Abstract
Deficiencies in the protein folding capacity of the endoplasmic reticulum (ER) in all eucaryotic cells lead to ER stress and triggers the unfolded protein response (UPR)1–3. ER stress is sensed by Ire1, a transmembrane kinase/endoribonuclease, which initiates the non-conventional splicing of the mRNA encoding a key transcription activator, Hac1 in yeast or XBP-1 in metazoans. In the absence of ER stress, ribosomes are stalled on unspliced HAC1 mRNA. The translational control is imposed by a base pairing interaction between the HAC1 intron and the HAC1 5′ untranslated region (5′UTR)4. After excision of the intron, tRNA ligase joins the severed exons5,6, lifting the translational block and allowing synthesis of Hac1 from the spliced HAC1 mRNA to ensue4. Hac1 in turn drives the UPR gene expression program comprising 7–8% of the yeast genome7 to counteract ER stress. We show here that upon activation, Ire1 molecules cluster in the ER membrane into discrete foci of higher-order oligomers, to which unspliced HAC1 mRNA is recruited by means of a conserved bipartite targeting element contained in the 3′ untranslated region (3′UTR). Disruption of either Ire1 clustering or of HAC1 mRNA recruitment impairs UPR signaling. The HAC1 3′UTR element is sufficient to target other mRNAs to Ire1 foci, as long as their translation is repressed. Translational repression afforded by the intron fulfills this requirement for HAC1 mRNA. Recruitment of mRNA to signaling centers provides a new paradigm for the control of eukaryotic gene expression.
In vitro studies suggest that the information required for HAC1 mRNA splicing is confined to the intron and the regions surrounding the splice junctions8. Surprisingly, in vivo splicing of HAC1 mRNA was greatly diminished when its 3′UTR was replaced by the 3′UTRs of other yeast mRNAs, such as of ACT1 (Fig. 1b) or PGK1 (data not shown). Consistent with this finding, cells bearing a chimeric hac1-3′act1 mutant gene expressed Hac1 protein at trace levels that were too low to mount a functional UPR and failed to grow in ER stress conditions (Fig. 1b). Thus, the HAC1 3′UTR harbors an element important for HAC1 mRNA splicing in vivo.
Figure 1. A conserved element in the 3’UTR of HAC1 mRNA is required for splicing in vivo, but not in vitro.
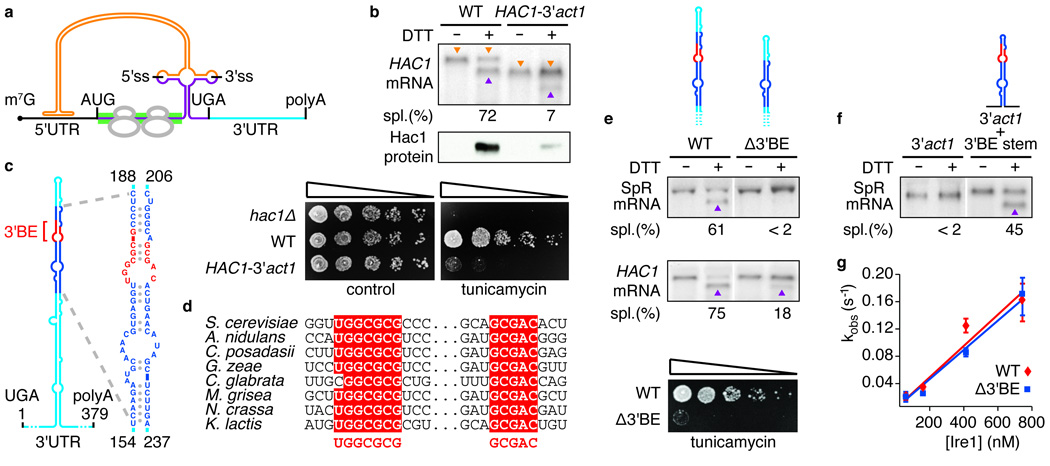
a, Schematic of HAC1 mRNA. The Hac1 ORF is divided into two exons (purple). The intron (orange) base pairs with the 5′UTR (black), causing ribosome stalling (grey). Ire1 cleaves the intron at the indicated splice sites (5′ss & 3′ss). The green bar depicts where the GFP ORF replaces the HAC1 sequence in the splicing reporter. The 3′UTR is indicated in light blue. The 5′cap (m7G), start codon (AUG), stop codon (UGA) and polyadenylation signal (polyA) are indicated. b, e, f, Northern blot of HAC1 or SpR mRNA variants before or after ER stress induction with DTT (10 mM) for 45 min. Purple triangles denote spliced mRNAs; orange triangles denote unspliced mRNAs (only in b). Percent mRNA splicing (“Spl. (%)“) is indicated. Yeast strains harbor a genomic HAC1 copy with its own (WT) or ACT1’s 3′UTR sequence (HAC1-3′act1) (b, top), a genomic copy of SpR (e, top) or HAC1 (e, middle) bearing either the wild-type (WT) or the Δ3′BE mutant 3′UTR, as depicted, or a genomic copy of SpR with the 3’UTR of ACT1 with (3′act1+3′BE stem) or without (3′act1) an insertion of the 64 nucleotide element — shown in expanded view in (c) — as depicted (f). b, middle, Western blot of HA-tagged Hac1 protein from lysates from strains as in (b, top). b,e, Viability assay by 1:5 serial dilutions of hac1Δ or strains as in (b, top) or (e, middle) spotted onto solid media with or without 0.2 µg/ml of the ER stress inducer tunicamycin. Plates were photographed after 3 days growing at 30°C. c, Schematic of the HAC1 3′UTR stem-loop structure with the 3’BE (red) in a region (dark blue) that is shown in expanded view to the right; positional numbering from UGA stop codon. d, alignment of the 3′BE in HAC1 homologues. g, An in vitro intron excision reaction was performed as described8 with Ire1 concentrations: 50 nM, 150 nM, 400 nM, 730 nM of wild-type (red diamonds) or Δ3′BE (blue squares) HAC1 mRNA as substrates.
Mutational probing experiments (not shown) indicate that the HAC1 3′UTR contains a prominent, extended stem-loop (Fig. 1c). Interestingly, two short sequence motifs within the stem-loop are highly conserved among all HAC1 orthologs identified; eight representatives are shown in Figure 1d. The sequence motifs map to opposite strands and are juxtaposed in the distal part of the stem, constituting a bipartite element (3′BE for 3′UTR Bipartite Element) (Fig. 1c, 3′BE in red).
To assess the importance of the 3′BE for HAC1 mRNA splicing in vivo, we employed a splicing reporter (SpR) in which we replaced the first 648 nucleotides of the HAC1 coding sequence in the first exon with that of GFP (Fig. 1a, green bar). This reporter allowed us to monitor the effect of 3′UTR mutations on mRNA splicing in cells that can mount a functional UPR, sustained by endogenous HAC1 mRNA. The SpR mRNA was efficiently spliced upon UPR induction. By contrast, splicing was significantly diminished when the 3′BE (Δ3′BE) was deleted (Fig. 1e). Consistent with these results, deletion of the 3′BE in HAC1 severely reduced HAC1 mRNA splicing and impaired cell survival under ER stress conditions (Fig. 1e). Only residual splicing of endogenous HAC1 mRNA occurred in the absence of the 3′BE, indicating that the 3′BE accounts in large part for the contribution of the 3′UTR to HAC1 mRNA splicing. Insertion of a 64-nucleotide 3′UTR fragment containing the central portion of the stem including the 3′BE (Fig. 1d, enlarged on right) into the SpR bearing the ACT1 3′UTR restored splicing significantly (Fig. 1f).
To test whether the 3′UTR affects the ability of Ire1 endonuclease to bind or catalyze the cleavage of HAC1 mRNA, we reconstituted the intron excision reaction in vitro. Ire1 cleaved HAC1 mRNA with the same rate in the presence or absence of the 3′BE (Fig. 1g). Thus, the 3′BE is not required for splicing in vitro.
The importance of the 3′BE for HAC1 mRNA splicing in vivo suggested it may serve to target the mRNA to sites in the cell where splicing takes place. To test this notion, we visualized Ire1 and HAC1 mRNA in vivo, using the imaging constructs depicted in Figure 2a. For Ire1, we inserted a GFP or mCherry into the cytosolic portion of Ire1 adjacent to its transmembrane region. For HAC1 mRNA, we inserted 16 copies of a U1A binding site into the 3′UTR downstream of the 3′BE. This mRNA can then be visualized by co-expression of a U1A-GFP fusion that docks to the U1A sites. Both Ire1 and HAC1 mRNA imaging constructs fully restored growth of ire1Δ and hac1Δ (Fig. 2b) cells under ER stress. In the absence of stress, Ire1-GFP co-localized with the ER marked by Sec63-mCherry (Fig. 2c). The majority of HAC1U1A mRNA displayed a grainy signal dispersed throughout the cytosol (Fig. 2d), with a fraction of HAC1 mRNA signal also found at the ER in agreement with previous observations9.
Figure 2. In response to ER stress HAC1 mRNA localizes to Ire1 foci in a 3’BE-dependent manner.
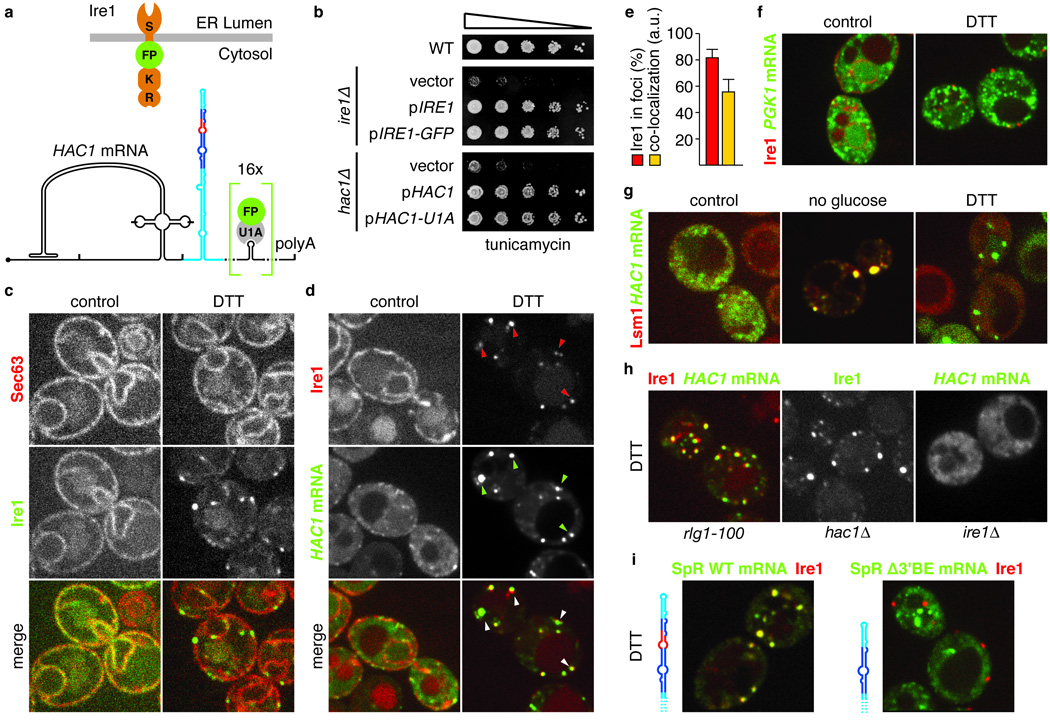
a, Schematic of Ire1 and HAC1 mRNA imaging constructs: Ire1 has an ER-luminal stress sensing domain (S) and a kinase (K) and endonuclease domain (E) at its cytosolic face. GFP or mCherry (FP) was inserted between the transmembrane region and the kinase domain (up). b, Viability assay under ER stress conditions (0.2 µg/ml tunicamycin) of wild-type or ire1Δ yeast complemented with either empty vector, or centromeric plasmids bearing a wild-type (pIRE1), or the GFP-tagged imaging copy of Ire1 (pIRE1-GFP) (top) or hac1Δ yeast complemented with either empty plasmid, or a 2-micron plasmid bearing a wild-type (pHAC1) or the U1A-tagged imaging copy of HAC1 (pHAC1-U1A) (bottom). c,d, Localization of Sec63-mCherry and Ire1-GFP (c) or Ire1-mCherry and HAC1U1A mRNA decorated with U1A-GFP (d) before (left panels, control) and after (right panels, DTT) induction of ER stress. Arrowheads in d, lower panels, denote Ire1/HAC1 mRNA foci. e, Histogram depicting the percentage of Ire1 signal in foci (red bar) and the C.I. for HAC1U1A mRNA recruitment into Ire1 foci expressed in arbitrary units (“a.u.”, yellow bar); means +/− s.e.m., n = 9. f, Localization of Ire1-mCherry and PGK1U1A mRNA decorated with U1A-GFP under normal (left panel, control) and ER stress (right panel, DTT) conditions. g, Localization of Lsm1-mCherry and HAC1U1A mRNA without stress (left panel, control), after nutrient starvation for 10 min (middle panel, no glucose), or after induction of ER stress (right panel, DTT). h,i, Localization of Ire1-mCherry, Ire1-GFP, or HAC1U1A or SpR having 16 U1A hairpins as HAC1U1A (SpRU1A) either with or without the Δ3′BE deletion after induction of ER stress (DTT). c–i, ER stress was induced with 10 mM DTT for 45 min; imaging was performed in ire1Δ cells, complemented with Ire1 imaging constructs, except in (h) cell were hac1Δ or rlg1-100.
Induction of ER stress dramatically altered localization of both Ire1 and HAC1 mRNA. The vast majority (82 ± 6%; see Methods) of Ire1 clustered into distinct foci localized both to the nuclear envelope and the cortical ER (Fig. 2c–e), in agreement with recent observations10. Excitingly, HAC1 mRNA strongly co-localized (co-localization index (“CI”): 56 ± 10; see Methods; Fig. 2e) with Ire1 in foci (Fig. 2d, arrowheads). This recruitment is specific, since control PGK1U1A mRNA remained dispersed in the cytosol under ER stress conditions (Fig. 2f).
Clustering of mRNAs in cytosolic foci is not unprecedented. Several stresses, such as nutrient starvation, cause aggregation of untranslated mRNAs into processing bodies (P-bodies) where they are stored and/or degraded11. The Ire1/HAC1 mRNA clusters however are distinct from P-bodies: Upon glucose depletion, Lsm1-mCherry clustered into P-bodies as reported12, and HAC1 mRNA co-localized with Lsm1-mCherry in these P-bodies. By contrast, under ER stress conditions the Lsm1-mCherry did not co-localize with HAC1 mRNA foci but remained dispersed throughout the cytosol (Fig. 2g). Thus, the Ire1/HAC1 mRNA foci constitute novel sites of mRNA clustering in the cytosol that are specific for the UPR.
We next determined the role of each of the three key UPR players, Ire1, HAC1 mRNA and tRNA ligase in organizing the foci. In rlg1-100 cells bearing mutant tRNA ligase defective in UPR signaling, co-clustering of Ire1 and HAC1 mRNA occurred normally (Fig. 2h). This result is consistent with the fact that cleavage of HAC1 mRNA by Ire1 is not dependent on the subsequent ligation step5. Likewise, HAC1 mRNA was not required for Ire1 clustering, as Ire1-GFP formed foci in hac1Δ cells (Fig. 2h and [ref 11]). Conversely, HAC1 mRNA failed to form foci in ire1Δ cells (Fig. 2h). Thus, clustering of Ire1 in response to ER stress is epistatic to HAC1 mRNA clustering.
Having established that HAC1 mRNA is targeted to Ire1 foci in an ER stress-driven manner, we assessed the role of HAC1 mRNA’s 3′UTR in the process. To this end, we added the U1A visualization module to the SpR used in Figure 1e. The SpRU1A mRNA containing a wild-type HAC1 3’UTR co-localized with Ire1-mCherry in foci (CI: 64 ± 20, Fig. 2i). By contrast, co-localization with Ire1 foci of the SpRU1A mRNA lacking the 3’BE was minimal (CI: 4 ± 6, Fig. 2i) at levels comparable to the control PGK1U1A mRNA (CI: 6 ± 4). Thus, the stem-loop structure in the 3′UTR of HAC1 mRNA – with the 3′BE at its core – indeed serves as a targeting element that guides HAC1 mRNA to Ire1 foci to allow splicing in vivo and cell survival under ER stress.
We next followed a time course of foci formation and downstream signaling upon induction of ER stress. Clustering of Ire1 into foci and recruitment into these foci of HAC1 mRNA (Fig. 3a,b) or of SpRU1A mRNA (Supplementary Fig. S1) correlated well with the onset of HAC1 mRNA splicing (Fig. 3) and Hac1 protein production (Fig. 3c). These findings show that Ire1 and HAC1 mRNA clustering is geared to rapidly transduce ER stress. Under conditions where ER stress builds up more gradually, the encounter of Ire1 and HAC1U1A mRNA in foci likewise paralleled the signaling response, but at a slower pace (Supplementary Fig. S2). The synchronicity of Ire1/HAC1 mRNA clustering and downstream signaling events underscores that the foci constitute functional mRNA splicing centers.
Figure 3. The HAC1 mRNA/Ire1 foci are functional UPR signaling centers.
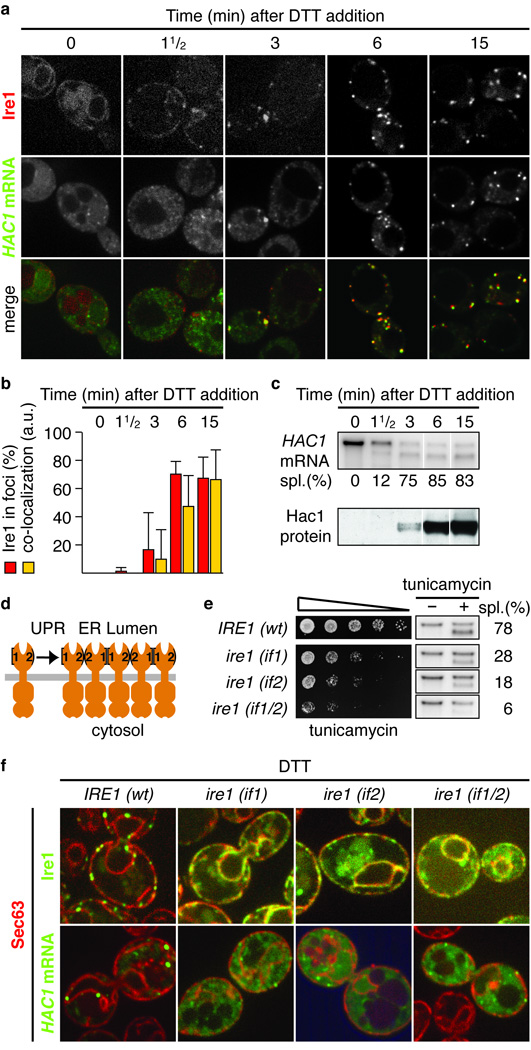
a, Localization of Ire1-mCherry and HAC1U1A mRNA decorated with U1A-GFP. b, Quantitation of the percentage of Ire1 signal in foci (red bars) and of the C.I. for HAC1U1A mRNA recruitment into Ire1 foci (yellow bars; means +/− s.e.m., n = 5). c, Northern blot of HAC1 mRNA (top) and Western blot of Hac1 protein (bottom). a–c, Samples were taken at indicated times after induction of ER stress with 10 mM DTT. d, Schematic of Ire1 oligomerization via interfaces 1 and 2. e, Viability assay under ER stress conditions (0.2 µg/ml tunicamycin) and Northern blot of HAC1 mRNA harvested from ire1Δ yeast complemented with wild-type Ire1 or interface mutants before or after treatment with 1 µg/ml tunicamycin for 1 h. f, Localization of Sec63-mCherry, Ire1-GFP, and HAC1U1A mRNA. Imaging was performed in ire1Δ yeast complemented with wild type or mutants of Ire1 that are defective in dimarization at interface 1 (if1), 2 (if2) or both (if1/2), either GFP-tagged (top) or untagged (bottom). ER stress was induced: 10 mM DTT 45 min. Separate channels are displayed in Supplementary Fig. S3.
Ire1 clusters in only few ~3–10 foci per cell. Because yeast contains ~200–300 molecules of Ire1 per cell13, the foci are composed of a few tens of Ire1 molecules each, suggesting that the foci harbor higher-order oligomers of Ire1. From the crystal structure of the Ire1 ER-luminal domain, we identified two separate dimerization interfaces that both are essential for optimal UPR signaling, suggesting that oligomerization is important for cells to mount a robust UPR14 (Fig. 3d). Accordingly, simultaneous disruption of both interfaces dramatically reduced HAC1 mRNA splicing and cell growth under ER stress conditions, while the single interface disruptions, which still can form Ire1 dimers via one interface, displayed intermediate splicing and growth phenotypes (Fig. 3e). Disruption of either interface prevented foci formation (Fig. 3f, Supplementary Fig. S3, and [ref 10]), indicating that Ire1 oligomerization is the organizing principle for UPR signaling foci. Importantly, Ire1’s inability to form foci impaired HAC1U1A mRNA recruitment (Fig. 3f and Supplementary Fig. S3). Thus, when Ire1 fails to oligomerize, HAC1 mRNA recruitment becomes rate limiting. In agreement, we found that artificially induced dimerization15 of Ire1 supported HAC1 mRNA splicing and cell survival under ER stress conditions only to the level of the single interface mutants and did not support Ire1 foci formation (Supplementary Fig. S4). We conclude that robust Ire1 oligomerization and HAC1 mRNA targeting serve to concentrate both key UPR components into foci to ensure efficient RNA processing and ER stress signaling.
HAC1 mRNA no longer is a substrate for Ire1 after removal of its intron, suggesting that the spliced HAC1 mRNA should disengage from Ire1 foci and not be recruited again. Accordingly, SpRU1A mRNA lacking the intron displayed reduced targeting to foci (CI: 17± 9) as compared to wild-type SpRU1A mRNA (Fig. 4a, b), although not as dramatically reduced as when the 3′BE was deleted (Fig. 2i and Fig. 4b). In further support, over-expression of SpRU1A mRNA containing the intron reduced splicing of endogenous HAC1 mRNA, presumably by competitively saturating Ire1 after being targeted there, but not so when SpRU1A mRNA lacked either the 3′BE or the intron (Fig. 4c). These observations suggest that the 3′BE alone is not sufficient for efficient targeting. In agreement, insertion of HAC1’s 3′UTR stem (Fig. 1c) into the 3′UTR of PGK1 could not facilitate recruitment of this heterologous mRNA to Ire1 foci (Fig. 4d). Thus, the intron and 3’BE cooperate to effect HAC1 mRNA targeting.
Figure 4. Translational repression is a prerequisite for mRNA targeting to Ire1 foci.
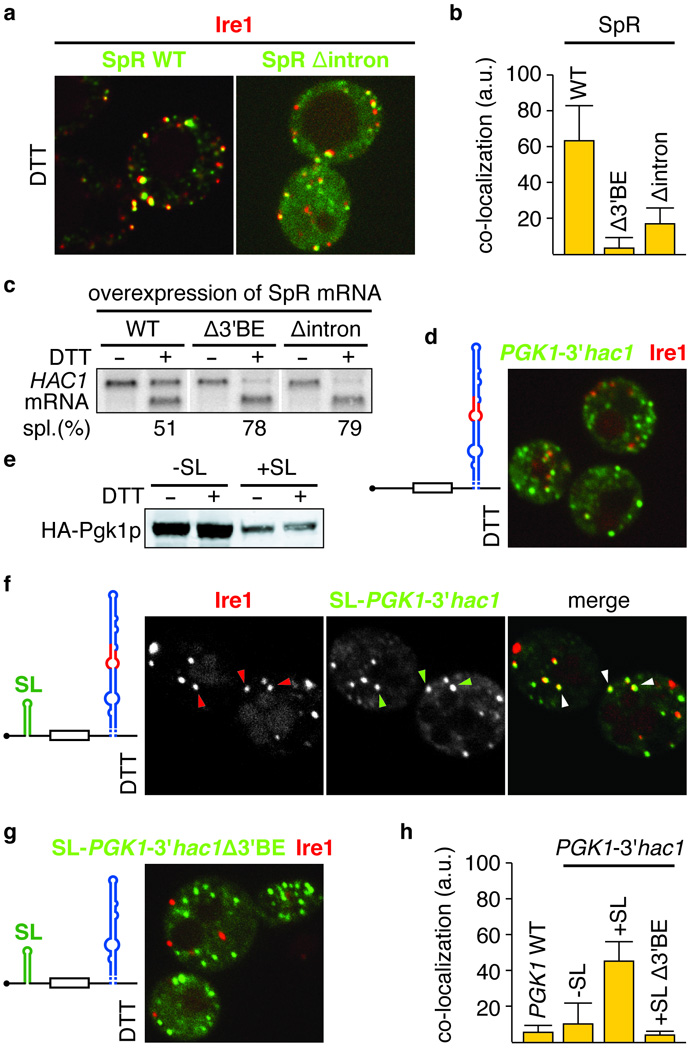
a,d,f–g, Localization of Ire1-mCherry, and U1A-GFP decorated mRNA of either the wild-type splicing reporter, as in Figure 2i (SpR WT), or an intron-less variant (SpR Δintron) (a), or of PGK1U1A, bearing either the wild-type (d,f) or mutant Δ3′BE (g) 3′UTR stem loop of HAC1 mRNA, in combination with (f,g) or without (d) a small stem loop (SL) that confers translational repression in its 5′UTR, as schematically depicted. b, C.I for mRNA recruitment of WT and Δintron SpR variants into Ire1 foci (means +/− s.e.m., n = 5); bar for the Δ3′BE mutant as depicted in Figure 2i is shown for comparison. c, Northern blot of HAC1 mRNA from yeast strains that over-expressed variants of the SpR, as indicated. e, Western blot of the variants of HA-tagged Pgk1 protein (HA-Pgk1p) bearing the 3′UTR from HAC1 with or without a 5′UTR SL a,c–g, ER stress was induced with 10 mM DTT for 45 min. h, C.I. for mRNA recruitment into Ire1 foci of PGK1U1A wild-type — see Figure 2f — or variants shown in d,f,g (means +/− s.e.m., n = 3–5).
The intron keeps HAC1 mRNA translationally silent (Fig. 1a), suggesting the possibility that translational repression may be key to HAC1 targeting as in other targeting mechanisms, as observed for ASH1 mRNA16. To test this hypothesis, we inserted a small stem loop into the 5′UTR of the PGK1U1A mRNA to repress its translation ([ref 17] and Fig. 4e). Remarkably, when we expressed PGK1U1A mRNA containing both the small stem loop in the 5′UTR and the HAC1 3′BE-containing stem in the 3′UTR, we found that this mRNA efficiently targeted to Ire1 foci (CI: 46 ± 11, Fig. 4f, h). Conversely, the corresponding mRNA lacking the 3′BE was not targeted (Fig. 4g, h). We conclude that the 3′BE-containing stem is both necessary and sufficient to target a heterologous mRNA to UPR-induced Ire1 foci, provided that its translation is on hold. Translational repression, therefore, is not only key to facilitate timely synthesis of Hac1 protein upon induction of the UPR, but is also integral to the targeting of HAC1 mRNA to ER stress signaling centers.
Our results describe a first example of mRNA targeting as a central feature in a signaling pathway. HAC1 mRNA is delivered to the site where it is processed as part of the main switch regulating the UPR. The mRNA guidance mechanisms characterized to date serve other goals, such as delivery of mRNA to sites of storage or degradation17,18, or restricted distribution of the proteins they encode19–21. HAC1 mRNA delivery to Ire1 foci has in common with other mRNA targeting mechanisms that it depends on a signal in the 3’UTR and on translational repression of the mRNA22. The mechanism of translational control of HAC1 mRNA serves both to prevent translation of a functional transcription factor, when the UPR is off, and to allow the mRNA access to the splicing machine, which removes the intron to allow its translation, when the UPR is on. In this way, the targeting signal is inactivated when translation of HAC1 mRNA resumes, even though the 3′BE remains present in the spliced mRNA.
The translational block in S. cerevisiae is exerted via a 16-base pairing interaction between sequences in the 252 nucleotide-long intron and the 5′UTR4. Most HAC1 or XBP1 orthologs bear introns that are shorter (~20–26 nucleotides) and show no sequence complementarity to support 5′UTR/intron based translational blocks. It is conceivable that other means of translational repression come into play. For instance, the PERK-mediated general translational attenuation in response to ER stress23 could serve a functionally similar role in XBP-1 mRNA targeting in metazoans.
Our findings emphasize the role of Ire1 oligomers, rather than dimers, in UPR signaling. Early co-immunoprecipitation studies already provided evidence for oligomerization24, and the identification of two functionally important interfaces that link Ire1 luminal domains into linear filaments in the crystal lattice supports an attractive model by which neighboring Ire1 molecules are ‘stitched’ together by the binding of unfolded proteins in the ER lumen14. This model and the epistasis data in Figure 2h suggest that Ire1 foci formation is governed by self-organization. Over-expression of Ire1 caused an enlargement of the foci, but did not increase their number (not shown), suggesting that there is a limited number of nucleation sites per cell and that foci may arise at such predisposed sites at the ER membrane. Since HAC1 mRNA recruitment occurs with amazing speed and efficiency (e.g., Fig. 3), one can further speculate that the 3’BE containing targeting signal may allow HAC1 mRNA to travel actively along cytoskeletal filaments to these pre-disposed sites, where Ire1 concentrates.
Clustering of activated signaling receptors occurs in many systems, such as the immunological synapse25 and in bacterial chemotaxis26, and the resulting local concentration of the signaling machinery can profoundly enhance the efficiency of signal transduction. Interestingly, we found that upon oligomerization in vitro the nuclease activity of the Ire1 kinase/nuclease domains vastly increases over the activity observed for Ire1 dimers27 (AVK & PW, submitted). Thus, by clustering into oligomers, Ire1 acquires enhanced avidity towards its substrate HAC1 mRNA and reaches full enzymatic activation at the same time. These mechanistic features converge into a signaling relay that provides the efficiency and timeliness required to combat ER stress.
METHODS SUMMARY
Microscopy data acquisition and analysis
Cells were visualized on a Yokogawa CSU-22 spinning disc confocal on a Nikon TE2000 microscope. Images of Ire1-mCherry and U1A-GFP decorated HAC1U1A, SpRU1A and PGK1U1A mRNAs and variants thereof were analyzed using a customized MatLab script to determine the fraction of Ire1-mCherry in foci and to score the recruitment of U1A-GFP decorated mRNA in Ire1 foci. The annotated MatLab script is available in the Supplementary Material. In brief, after background subtraction we defined the fraction of Ire1-mCherry in foci as the ratio between the integrated fluorescence intensity of pixels with a signal greater than a threshold value and the total integrated fluorescence intensity. The threshold was empirically defined such that under non-stress conditions no signal was scored as “foci”. Similarly, RNA foci were defined as pixels exceeding twofold the mean intensity in the RNA channel. A “co-localization index” was then defined as the integrated intensity of the pixels within the RNA foci that had pixels in common with Ire1 foci divided by the total RNA intensity and expressed in arbitrary units in a range of 0 to 100. Per condition, the percentage of Ire1-mCherry in foci and the co-localization index for the mRNA recruited to the foci was determined for 3–9 individual cells. Values and the standard error of the mean are given in histograms in Figure 2–Figure 4. Since, in contrast to the covalently fluorescently tagged Ire1, we do not know what fraction of U1A-GFP is bound to mRNAs containing U1A binding sites, background subtraction for U1A-GFP was arbitrary. Therefore, we quantified the data by the co-localization index rather an absolute % co-localization measure. The co-localization index robustly scores the differences in mRNA recruitment we observed qualitatively in the fluorescent micrographs.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Acknowledgements
We thank Martin Jonikas and Benoît Kornmann for their help with the MatLab scripts; Roy Parker for the pPS2037 and pRP1187 plasmids; Kurt Thorn for the pKT127 plasmid and for his invaluable assistance with microscopy at the Nikon Imaging Center at UCSF; and Christine Guthrie, Raúl Andino, John Gross, and Walter lab members for discussion and comments on the manuscript. T.A. was supported by the Basque Foundation for Science and the Howard Hughes Medical Institute; E.v.A. by the Netherlands Organization for Scientific Research (NWO); D.P. and C.R. by the National Science Foundation; C.R. by the President’s Dissertation Year Fellowship; A.V.K. by the Jane Childs Memorial Fund for Medical Research. P.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.van Anken E, Braakman I. Endoplasmic reticulum stress and the making of a professional secretory cell. Crit. Rev. Biochem. Mol. Biol. 2005;40:269–283. doi: 10.1080/10409230500315352. [DOI] [PubMed] [Google Scholar]
- 4.Rüegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 5.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 6.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 7.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez TN, Sidrauski C, Dörfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 10.Kimata Y, et al. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 14.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl Acad. Sci. USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollock R, Rivera VM. Regulation of gene expression with synthetic dimerizers. Methods Enzymol. 1999;306:263–281. doi: 10.1016/s0076-6879(99)06017-6. [DOI] [PubMed] [Google Scholar]
- 16.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 17.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi SB, et al. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- 21.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 22.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 24.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 25.Bromley SK, et al. The immunological synapse. Annu. Rev. Immunol. 2001;19:375–396. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- 26.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 27.Lee KP, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature


