Summary
As is typical for S100-target protein interactions, a Ca2+-dependant conformational change in S100A1 is required to bind to a 12-residue peptide (TRTK12) derived from the actin capping protein, CapZ. In addition, the Ca2+-binding affinity of S100A1 is found to be tightened (> 3-fold) when TRTK12 is bound. To examine the biophysical basis for these observations, the solution NMR structure of TRTK12 in a complex with Ca2+-loaded S100A1 was determined. When bound to S100A1, TRTK12 forms an amphipathic helix (residues N6 to S12) with several favorable hydrophobic interactions observed between W7, I10, and L11 of the peptide and a well-defined hydrophobic binding pocket in S100A1 that is only present in the Ca2+-bound state. Next, the structure of S100A1-TRTK12 was compared to that of another S100A1-target complex (i.e. S100A1-RyRP12), which illustrated how the binding pocket in Ca2+-S100A1 can accommodate peptide targets with varying amino acid sequences. Similarities and differences were observed when comparing the structures of S100A1-TRTK12 and S100B-TRTK12, providing insights regarding how more than one S100 protein can interact with the same peptide target. Such comparisons, including those to other S100-target and S100-drug complexes, provide the basis for designing novel small molecule inhibitors that could be specific for blocking one or more S100-target protein interaction(s).
Keywords: S100A1, target binding, CapZ, S100 proteins, NMR
Introduction
S100A1 is a dimeric Ca2+-binding protein (10.5 kDa per subunit) in the S100 protein family and has two EF-hands per subunit. In the N-terminus, each S100A1 subunit has a 14-residue EF-hand (EF1; residues 19-32) termed the “S100-hand” or the “pseudo EF-hand”. EF1 typically binds Ca2+ weakly via several backbone carbonyl ligands and a bidendate ligand from a glutamate (E32) at position 14 of the Ca2+-binding domain. In the C-terminus, each S100A1 subunit has a canonical 12 residue EF-hand (EF2; residues 62–73) that coordinates Ca2+ in a manner identical to that of other proteins in the EF-hand super family such as calmodulin (CaM) and troponin C (TnC) 1. As with most dimeric S100 proteins, only EF2 of S100A1 undergoes a large structural rearrangement upon binding Ca2+, and it is this conformational change that allows S100A1 to exert its biological function via interaction with and modulation of cellular targets. This conformational change in S100A1 involves the rotation of the entering helix of EF2 (helix 3) by ~90° rather than the exiting helix (helix 4) as found for most other EF-hand containing proteins, and is the defining characteristic of an “S100 Ca2+ switch” 2; 3; 4. Such a conformational change is unique to S100 proteins because the exiting helix (helix 4 of EF2) is typically held in place via an X-type four helix bundle that comprises the tight dimer interface; whereas, in other EF-hand containing proteins, the exiting helix is free to rotate.
In general, proteins in the S100 family are distributed in a cell-specific manner and interact with a diverse set of molecules 5; 6; 7. For S100A1, at least 20 known protein targets have been identified including Ca2+ signaling proteins (ryanodine receptors 1 & 2, Serca2a, phospholamban), neurotransmitter release proteins (synapsins I & II), cytoskeletal and filament associated proteins (CapZ, microtubules, intermediate filaments, tau, microfilaments, desmin, tubulin, F-actin, titin, and the glial fibrillary acidic protein GFAP), transcription factors and their regulators (myoD, p53), enzymes (aldolase, phosphoglucomutase, malate dehydrogenase, glycogen phosphorylase, photoreceptor guanyl cyclases, adenylate cyclases, glyceraldehyde-3-phosphate dehydrogenase, twitchin kinase, Ndr kinase, and F1 ATP synthase), and other Ca2+-activated proteins (annexins V & VI, S100B, S100A4, S100P, and other S100 proteins) (reviewed in 6; 7). It is also the case that several S100 protein family members bind to the same protein target 6; 7. For example, S100A1 and S100B, two of the earliest discovered S100 proteins, both interact with several of the same protein targets in a Ca2+-dependent manner including the ryanodine receptor, microtubules, GFAP, p53, NDR kinase, phosphoglucomutase, CacyBP/Sip1, and annexin A6 6; 8; 9; 10; 11; 12; 13, so it is possible that some S100 proteins may function redundantly. Furthermore, using phage display techniques, Dimlich et al. identified a 12 amino acid peptide (TRTKIDWNKILS) derived from the actin capping protein (CapZ) that bound to S100B in a Ca2+-dependent manner at low micromolar concentrations 14. Subsequently, S100A1 and several other S100 proteins (i.e. S100A1, S100A4, and S100A5) have also been shown to bind this same peptide, termed TRTK12 (unpublished data) 5. As a result, the TRTK12 peptide is considered a general S100 protein consensus binding sequence ((K/R)(L/I)XWXXIL).
To examine in detail how two S100 proteins can bind to the same protein target, we have determined the solution structure of S100A1-TRTK12, and compared it to the previously determined structure of S100B-TRTK12 15. NMR chemical shift perturbation data was suggestive that TRTK12 interacts with the hydrophobic pocket that is common to both Ca2+-S100A1 and Ca2+-S100B 4; 5. However, when these two structures were compared here at atomic resolution, it was found that the specific positioning of the TRTK12 peptide in the target binding pocket was not conserved in S100A1 and S100B, and that interactions between the peptide and the two S100 protein side chains were considerably different. This structure also leads to salient insights into the necessary attributes of productive S100A1-target protein interactions, some of which may be common in several S100-target protein interactions, including those for S100B.
Results
Chemical shift and NOE assignments for the S100A1-TRTK12 complex
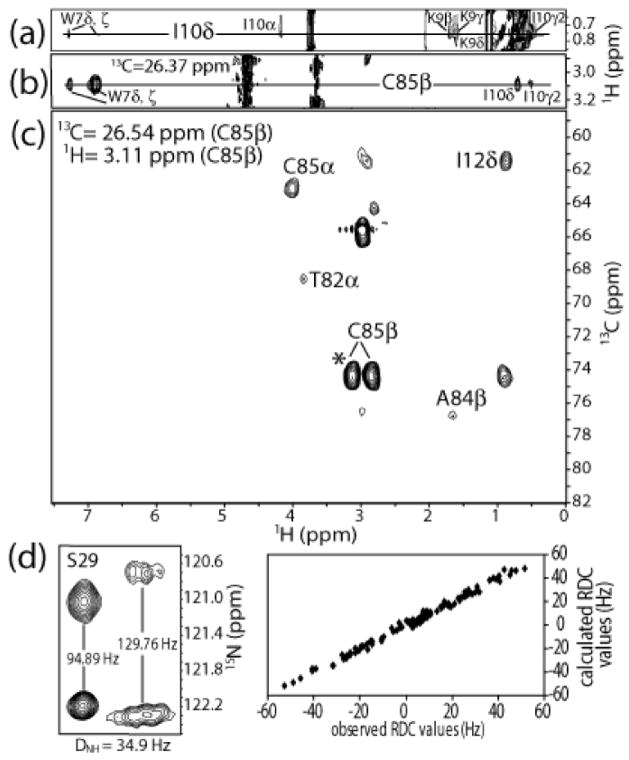
The first step in solving the high-resolution structure of S100A1-TRTK12 (24 kDa) by NMR was to unambiguously assign the resonances and NOE correlations for the complex using data from a series of heteronuclear multidimensional NMR experiments. The 1H, 13C, and 15N chemical shift assignments for all observable backbone and sidechain resonances of 13C, 15N-labeled S100A1 bound to TRTK12 were completed a priori using standard NMR through-bond experiments as described in Wright et al. 2005 5. Unambiguous resonance and NOE assignments for protons of the unlabeled TRTK12 peptide bound to 13C, 15N-labeled S100A1 were then made using 2D 12C-filtered spectra (NOESY, TOCSY in H2O and D2O), as previously described for other protein-peptide complexes 15; 16; 17; 18. Representative NOE data from a region of a two-dimensional 12C-filtered NOESY collected in D2O is illustrated (Fig. 1a), which show NOE correlations for bound TRTK12 between I10δ and other protons of I10 (I10α, I10γ2) as well as to protons of K9 (K9β, γ, δ) and W7 (W7δ,ξ). That W7 was proximal to I10 also provided an early indication that the TRTK12 peptide was helical when bound to Ca2+-S100A1 (Fig. 1a). In addition, proton resonances for I10 and W7 of TRTK12 (i.e. I10γ2, I10δ, W7δ, ξ) were found to be proximal to the β-protons of C85 of 13C, 15N-labeled S100B in a 3D 13C edited, 12C filtered NOESY experiment (Fig. 1b). Intermolecular NOE data such as these were critically important for the structure determination of the S100A1-TRTK12 complex as well as for validating proton assignments on unlabeled TRTK12 bound to S100A1 (Fig. 1b). In summary, the observable 1H resonances of TRTK12 together with the 1H, 13C, and 15N resonances of S100A1 in the S100A1-TRTK12 complex were assigned unambiguously and deposited into the BioMagResBank database (http://www.bmrb.wisc.edu) under the BMRB Accession number 16050.
Figure 1.
NOE data used to determine the structure of Ca2+-S100A1 bound to TRTK12 at 37 °C, pH 7.2. (a) Region of the 12C filtered NOESY experiment, showing NOE correlations between protons of Trp-7 and Lys-9 to Ile-10δ of TRTK12 when bound to Ca2+-S100A1. These NOE correlations are not present in spectra of samples containing the TRTK12 peptide alone. (b) Strip of the 3D 13C edited, 12C filtered NOESY spectrum, demonstrating NOE correlations between C85β of S100A1 to several protons of both Trp-7 and Ile-10 of TRTK12. (c) Plane of the 4D 13C, 13C-edited NOESY, showing medium and long range NOE correlations from C85β of S100A1. Each of these spectra was collected on samples containing 13C, 15N-labeled S100A1 and unlabeled TRTK12 peptide. (d) Residual dipolar coupling (RDC) data from the amide of S29 in isotropic (left) and aligned (right) media, illustrating typical N-HN splittings. On the right, a plot of expected RDCs versus observed RDCs, showing that the data fit well into structure calculations.
NOE assignments were made using data from 3D 15N-edited NOESY, 3D 13C-edited NOESY, 4D 15N, 13C edited NOESY and 4D 13C, 13C-edited NOESY experiments (Fig. 1c). As found in all other dimeric S100 protein structures, it was clear from NOE data that helices 1 and 4 were an integral part of the S100A1 dimer interface in the S100A1-TRTK12 complex 19. For example, early in the NOE assignment and structure determination process, several NOE correlations were observed between residues at the N- and C-terminus of helix 1 (i.e. L4δ1 to F15HN and several others). Because of the physical impossibility of having two residues at opposite ends of a helix being proximal in space, such NOE correlations were assigned as inter-subunit between helices 1 and 1′ of the S100A1 dimer. Similarly, the assignment of intermolecular NOEs could be made for residues at the N- and C-terminus of helices 4 and 4′ due to the antiparallel alignment of these helices (i.e. F71HN to V83γ1, and several others). As expected, such NOE data for S100A1 in the S100A1-TRTK12 complex were fully consistent with the antiparallel alignment of helices 1, 1′, 4, and 4′ into an X-type four-helix bundle at the dimer interface as found for other S100 proteins. It was then relatively straightforward to continue assigning both intra- and intersubunit NOE correlations in an iterative manner using preliminary structural models as a guide. For example, an NOE correlation observed between the Hβ protons of C85 and Hδ protons of I12 (Fig. 1c) was assigned as an intermolecular NOE since these two protons were more than 20 Å apart within the same subunit. In total, 130 intermolecular NOE constraints were assigned unambiguously at the dimer interface for residues in helix 1 (E3, L4, E5, T6, A7, M8, T10, L11, I12, V14, F15), loop 2 or ‘hinge’ (E40, L41), and helix 4 (F71, F74, V75, V76, A80, T82, V83, C85). An additional 38 NOE correlations remained ambiguous and were allowed to satisfy inter- and/or intramolecular distance constraints even in the final structure calculation (table 1), as previously described for other symmetric dimers 20.
Table 1.
NMR-derived restraints and statistics of 20 NMR structures1
| <20> | best | |
|---|---|---|
| rmsd from distance constraints (Å)2 | ||
| total (3382) | 0.037 ± 0.002 | 0.036 |
| intraresidue (570) | 0.009 ± 0.004 | 0.010 |
| sequential ( |i − j| = 1) (962) | 0.029 ± 0.003 | 0.028 |
| medium range (1 < |i − j| ≤1) (848) | 0.036 ± 0.003 | 0.035 |
| long range ( |i − j| > 5) (446) | 0.045 ± 0.005 | 0.045 |
| intermolecular for dimer interface (130) | 0.044 ± 0.006 | 0.034 |
| ambiguous intra- or inter- S100A1 subunit (38) | 0.012 ± 0.010 | 0.005 |
| TRTK12 peptide (seq, med) (90) | 0.028 ± 0.009 | 0.041 |
| Intermolecular S100A1 to TRTK12 peptide (96) | 0.060 ± 0.011 | 0.062 |
| calcium ligand (18) | 0.025 ± 0.010 | 0.029 |
| hydrogen bonds (184) | 0.069 ± 0.005 | 0.064 |
| rmsd from exptl dihedral constraints (°) | ||
| Φ,Ψ (276) | 0.516 ± 0.120 | 0.502 |
| rmsd from dipolar coupling restraints (Hz) | ||
| DNH (112) | 1.652 ± 0.093 | 1.606 |
| DCH (136) | 3.296 ± 0.171 | 3.261 |
| rmsd from exptl13C chemical shifts | ||
| 13Cα (ppm) | 1.261 ± 0.041 | 1.253 |
| 13Cβ (ppm) | 1.054 ± 0.039 | 1.020 |
| rmsd from idealized geometry | ||
| bonds (Å) | 0.006 ± 0.001 | 0.006 |
| angles (°) | 0.978 ± 0.018 | 0.959 |
| impropers (°) | 1.841 ± 0.010 | 1.844 |
| Lennard-Jones potential energy (kcal/mol)3 | −889 ± 28 | −917 |
| Q-factor4 | 0.26 ±0.04 | 0.25 |
| % of residues in the most favorable region of the Ramachandran plot5 | 87.6 ± 2.4 | 91.3 |
| Rmsd to the mean structure (Å)6 | ||
| all backbone atoms in S100A1 (3–88) | 0.518 ± 0.088 | 0.339 |
| all heavy atoms in S100A1 (3–88) | 1.097 ± 0.128 | 0.943 |
| all ordered backbone (S100A1 3–88, TRTK12 4–12) | 0.565 ± 0.089 | 0.476 |
| all heavy atoms (S100A1 3–88, TRTK12 4–12) | 1.118 ± 0.118 | 0.969 |
The 20 ensemble structures, <20>, are the results of simulated annealing calculations. The best structure is the closest to the average structure. The values shown for the <20> are the mean ± standard deviation.
All four subunits of the protein complex are included in all of these values. None of the 20 structures has a distance violation > 0.4 Å or a dihedral angle violation of > 5°. The force constants used in the SA calculations are as follows: 1000 kcal mol −1 Å2 for bond length, 500 kcal mol−1 rad−2 for angles and improper torsions, 4 kcal mol−1 Å−4 for the quartic van der Waals (vdw) repulsion term (hard-sphere effective vdw set to 0.8 times their values in CHARMm parameters), 50 kcal mole−1 Å−2 for experimental distance constraints, 100 kcal mol−1 Å−2 for non-crystallographic symmetry, 1kcal mol−1 Å−2 for distance symmetry constraints, 0.5 kcal mol−1 ppm−2 for the 13C chemical shift constraints, and 1.0 for the conformational database potential. The force constants (in kcal Hz−2) used for dipolar coupling restraints were as follows: 0.40 for 15N-1HN and 0.2 for 13Cα-1Hα.
Lennard-Jones van der Waals energies were calculated using CHARMm parameters and were not used in any stage of the structure determination
Q-values were determined by randomly removing 10% of all RDC values. To ensure accuracy, an ensemble of structures with a second randomly removed subset of RDCs was also run. The Q-value of this second set was 0.26.
PROCHECK was utilized to generate the Ramachandran plot.
Backbone calculations include Cα, N, and C′ atoms. Only residues 3–88 are included since no long-range NOE correlations were observed for residues 1–2 and 89–93 in S100A1 or residues 1–3 in the TRTK peptide.
Next, the structure and positioning of the TRTK12 peptide in the S100A1-TRTK12 complex was examined in detail. Important for these analyses was a 3D 13C-edited, 12C-filtered NOESY experiment, which provided 96 intermolecular protein-peptide distance constraints between 13C, 15N-labeled S100A1 and unlabeled TRTK12 peptide (Fig. 1b). For example, W7, I10, and L11 of TRTK12 exhibited intermolecular NOE correlations to C85 on S100A1. In general these three hydrophobic residues of TRTK12 provided a large number of NOE correlations to hydrophobic residues in helix 4. This periodicity of recurring intermolecular NOE correlations, combined with (i, i+3) intrapeptide NOE correlations from the 12C-filtered NOESY experiment (Fig. 1a), allowed for the precise orientation of TRTK12 into the hydrophobic binding site in Ca2+-S100A1, and further confirmed that TRTK12 folds as a helix when bound to S100A1. As was also found for the S100B-TRTK12 complex, all five residues defining the S100A1/S100B consensus binding sequence of TRTK12 peptide (K/R)(L/I)XWXXIL) including K4, I5, W7, I10, and L11 were found to be at the S100A1-TRTK12 binding interface 14. In total, protons from eight residues of the TRTK12 peptide (K4, I5, W7, N8, K9, I10, L11, and S12) gave intermolecular NOE correlations to protons from fourteen residues on S100A1 (F44, L45, K49, D50, D52, A53, K56, I57, E60, L77, L81, A84, C85, and F88) consistent with the helical TRTK12 peptide oriented nearly parallel to helix 3 on S100A1, spanning from the hinge region (loop 2) to the middle of helices 3 and 4.
Residual dipolar coupling measurements
In an effort to both improve and independently verify the NOE-based structure, N-HN and Cα-Hα residual dipolar coupling (RDC) data were measured using a radially compressed polyacrylamide gel to partially align the protein (Fig. 1d), as previously described 21. In the absence of decoupling, the observed splitting between two nuclei P and Q corresponds to the sum of the scalar and dipolar interactions, JPQ and DPQ. JPQ is independent of alignment, and DPQ is simply obtained from the difference between the P–Q splitting measured in conditions where the sample is partially aligned (i.e. in radially compressed gels) versus conditions with isotropic tumbling 22. Here, 112 1DN-H values for S100A1 in the S100A1-TRTK12 complex were recorded from a [15N-1H]-2D correlation spectrum in the absence of 1H decoupling during the t1 evolution period, and the upfield and downfield components of the 15N-{1HN} doublets were separated into two spectra in an interleaved manner as described previously 22 (Fig. 1d). Additionally, 1DCα-Hα dipolar couplings were obtained under the same alignment conditions as those for 1DN-H RDCs. These data were collected using a 3D (H)CA(CO)NH experiment recorded in the absence of 1H decoupling during the Cα evolution period permitting accurate measurement of 136 1DCα-Hα residual dipolar couplings, as described previously 23. Because the 1DNH and 1DCαHα data were collected under identical alignment conditions, they could be fit simultaneously in structure calculations to enforce a single alignment tensor (Aa ~ 0.6 × 10−3).
Structure calculations, structural statistics and description of the S100A1-TRTK12 complex solved in solution by NMR spectroscopy
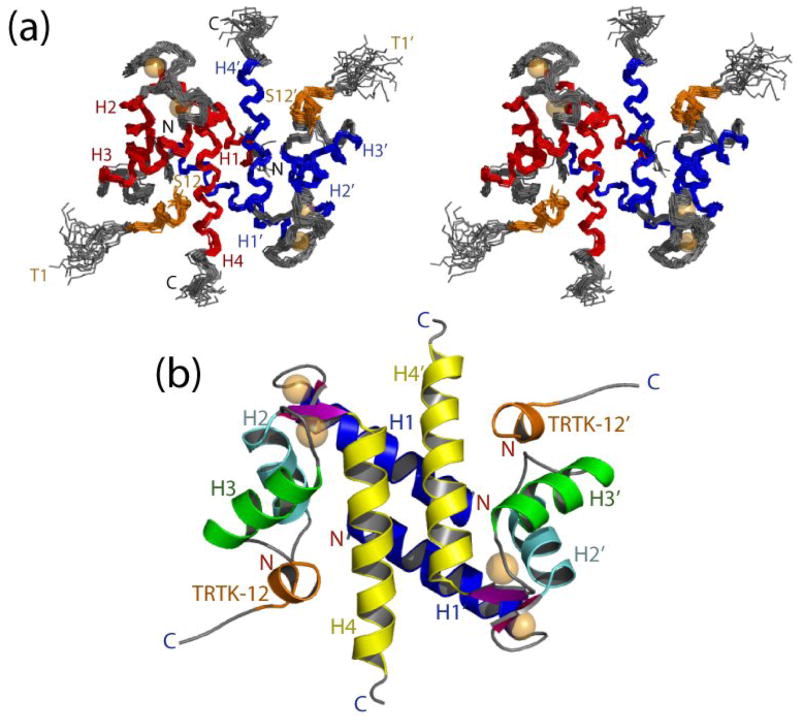
In total, 3,382 experimental distance constraints, 276 dihedral angle constraints, and 248 residual dipolar coupling constraints (RDCs) were used to calculate the solution structure of the dimeric Ca2+-S100A1-TRTK12 complex (>18 constraints/residue) (Fig. 2). The first three residues of the TRTK12 peptide showed no intermolecular NOE correlations, while the last nine residues showed more than 10 NOE correlations per residue on average. The large number of assigned NOE correlations allowed for an accurate residue-by-residue examination of the S100A1-TRTK12 binding interface, although the N-terminal tail of TRTK12 remains undefined and likely is dynamic in solution. A family of the twenty lowest energy structures of S100A1-TRTK12 is depicted in stereoview in figure 2a. These structures all have low Q-factors (0.26), no dihedral violations greater than 5°, no NOE violations greater than 0.4 Å, and no residues in the unfavorable portion of the Ramachandran plot (table 1). The backbone atoms in each of the twenty structures are well-defined with an RMSD of 0.57 for all ordered residues and an RMSD of 0.52 for just the S100A1 backbone alone (table 1). For the bound TRTK12 peptide, the backbone is slightly less well defined, with an RMSD of 0.81 for all ordered residues. No long-range NOE correlations were observed for residues 1–2, and 89–93 in Ca2+-S100A1 or for residues 1–3 of the TRTK12 peptide, and thus these were removed from RMSD calculations (table 1).
Figure 2.
The three-dimensional structure of the S100A1-TRTK12 complex. (a) Stereoview of the 20 lowest-energy structures with the RMSD for backbone atoms found to be 0.672 (residues 3–87 of S100A1 and residues 4–12 of TRTK12). The two subunits are colored red and blue for S100A1, respectively, and the TRTK12 peptide is colored orange. (b) Ribbon diagram illustrating the fold of the S100A1-TRTK12 complex.
Overall, the backbone atoms of S100A1 do not significantly deviate in position between the Ca2+-bound state and the peptide bound state, with the RMSD for Cα carbons between the two states being 1.31Å (table 2, 3). This maintenance of the classical S100 fold upon target protein binding was also observed when TRTK12 bound to Ca2+-S100B as well as when a peptide derived from the ryanodine receptor (RyRP12) bound to Ca2+-S100A1 (table 2, 3); thus, the S100A1 structure reported here is likely a conserved feature of Ca2+-dependant S100A1-target protein interactions. In the absence of S100A1, the TRTK12 peptide exists as a random coil, as judged by a lack of NOE correlations and a narrow range of NMR spectral data 15. However, when bound to Ca2+-S100A1 the C-terminal region of TRTK12 adopts a helical conformation, as judged by both characteristically α-helical interpeptide NOEs (Fig. 1a) and intrasubunit NOE correlations (Fig. 1b, 3a). Interestingly, the observation that S100A1 interacts with helical portions of target proteins is another feature that is observed in nearly all of the S100-target protein structures reported thus far, and is likely another general feature for S100-target protein interactions.
Table 2.
Interhelical angles of S100A1 and S100B complexes
| Helices7 | Apo-S100A18 | Ca2+-S100A19 | S100A1-Ca2+- RyRP1210 | S100A1-Ca2+- TRTK12 | Ca2+-S100B11 | S100B-Ca2+- TRTK1212 |
|---|---|---|---|---|---|---|
| I–II | 120 ± 3 | 132 ± 1 | 136 ± 2 | 131 ± 1 | 137 ± 5 | 132 ± 2 |
| I–III | −45 ± 2 | −102 ± 2 | −94 ± 2 | −106 ± 2 | −118 ± 5 | −118 ± 3 |
| I–IV | 107 ± 2 | 131 ± 2 | 125 ± 2 | 126 ± 2 | 128 ± 4 | 128 ± 1 |
| II–III | 148 ± 2 | 125 ± 2 | 126 ± 1 | 121 ± 2 | 104 ± 3 | 109 ± 2 |
| II–IV | 46 ± 1 | −29 ± 1 | −35 ± 2 | −37 ± 2 | −35 ± 4 | −33 ± 2 |
| III–IV | −150 ± 1 | 121 ± 2 | 136 ± 2 | 120 ± 2 | 106 ± 4 | 108 ± 4 |
| I-I′ | −165 ± 3 | −157 ± 3 | −153 ± 2 | −155 ± 2 | −155 ± 1 | −148 ± 2 |
| IV-IV′ | 176 ± 2 | 152 ± 3 | 156 ± 2 | 156 ± 2 | 159 ± 5 | 146 ± 3 |
| I-peptide | - | - | 88 ± 7 | −106 ± 4 | - | −63 ± 11 |
| II-peptide | - | - | −49 ± 7 | 103 ± 3 | - | 164 ± 11 |
| III-peptide | - | - | −163 ± 4 | 38 ± 2 | - | 55 ± 11 |
| IV-peptide | - | - | 49 ± 7 | 127 ± 4 | - | 143 ± 7 |
Interhelical angles (Ω) range from −180° to 180° and are classified as either parellel (||) for 0° |Ω| 40°and 140°|Ω|180° or as perpendicular (⊥) for 40°| Ω|140°as described 54.
Taken from the NMR structure (PDB code 1K2H) 55.
Taken from the NMR structure (PDB code 1ZFS) 5.
Taken from the NMR structure (PDB code 2J2K) 17.
Taken from the NMR structure (PDB code 1QLK) 4.
Taken from the NMR structure (PDB code 1MWN) 15.
Table 3.
EF-hand angles of S100 proteins based on the VGM method
| EF-hand | N-terminal coordinate of second helix | θ (deg.) | φ (deg) |
|---|---|---|---|
| Pseudo-EF-hand | |||
| S100A1-Ca2+-TRTK1213 | (8.659, 2.341, −5.416) | 53 ± 2 | 80 ± 3 |
| S100A1-Ca2+-RyRP1214 | (9.236, −1.916, −5.779) | 45 ± 2 | 84 ± 5 |
| Ca2+-S100A115 | (9.237, −2.133, −4.516) | 51 ± 2 | 82 ± 2 |
| Ca2+-S100B-TRTK1216 | (0.927, −0.838, −7.262) | 55 ± 2 | 95 ± 3 |
| Ca2+-S100B17 | (11.199, −3.856, −6.578) | 53± 4 | 79 ± 5 |
| Apo-S100A118 | (11.843, −4.311, −5.724) | 68 ± 4 | 85 ± 9 |
| Typical EF-hand | |||
| S100A1-Ca2+-TRTK12a | (9.191, 2.880, −4.310) | 60 ± 2 | 115 ± 5 |
| S100A1-Ca2+-RyRP12b | (9.919, 1.444, 5.583) | 45 ± 2 | 83 ± 2 |
| Ca2+-S100A1c | (8.928, −2.392, −3.789) | 58 ± 2 | 108 ± 3 |
| Ca2+-S100B-TRTK12d | (9.070, −1.009, −5.863) | 70 ± 4 | 112 ± 3 |
| Ca2+-S100Be | (9.073, −2.602, −6.819) | 76 ± 5 | 95 ± 7 |
| Apo-S100A1f | (5.246, 13.667, −0.067) | 35 ± 2 | −90 ± 7 |
Vector geometry mapping results were obtained using the program VGM as described 54. Apo-calmodulin of Zhang et al. (PDB code 1DMO) was used as the reference for these calculations as previously described 56.
Taken from the NMR structure (PDB code 2J2K) 17.
Taken from the NMR structure (PDB code 1ZFS) 5.
Taken from the NMR structure (PDB code 1MWN) 15.
Taken from the NMR structure (PDB code 1QLK) 4.
Taken from the NMR structure (PDB code 1K2H) 55.
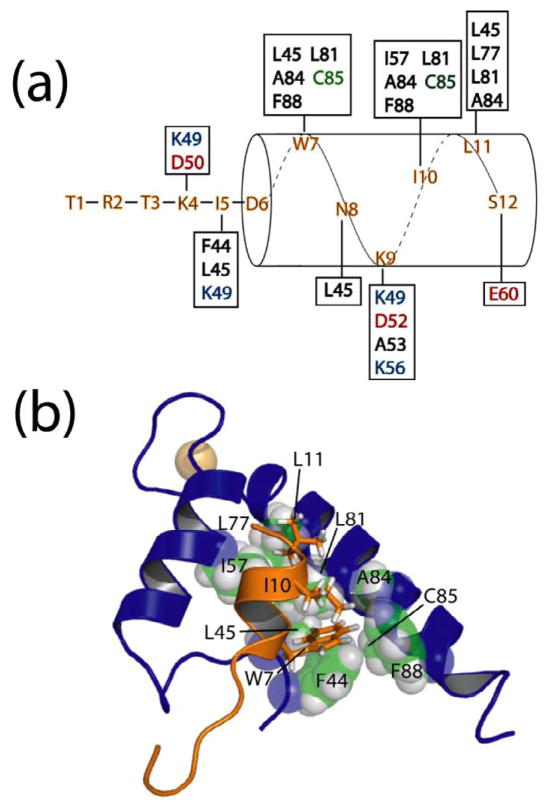
Figure 3.
Hydrophobic residues of TRTK12 interact with a hydrophobic binding pocket on Ca2+-S100A1. (a) Residues that give intermolecular NOE correlations between the TRTK12 peptide and residues on S100A1 (in boxes) are illustrated. S100A1 residues are colored black for hydrophobic, green for polar, red for negatively charged, and blue for positively charged residues. (b) Residues of TRTK12 including W7, I10, and L11 are packed nearby hydrophobic residues from the hinge region (F44, L45), helix 3 (I57), and helix 4 (L77, L81, A84, C85, and F88) of S100A1. In particular, W7 fits into a well-formed hydrophobic pocket consisting of F44, L45, L81, A84, C85, and F88 of Ca2+-S100A1.
The Ca2+-dependence of the S100A1-TRTK12 interaction
Upon the addition of Ca2+ to dimeric apo-S100A1, helix 3 of both subunits reorients by ~90° relative to helix 4 5. As a consequence of this conformational change, a hydrophobic pocket defined by amino acid residues in the hinge (loop 2; F44, L45), helix 3 (I57), and helix 4 (L77, L81, A84, C85, and F88), which were buried in the apo-state, are exposed when Ca2+ binds to S100A1 (Fig. 3). These same residues form multiple contacts with W7, I10, and L11 of TRTK12 (Fig. 3). The most prominent TRTK12 sidechain involved in an interaction with S100A1 is the large hydrophobic sidechain of W7, which extends into a well-formed hydrophobic groove formed by residues F44, L45, L81, A84, C85, and F88 of Ca2+-S100A1. Additionally, ring stacking between F88 and F44 on S100A1 and W7 on TRTK12 is observed, which is consistent with experiments in which S100A1 is truncated at F88 and binding to TRTK12 is abrogated 8. The hydrophobic residues I10 and L11 of TRTK12 also interact with numerous hydrophobic residues on helix 3 and helix 4 of S100A1 and are peripherally associated with the deep W7 binding pocket, as judged by a large number of NOE correlations to S100A1 between these residues and hydrophobic residues in the S100A1 binding pocket (Fig. 3). Thus, it is concluded that TRTK12 forms an amphipathic helix and that substantial hydrophobic interactions, most notably those between W7 on TRTK12 and a series of residues on S100A1, serve to stabilize the S100A1-TRTK12 complex.
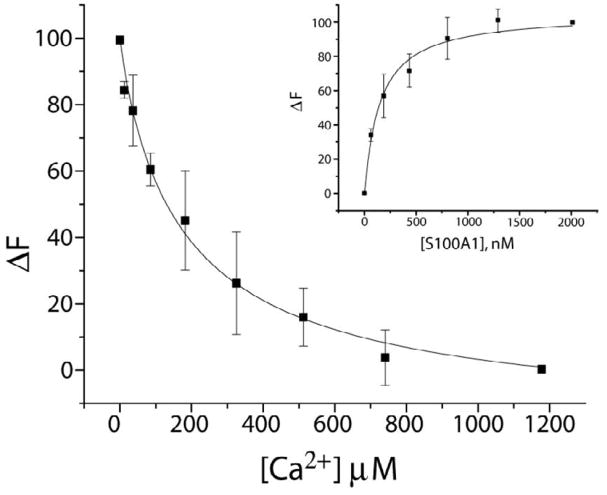
The TRTK12-S100A1 interaction, driven by the formation of an interprotein hydrophobic binding site, stabilizes the canonical EF-hand of S100A1 (EF2). This is experimentally manifested through an increase in the Ca2+-binding affinity of S100A1 in the presence of peptide (Fig. 4). Here, Tb3+ binding and kickoff experiments were performed with S100A1 in the presence of TRTK12, as previously described 5. Changes in Tb3+ luminescence intensity were measured upon binding to the tight site (EF2) of S100A1 (TbKD=149 ± 3 nM; Fig. 4, inset), and Ca2+-binding to S100A1 was then determined in direct competition studies with Tb3+. As observed in figure 4, S100A1 binds Ca2+ at 8 ± 3 μM in the presence of TRTK12, which is more than 3-fold tighter than when no peptide is present (27 μM). Such an increase in the Ca2+ affinity is well documented among S100 proteins and other EF-hand containing proteins in general when target proteins are bound 24; 25; 26; 27; 28; 29; 30; 31.
Figure 4.
Ca2+ and Tb3+ binding to S100A1-TRTK12 as monitored via Tb3+ luminescence. Displacement of Tb3+ by Ca2+ from a Tb3+-S100A1-TRTK12 complex as monitored by the decrease in Tb3+ luminescence. The sample conditions included 50μM TRTK12, 20 mM HEPES buffer, pH 7.0, and 20 mM DTT at 25 °C in D2O. The CaKD calculated from this competition experiment (CaKD=8 ± 3 μM) relied on the dissociation of Tb3+ from the Tb3+-S100A1-TRTK12 complex in the same buffer (TbKD = 149 ± 3 nM; inset). In this titration (inset), S100A1 was titrated into a solution where [Tb3+] was kept constant (3 μM) and [TRTK12] was kept at a concentration (50μM) where Tb3+-S100A1 is fully saturated with TRTK12 peptide at all points in the titration.
The drug pentamidine inhibits S100A1-TRTK12 complex formation
There is a growing interest in developing inhibitors of S100A1 since they could be beneficial in treating a variety of human diseases including neurological diseases, diabetes mellitus, heart failure, and several types of cancer 32; 33; 34; 35. The absence of significant phenotypes in S100A1 knockout mice suggests that an S100A1 antagonist would have minimal side effects in normal tissues 36; 37. In the brain, increased S100A1 expression contributes to pathologies related to neurological diseases, which suggests that S100A1 inhibitors could reverse such processes. For example, neuronal PC12 cells that do not express S100A1 are more resistant to Aβ-induced cell death than cells that express normal levels of S100A1 32, indicating that S100A1 antagonists would be beneficial in treating Alzheimer’s disease. In addition, extracellular S100A1 is cytotoxic to PC12 cells 38; 39 and injection of anti-S100A1 antibodies were found to lessen learning and memory deficits 40. Other S100A1-regulated intracellular processes are also consistent with S100A1 augmentation of Alzheimer’s disease pathology. These include S100A1-linked altered APP expression, destabilization of Ca2+ homeostasis, S100A1-mediated increased cell growth, decreased dendritic arborization, and decreased tubulin polymerization/stability 32; 41.
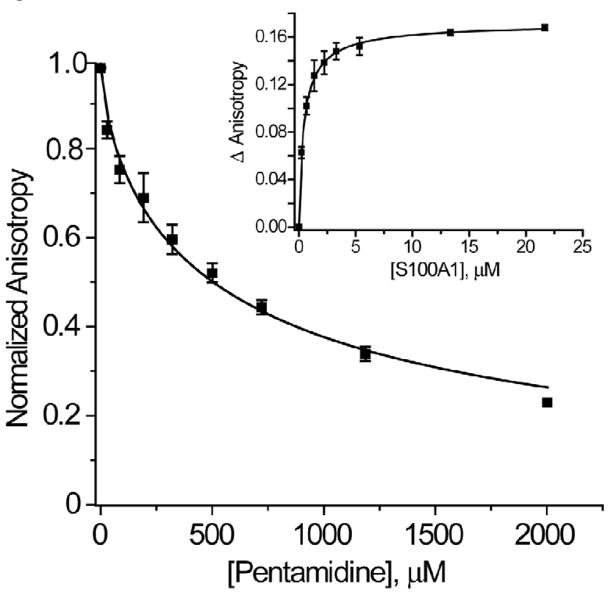
As previously found for S100B, the FDA-approved drug pentamidine (Pnt) has been found to bind S100A1 and block its interaction with TRTK12. Specifically, a fluorescently labeled TAMRA-TRTK peptide was found to bind (KD= 1.0 ± 0.6 μM) to Ca2+-S100A1 (Fig. 5; inset). This affinity of TAMRA-TRTK12 is ~20 fold tighter than for unlabeled TRTK12 (KD= 23 ± 6 μM), which is likely due to additional interactions between the protein and the hydrophobic TAMRA moiety. Furthermore, increasing amounts of Pnt were sufficiently able to displace the TAMRA-labeled peptide from Ca2+-S100A1, as judged by a decrease in fluorescence anisotropy values (Fig. 5). Using the equations employed by Nikolovska-Coleska et al. 42, the calculated KD of Pnt for S100A1 is 130 ± 60μM (as compared to a KD of 39 ± 5 μM for the S100B-Pnt complex) 43 and provides evidence that Pnt is a potential lead compound for developing S100A1 inhibitors. This competitive binding data also indicates that Pnt binds to S100A1 in the TAMRA-TRTK12 binding pocket, and could represent a starting point for developing drugs that block S100A1 through at least partial occlusion of this site. This may eventually lead to the development of novel high affinity molecules that specifically inhibit S100A1 and can be tested in vivo.
Figure 5.
Pentamidine (Pnt) binding to Ca2+-S100A1. Displacement of TAMRA-labeled TRTK12 bound to Ca2+-S100A1 by the FDA approved drug pentamidine as monitored by a decrease in TAMRA-TRTK12 fluorescence anisotropy. The sample conditions included 3.5 μM S100A1, 50 nM TAMRA-labeled TRTK12, 25 mM NaCl, 1.0 mM CaCl2, 1 mM DTT, 5 mM HEPES buffer, pH 7.5, at 25 °C. The PntKD calculated from this competition experiment (PntKD=120 ± 60 μM) relied on the dissociation of TAMRA-labeled TRTK12 from Ca2+-S100A1 (TAMRA-TRTKKD = 1.0 ± 0.6 μM; inset). In this titration (inset), S100A1 was titrated into a solution where [TAMRA-TRTK] was kept constant (50 nM) with all other buffer conditions the same as in the competition experiment.
Discussion
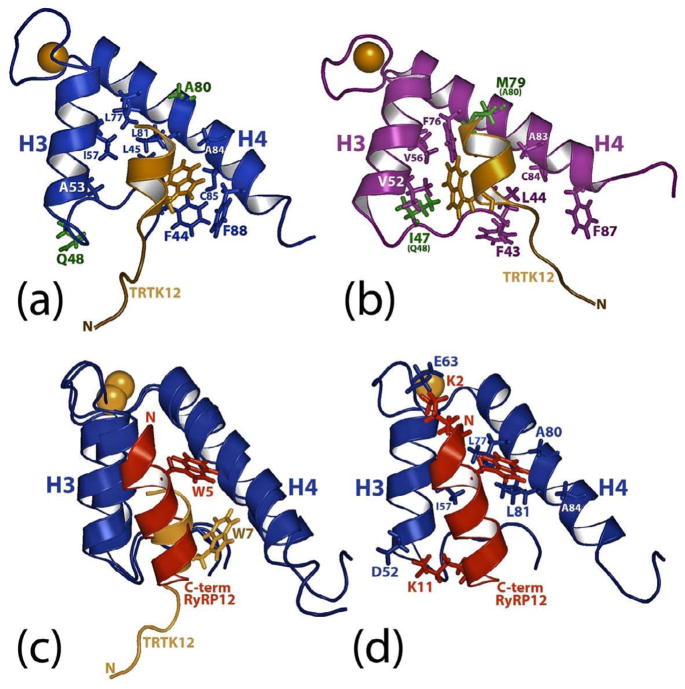
S100A1 and S100B, two of the earliest S100 proteins discovered, both have been reported to bind the ryanodine receptor, microtubules, GFAP, p53 (also binds S100A2, S100A4), NDR kinase, phosphoglucomutase, CacyBP/Sip1 (also binds S100A6, S100A12), and Annexin A6 (also binds S100A6) (reviewed in 7,17, unpublished results). Likewise, other target proteins have also been shown to bind multiple S100 proteins including annexin A5 (S100A1, S100A6, S100A12), caldesmon (S100A1, S100A6), and Sgt1 (S100A6, S100P). The completion of this structure represents a chance to study in detail how two S100 proteins, S100A1 and S100B, interact with the same peptide, TRTK12 15. Specifically, both S100A1 and S100B bind to TRTK12 at μM concentrations (S100B, KD= 7 ± 1 μM 6; S100A1, KD= 23 ± 6 μM), and upon comparison of chemical shift perturbation data, TRTK12 was thought to be oriented in a similar manner relative to helix 3 and helix 4 in the S100B-TRTK12 and S100A1-TRTK12 complexes 5. However, a comparison of the two NMR structures shows sidechain interactions that stabilize the peptide-protein interface differ substantially between S100A1 and S100B (Fig. 6a, 6b), which could provide an explanation for the ~3-fold difference in affinity observed. Specifically, W7 in TRTK12 does not extend close to helix 3 in Ca2+-S100A1, as observed in Ca2+-S100B, but instead is located near another hydrophobic pocket close to the C-terminus of S100A1 (Fig. 3b, 6a, 6b). This change in tryptophan position for TRTK12 has the consequence of rotating the entire peptide so that the hydrophobic face of the helical peptide faces helix 4 of S100A1 more directly than it does helix 4 of S100B. Specifically, the hydrophobic tryptophan residue of TRTK12 interacts with a binding pocket in S100A1 that comprises residues from loop 2 termed the ‘hinge’ (F44, L45) and helix 4 (L81, A84, C85, and F88). Comparatively, different residues in S100B from the ‘hinge’ (I47), helix 3 (V52, V56), and helix 4 interact with W7 on TRTK12 (Fig. 6b). That this pocket is so different on S100A1 versus S100B is likely the result of differences in protein sequence, most notably the nonconservative changes of an isoleucine (I47 in S100B) for a glutamine (Q48 in S100A1) and a methionine (M79 in S100B) for an alanine (A80 in S100A1; Fig. 6a, b; residues colored green). Another more subtle change in the sequences is a valine residue (V52 in S100B) for an alanine residue (A53 in S100A1). These and other differences in sequence result in a deep hydrophobic tryptophan-binding site in S100B nearby helix 3, which does not exist in S100A1. That S100A1 can form a different hydrophobic tryptophan-binding pocket adjacent to helix 4 is also likely facilitated by the fact that S100A1 has an alanine residue at position 80 rather than a methionine (i.e. M79). This allows W7 of TRTK12 to more readily approach the other large hydrophobic residues (i.e. F44, L45, L81, F88) comprising the S100A1 TRTK12 binding site (Fig. 6).
Figure 6.
Comparison of the S100B-TRTK12, S100A1-RyRP12, and S100A1-TRTK12 structures. (a) Ribbon diagram illustrating TRTK12 (tan) bound to Ca2+-S100A1 (blue). Residues that are not similar between the S100A1 and S100B sequences are shown in green. (b) Ribbon diagram illustrating TRTK12 (tan) bound to Ca2+-S100B (purple). As in (a), residues that are very dissimilar between the S100A1 and S100B sequences are shown in green. (c). Overlay of ribbon diagrams of TRTK12 (tan) and RyRP12 (red) bound to Ca2+-S100A1 (blue). In this panel, the location of the tryptophan residue in each peptide (W5 for RyRP12; W7 for TRTK12) is illustrated. (d). Ribbon diagram illustrating RyRP12 (red) bound to Ca2+-S100A1 (blue) illustrating the sidechains of S100A1 that interact with the RyRP12 peptide.
Our lab recently published a structure of S100A1 bound to a peptide, RyRP12, derived from ryanodine receptor (KKAVWHKLLSKQ)17; 36. Like TRTK12, this peptide closely resembles the consensus binding sequence of S100 proteins. However, the peptide binding sites on Ca2+-S100A1 were found to be quite different when the NMR structures of these two complexes were compared (Fig. 6c, 6d). For example, both RyRP12 and TRTK12 form amphipathic helices and bind a hydrophobic binding site exposed in Ca2+-S100A1, but the two peptides were oriented differently in the pocket by nearly 180o. For example, RyRP12 aligns parallel to helix 4 of S100A1, with the tryptophan residue on this peptide (W5) located in a hydrophobic binding pocket slightly higher in the Ca2+-S100A1 binding pocket than the tryptophan residue (W7) of TRTK12 (Fig. 6c). Furthermore, in RyRP12, W5 is stabilized by different residues in helix 3 (I57) and helix 4 (L77, A80, L81) than W7 in TRTK-12, which interacts with residues in the hinge (F44, L45) and helix 4 (L81, A84, C85, F88). Examination of residue-by-residue contacts between both complexes also reveals that the RyRP12 and TRTK12 peptides interact with S100A1 via leucine/isoleucine residues (I10 and L11 in TRTK12 and L8 and L9 in RyRP12) in addition to the aromatic tryptophan residue. However, unlike what is found in the S100A1-TRTK12 complex, two lysine residues in RyRP12 (K2, K11) likely form electrostatic interactions with S100A1 (E63 and D52 in S100A1), which could contribute to the orientation of RyRP12 in the S100A1 binding site 44. The TRTK12 peptide has no such electrostatic interactions with S100A1, and is oriented in the hydrophobic pocket based on Van der Waals and hydrophobic interactions alone. From these two structures, it appears likely that there is no one preferred orientation of binding for S100A1 targets, and that each S100A1 binding partner may orient differently using different key residues in the hydrophobic pockets nearby helices 3, 4 and the hinge.
Examination of other S100-target peptide complexes reveals that while tryptophan is frequently used in S100-target interactions, other residues can substitute; in both the S100A11-annexin 1 complex and the S100A6-Siah 1 structure, tryptophan is replaced with leucine 45; 46. Such variability in a residue position that has been assumed to be fairly well-conserved amongst S100 binding motifs suggests that other previously unsuspected sequences could be high affinity targets to various members of the S100 family. The fact that S100 proteins can bind to more than one target (i.e. KD of S100A1 for TRTK12 = 23 ± 6 μM while the KD for RyRP12= 8 μM) 36; 47, and that different S100 proteins can bind to the same target, suggests that many S100 proteins interact with their targets via the ‘selected fit’ mechanism of binding. Using this model, Ca2+-S100A1 exists as an ensemble of equilibrium structures, with the protein binding pocket sampling multiple similar but not identical conformations. Productive S100A1-target complex formation occur when S100A1 attains the conformation(s) most compatible for that particular target protein 48. Thus unique targets can select for particular conformations, leading to a diverse set of potential interactions. Evidence supporting this hypothesis is seen in Ca2+-S100B, where regions comprising the target binding pocket experience both fast and slow timescale dynamics 49. This mechanism of binding may be a common feature in the S100 family of proteins, since most S100 proteins are reported to bind multiple protein targets.
Conclusions
Here we present the solution structure of Ca2+-S100A1 bound to the 12-residue peptide TRTK12, which comprises an exact match of the S100 consensus binding sequence. A comparison of the apo-, Ca2+-, and Ca2+-S100A1-TRTK12 structures clearly reveals why this interaction is Ca2+-dependant. TRTK12 binds S100A1 as an amphipathic helix, with its hydrophobic face in close contact with the hydrophobic binding pocket of S100A1. Close examination of this interaction reveals that several residues, most notably W7 on the TRTK12 peptide, are fully buried in a well-formed hydrophobic cleft on S100A1. TRTK12 is uniquely oriented in the S100A1 binding site, as compared to the similar peptide RyRP12. Additionally, S100B and S100A1 both bind TRTK12 differently. A detailed examination of the S100A1 and S100B structures furthers our understanding of how these similar proteins interact with their respective target molecules and elicit distinct cellular functions. This information in turn forms the underpinning for specific rational drug design efforts towards inhibiting one or both of these S100 proteins when they are elevated in a disease state.
Materials and Methods
Sample preparations
A synthetic peptide (ac-TRTKIDWNKILS-am) derived from the α subunit of CapZ (residues 267-276) was chemically synthesized with its N-terminus acetylated (-ac) and C-terminus amidated (-am) and prepared for NMR as described previously 10. The TAMRA-TRTK peptide is synthesized with the TAMRA fluorescent covalently labeled at the N-terminus with the C-terminus amidated (-am). Recombinant 15N- and 13C, 15N-labeled rat S100A1 protein (10.5 kDa per subunit) was purified after overexpression in Escherichia coli (HMS174(DE3)) as described 5. NMR samples contained 15 mM d11-Tris-HCl, pH 7.2, 15 mM dithiothreitol (DTT), 10 mM CaCl2, 0.34 mM NaN3, 20 mM NaCl, 10% 2H2O, TRTK12 peptide (2–6 mM) and S100A1 (1–3 mM; subunit concentration). The acrylamide solutions used in the dipolar coupling experiments were prepared fresh at 30% acrylamide (1:20 bis) concentration. Compressed gels contained 5% acrylamide solution, 0.015% TEMED, and 0.015% ammonium persulfate (APS), and were prepared as previously described 5.
NMR spectroscopy
NMR spectra were collected at 37 °C with a Bruker DMX600 NMR spectrometer (600.13 MHz for protons) and a Bruker AVANCE 800 NMR spectrometer (800.27 MHz for protons) each equipped with four frequency channels and 5mm triple-resonance z-axis gradient cryogenic probeheads. Sequential backbone and side chain assignments of S100A1 in the TRTK12 peptide complex were obtained using standard NMR spectroscopy methods as described 5. The sequential assignments for the unlabeled TRTK12 peptide bound to 13C, 15N-labeled Ca2+-S100A1 were based on correlations recorded in 12C-filtered TOCSY and 12C-filtered NOESY experiments 50; 51; 52. The filtered TOCSY spinlock time (75 ms) and the filtered NOESY mixing time (200 ms) were sufficient for the collection of high-quality proton NMR data for TRTK12 in a protein complex of this molecular weight (24.2 kDa) 15; 18. 4D 13C, 15N -edited NOESY-HSQC 53, and 4D 13C, 13C -edited NOESY HSQC spectra, both with a mixing times of 130 ms, were collected on sample containing 13C, 15N S100A1 bound to unlabeled TRTK12. A 3D 12C filtered, 13C edited NOESY with a mixing time of 200 ms was also collected for recording intermolecular NOE correlations at the peptide-protein interface, as previously described 15; 36.
Structure calculation
Interproton distance constraints were derived from 2D, 3D, and 4D NOESY experiments (2D NOESY, 12C-filtered 2D NOESY, 15N-edited 3D NOESY, 12C filtered, 13C-edited 3D NOESY, 15N, 13C-edited 4D NOESY, and 13C, 13C-edited 4D NOESY) as described previously 5. Dihedral constraints φ ± 20 and ψ ± 15° for α-helix and φ± 40 and ψ± 40 for β-sheet were included based on 3JNH-Hα coupling constants, hydrogen exchange rates, and the chemical shift index of 1Hα and 13Cα resonances. Distance constraints of 2.0–2.8 Å between Ca2+ and protein ligands were included based on the EF-hand model for a typical and S100-type Ca2+-binding domain, respectively. An ensemble of structures, calculated without these Ca2+ restraints, had an RMSD of 0.625, demonstrating that the inclusion of such restraints had no effect on the overall structure of the complex. Hydrogen bond constraints of rHN-O = 1.5–2.8 Å and rN-O = 2.4–2.5 Å were included in the final stage of structure calculations. Structures calculated without hydrogen bonds had an RMSD of 0.653 when compared to those calculated with hydrogen bonds, indicating that inclusion of these constraints also had little or no influence on the overall structure. Pseudopotentials for secondary 13Cα and 13Cβ chemical shifts and a conformational data base potential were included in the final simulated annealing refinements using the computer program XPLOR 54. The internuclear dipolar coupling (in Hz) were determined from the difference in J splitting between isotropic and radially compressed polyacrylamide-aligned phases using both a 2D IPAP 1H-15N HSQC to record N-HN splittings and a 3D (H)CA(CO)NH experiment without Hα decoupling during Cα cquisition in t2 to record C α -H α splittings, and were incorporated into the final structure calculation as previously described 5; 17. Q-factors were calculated by randomly removing ~10% of both the N-HN and C α -H α RDC data, and then comparing these values to those back-calculated from the structure. The final 20 structures were selected (from 200) based on lowest energy and were of high quality based on the statistical criteria listed in table 1. The coordinates of the S100A1-TRTK12 structure have been deposited in the Protein Data Base (PDB accession number 2kbm) and chemical shift assignments for the S100A1-TRTK12 complex have been deposited in the BioMagResBank (accession numbers 16050).
Luminescence Spectroscopy
All experiments used to determine binding constants were performed on an Aminco Bowman series 2 luminescence spectrophotofluorometer with a temperature probe (quartz cuvette) maintained at 25 °C. The binding constants were obtained from titrations, repeated in triplicate and performed with at least two different concentrations of Tb3+. Typical conditions for S100A1 titrations included 20 mM HEPES buffer pH 7.0 and 20 mM DTT, 100 μM TRTK12, and 1–5 μM Tb3+. These samples were lyophilized and resuspended in the appropriate volume of D2O. Competition experiments, repeated in triplicate, used the same sample as the Tb3+ binding experiments, with the addition of 1–5 μM S100A1. Ca2+ KD values were calculated using KD = K′/(1 + [Tb3+]/TbKD, where K′ is from the best-fit curve of the Ca2+ titration data, and the TbKD term is from the best-fit curve of the Tb3+ titration data.
Fluorescence Anisotropy
All experiments were performed using fluorescence spectroscopy techniques on an SLM-Aminco Bowman series 2 (Thermo, Asheville, NC) fluorescence spectrophotometer with an excitation wavelength of 542 nm and an emission wavelength of 590 nm. All experiments were repeated three or more times, and all samples were studied at 25 °C with sample conditions consisting of 5mM HEPES, pH 7.5, 1 mM DTT, 25 mM NaCl, 1 mM CaCl2, and 50 nM TRTK12-TAMRA. Fluorescence anisotropy measurements during Pnt competition binding studies were used to monitor the binding of TRTK-TAMRA to S100A1 using the same set of conditions with the addition of 3.5 μM S100A1. The binding data were fit using a single site binding model with Origin software (OriginLab Corp., Northampton, MA) and one peptide and one Pnt molecule bound per S100A1 subunit. An equation derived by Nikolovska-Coleska et al. was used for determination of the KD, as previously described 42 using:
Where [I]50 is the concentration of pentamidine at 50% inhibition, [L]50 is the concentration of the free TAMRA-TRTK peptide at 50% inhibition, [P]0 is the concentration of the free protein at 0% inhibition, and TAMRA-TRTKKD is the dissociation constant of the S100A1-TAMRA-TRTK12.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM58888; CA107331 to DJW). The NMR spectrometers used in these studies were purchased, in part, with funds from shared instrumentation grants from the NIH (S10 RR10441; S10 RR15741; S10 RR16812; S10 RR23447 to DJW) and from the NSF (DBI 0115795 to DJW). NTW was partially supported by NIAMS training grant T32 AR007592 to the Interdisciplinary Program in Muscle Biology, University of Maryland School of Medicine, and by an American Heart Association training grant 0615343U. We would also like to thank Thomas Charpentier for his assistance in editing the manuscript and for helping to prepare figures.
Footnotes
ACCESSION NUMBERS: Coordinates and structure factors for the S100A1-TRTK12 complex have been deposited in the Protein Data Bank with accession number 2kbm.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strynadka NCJ, James MNG. Crystal structures of the helix-loop-helix calcium- binding proteins. Ann Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 2.Ikura M, Yap KL. Where cancer meets calcium--p53 crosstalk with EF-hands. Nat Struct Biol. 2000;7:525–7. doi: 10.1038/76721. [DOI] [PubMed] [Google Scholar]
- 3.Smith SP, Shaw GS. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure. 1998;6:211–22. doi: 10.1016/s0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 4.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37:2729–40. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 5.Wright NT, Varney KM, Ellis KC, Markowitz J, Gitti RK, Zimmer DB, Weber DJ. The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. J Mol Biol. 2005;353:410–426. doi: 10.1016/j.jmb.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763:1284–97. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 7.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;296:201–14. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbuglia M, Verzini M, Rustandi RR, Osterloh D, Weber DJ, Gerke V, Donato R. Role of the C-terminal extension in the interaction of S100A1 with GFAP, tubulin, the S100A1- and S100B-inhibitory peptide, TRTK-12, and a peptide derived from p53, and the S100A1 inhibitory effect on GFAP polymerization. Biochem Biophys Res Commun. 1999;254:36–41. doi: 10.1006/bbrc.1998.9881. [DOI] [PubMed] [Google Scholar]
- 9.Garbuglia M, Verzini M, Donato R. Annexin VI binds S100A1 and S100B and blocks the ability of S100A1 and S100B to inhibit desmin and GFAP assemblies into intermediate filaments. Cell Calcium. 1998;24:177–91. doi: 10.1016/s0143-4160(98)90127-0. [DOI] [PubMed] [Google Scholar]
- 10.Rustandi RR, Baldisseri DM, Drohat AC, Weber DJ. Structural changes in the C-terminus of Ca2+-bound rat S100B (beta beta) upon binding to a peptide derived from the C-terminal regulatory domain of p53. Protein Sci. 1999;8:1743–51. doi: 10.1110/ps.8.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landar A, Caddell G, Chessher J, Zimmer DB. Identification of an S100A1/S100B target protein: phosphoglucomutase. Cell Calcium. 1996;20:279–85. doi: 10.1016/s0143-4160(96)90033-0. [DOI] [PubMed] [Google Scholar]
- 12.Arcuri C, Giambanco I, Bianchi R, Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience. 2002;109:371–88. doi: 10.1016/s0306-4522(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 13.Filipek A, Jastrzebska B, Nowotny M, Kuznicki J. CacyBP/SIP, a calcyclin and Siah-1-interacting protein, binds EF-hand proteins of the S100 family. J Biol Chem. 2002;277:28848–52. doi: 10.1074/jbc.M203602200. [DOI] [PubMed] [Google Scholar]
- 14.Ivanenkov VV, Jamieson GA, Jr, Gruenstein E, Dimlich RV. Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J Biol Chem. 1995;270:14651–8. doi: 10.1074/jbc.270.24.14651. [DOI] [PubMed] [Google Scholar]
- 15.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;324:1003–14. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 16.Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–24. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 17.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J Biol Chem. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nat Struct Biol. 2000;7:570–4. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 19.Weber DJ, Rustandi RR, Carrier F, Zimmer DB. Interaction of dimeric S100B(ββ) with the tumor suppressor protein: A model for Ca-dependent S100-target protein interactions. In: Pochet R, editor. The molecular basis of calcium action in biology and medicine. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. [Google Scholar]
- 20.Nilges M. A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins. 1993;17:297–309. doi: 10.1002/prot.340170307. [DOI] [PubMed] [Google Scholar]
- 21.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 22.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–8. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 23.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol. 1997;4:732–8. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 24.Markowitz J, Rustandi RR, Varney KM, Wilder PT, Udan R, Wu SL, Horrocks WD, Weber DJ. Calcium-Binding Properties of Wild-Type and EF-Hand Mutants of S100B in the Presence and Absence of a Peptide Derived from the C-Terminal Negative Regulatory Domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 25.Johnson JD, Snyder C, Walsh M, Flynn M. Effects of myosin light chain kinase and peptides on Ca2+ exchange with the N- and C-terminal Ca2+ binding sites of calmodulin. J Biol Chem. 1996;271:761–7. doi: 10.1074/jbc.271.2.761. [DOI] [PubMed] [Google Scholar]
- 26.Olwin BB, Edelman AM, Krebs EG, Storm DR. Quantitation of energy coupling between Ca2+, calmodulin, skeletal muscle myosin light chain kinase, and kinase substrates. J Biol Chem. 1984;259:10949–55. [PubMed] [Google Scholar]
- 27.Persechini A, White HD, Gansz KJ. Different mechanisms for Ca2+ dissociation from complexes of calmodulin with nitric oxide synthase or myosin light chain kinase. J Biol Chem. 1996;271:62–7. doi: 10.1074/jbc.271.1.62. [DOI] [PubMed] [Google Scholar]
- 28.Yazawa M, Vorherr T, James P, Carafoli E, Yagi K. Binding of calcium by calmodulin: influence of the calmodulin binding domain of the plasma membrane calcium pump. Biochemistry. 1992;31:3171–6. doi: 10.1021/bi00127a018. [DOI] [PubMed] [Google Scholar]
- 29.Durussel I, Méhul B, Bernard D, Schmidt R, Cox JA. Cation- and peptide-binding properties of human calmodulin-like skin protein. Biochemistry. 2002;41:5439–48. doi: 10.1021/bi016062z. [DOI] [PubMed] [Google Scholar]
- 30.Stemmer PM, Klee CB. Dual calcium ion regulation of calcineurin by calmodulin and calcineurin B. Biochemistry. 1994;33:6859–66. doi: 10.1021/bi00188a015. [DOI] [PubMed] [Google Scholar]
- 31.Peersen OB, Madsen TS, Falke JJ. Intermolecular tuning of calmodulin by target peptides and proteins: differential effects on Ca2+ binding and implications for kinase activation. Protein Sci. 1997;6:794–807. doi: 10.1002/pro.5560060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer DB, Chaplin J, Baldwin A, Rast M. S100-mediated signal transduction in the nervous system and neurological diseases. Cell Mol Biol (Noisy-le-grand) 2005;51:201–14. [PubMed] [Google Scholar]
- 33.Zimmer DB, Chessher J, Wilson GL, Zimmer WE. S100A1 and S100B expression and target proteins in type I diabetes. Endocrinology. 1997;138:5176–83. doi: 10.1210/endo.138.12.5579. [DOI] [PubMed] [Google Scholar]
- 34.Li G, Barthelemy A, Feng G, Gentil-Perret A, Peoc’h M, Genin C, Tostain J. S100A1: a powerful marker to differentiate chromophobe renal cell carcinoma from renal oncocytoma. Histopathology. 2007;50:642–7. doi: 10.1111/j.1365-2559.2007.02655.x. [DOI] [PubMed] [Google Scholar]
- 35.Pelc P, Vanmuylder N, Lefranc F, Heizmann CW, Hassid S, Salmon I, Kiss R, Louryan S, Decaestecker C. Differential expression of S100 calcium-binding proteins in epidermoid cysts, branchial cysts, craniopharyngiomas and cholesteatomas. Histopathology. 2003;42:387–94. doi: 10.1046/j.1365-2559.2003.01588.x. [DOI] [PubMed] [Google Scholar]
- 36.Prosser BL, Wright NT, Hernandez-Ochoa E, Varney KM, Liu Y, Olojo RO, Zimmer DB, Weber DJ, Schneider MF. S100A1 binds to the calmodulin binding site of ryanodine receptor and modulates skeletal muscle EC coupling. J Biol Chem. 2008 doi: 10.1074/jbc.M709231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du SJ, Cole JJ, Tenis N, Gao XM, Kontgen F, Kemp BE, Heierhorst J. Impaired cardiac contractility response to hemodynamic stress in S100A1-deficient mice. Mol Cell Biol. 2002;22:2821–2829. doi: 10.1128/MCB.22.8.2821-2829.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulle S, Mariggio MA, Belia S, Petrelli C, Ballarini P, Guarnieri S, Fano G. Rapid desensitization of PC12 cells stimulated with high concentrations of extracellular S100. Neurosci. 1999;89:991–997. doi: 10.1016/s0306-4522(98)00386-8. [DOI] [PubMed] [Google Scholar]
- 39.Mariggio MA, Fulle S, Calissano P, Nicoletti I, Fano G. The brain protein S-100ab induces apoptosis in PC12 cells. Neuroscience. 1994;60:29–35. doi: 10.1016/0306-4522(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 40.O’Dowd BS, Zhao WQ, Ng KT, Robinson SR. Chicks injected with antisera to either S-100 alpha or S-100 beta protein develop amnesia for a passive avoidance task. Neurobiol Learn Mem. 1997;67:197–206. doi: 10.1006/nlme.1997.3766. [DOI] [PubMed] [Google Scholar]
- 41.Zimmer DB, Cornwall EH, Reynolds PD, Donald CM. S100A1 regulates neurite organization, tubulin levels, and proliferation in PC12 cells. J Biol Chem. 1998;273:4705–11. doi: 10.1074/jbc.273.8.4705. [DOI] [PubMed] [Google Scholar]
- 42.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–73. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 43.Charpentier TH, Wilder PT, Liriano MA, Varney KM, Pozharski E, Mackerell AD, Jr, Coop A, Toth EA, Weber DJ. Divalent Metal Ion Complexes of S100B in the Absence and Presence of Pentamidine. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.06.047. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J Biol Chem. 2008;283:26676–83. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dempsey AC, Walsh MP, Shaw GS. Unmasking the annexin I interaction from the structure of Apo-S100A11. Structure (Camb) 2003;11:887–97. doi: 10.1016/s0969-2126(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 46.Lee YT, Dimitrova YN, Schneider G, Ridenour WB, Bhattacharya S, Soss SE, Caprioli RM, Filipek A, Chazin WJ. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protein: a novel mode for S100 protein target recognition. Biochemistry. 2008;47:10921–32. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright NT, Margolis JW, Margolis FL, Weber DJ. Refinement of the solution structure of rat olfactory marker protein (OMP) J Biomol NMR. 2005;33:63–8. doi: 10.1007/s10858-005-1281-7. [DOI] [PubMed] [Google Scholar]
- 48.Wang C, Karpowich N, Hunt JF, Rance M, Palmer AG. Dynamics of ATP-binding cassette contribute to allosteric control, nucleotide binding and energy transduction in ABC transporters. J Mol Biol. 2004;342:525–37. doi: 10.1016/j.jmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Wright NT, Inman KG, Levine JA, Cannon BR, Varney KM, Weber DJ. Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B. J Biomol NMR. 2008;42:279–86. doi: 10.1007/s10858-008-9282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikura M, Bax A. Isotope-filtered 2D NMR of a protein-peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J Am Chem Soc. 1992;114:2433–2440. [Google Scholar]
- 51.Bax A, Grzesiek S, Gronenborn AM, Clore GM. Isotope-Filtered 2D HOHAHA Spectroscopy of a Peptide-Protein Complex Using Heteronuclear Hartmann-Hahn Dephasing. J Magn Reson A. 1994;106:269–73. [Google Scholar]
- 52.Vuister GW, Kim SJ, Orosz A, Marquardt J, Wu C, Bax A. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol. 1994;1:605–14. [PubMed] [Google Scholar]
- 53.Muhandiram DR, Guang YX, Kay LE. An enhanced-sensitivity pure absorption gradient 4D 15N,13C-edited NOESY experiment. J Biomol NMR. 1993;3:463–470. [Google Scholar]
- 54.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]