Abstract
Background
Uteroglobin-related protein 1 (UGRP1) is a secretory protein expressed in the airways and is speculated to have anti-inflammatory activity. In the mouse, its gene expression is down-regulated by interleukin (IL)-5 and -9, and up-regulated by IL-10. However, the precise role of UGRP1 in human inflammatory airway diseases such as asthma has not been clarified. The objectives of this study were to establish an ELISA system to quantify UGRP1 protein, and to examine whether plasma UGRP1 levels are associated with the G-112A polymorphism, asthma susceptibility, and its severity.
Methods
152 asthma patients and 103 normal controls were involved in this study. Mice were immunized with recombinant UGRP1 and hybridoma cell lines were established. A sandwich ELISA system was established by using two monoclonal antibodies recognizing different epitopes. Plasma UGRP1 levels were measured with the ELISA system and the G-112A allele was detected by using real-time PCR.
Results
An ELISA system was established that allowed determination of UGRP1 levels within the range of 9.6–1250 pg/ml. The mean plasma UGRP1 levels for subjects with -112A allele were significantly lower than those without it (p = 0.025). Although there was no significant difference in the plasma UGRP1 levels between asthma patients and controls (p = 0.13), severe asthma patients without oral corticosteroid had significantly lower plasma UGRP1 levels compared to mild- or moderate- asthma patients and controls (p = 0.004, 0.03 and 0.003, respectively).
Conclusions
The ELISA system for quantifing UGRP1 protein was established, and plasma UGRP1 levels were associated with the G-112A UGRP1 gene promoter polymorphism and the severity of asthma.
Keywords: asthma, genetic polymorphism, plasma levels, severity, uteroglobin-related protein 1
INTRODUCTION
The uteroglobin-related protein 1 (UGRP1) gene, officially named secretoglobin 3A2 (SCGB3A2) encodes a homodimeric secretory protein of 10 kDa.1 It was originally identified as a downstream target gene for the homeodomain transcription factor, thyroid-specific enhancer-binding protein (T/EBP), also called thyroid transcription factor 1 (TTF1) or NKX 2.1.2 T/EBP regulates the expression of thyroid- and lung-specific genes, the latter of which includes surfactant proteins A, B, and C, and Clara cell secretory protein (CCSP).1,3–6 Mice homozygous for the disrupted gene for T/ebp were born dead and lacked lung parenchyma, demonstrating that T/EBP plays an essential role in lung development.7 In murine lung, UGRP1 is expressed mainly in the airway epithelial cells and is an early molecular marker for Clara cells.8 The spatial distribution of UGRP1 is similar between humans and mice although in humans the expression is primarily found in serous-like cells of bronchi and associated glands.8 These findings suggest that UGRP1 may play a significant physiological role in the lung. However, little is known about the function of UGRP1.
The amino acid sequence of human UGRP1 has 25% identity to human CCSP (SCGB1A1), a prototypical member of the SCGB gene super-family, that is believed to function as an anti-inflammatory protein.2 The expression pattern of UGRP1 is similar to that of CCSP although UGRP1 expression is found in the epithelial cells of the trachea where CCSP is not expressed.2 The CCSP family (SCGB1A) proteins are characterized by the presence of two conserved cysteine residues at the N- and C-terminal regions of polypeptides separated by a lysine that are required to form a dimmer.9 The area containing the conserved lysine residue is called antiflammin. 9 The antiflammin domain exhibits potent anti-inflammatory and immunomodulatory activities and appears to be responsible for the PLA2-inhibitory activity of CCSP.9,10 The amino acid sequence similarities between UGRP1 and the CCSP family proteins are significant in the areas of the signal peptide and antiflammin,2 These findings suggest that UGRP1 may also have an anti-inflammatory function.
Recently, it was reported that allergen-induced inflammation in allergen-sensitized mice was associated with a reduction in UGRP1 mRNA expression in lung as compared with naïve animals, and the expression returned to normal with dexamethasone treatment.2 Further, the possible involvement of interleukin (IL)-5 and -9 in the decreased airway UGRP1 expression in allergic airway inflammation was demonstrated.11,12 On the other hand, IL-10, known as an anti-inflammatory cytokine, induced Ugrp1 gene expression in lung epithelial cells, suggesting that UGRP1 might be a target for the anti-inflammatory activities of IL-10.13
The human UGRP1 gene is located on chromosome 5q31–32, the area containing one or more genes that may potentially play a role in airway inflammation associated with atopic asthma. These genes include a number of proinflammatory cytokines such as IL-3, -4, -5, -9, and -13.14–18 We previously reported the presence of a G to A polymorphism at -112 bp in the human UGRP1 gene promoter and that the -112A allele is responsible for a 24% reduction in the promoter activity as compared to the wild-type -112G allele. 19 This polymorphism exhibited a significant association with asthma phenotype in an adult Japanese population.19
In the present study, we established an ELISA assay to measure the concentration of human UGRP1 in plasma. Using this ELISA system, we successfully demonstrated an association of UGRP1 levels to the G-112A polymorphism and the severity of asthma.
METHODS
SUBJECTS
A total of 255 Japanese subjects, 152 asthma patients and 103 controls, were enrolled in this study. Subjects with bronchial asthma were recruited from the outpatient clinic at the Fukushima Medical University Hospital according to the following criteria: First, the presence of at least two of the following symptoms was examined: recurrent cough, wheezing or dyspnea. Second, we examined for increased airway responsiveness to methacholine or the presence of reversible airflow limitation; the latter refers to 15% variability in the forced expiratory volume in 1 second (FEV1), or in the peak expiratory flow rate with or without an inhaled short-acting β2-agonist. The third criterion was the absence of any other pulmonary diseases. Control subjects were normal volunteers who had no symptoms, or past history of asthma, or other airway or allergic diseases. All asthma patients had been treated according to the Japanese Asthma Prevention and Management Guideline20 and were at a clinically stable phase. Lung function, serum nonspecific IgE, and antigen-specific IgE for 10 common inhalant antigens including house-dust mites, molds, pollens, and animal dander (cat and dog) were examined in both asthma patients and controls. Concentrations of antigen-specific IgE of more than 0.69 UA/ml were considered as positive. Subjects were characterized as “atopic” if they had at least one positive antigen-specific IgE or a nonspecific IgE level of 250 IU/ml or greater. The severity of asthma was classified into mild, moderate and severe, according to the Global Initiative for Asthma (GINA) guideline.21 DNA was extracted from peripheral blood cells. All of the participants gave informed consent and the study was approved by the ethical committee of the Fukushima Medical University.
EXPRESSION AND PURIFICATION OF HUMAN UGRP1 PROTEIN
Expression of recombinant human UGRP1 (hUGRP1) was performed using the pET Expression System (Novagen, EMD Biosciences, Dermstdt, Germany). A cDNA encoding UGRP1 mature sequence was sub-cloned in-frame into pET32a (+), which was transformed into E. coli OrigamiDE3. A scaled-up culture was performed in LB medium containing 0.1 mg/ml ampicillin and hUGRP1 expression was induced by the addition of 1 mM IPTG. The cultured medium was centrifuged at 8,000 rpm for 15 minutes and pellets were suspended in binding buffer (20 mM Tris-HC1 buffer, pH 8.0 containing 0.5 M NaCl and 5 mM imidazole). The suspension was sonicated and centrifuged at 18,000 rpm for 15 minutes, and supernatants were collected. hUGRP1 in the supernatants was purified with a Ni2+-charged HiTrap Chelating HP column equipped with ÄKTA prime purification system (Amersham Biosciences, Buckinghamshire, UK). Briefly, samples were loaded onto the column equilibrated with binding buffer. The column was washed with the binding buffer and elution was carried out with a linear imidazole gradient from 5 to 500 mM. Fractions containing recombinant protein were collected and dialyzed against 20 mM Tris-HCl buffer, pH 7.4, resulting in purified hUGRP1. The N-terminal His-tag of hUGRP1 was then removed by cleavage with thrombin using the Thrombin Cleavage Capture Kit (Novagen). The cleaved His-tag peptide was removed by incubation with Ni-NTA agarose (Qiagen, Hilden, Germany). The hUGRP1 (with His-tag) and mature UGRP1 (without His-tag) were used as a standard for ELISA and a protein antigen for immunization, respectively.
MONOCLONAL ANTIBODIES
Monoclonal antibodies against hUGRP1 were obtained by immunizing BALB/c mice with mature hUGRP1 prepared as above. Briefly, spleen cells from immunized mice were fused with myeloma-cells (SP2) by using polyethylene glycol to produce hybridoma cell lines. Five hybridoma-cell lines that survived the hypoxanthine-aminopterin-thymidine (HAT) selection were chosen for further studies. Each hybridoma-cell line was confirmed for its appropriate antibody titer, and the class of monoclonal antibody was determined as IgG1K for all five hybridoma-cell lines. Each of the cloned hybridoma-cell lines were grown as tumors in mouse abdominal cavities. Each monoclonal antibody was collected from their ascites fluid and purified with a protein G column. Specificity of five antibodies was confirmed by examining specific findings to hUGPR1 by Western blotting. Two monoclonal antibodies (MoAb 4 G10 and 5B1), recognizing different epitopes of hUGRP1, were selected for the sandwich ELISA system. MoAb 4G10 was prepared as a capture antibody. MoAb 5B1 was used as a detection antibody after labeling with horseradish peroxidase using a Peroxidase Labeling Kit (Roche, Grenzacherstrasse, Switzerland) according to the manufacturer’s instruction. The protocols to produce monoclonal antibodies were approved by the Institutional Animal Care and Use Committee.
QUANTITATION OF UGRP1
UGRP1 concentrations were measured with ELISA. Briefly, SUMIRON Multi Well Plates for ELISA (Sumitomo Bakelite Co., Ltd, Tokyo, Japan) were coated overnight at 4°C with MoAb 4G10 at a concentration of 1 µg/ml in Tris-buffered saline (TBS), pH 7.4. After unbound antibody was removed, the plate was blocked with 300 µl of 1% bovine serum albumin in TBS (BSA-TBS). Standards or samples (100 µl) were added and allowed to react with the coated antibody at 37°C for 60 minutes. Unbound antigen was removed, and then 100 µl of horseradish peroxidase-conjugated MoAb 5B1 diluted 1 in 500 in BSA-TBS was added, followed by incubation at 37°C for 60 minutes. After repeating a wash with 0.05% Tween 20 in TBS, 100 µl substrate solution (prepared according to the manufacturer’s instruction) was added to each well. Enzyme activity was measured at the absorbance of 450 nm after terminating the reaction by the addition of phosphoric acid (1M, 100 µl). A linear standard curve was obtained between 9.6 and 1250 pg/ml of UGRP1. All assays were performed in duplicate.
GENOTYPING
For each individual, the -112G/A polymorphism in the human UGRP1 gene promoter was determined by real-time polymerase chain reactions (PCR). Allelic discrimination assays were performed by a Taqman 7000 platform (Applied Biosystems, USA) according to the manufacturer’s instruction. Briefly, the PCR reaction contained 20 ng genomic DNA, 2.5 µL Taqman master mix and 0.125 µL of 40X assay mix which contains primers (forward; 5’ GCT GTA CTG TAG AGC TTT GTT TCT CA 3’, reverse 5’ CCC CAC CAA AGA AAG GGA TAA ATG T 3’) and probes (wild type; VIC-CAA ATT GTT TGG TGA GAA A, mutant type; FAM-TCC AAA TTG TTT AGT GAG AAA). PCR was performed using 96 well plates in a real-time PCR system (ABI PRISM 7000) with reaction conditions of 95°C for 10 minutes followed by 40 cycles of 92°C for 15 seconds and 60°C for 1 minute.
STATISTICAL ANALYSIS
Genotype distributions were analyzed by using the SNP Analyze software (Dynacom, Kanagawa, Japan). Statistical analyses were performed with SPSS II for Windows. Values for UGRP1 and nonspecific IgE were log transformed for statistical analysis, since these values were log normally distributed in the population. The Mann-Whitney U-test was used to compare unpaired sets of data. The Chi square test was used to analyze the relationship between the existence of the -112A allele and asthma status. Oneway analysis of variance test was used to analyze the relationship between plasma UGRP1 levels and the severity of asthma. Correlations were examined by using Spearman rank analysis. The level of critical significance was assigned at p values less than 0.05.
RESULTS
CHARACTERISTICS OF SUBJECTS
The numbers of control subjects and asthma patients enrolled in this study were 103 and 152, respectively (Table 1). There were significant differences between the cases and control group with respect to age and male/female ratio. Asthma patients exhibited significantly higher total serum IgE levels and atopic/non-atopic ratio, and reduced pulmonary functions as compared with the control group. Within asthma patients, no significant differences were found in these parameters among mild, moderate and severe asthma patients (p = 0.167).
Table 1.
Characteristics of the subjects
| Control (n = 103) | Asthma (n = 141) |
||||
|---|---|---|---|---|---|
| Mild (n = 35) | Moderate (n = 87) | Severe (n = 19) | Total | ||
| Age (Y) | 30.5 (29.1–31.9)* | 43.9 (40.0–47.8) | 47.3 (44.2–50.4) | 43.5 (37.7–49.2) | 45.9 (43.7–48.2) |
| Sex, M/F (male%) | 62/41 (60.2%)† | 16/19 (45.7%) | 41/46 (47.1%) | 7/12 (36.8%) | 64/77 (45.4%) |
| Total IgE (UA/Ml) | 71.5 (55.2–92.5)* | 228.2 (146.6–355.3) | 215.2 (159.1–291.1) | 158.6 (57.8–435.2) | 209.4 (163.3–268.5) |
| A/N (Atopic%) | 25/78 (24.3%)* | 30/5 (85.7%) | 67/20 (77.0%) | 12/7 (63.2%) | 109/32 (77.3%) |
| FEV1 (L) | 3.05 (2.72–3.39)* | 2.61 (2.37–2.85) | 2.38 (2.19–2.56) | 2.46 (2.08–2.83) | 2.45 (2.31–2.58) |
| FEV1% (%) | 86.1 (83.5–88.8)* | 75.8 (72.4–79.2) | 71.1 (68.0–74.1) | 75.3 (68.0–82.6) | 72.8 (70.6–75.1) |
Data are expressed as geometric means and 95% Cls. Total IgE: total immunoglobulin E. M/F: Male/Female. A/N: atopic/non-atopic. FEV1: forced expiratory volume in 1 second. FEV1% percentage of FEV1 in forced vital capacity.
p < 0.01
p < 0.05 Compared with asthma patients.
QUANTIFICATION OF PLASMA UGRP1 LEVELS
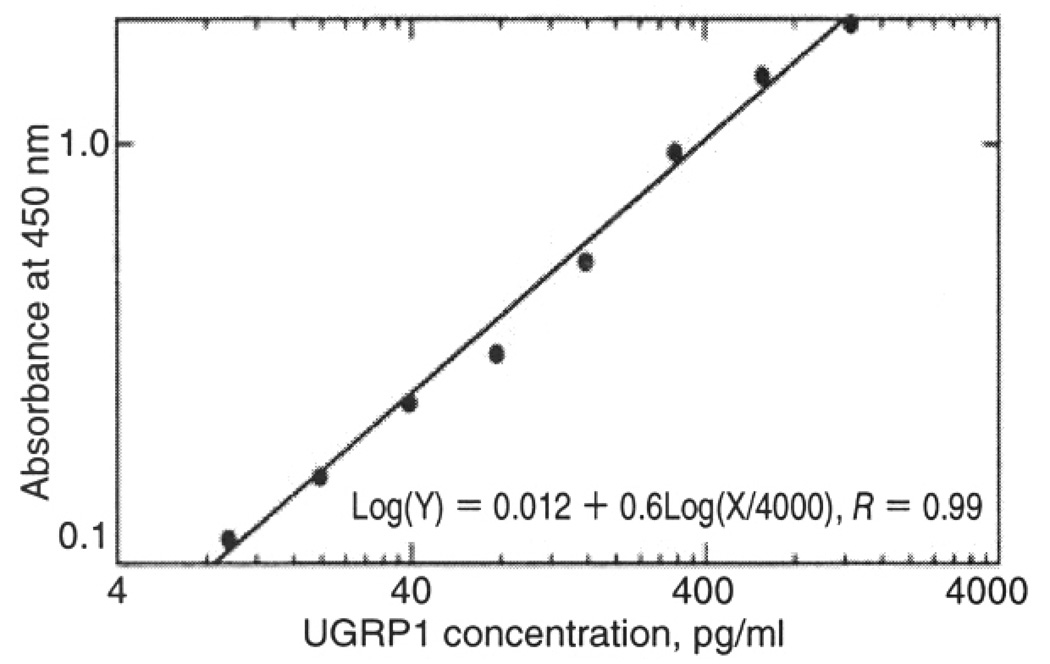
The standard curve (log-log fit) for UGRP1 demonstrated good linearity within 9.6–1250 pg/ml of UGRP1 (r = 0.99), suggesting that UGRP1 levels can be estimated within this range (Fig. 1). In only one asthmatic subject, the level was over 1250 pg/ml; this sample was diluted in assay buffer and re-measured. Age and sex distributions did not affect plasma UGRP1 levels (data not shown). There was no significant correlation between nonspecific IgE and plasma UGRP1 levels (p = 0.37), and no significant difference in plasma UGRP1 levels between atopic and non-atopic subjects (p = 0.88).
Fig. 1.
The standard curve (log-log fit) for measuring UGRP1 levels. UGRP1 concentration can be accurately determined between 9.6–1250 pg/ml.
UGRP1 PROMOTER G-112A POLYMORPHISM, ASTHMA PHENOTYPE AND PLASMA UGRP1 LEVELS
The frequencies of G-112A polymorphism were determined in control and asthmatic subjects (Table 2). These genotype distributions were confirmed to fit to Hardy-Weinberg distribution. Thirty-seven out of 103 normal control subjects (35.9%) had the -112A allele, while 69 of 152 asthmatic subjects (44.3%) had the -112A allele, suggesting a weak trend of a more frequent presence of the -112A allele in asthma patients as compared with control subjects although no statistical significance was obtained. When asthma patients were classified by the severity of their asthma, the -112A allele was more frequently found in severe asthma patients; although there was no statistically significant difference.
Table 2.
Distribution of UGRP1 G-112A genotype in asthmatic patients and controls
| Control | Asthma |
||||
|---|---|---|---|---|---|
| Mild | Moderate | Severe | Total | ||
| Genotype | |||||
| G/G | 66 (64.1%) | 20 (57.1%) | 49 (56.3%) | 7 (36.8%) | 76 (53.9%) |
| G/A or A/A | 37 (35.9%) | 15 (42.9%) | 38 (43.7%) | 12 (63.2%) | 65 (46.1%) |
| A allele frequency (%) | 20.4 | 25.7 | 25.3 | 39.5 | 27.3 |
| Total subjects | 103 (100%) | 35 (100%) | 87 (100%) | 19 (100%) | 141 (100%) |
There was no significant different between each group.
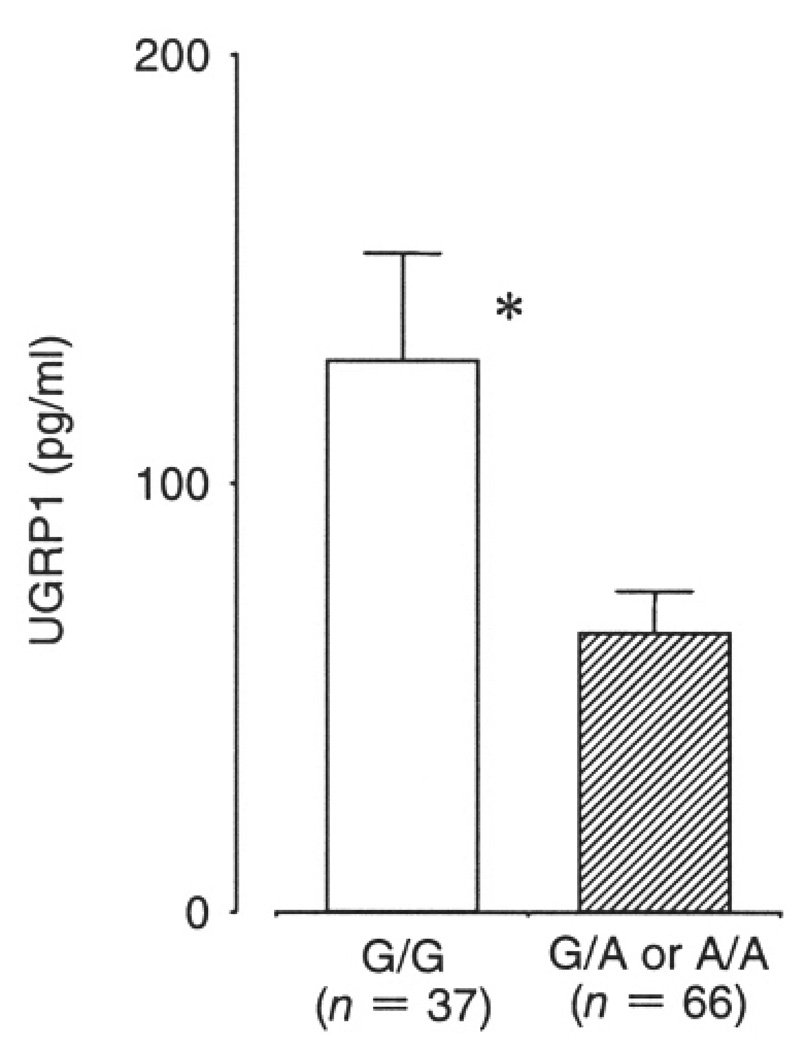
When plasma UGRP1 levels were examined, those having at least one -112A allele (G/A or A/A genotype) had a mean plasma UGRP1 level of 70.6+/−6.8 (SE) pg/ml that was significantly lower than that of wild-type subjects (G/G genotype) (123.0+/−18.0 pg/ml, p = 0.007). Significantly reduced plasma UGRP1 levels were also found for healthy control subjects with A-allele (G/A or G/G genotype, 65.5+/−9.6 pg/ ml) as compared with those without A-allele (G/G genotype, 128.7+/−24.7 pg/ml) (p = 0.02) (Fig. 2).
Fig. 2.
Comparison of plasma UGRP1 levels between subjects with (G/A or A/A) and without (G/G) -112A allele. The results for control subjects are shown. The total number of subjects with -112A allele was 37 and those without A allele was 66. Data are means +/− SE. * p = 0.02 compared to control.
ASTHMA PHENOTYPE AND PLASMA UGRP1 LEVELS
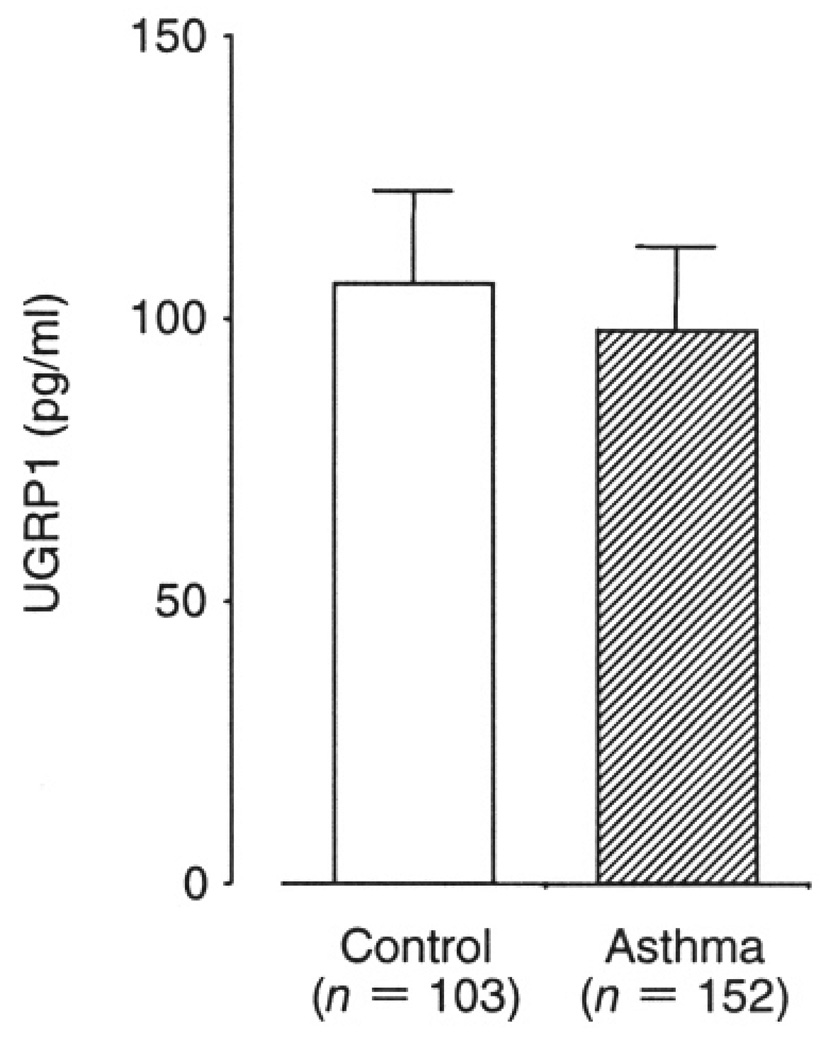
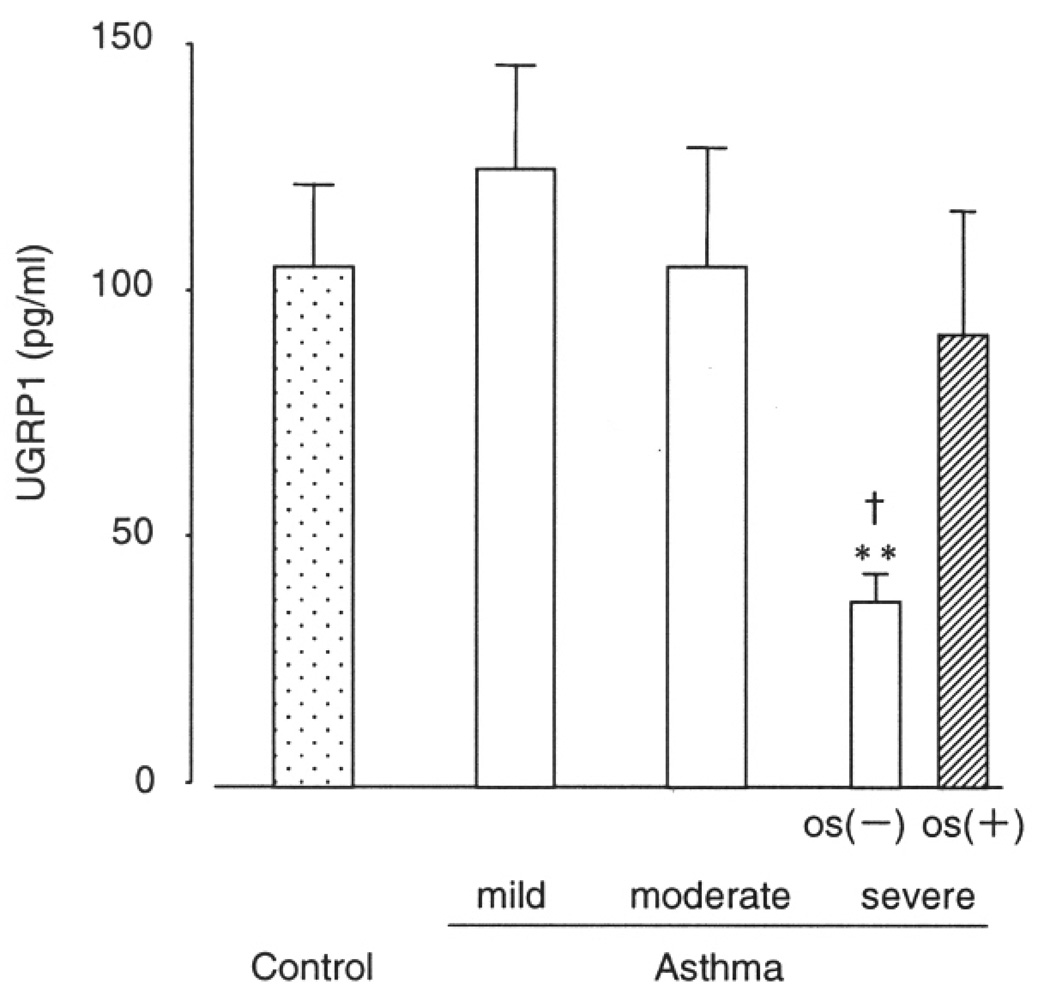
The average plasma UGRP1 level was 98.0+/−14.7 (SE) pg/ml in asthma patients, and 106.0+/−16.4 pg/ml in control subjects. Although asthma patients had lower plasma UGRP1 levels as compared to controls, the difference was not statistically significant (p = 0.12, Fig. 3). Among asthma patients, the average plasma UGRP1 levels decreased in the order of asthma severity; 112.7+/−21.3 pg/ml, 106.0+/−23.9 pg/ml and 57.6+/−10.8 pg/ml for mild, moderate and severe asthma patients, respectively, and severe asthma patients tended to have lower plasma UGRP1 (p = 0.06). However, 11 of 19 severe asthma patients were treated with systemic corticosteroids that have been known to increase UGRP1 expression. For this reason, this group was divided into two groups, subjects treated with or without oral corticosteroid (OS). Mean plasma UGRP1 levels for severe asthma patients with- and without-OS were 92.0+/−25.3 pg/ml and 37.7+/−5.7 pg/ml, and subjects with OS had significantly higher plasma UGPR1 levels compared to those without it (p = 0.013). In addition, as shown in Figure 4, severe asthma patients without OS had significantly lower plasma UGRP1 levels compared to controls (P = 0.003) and mild and moderate asthma patients (p = 0.004 and 0.03, respectively).
Fig. 3.
Comparison of plasma UGRP1 levels between control and asthma patients. Data are means +/− SE. There was no statistically significant difference.
Fig. 4.
Comparison of plasma UGRP1 levels among healthy control and mild-severe asthmatic with or without oral corticosteroid (OS). Data are means +/− SE. ** p < 0.05 compared to control, mild, and moderate asthma. † p = 0.013 compared to severe asthma with OS.
DISCUSSION
In this study, we established a sandwich ELISA system to measure human UGRP1 protein concentrations using two monoclonal antibodies produced against human UGRP1. Plasma UGRP1 levels could be determined by this ELISA system within the range of 9.6–1250 pg/ml. Since in humans, expression of UGRP1 is restricted to the airways, plasma UGRP1 is likely to be derived from the respiratory tracts.2
By using this ELISA system, we demonstrated that plasma UGRP1 levels are affected by the G-112A polymorphism of the UGRP1 gene promoter. We previously reported the functional significance of the G-112A polymorphism in the promoter of UGRP1 gene.19 By luciferase reporter assays, a 24% reduction of the promoter activity was observed for the -112A allele as compared with the -112 G allele. Electrophoretic mobility shift analysis revealed that this was due to decreased affinity of a particular nuclear protein to the -112A binding site.19 In the present study, the mean plasma UGRP1 levels for subjects having at least one -112A allele had 43% lower plasma UGRP1 levels than those having -112 G allele (70.6 and 123.0 pg/ml, respectively, p = 0.007). Previously, we also demonstrated that the G-112A polymorphism of the UGRP1 gene promoter is associated with asthma phenotypes.19 Later, two groups failed to find the same association in populations of childhood asthma.22 This study was not designed to determine the association between asthma phenotype and genotype, meaning that the number of subjects in this study was not large enough to analyze this relation. In spite of such a small subject number, the trend was similar to the previous study in that subjects with the -112A allele were more likely to have asthma than those without (Table 2).
The most important finding in the present study is that a significant association was observed between plasma UGRP1 levels and the severity of asthma when the subjects with severe asthma were grouped according to oral corticosteroid use. Subjects treated with oral corticosteroid had significantly higher plasma UGRP1 levels. This phenomenon has already been demonstrated in the mouse asthma model.2 The reduced plasma UGRP1 levels in severe asthma patients without oral corticosteroid could be due to genetic or acquired mechanisms. This study demonstrated that the UGRP1 gene promoter polymorphism affects plasma UGRP1 levels and that the -112A allele is more likely found in asthma patients especially in severe asthma patients, although there was no statistical significance. This suggests the possibility that the G-112A genotype could be an independent determinant of plasma UGRP1 levels and that UGRP1 might be one of the genetic determinants of asthma severity.
Currently, only limited information is available for the functional role of UGRP1 in lung. However, its similarity to CCSP, especially in the antiflammin domain that exhibits major anti-inflammatory and immunomodulatory activities, suggests a possible anti-inflammatory function for UGRP1. In addition, the suppression of allergic airway inflammation by an over-expression of UGRP1 in the airways has been reported in mice.23 A recent study also demonstrated that the anti-inflammatory effect of IL-10 may be mediated by UGRP1.13 In general, IL-10 is associated with resolution of the inflammatory response process24 and is known to block allergic inflammation in animal models.25,26 IL-10 is up-regulated during acute exacerbation of asthma induced by viral infection, especially in its recovery phase.24,27–29 If the reduction of UGRP1 levels is related to the G-112A UGRP1 promoter polymorphism, the anti-inflammatory action of IL-10 could be reduced in individuals with the -112A allele. A defect in such anti-inflammatory functions in the asthmatic airways might be related to the severity of asthma.
Another possible mechanism for reduced plasma UGRP1 levels in severe asthma is decreased UGRP1 production by remodeling of the small airways. In human airways, the major source of UGRP1 is non-ciliated cells (such as Clara cells), in common with CCSP.8 A decreased number of CCSP positive cells in asthmatic airways have been also demonstrated.30 Further in asthma patients, a significant decrease of CCSP was reported both in their bronchoalveolar lavage fluid31 and serum32 compared to control subjects. Asthmatic patients with a long duration of the disease (>/=10 years) had significantly lower serum CCSP levels than those with a short duration of the disease (<10 years), which may reflect decreased production of CCSP caused by remodeling of the small airways in asthma.32 Since UGRP1 is also expressed mainly in non-ciliated cells (such as Clara cells), a similar mechanism could be estimated for UGRP1.8
In our previous study with a mouse model of allergic airway inflammation, the expression of UGRP1 mRNA in lungs was reduced by antigen inhalation and recovered by dexamethasone treatment. 2 In addition, recent studies demonstrated that IL-5 and -9 reduce UGRP1 expression while IL-10 increases UGRP1 expression in mouse airways.11–13 These results suggest a possible reduction of UGRP1 expression in human asthmatic airways because IL-5 and -9 are known to be elevated in asthmatic airways.33 In this study, the mean plasma UGRP1 levels for asthma patients were close to that of controls (98.0 pg/ml and 106 pg/ml, respectively). One possible explanation for this discrepancy is that plasma UGRP1 levels are associated with asthma severity independent of asthma phenotype as shown in Figure 4, in which plasma UGRP1 levels of mild asthma patients are similar or slightly higher than those of controls.
Another explanation for this discrepancy might be due to a difference in mechanisms of asthma between humans and mice. Th2 dominated immune responses have been recognized to cause allergic diseases including asthma in mice, 34,35 whereas in humans the pattern of T-cell immunity is thought to be more heterogenous.36 Some examples include production of IL-10, which is a Th2 marker for mice but is secreted by human Th1 and Th2 clones,37 and the production of Th2-depedent IgG4 in non-atopies in the absence of IgE.38 Since the subjects of the latter study were adult asthma patients including both atopic and non-atopic types, heterogeneity of immune responses are expected to be large. Asthmatic individuals with dominant expression of 1L-5 and/or -9 compared with IL-10 might have reduced UGRP1, and those with dominant expression of IL-10 might have increased UGRP1 in their lungs. It is likely that UGRP1 gene expression may be under more complex regulation and may be heterogeneous in human asthmatic airways as compared to mice.
Finally, the effect of inhaled corticosteroid (ICS) in asthma patients should be considered. A previous mice study clearly demonstrated that systemic corticosteroids reversed the reduced expression of UGRP1 in the airways.2 In this study, oral corticosteroid was found to increase plasma UGRP1 levels in severe asthma patients. This may raise the possibility that asthma patients treated with inhaled corticosteroid (ICS) might have reduced plasma UGRP1 levels. In our study, 94.3% of asthma patients were treated with ICS. Even in mild asthma patients, a major part (77%) is treated with ICS. ICS might up-regulate the expression of UGRP1 in their airways and increase plasma UGRP1 levels in asthma patients, which masks the difference between asthma patients and controls.
In conclusion, UGRP1 levels in human plasma can be determined with the newly established ELISA system. Plasma UGRP1 levels are associated with the G-112A UGRP1 promoter polymorphism and the severity of asthma.
REFERENCES
- 1.Niimi T, Copeland NG, Gilbert DJ, et al. Cloning, expression, and chromosomal localization of the mouse gene (Scgb3a1, alias Ugrp2) that encodes a member of the novel uteroglobin-related protein gene family. Cytogenet. Genome. Res. 2002;97:120–127. doi: 10.1159/000064067. [DOI] [PubMed] [Google Scholar]
- 2.Niimi T, Keek-Waggoner CL, Popescu NC, et al. UGRP1, a uteroglobin/Clara cell secretory protein-related protein, is a novel lung-enriched downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor. Mol. Endocrinol. 2001;15:2021–2036. doi: 10.1210/mend.15.11.0728. [DOI] [PubMed] [Google Scholar]
- 3.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol. Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno MD, Bohinski RJ, Huelsman KM, Whitsett JA, Korfhagen TR. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J. Biol. Chem. 1995;270:6531–6536. doi: 10.1074/jbc.270.12.6531. [DOI] [PubMed] [Google Scholar]
- 5.Kelly SE, Bachurski CJ, Burhans MS, Glasser SW. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol. Chem. 1996;271:6881–6888. doi: 10.1074/jbc.271.12.6881. [DOI] [PubMed] [Google Scholar]
- 6.Ray MK, Chen CY, Schwartz RJ, DeMayo FJ. Transcriptional regulation of a mouse Clara cell-specific protein (mCC10) gene by the NKx transcription factor family members thyroid transciption factor 1 and cardiac muscle-specific homeobox protein (CSX) Mol. Cell Biol. 1996;16:2056–2064. doi: 10.1128/mcb.16.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura S, Hara Y, Pineau T, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes. Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds SD, Reynolds PR, Pryhuber GS, Finder JD, Stripp BR. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am. J. Respir. Crit. Care Med. 2002;166:1498–1509. doi: 10.1164/rccm.200204-285OC. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee AB, Chilton BS. The uteroglobin/Clara cell protein family. Ann. NY. Acad. Sci. 2000;923:147–155. [Google Scholar]
- 10.Miele L, Cordella-Miele E, Facchiano A, Mukherjee AB. Novel anti-inflammatory peptides from the region of highest similarity between uteroglobin and lipocortin I. Nature. 1988;335:726–730. doi: 10.1038/335726a0. [DOI] [PubMed] [Google Scholar]
- 11.Chiba Y, Srisodsai A, Supavilai P, Kimura S. Interleukin-5 reduces the expression of uteroglobin-related protein (UGRP) 1 gene in allergic airway inflammation. Immunol. Lett. 2005;97:123–129. doi: 10.1016/j.imlet.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba Y, Kusakabe T, Kimura S. Decreased expression of uteroglobin- related protein 1 in inflamed mouse airways is mediated by IL-9. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;287:L1193–L1198. doi: 10.1152/ajplung.00263.2004. [DOI] [PubMed] [Google Scholar]
- 13.Srisodsai A, Kurotani R, Chiba Y, et al. Interleukin-10 induces uteroglobin-related protein (UGRP) 1 gene expression in lung epithelial cells through homeodomain transcription factor T/EBP/NKX2.1. J. Biol. Chem. 2004;279:54358–54368. doi: 10.1074/jbc.M405331200. [DOI] [PubMed] [Google Scholar]
- 14.Ober C, Moffatt MF. Contributing factors to the pathobiology. The genetics of asthma. Clin. Chest Med. 2000;21:245–261. doi: 10.1016/s0272-5231(05)70264-1. [DOI] [PubMed] [Google Scholar]
- 15.Postma DS, Bleecker ER, Amelung PJ, et al. Genetic susceptibility to asthma—bronchial hyperresponsiveness coinherited with a major gene for atopy. N. Engl. J. Med. 1995;333:894–900. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 16.Ruffilli A, Bonini S. Susceptibility genes for allergy and asthma. Allergy. 1997;52:256–273. doi: 10.1111/j.1398-9995.1997.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 17.Bleecker ER. Mapping susceptibility genes for asthma and allergy. Clin. Exp. Allergy. 1998;28 Suppl 5:6–12. doi: 10.1046/j.1365-2222.1998.028s5006.x. discussion 26–28. [DOI] [PubMed] [Google Scholar]
- 18.Cookson WO, Moffatt MF. Genetics of asthma and allergic disease. Hum. Mol. Genet. 2000;9:2359–2364. doi: 10.1093/hmg/9.16.2359. [DOI] [PubMed] [Google Scholar]
- 19.Niimi T, Munakata M, Keck-Waggoner CL, et al. A polymorphism in the human UGRP1 gene promoter that regulates transcription is associated with an increased risk of asthma. Am. J. Hum. Genet. 2002;70:718–725. doi: 10.1086/339272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asthma prevention and management guidelines. Ministry of Health and Welfare, Japan. Int. Arch. Allergy Immunol. 2000;121 Suppl 1:I–VIII. 1–77. doi: 10.1159/000053608. [DOI] [PubMed] [Google Scholar]
- 21.Global Initiative for Asthma (GINA2002) Global Strategy for Asthma Management and Prevention. National Institute of Health, National Heart, Lung, and Blood Institute; 2002. [Google Scholar]
- 22.Jian Z, Nakayama J, Noguchi E, Shibasaki M, Arinami T. No evidence for association between the -112G/A polymorphism of UGRP1 and childhood atopic asthma. Clin. Exp. Allergy. 2003;33:902–904. doi: 10.1046/j.1365-2222.2003.01703.x. [DOI] [PubMed] [Google Scholar]
- 23.Chiba Y, Kurotani R, Kusakabe T, et al. Uteroglobin-related protein 1 expression suppresses allergic airway inflammation in mice. Am. J. Respir. Crit. Care Med. 2006;173:958–964. doi: 10.1164/rccm.200503-456OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawrylowicz CM, O’Garra A. Potential role of interleukin-10-secreting regulatory T cells in allergy and asthma. Nat. Rev. Immunol. 2005;5:271–283. doi: 10.1038/nri1589. [DOI] [PubMed] [Google Scholar]
- 25.Stampfli MR, Cwiartka M, Gajewska BU, et al. Interleukin-10 gene transfer to the airway regulates allergic mucosal sensitization in mice. Am. J. Respir. Cell Mol. Biol. 1999;21:586–596. doi: 10.1165/ajrcmb.21.5.3755. [DOI] [PubMed] [Google Scholar]
- 26.Oh JW, Seroogy CM, Meyer EH, et al. CD4 T-helper ceils engineered to produce IL-10 prevent allergen-induced airway hyperreactivity and inflammation. J Allergy Clin. Immunol. 2002;110:460–468. doi: 10.1067/mai.2002.127512. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson KG, Kent J, Ireland DC. Respiratory viruses and exacerbations of asthma in adults. BMJ. 1993;307:982–986. doi: 10.1136/bmj.307.6910.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston SL, Pattemore PK, Sanderson G, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busse WW, Gern JE. Is interleukin-10 a “10” in virus-provoked asthma? Am. J. Respir. Crit. Care Med. 2005;172:405–406. doi: 10.1164/rccm.2506005. [DOI] [PubMed] [Google Scholar]
- 30.Shijubo N, Itoh Y, Yamaguchi T, et al. Clara cell protein-positive epithelial cells are reduced in small airways of asthmatics. Am. J. Respir. Crit. Care Med. 1999;160:930–933. doi: 10.1164/ajrccm.160.3.9803113. [DOI] [PubMed] [Google Scholar]
- 31.Van Vyve T, Chanez P, Bernard A, et al. Protein content in bronchoalveolar lavage fluid of patients with asthma and control subjects. J. Allergy. Clin. Immunol. 1995;95:60–68. doi: 10.1016/s0091-6749(95)70153-2. [DOI] [PubMed] [Google Scholar]
- 32.Shijubo N, Itoh Y, Yamaguchi T, et al. Serum levels of Clara cell 10-kDa protein are decreased in patients with asthma. Lung. 1999;177:45–52. doi: 10.1007/pl00007626. [DOI] [PubMed] [Google Scholar]
- 33.Renauld JC. New insights into the role of cytokines in asthma. J. Clin. Pathol. 2001;54:577–589. doi: 10.1136/jcp.54.8.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtzman MJ, Morton JD, Shornick LP, et al. Immunity, inflammation, and remodeling in the airway epithelial barrier: epithelial-viral-allergic paradigm. Physiol. Rev. 2002;82:19–46. doi: 10.1152/physrev.00020.2001. [DOI] [PubMed] [Google Scholar]
- 35.Umetsu DT. Revising the immunological theories of asthma and allergy. Lancet. 2005;365:98–100. doi: 10.1016/S0140-6736(05)17714-9. [DOI] [PubMed] [Google Scholar]
- 36.Heaton T, Rowe J, Turner S, et al. An immunoepidemiological approach to asthma: identification of in-vitro T-cell response patterns associated with different wheezing phenotypes in children. Lancet. 2005;365:142–149. doi: 10.1016/S0140-6736(05)17704-6. [DOI] [PubMed] [Google Scholar]
- 37.Del Prete G, De Carli M, Almerigogna F, et al. Human IL-10 is produced by both type 1 helper (Thl) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J. Immunol. 1993;150:353–360. [PubMed] [Google Scholar]
- 38.Platts-Mills T, Vaughan J, Squillace S, Woodfolk J, Sporik R. Sensitisation, asthma, and a modified Th2 response in children exposed to cat allergen: a population-based cross-sectional study. Lancet. 2001;357:752–756. doi: 10.1016/S0140-6736(00)04168-4. [DOI] [PubMed] [Google Scholar]