activation of full-length Csk. These analyses provide a model for the differential regulations of Csk and Src by domain-domain interactions. It is as if the Src N-lobe contains a fully functional “ON” switch, and the Csk N-lobe contains an incomplete “ON” switch that needs to be complemented by certain structural elements from the regulatory domain for Csk activation.
Figure 7. Ribbon structures of the N-lobes of Src and Csk and their interactions.
A. The structure of the N-lobe of Src. The identified motifs that are important in activating the Src catalytic domain are indicated. B. The structure of the N-lobe of Csk. In addition to the N-lobe motifs, the SH2 domain is also shown to illustrate the interaction between the SH2 domain and the N-lobe.
The most compelling evidence for the correct functional assignment of these motifs is that grafting these structural elements from Src to the catalytic domain of Csk results in an active Csk catalytic domain, while missing any of these elements would result in an inactive catalytic domain. This result indicates that the interactions among these structural elements are indeed sufficient to activate an otherwise inactive catalytic domain. It is unlikely that any catalytic functionality is gained from such grafting, as the Csk catalytic domain is capable of being active when properly activated. We speculate that correct positioning of the α-helix C is likely the key consequence of these grafted interactions.
It is not clear how the interactions that activate the Src catalytic domain are related to the inactivation of Src. Src is inactivated by phosphorylation of a C-terminal Tyr residue, Tyr527 (9, 10). The phosphorylated Tyr527 binds to the SH2 domain, which leads to interactions between the SH3 domain and the SH2-catalytic domain linker (6). Because the identified motifs and their interactions are the key to the Src activation, any conformational changes that disrupt these interactions could result in Src inactivation. Thus, it is conceivable that the binding between phospho-Tyr527 to the SH2 domain may lead to disruption of the activating interactions among the identified motifs. A comparison of the active and inactive Src structures does not reveal any major differences in the conformations and interactions of these motifs. Alternatively, there may be a separate mechanism that disrupts the active conformation. It is interesting to note that the mutation of several residues activates Src, suggesting that these residues may be involved in suppressing Src kinase activity (Table 1). Further investigation is required to determine the detailed mechanisms of Src inactivation.
The regulation model developed in this study may apply to other protein tyrosine kinases or protein kinases in general. Accumulating evidence indicates that the N-lobe is a crucial region that determines the regulation of protein kinases (33, 34). Molecular dynamics free energy calculations indicate that the N-terminal end of the catalytic domain of Hck is a conformational switch in the allosteric regulation, although the structural basis for the conformational switch is not determined (33). Several structural studies also point to the same surface area in the N-lobe as key regulatory structures in other protein kinases. For example, similar to the catalytic domain of Csk, all cyclin-dependent kinases contain a catalytic domain that is inactive by itself. Activation is achieved by the binding to a cyclin molecule among other events (38). Among other interactions, the cyclin directly interacts with the β-4.5 turn, the C-loop, and the α-helix C of the cyclin-dependent kinase 2 (39, PDB ID: 1JTS). This regulation is highly analogous to the activating interaction between the Csk catalytic domain and the regulatory domains. Another example is the receptor-type protein tyrosine kinase EGFR, which is activated by the formation of an asymmetric homodimer between two EGFR molecules (40). In the asymmetric dimer, the C-terminal lobe of one kinase domain binds to the β-4.5 turn, C-loop and the α-helix C region of another kinase molecule, analogous to that of cyclin in activated CDK/cyclin complexes. In a recently published structure of v-Fps (3), in which the catalytic domain is activated by the SH2 domain, the SH2 domain also interacts with the α-helix C and the β-4.5 turn region in a way generally similar to that in Csk. These structural observations suggest that β-4.5 turn and its interactions with the C-loop and other motifs may be a focal point of regulatory interactions in many different protein kinases.
Materials and Methods
Reagents and chemicals
All reagents used for bacterial culture and protein expression were purchased from Fisher. The chromatographic resin, iminodiacetic acid-Agarose, was purchased from Sigma, Amylose-Agarose was purchased from New England Biolabs. DNA primers were synthesized by Integrated DNA Technologies. [γ-32P]ATP (6,000 Ci mol−1) was purchased from PerkinElmer.
Plasmid construction and protein expression
Src mutants were expressed as a fusion protein with maltose-binding protein (MBP) using pMAL-c2x expression vector (New England Biolabs) or as (His)6-tagged protein using the pRSET-a vector as indicated. The MBP fusion proteins were purified as described previously (41). The (His)6-tagged Src mutants from the pRSET plasmids were coexpressed with GroES/EL chaperone (pREP4groESL) and protein tyrosine phosphatase 1B (pPCDF-PTP1B). The chaperone helps with the correct folding and increases the solubility of the protein, and the PTP 1B helps reduce toxicity due to Src overexpression. The (His)6-fusion proteins were purified using immobilized Ni2+-iminodiacetic acid-Agarose as described previously (29). Mutant plasmids were constructed using Quikchange from Stratagene, and confirmed by DNA sequencing. Upon confirmation, the plasmids were transformed into appropriate host E. coli cells for protein expression and purification. After each mutant enzyme was obtained, the concentration and purity of the enzyme was determined by Bradford assay and SDS-PAGE, respectively. The purified proteins were stored in 50% glycerol and 25 mM Tris, pH 8.0 at −20 °C.
Kinase assays
For quantifying PTK activities, we measured the phosphorylation of polyE4Y or kdSrc (kinase-defective Src) using the acid precipitation assay (31). PolyE4Y is a random copolymer of Glu and Tyr in the ratio of 4:1 (Sigma). A standard assay reaction of 50 μl contained the kinase and the substrates (1 mg ml−1 polyE4Y, and 200 μM 32P-ATP [~1,000 dpm/pmol]) in the kinase assay buffer (75 mM 4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid [EPPS], pH 8.0, 12 mM MgCl2, 5% glycerol, 0.005% Triton X-100). After the reaction at 30°C for 30 min, 35 μl reaction mixtures were spotted onto filter paper strips (1×2 cm), which were then washed in 5% trichloroacetic acid at approximately 65°C 3-times for approximately 10 min each. Phosphorylated and unphosphorylated polyE4Y was precipitated onto the filter paper and the amount of phosphate incorporated into polyE4Y on the filter paper was determined by liquid scintillation counting. For each mutant enzyme, the activity was first determined at a series of enzyme concentrations to find the concentration range that gave a reasonable signal and a linear relationship between the signal and the enzyme concentration. Then the activity was determined at least three times using enzyme concentrations in the linear range. The specific activity of a mutant enzyme, expressed as the mole of phosphate transferred by each mole of enzyme within a min (min−1) under the standard assay conditions, was calculated from each determination. The average activity and the standard error were calculated from at least three such determinations for each mutant enzyme.
A kinase-defective mutant of Src (kdSrc) was used as a substrate to determine the ability of certain mutants to phosphorylate a physiological substrate of Csk. KdSrc differs from the active Src by only a single residue substitution (K295M), and retains the structural and functional specificity as a substrate for Csk. KdSrc was prepared as described previously (29). The level of kdSrc phosphorylation was determined as described for polyE4Y phosphorylation above except the concentration of kdSrc used was 10 μM. To determine the steady state kinetic parameters of Csk and mutants phosphorylating kdSrc, a range of kdSrc concentrations (2–10 μM) were used, and the kcat and Km values were calculated by Lineweaver-Burk plot. The reported values were based on three independent determinations.
Footnotes
This work was supported by grants from the American Cancer Society (RSG-04-247-01-CDD) and NIH (1RO1CA111687). DNA sequencing was performed at University of Rhode Island Genomics and Sequencing Center.
Abbreviations used: C-lobe, the C-terminal lobe of the catalytic domain of a protein tyrosine kinase; kdSrc, kinase-defective Src; N-lobe, the N-terminal lobe of the catalytic domain of a protein tyrosine kinase; PTKs, protein tyrosine kinases.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sondhi D, Cole PA. Domain interactions in protein tyrosine kinase Csk. Biochemistry. 1999;38:11147–11155. doi: 10.1021/bi990827+. [DOI] [PubMed] [Google Scholar]
- 2.Sun G, Budde RJ. Mutations in the N-terminal regulatory region reduce the catalytic activity of Csk, but do not affect its recognition of Src. Arch Biochem Biophys. 1999;367:167–172. doi: 10.1006/abbi.1999.1253. [DOI] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, Kofler M, Hantschel O, Gish GD, Grebien F, Salah E, Neudecker P, Kay LE, Turk BE, Superti-Furga G, Pawson T, Knapp S. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weijland A, Neubauer G, Courtneidge SA, Mann M, Wierenga RK, Superti-Furga G. The purification and characterization of the catalytic domain of Src expressed in Schizosaccharomyces pombe. Comparison of unphosphorylated and tyrosine phosphorylated species Eur J Biochem. 1996;240:756–764. doi: 10.1111/j.1432-1033.1996.0756h.x. [DOI] [PubMed] [Google Scholar]
- 5.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 7.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 8.Smith KM, Yacobi R, Van Etten RA. Autoinhibition of Bcr-Abl through its SH3 domain. Mol Cell. 2003;12:27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper JA, Howell B. The when and how of Src regulation. Cell. 1993;73:1051–1054. doi: 10.1016/0092-8674(93)90634-3. [DOI] [PubMed] [Google Scholar]
- 10.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 11.Cooper JA, Gould KL, Cartwright CA, Hunter T. Tyr527 is phosphorylated in pp60c-src: implications for regulation. Science. 1986;231:1431–1434. doi: 10.1126/science.2420005. [DOI] [PubMed] [Google Scholar]
- 12.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 13.Muthuswamy SK, Muller WJ. Activation of Src family kinases in Neu-induced mammary tumors correlates with their association with distinct sets of tyrosine phosphorylated proteins in vivo. Oncogene. 1995;11:1801–1810. [PubMed] [Google Scholar]
- 14.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 15.Sun G, Ramdas L, Wang W, Vinci J, McMurray J, Budde RJ. Effect of autophosphorylation on the catalytic and regulatory properties of protein tyrosine kinase Src. Arch Biochem Biophys. 2002;397:11–17. doi: 10.1006/abbi.2001.2627. [DOI] [PubMed] [Google Scholar]
- 16.Sun G, Sharma AK, Budde RJ. Autophosphorylation of Src and Yes blocks their inactivation by Csk phosphorylation. Oncogene. 1998;17:1587–1595. doi: 10.1038/sj.onc.1202076. [DOI] [PubMed] [Google Scholar]
- 17.Ma YC, Huang J, Ali S, Lowry W, Huang XY. Src tyrosine kinase is a novel direct effector of G proteins. Cell. 2000;102:635–646. doi: 10.1016/s0092-8674(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, Wang Y, Ahmadibeni Y, Parang K, Sun G. Structural basis for domain-domain communication in a protein tyrosine kinase, the C-terminal Src kinase. J Mol Biol. 2006;357:1263–1273. doi: 10.1016/j.jmb.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 19.Wong L, Lieser S, Chie-Leon B, Miyashita O, Aubol B, Shaffer J, Onuchic JN, Jennings PA, Woods VL, Jr, Adams JA. Dynamic coupling between the SH2 domain and active site of the COOH terminal Src kinase, Csk. J Mol Biol. 2004;341:93–106. doi: 10.1016/j.jmb.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 20.Wong L, Lieser SA, Miyashita O, Miller M, Tasken K, Onuchic JN, Adams JA, Woods VL, Jr, Jennings PA. Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J Mol Biol. 2005;351:131–143. doi: 10.1016/j.jmb.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 21.Kawabuchi M, Satomi Y, Takao T, Shimonishi Y, Nada S, Nagai K, Tarakhovsky A, Okada M. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature. 2000;404:999–1003. doi: 10.1038/35010121. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi S, Takayama Y, Ogawa A, Tamura K, Okada M. Transmembrane phosphoprotein Cbp positively regulates the activity of the carboxyl- terminal Src kinase, Csk. J Biol Chem. 2000;275:29183–29186. doi: 10.1074/jbc.C000326200. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa A, Takayama Y, Sakai H, Chong KT, Takeuchi S, Nakagawa A, Nada S, Okada M, Tsukihara T. Structure of the carboxyl-terminal Src kinase, Csk. J Biol Chem. 2002;277:14351–14354. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- 24.Lamers MB, Antson AA, Hubbard RE, Scott RK, Williams DH. Structure of the protein tyrosine kinase domain of C-terminal Src kinase (CSK) in complex with staurosporine. J Mol Biol. 1999;285:713–725. doi: 10.1006/jmbi.1998.2369. [DOI] [PubMed] [Google Scholar]
- 25.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 26.Williams JC, Weijland A, Gonfloni S, Thompson A, Courtneidge SA, Superti-Furga G, Wierenga RK. The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions. J Mol Biol. 1997;274:757–575. doi: 10.1006/jmbi.1997.1426. [DOI] [PubMed] [Google Scholar]
- 27.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Huang XY, Cole PA. Molecular determinants for Csk-catalyzed tyrosine phosphorylation of the Src tail. Biochemistry. 2001;40:2004–2010. doi: 10.1021/bi002342n. [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Ayrapetov MK, Kemble DJ, Parang K, Sun G. Docking-based substrate recognition by the catalytic domain of a protein tyrosine kinase, C-terminal Src kinase (Csk) J Biol Chem. 2006;281:8183–8189. doi: 10.1074/jbc.M508120200. [DOI] [PubMed] [Google Scholar]
- 31.Lee S, Lin X, Nam NH, Parang K, Sun G. Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase. Proc Natl Acad Sci U S A. 2003;100:14707–14712. doi: 10.1073/pnas.2534493100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levinson NM, Seeliger MA, Cole PA, Kuriyan J. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134:124–134. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banavali NK, Roux B. The N-terminal end of the catalytic domain of SRC kinase Hck is a conformational switch implicated in long-range allosteric regulation. Structure. 2005;13:1715–1723. doi: 10.1016/j.str.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Wang YH, Huang K, Lin X, Sun G. Subdomain switching reveals regions that harbor substrate specificity and regulatory properties of protein tyrosine kinases. Biochemistry. 2007;46:10162–10169. doi: 10.1021/bi7007257. [DOI] [PubMed] [Google Scholar]
- 35.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 36.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 37.Sondhi D, Xu W, Songyang Z, Eck MJ, Cole PA. Peptide and protein phosphorylation by protein tyrosine kinase Csk: insights into specificity and mechanism. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- 38.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 39.Russo AA, Jeffrey PD, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Wang YH, Ayrapetov MK, Lin X, Sun G. A new strategy to produce active human Src from bacteria for biochemical study of its regulation. Biochem Biophys Res Commun. 2006;346:606–611. doi: 10.1016/j.bbrc.2006.05.180. [DOI] [PubMed] [Google Scholar]