Abstract
Frontotemporal dementia (FTD) is a clinical syndrome with a heterogeneous molecular basis. The neuropathology associated with most FTD is characterized by abnormal cellular aggregates of either transactive response DNA-binding protein with Mr 43 kDa (TDP-43) or tau protein. However, we recently described a subgroup of FTD patients, representing around 10%, with an unusual clinical phenotype and pathology characterized by frontotemporal lobar degeneration with neuronal inclusions composed of an unidentified ubiquitinated protein (atypical FTLD-U; aFTLD-U). All cases were sporadic and had early-onset FTD with severe progressive behavioural and personality changes in the absence of aphasia or significant motor features. Mutations in the fused in sarcoma (FUS) gene have recently been identified as a cause of familial amyotrophic lateral sclerosis, with these cases reported to have abnormal cellular accumulations of FUS protein. Because of the recognized clinical, genetic and pathological overlap between FTD and amyotrophic lateral sclerosis, we investigated whether FUS might also be the pathological protein in aFTLD-U. In all our aFTLD-U cases (n = 15), FUS immunohistochemistry labelled all the neuronal inclusions and also demonstrated previously unrecognized glial pathology. Immunoblot analysis of protein extracted from post-mortem aFTLD-U brain tissue demonstrated increased levels of insoluble FUS. No mutations in the FUS gene were identified in any of our patients. These findings suggest that FUS is the pathological protein in a significant subgroup of sporadic FTD and reinforce the concept that FTD and amyotrophic lateral sclerosis are closely related conditions.
Keywords: frontotemporal lobar degeneration, frontotemporal dementia, FUS, fused in sarcoma, TLS, translocated in liposarcoma
Introduction
Frontotemporal dementia (FTD) is a clinical syndrome characterized by progressive changes in behaviour, personality and/or language with relative preservation of memory (Neary et al., 1998; McKhann et al., 2001). The neuropathology associated with clinical FTD is heterogeneous, with the common feature being relatively selective degeneration of the frontal and temporal lobes (frontotemporal lobar degeneration, FTLD) (Trojanowski and Dickson, 2001; Cairns et al., 2007a). As with many other neurodegenerative conditions, the pathology in most cases of FTLD also includes the presence of abnormal intracellular protein aggregates. This feature is the basis of recently published consensus recommendations for the nomenclature of FTLD in which classification is based on the molecular defect that is presumed to be pathogenic or most characteristic (Mackenzie et al., 2009). Approximately half of cases show accumulation of hyperphosphorylated tau protein in neurons and glia (FTLD-tau). The majority of tau-negative cases have neuronal inclusions that were originally identified by their immunoreactivity for ubiquitin (FTLD-U) (Jackson et al., 1996; Josephs et al., 2004; Johnson et al., 2005; Mackenzie et al., 2006). Recently, the transactive response (TAR) DNA-binding protein with Mr 43 kDa (TDP-43) was identified as the pathological protein in both FTLD-U (now referred to as FTLD-TDP) and amyotrophic lateral sclerosis (ALS) (Arai et al., 2006; Neumann et al., 2006). This finding has provided strong evidence that FTD with FTLD-TDP pathology, ALS with dementia and classical ALS are all part of a clinicopathological spectrum of disease.
Although TDP-43 was initially thought to be the pathological protein in all cases of FTLD-U and ALS (Arai et al., 2006; Neumann et al., 2006; Davidson et al., 2007), subsequent studies identified some important exceptions (Cairns et al., 2007b; Holm et al., 2007; Mackenzie et al., 2007; Josephs et al., 2008; Pikkarainen et al., 2008). In two recent papers, we described a subgroup of patients with sporadic FTD and FTLD-U pathology that was negative for TDP-43, accounting for 10%–20% of our respective FTLD-U series (Mackenzie et al., 2008a; Roeber et al., 2008). The unusual and highly consistent clinical and pathological phenotype suggested to us that these cases represent a specific disease entity, which we referred to as ‘atypical’ FTLD-U (aFTLD-U). Identification of this group indicated that there was at least one additional FTD-related protein yet to be discovered.
Recently, two studies identified mutations in the gene encoding the fused in sarcoma (FUS) protein (also known as translated in liposarcoma, TLS), as the cause of familial ALS (FALS) type 6 (Kwiatkowski et al., 2009; Vance et al., 2009). These studies reported that a total of 14 different mutations were found in 26 unrelated families (∼4% of FALS in these combined series). Most were missense mutations, affecting highly conserved regions in exon 15 that encodes the C-terminus. With the exception of one family with autosomal recessive disease, caused by the c.1551C>G mutation (Kwiatkowski et al., 2009), all other mutations produced autosomal dominant ALS, although with incomplete penetrance. No mutations were found in 293 sporadic ALS (SALS) cases screened in one study (Kwiatkowski et al., 2009). The clinical phenotype was classical ALS, with a mean age of onset of 46 years and mean disease duration of 33 months. There was no associated cognitive dysfunction. Post-mortem pathology was described in four patients and included degeneration of both upper and lower motor neurons. One study reported only increased neuronal cytoplasmic FUS-immunoreactivity in a single affected individual (Kwiatkowski et al., 2009), while the other described FUS-ir dystrophic neurites (DN) and globular neuronal cytoplasmic inclusions (NCI) in lower motor neurons, in the absence of TDP-43 pathology (Vance et al., 2009). In vitro experiments from both groups suggested increased cytoplasmic FUS localization in cells expressing mutations and one study reported increased levels of insoluble FUS (Kwiatkowski et al., 2009).
The FUS gene, located on chromosome 16, consists of 15 exons that encode a 526 amino-acid protein (Aman et al., 1996). The C-terminus region contains multiple domains involved in RNA–protein interactions, while the N-terminus functions in transcriptional activation (Prasad et al., 1994). FUS is a ubiquitously expressed protein (Aman et al., 1996; Andersson et al., 2008) that binds to RNA (Crozat et al., 1993; Zinszner et al., 1997) and DNA (Perrotti et al., 1998) and is involved in diverse cellular processes including cell proliferation (Bertrand et al., 1999), DNA repair (Baechtold et al., 1999), transcription regulation, RNA splicing (Yang et al., 1998) and the transport of RNA between intracellular compartments (Zinszner et al., 1997). In most cell types, FUS is present in both the nucleus and cytoplasm, however in neurons there is proportionally more in the nucleus and expression in glia is exclusively nuclear (Andersson et al., 2008). FUS may be involved in neuronal plasticity and maintenance of dendritic integrity by transporting mRNA, including those that encode actin-related proteins, to dendritic spines for local translation in response to synaptic stimulation (Fujii et al., 2005a, b). In contrast, FUS deficient neurons show decreased spine arborization and morphology (Fujii et al., 2005a). Chromosomal translocation of the 5′ portion of FUS results in several fusion oncogenes that are each associated with specific types of human cancer, including myxoid liposarcoma, Ewing's sarcoma and acute myeloid leukemia (Law et al., 2006). FUS knock-out mice show perinatal mortality (Hicks et al., 2000). The finding that FUS mutations cause FALS is the first association between this protein and a neurodegenerative condition.
The recognized clinical, genetic and pathological overlap between ALS and FTD, and the high degree of functional homology between FUS and another ALS/FTD-related protein (TDP-43) (Lagier-Tourenne and Cleveland, 2009), led us to speculate that FUS might also be the pathological protein in some cases of tau/TDP-43-negative FTLD. In this study, we investigate the possible role of FUS in our aFTLD-U cases.
Materials and methods
Cases
All of the 15 cases of aFTLD-U from our previous two studies (Mackenzie et al., 2008a; Roeber et al., 2008) were evaluated for FUS pathology. These cases had previously been characterized as having pathological inclusions that were immunoreactive for ubiquitin and the ubiquitin proteasome system associated sequestosome p62 (p62) but not for tau, TDP-43, neuronal intermediate filaments or α-synuclein. Neurological control cases included FTLD-TDP [n = 12; including two each of sporadic type 1, sporadic type 2, sporadic type 3, familial with granulin gene (GRN) mutations, familial with valosin-containing protein (VCP) mutations and familial linked to chromosome 9p] (Cairns et al., 2007b); FTLD-tau [n = 8; including two each of Pick's Disease (PiD), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD) and argyrophilic grain disease (AGD)]; Alzheimer's disease (AD; n = 2); Parkinson's disease combined with dementia with Lewy bodies (n = 2); multiple system atrophy (MSA; n = 2), Huntington's disease (HD; n = 2) and ALS (n = 6; including two each of SALS, FALS with superoxide dismutase (SOD)1 mutations and FALS with SOD1 mutations excluded). Normal control tissue was from two elderly patients with no history of neurological disease.
FUS antibodies
We tested a number of commercially available anti-FUS antibodies, each of which recognizes a different epitope. Results are summarized in Table 1. Immunohistochemistry (IHC) using three of the four antibodies demonstrated the normal physiological pattern of staining and also labelled the pathological lesions. One of these (Santa Cruz sc-47 711) only worked on frozen sections. The other two showed similar results on sections of formalin fixed, paraffin embedded material. The polyclonal antibody from Sigma-Aldrich was used for all subsequent IHC.
Table 1.
Anti-FUS antibodies tested
| Company | Product no. | Type | Epitope (aa 1–526) | Paraffin section IHC (dilution) | Frozen section IHC (dilution) | Immunoblot |
|---|---|---|---|---|---|---|
| Bethyl Laboratories | A300-302A | RP | N-terminus (aa 1–50) | Yes | Yes | Yes |
| (1:200–500) | (1:5000–10 000) | (1:20 000) | ||||
| Sigma-Aldrich | HPA008784 | RP | mid region (aa 86–213) | Yes | Not tested | Yes |
| (1:25–500) | (1:500) | |||||
| Bethyl Laboratories | A300-292A | RP | mid region (aa 200–250) | No | No | Yes |
| (1:2500) | ||||||
| Santa Cruz Biotechnology | sc-47711 | MM | C-terminus | No | Yes | Yes |
| (1:50–200) | (1:1000) |
MM = mouse monoclonal; RP = rabbit polyclonal.
Immunohistochemistry
Cases of aFTLD-U had previously been immunostained with antibodies against ubiquitin, p62, TDP-43, hyperphosphorylated tau, α-synuclein, Aβ, α-internexin, non-phosphorylated neurofilament (NF), phosphorylated neurofilament (pNF) and expanded polyglutamine repeat regions, as described (Mackenzie et al., 2008a; Roeber et al., 2008). In these cases, FUS IHC was performed on sections of frontal and temporal neocortex, hippocampus, basal ganglia, midbrain and either medulla or spinal cord. Ubiquitin, p62 and TDP-43 IHC was repeated on selected sections. For control cases, the region of maximal pathology was evaluated with FUS IHC.
All IHC was performed on 5 μm thick sections of formalin fixed, paraffin embedded tissue using the Ventana BenchMark® XT automated staining system (Ventana, Tuscon, AZ) and developed with aminoethylcarbizole (AEC). The primary antibodies employed recognized FUS (Sigma-Aldrich anti-FUS; 1:25–1:200 with initial overnight incubation at room temperature, following microwave antigen retrieval), ubiquitin (DAKO anti-ubiquitin; 1:500, following microwave antigen retrieval), p62 (BD Transduction Laboratories p62 Lck ligand; 1:500 following microwave antigen retrieval) and TDP-43 (ProteinTech Group anti-TARDBP; 1:1000 following microwave antigen retrieval). Based on the amount of normal physiological staining, it was apparent that the anti-FUS sensitivity was greatly influenced by the degree of tissue fixation and that this was only partially reversed by antigen retrieval. Therefore, the dilution of the primary antibody was adjusted in each case (from 1:25 to 1:200) to allow for faint physiological staining that ensured sensitivity (internal positive control) but did not compromise visualization of the pathology.
Immunofluorescence
Double-label immunofluorescence was performed on selected cases of aFTLD-U and FTLD-TDP, using a mouse monoclonal anti-ubiquitin antibody (Chemicon 1510; 1:20 000) and a rabbit polyclonal (RP) anti-FUS antibody (Sigma-Aldrich anti-FUS; 1:25). The secondary antibodies were Alexa Fluor 594 conjugated anti-rabbit and Alexa Fluor 488 conjugated anti-mouse (Molecular Probes; 1:500). 4′-6-diamidino-2-phenylindol (DAPI) was used for nuclear counterstaining.
Biochemical fractionation and immunoblot analysis
Fresh-frozen post-mortem frontal cortical tissue from aFTLD-U (n = 6), FTLD-TDP (n = 6) and normal controls (n = 7) was used for the sequential extraction of proteins with buffers of increasing stringency using a protocol commonly used for the sequential extraction of tau (Zhukareva et al., 2002). Briefly, grey matter was extracted at 2 ml/g (v/w) by repeated homogenization and centrifugation steps (120 000 × g, 30 min, 4°C) with high-salt (HS) buffer (50 mM Tris–HCl, 750 mM NaCl, 10 mM NaF, 5 mM EDTA, pH 7.4), 1% Triton-X 100 (TX) in high-salt buffer, radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] and 2% SDS buffer. To prevent carry over, each extraction step was performed twice. Only supernatants from the first extraction steps were analysed while supernatants from the second wash steps were discarded. The 2%-SDS insoluble pellet was extracted in 70% formic acid (FA) at 0.5 ml/g (v/w). Formic acid was evaporated in a SpeedVac system and the dried pellet was resuspendend in sample buffer and the pH was adjusted to neutral with NaOH. Protease inhibitors were added to all buffers prior to use. For immunoblot analysis, equal volumes of fractions from different samples (10 µl of high-salt buffer and TX, 20 µl of RIPA and SDS, 25 µl of formic acid) were resolved by 7.5% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). Following transfer, membranes were blocked with Tris buffered saline (TBS) containing 5% powdered milk and probed with anti-FUS antibodies (see Table 1 for dilutions). Primary antibodies were detected with horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG (Jackson ImmunoResearch Europe, UK) and signals were visualized by an HRP-based chemiluminescent reaction (Pierce, Rockford, IL) and the Chemiluminescence Imager Stella 3200 (Raytest, Switzerland). The intensity of the FUS bands in the soluble (HS and TX) and insoluble (SDS) fractions were measured and the ratio calculated.
Molecular genetic analysis
Molecular genetic analysis was performed on six cases of aFTLD-U and six pathologically normal controls where fresh-frozen post-mortem cerebellar tissue was available.
Genomic DNA sequencing
Genomic DNA (gDNA), prepared using standard automation protocols with the AutoGenprep 245T (Autogen, Holliston, MA), was used as a template to amplify the 15 exons of FUS by polymerase chain reaction (PCR). Primers designed to flanking intronic regions were used for both PCR and sequencing reactions (primer sequences available on request). Twenty microlitres of PCR product for each exon (Qiagen, Valencia, CA) was purified using the Ampure system (Agencourt Bioscience Corporation, Beverly, MA), then sequenced in both directions using Big Dye chemistry (Applied Biosystems, Foster City, CA). Sequencing products were purified using the CleanSeq method (Agencourt Bioscience Corporation, Beverly, MA) and analysed on an ABI3700.
Complementary DNA analysis
Total RNA was extracted using Trizol and the Pure Link system (Invitrogen, Carlsbad, CA), and the quality and quantity assessed on the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). First-strand synthesis was carried out using the Superscript III system (Invitrogen, Carlsbad, CA), with 200 ng of total RNA as the template and a gene-specific primer, designed to the 3′ untranslated region (UTR) of FUS (CTTGGGTGATCAGGAATTG). The resultant complementary DNA (cDNA) was diluted 1:5 and used as the template for RT–PCR reactions (Qiagen, Valencia, CA) using the following primer pairs:
c.1F:CATGGCCTCAAACGATTATAC and c.6:ATGGAGGATTGATCTTGGC,
c.5F:GCAGAACCAGTACAACAGC and c.10R:CTTCAGCTTGCCAGTTTC,
c.9F:CAATTGAGTCTGTGGCTGATTAC and c.14R:CACCACGACGATCATCCC
and c.12F:GTCCTAATCCCACCTGTGAG and c.utrR:CTTGGGTGATCAGGAATTG.
RT–PCR reactions were denatured for 3 min at 94°C, then cycled at 60–50°C touchdown (30″, 30″, 45″) for 35 cycles. RT–PCR products were visualized by electrophoresis on 2% agarose gel and sequenced in both directions.
mRNA expression analysis
Total RNA samples were normalized to 50 ng/μl and using 200 ng as the template, a reverse transcription reaction was performed using a 1:1 mix of random hexamers and oligo(dT) primers, and the SuperScript III system (Invitrogen, Carlsbad, CA). Gene expression assays were ordered from Applied Biosystems for FUS (Hs00192029_m1), and for the endogenous controls GAPDH (Hs00266705_g1), YWHAZ (Hs00852925_sH) and HPRT1 (Hs99999909_m1). Real-time PCR was performed on an ABI 7900 using the TaqMan method. Reactions contained 1 μl of cDNA amplified with 0.25 μl primer/probe mix and 2.5 μl TaqMan 2 × Universal PCR Master Mix (Applied Biosystems, Foster City, CA). The cycling parameters recommended by the manufacturer were followed; 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s/60°C for 1 min. All samples were run in triplicate and normalized to GAPDH (or other controls where necessary). The carboxyfluorescein (FAM)-fluorescent signal was analysed using SDS2.2.2 software, and relative quantities of FUS mRNA were determined using the ΔΔct method.
Results
Clinical summary
A detailed clinical description of these cases has been published previously (Mackenzie et al., 2008a; Roeber et al., 2008). In brief, the 15 subjects included nine females and six males. The mean age of disease onset was 38 years (range 28–55 years) and the mean duration was 7 years (range 4–15 years). All fulfilled clinical criteria for FTD and presented with severe progressive changes in personality and behaviour. Common features included lack of insight, poor judgement, decline in personal hygiene, hyperorality, poor attention, emotional blunting, disinhibition with inappropriate interpersonal conduct and antisocial behaviour that was sometimes aggressive and even criminal. Language remained fluent but eventually showed features of frontal dysfunction with reduced output, aspontaneity, stereotypy, perseveration and eventual mutism. Memory was only affected late in the disease course. Mild Parkinsonism was present in some but none showed clinical evidence of pyramidal system dysfunction. None of the subjects had a family history of similar disease.
Neuropathology
General findings
All cases showed symmetric atrophy of the frontal and temporal lobes and caudate nucleus. Histological evidence of chronic degeneration was present in the frontal and temporal neocortex, hippocampal CA1 sector and subiculum (hippocampal sclerosis), striatum, globus pallidus and substantia nigra. As previously described, no pathological changes were demonstrated with silver impregnation methods (Bielschowsky or Gallyas stains) or IHC for Aβ, tau, α-synuclein, neurofilament proteins, α-internexin, TDP-43 or expanded polyglutamine repeats (Mackenzie et al., 2008a, Roeber et al., 2008).
Ubiquitin/p62 IHC
All cases showed similar morphology and anatomical distribution of pathology. Variable numbers of well-defined, round, oval or crescentic neuronal cytoplasmic inclusions (NCI) were present in middle and deeper layers of neocortex, along with occasional short dystrophic neurites. Similar NCI were moderate to numerous in the dentate granule cells and less abundant in hippocampal pyramidal neurons and subcortical regions including the striatum, nucleus basalis, hypothalamus, substantia nigra and periaqueductal grey matter. Despite there being no obvious neuronal loss or degenerative changes, ub-ir inclusions were present in lower motor neurons (LMN) of the hypoglossal nucleus and/or spinal cord in 7 of the 12 (58%) cases available for evaluation.
In addition to cytoplasmic inclusions, ubiquitin IHC demonstrated a unique type of neuronal intranuclear inclusion (NII) in all cases. These appeared as a single straight, curved or twisted (vermiform), thick filament. Neuronal intranuclear inclusions were most numerous in the dentate granules cells but also found in pyramidal neurons of the neocortex and hippocampus and some subcortical regions. Unlike the neuronal cytoplasmic inclusions and dystrophic neurites, neuronal intranuclear inclusions were only immunoreactive for ubiquitin and not p62.
FUS IHC/immunofluorescence
The normal physiological staining pattern of FUS was equally well demonstrated in normal controls, neurological controls and aFTLD-U cases. This consisted of strong immunoreactivity of neuronal nuclei, weaker but consistent staining of neuronal cytoplasm and more variable reactivity of glial nuclei (Fig. 1A) (Andersson et al., 2008). In both the nuclei and cytoplasm, the normal staining pattern was generally diffuse but with occasional small granules.
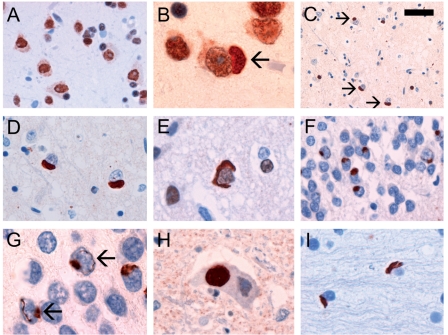
Figure 1.
FUS IHC performed on sections of post-mortem brain tissue from normal control (A) and aFTLD-U subjects (B–I). The normal physiological staining pattern, consisting of strong immunoreactivity of neuronal nuclei, weaker but consistent staining of neuronal cytoplasm and more variable reactivity of glial nuclei was demonstrated in all cases, including normal controls (A), neurological controls and aFTLD-U subjects (B). In patients with aFTLD-U, neurons with inclusions (arrow) retained at least some of the normal nuclear and cytoplasmic staining (B). Neuronal cytoplasmic inclusions (NCI) were most numerous in the middle and deeper layers of neocortex (C–E) and dentate granule cells of the hippocampus (F) and ranged in morphology from round or oval (D and F) to crescentic (E). Dentate granule cells with vermiform neuronal intranuclear inclusions (arrows) often also had NCI (G). Globular NCI were present in lower motor neurons (H). Flame-shaped glial cytoplasmic inclusions were present in white matter (I). FUS IHC with concentration of primary antibody adjusted to demonstrate normal physiological staining (A and B) or optimize visualization of pathological inclusions (C–I). Scale bar: A and H, 20 μm; B, E, G and I, 10 μm; C, 50 μm; D, 15 μm; F, 30 μm.
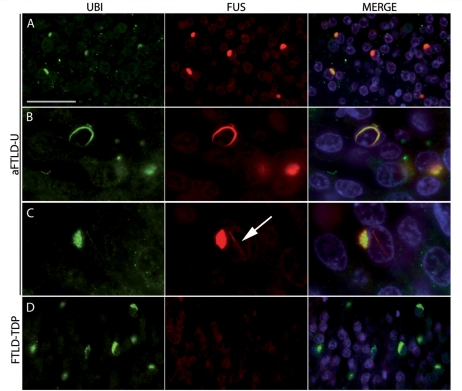
In cases of aFTLD-U, FUS IHC labelled neuronal cytoplasmic inclusions, neuronal intranuclear inclusions and dystrophic neurites of similar morphology, number and anatomical distribution as were demonstrated with ubiquitin and p62 antibodies (Fig. 1B–H). Some types of inclusions were even more apparent with FUS IHC. For instance, all cases in which medulla or spinal cord tissue was available had at least some large globular FUS-ir neuronal cytoplasmic inclusion in LMN (Fig. 1H, Table 2), compared to only 58% of cases with ub-ir NCI in this location (see above). FUS IHC also demonstrated moderate numbers of oval or flame-shaped glial cytoplasmic inclusions in the white matter that were not evident with ubiquitin IHC (Fig. 1I). On average, the burden of FUS pathology tended to be greatest in the hippocampal dentate fascia, moderate in the frontal and temporal neocortex and striatum, while other subcortical regions were less consistently involved and to a milder degree (Table 2). The co-localization of FUS and ubiquitin in NCI and NII was confirmed with double label immunofluorescence (Fig. 2 ). The pathological changes were immunoreactive with different FUS antibodies that recognize the N-terminus, mid-region and C-terminus of the protein, respectively (Table 1). Although the variation in staining intensity prevented quantitation, it was evident that neurons harbouring inclusions (either NCI or NII) still retained at least some of the normal distribution of nuclear and cytoplasmic staining (Figs 1B and 2).
Table 2.
Anatomical distribution and severity of FUS-immunoreactive pathology in aFTLD-U cases
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | Case 11 | Case 12 | Case 13 | Case 14 | Case 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frontotemporal neocortex | ++ | ++ | ++ | ++ | ++ | +++ | + | + | ++ | + | +++ | + | + | ++ | + |
| Hippocampus | +++ | NA | ++ | ++ | +++ | +++ | ++ | + | ++ | ++ | +++ | ++ | +++ | ++ | +++ |
| Striatum | ++ | ++ | + | ++ | ++ | ++ | + | + | ++ | + | ++ | NA | + | NA | ++ |
| Lower motor neurons | + | ++ | ++ | NA | ++ | + | + | NA | NA | + | + | NA | ++ | + | NA |
Grading: −= absent; += mild; ++ = moderate; +++ = severe; NA = not available.
Score is aggregate of all FUS-positive pathology, including neuronal cytoplasmic inclusions, neuronal intranuclear inclusions, glial inclusions and dystrophic neurites.
Figure 2.
Double-label immunofluorescence of inclusions in aFTLD-U and FTLD-TDP. Neuronal cytoplasmic inclusions (A) and neuronal intranuclear inclusions (B and C) in aFTLD-U cases showed colocalization of ubiquitin (green) and FUS (red). Note the presence of a cytoplasmic and intranuclear inclusion (arrow) in a single cell (C). No obvious difference in the intensity of FUS nuclear staining was observed between neurons with and without inclusions (A). In sharp contrast, ubiquitin-positive inclusions in FTLD-TDP were not stained with anti-FUS antibodies (D). Scale bar: A and D, 50 µm; B, 20 µm; C, 16.5 µm.
With one exception, none of the normal or neurological controls showed any FUS-ir pathology. Specifically, FUS IHC did not label senile plaques, neurofibrillary tangles, dystrophic neurites, Lewy bodies, Lewy neurites, Pick bodies, ballooned neurons, neuronal inclusions in ALS or FTLD with TDP pathology (Fig. 2) or glial inclusions in tauopathies or MSA. The exception was Huntingdon's disease in which the characteristic NII were strongly FUS-ir, a finding that has been reported previously (Doi et al., 2008).
FUS immunoblot analysis
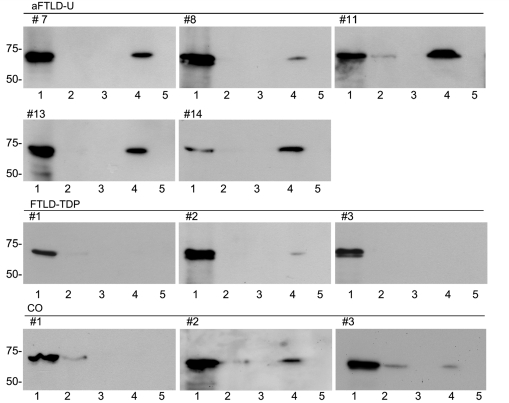
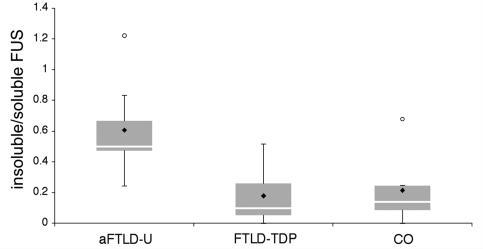
To characterize FUS biochemically, protein was sequentially extracted from fresh-frozen post-mortem brain tissue from aFTLD-U, FTLD-TDP and controls, using buffers containing increasingly strong detergents or acids. Fractions were then separated by SDS–PAGE and analysed by anti-FUS immunoblotting. FUS was consistently detected as a major ∼73-kDa band in the soluble high salt fraction from aFTLD-U as well as from normal or neurological controls (Fig. 3). Some cases, independent of the diagnosis, also showed a weak band in the soluble TX fraction. Although the amount of FUS in the SDS fraction, which is enriched for more insoluble proteins, varied within each group of patients, aFTLD-U cases tended to show stronger bands compared to both normal and FTLD-TDP controls. Within the aFTLD-U group, the amount of this insoluble FUS species roughly correlated with the severity of FUS pathology as detected by IHC (Table 2). The shift of FUS towards the insoluble fraction in aFTLD-U was confirmed by quantitative analysis of the intensity of the FUS bands in the soluble (HS and TX) and insoluble (SDS) fractions and calculation of insoluble:soluble ratios (Fig. 4). While there was some overlap between the aFTLD-U and control groups, statistical analysis revealed significantly higher ratios for aFTLD-U (mean = 0.62 ± 0.25) compared to FTLD-TDP (mean = 0.17 ± 0.19) or controls (mean = 0.21 ± 0.22) (P < 0.05; Student's t-test). Despite the shift of FUS to the insoluble fraction in aFTLD-U, we found no other evidence of biochemical abnormality; specifically, immunoblot analysis using four antibodies that each recognize different epitopes across the entire FUS protein (Table 1) did not identify any additional protein bands of higher or lower molecular weight.
Figure 3.
Biochemical analysis of FUS. Proteins were sequentially extracted from aFTLD-U, FTLD-TDP and control (CO) brains. High salt (lane 1), Triton X-100 (lane 2), RIPA (lane 3), 2% SDS (lane 4) and formic acid (lane 5) fractions were separated by 7.5% SDS–PAGE and immunoblotted with anti-FUS antibody (RP A300-302A). All cases showed a strong ∼73-kDa band in the soluble high-salt fraction (lane 1). Although the amount of SDS–soluble FUS (lane 4) was variable within each group, aFTLD-U cases always showed a strong band that was greater than that seen for most of the controls.
Figure 4.
Ratio of insoluble to soluble FUS. Band intensities of FUS in insoluble (SDS fraction) and soluble (high salt and Triton-X100 fractions) were analysed and the ratio calculated. Ratios are depicted as a box and whiskers blot that shows the range of values, with the box being subdivided into the 25 and 75% quartiles by the median; circles represent outliers, filled rhombus represent the mean. Although there is some overlap, the aFTLD-U group showed significantly higher ratios compared to both the FTLD-TDP and control groups (P < 0.05).
Molecular genetic analysis of FUS
Sequencing of all exons and flanking intronic regions of FUS gDNA did not identify any mutations in the six aFTLD-U cases analysed. In addition, complete FUS cDNA analyses, using four overlapping fragments, did not show aberrant transcripts by agarose gel-electrophoresis. cDNA sequencing further excluded mutations but confirmed the known alternative splicing at the start of exon 4, resulting in two transcripts with a 3-bp difference (Morohoshi et al., 1998). Finally, FUS mRNA expression in the aFTLD-U cases did not show any significant increase or decrease in expression, compared to normal controls.
Discussion
The findings of our study provide strong support for FUS being the pathological protein in an important new subtype of FTD. We found that FUS co-localized with all the ub-ir pathological inclusions in our aFTLD-U cases, including dystrophic neurites, neuronal cytoplasmic inclusions and, perhaps most importantly, the unusual vermiform NII that is the most unique feature of these cases. Moreover, FUS immunohistochemistry demonstrated additional inclusions, in glial cells, that were not seen with ubiquitin staining. The pathology was demonstrated with multiple antibodies that recognize different epitopes of the FUS protein but not with antibodies against the other proteins commonly associated with neurodegenerative disease (tau, α-synuclein, TDP-43 and intermediate filament proteins). FUS immunoreactivity was specific for this group of cases and did not label the characteristic pathological lesions of FTLD-tau, FTLD-TDP, ALS or most of the other neurodegenerative conditions we examined. Although the only evidence we found for a disease-associated biochemical modification of FUS in aFTLD-U was a relative change in solubility (higher mean ratio of insoluble versus soluble FUS in aFTLD-U compared with controls), this none-the-less suggests that FUS accumulation is a primary rather than a secondary phenomenon in this condition (i.e. normal FUS is not simply becoming entrapped in pre-existing inclusions made of some other protein). The recent recognition that FUS plays a pathogenic role in a subgroup of ALS supports its potential to cause FTD, which is considered to be a closely related neurodegenerative syndrome. Finally, the highly unusual and stereotypic clinical phenotype of our aFTLD-U subjects is consistent with these cases representing a distinct entity, which should have a novel and consistent pathology. In accordance with the recently proposed system of FTLD nomenclature (Mackenzie et al., 2009), this newly recognized pathology should be designated as FTLD-FUS.
It is perhaps not surprising to discover that FUS is also the pathological protein in a subgroup of FTD, given the high degree of functional homology it shares with another ALS/FTLD related protein, TDP-43 (recently reviewed in Lagier-Tourenne and Cleveland, 2009). Both TDP-43 and FUS are ubiquitously expressed DNA/RNA-binding proteins involved in multiple aspects of gene expression, transcription regulation, RNA splicing, transport and translation (Crozat et al., 1993; Prasad et al., 1994; Aman et al., 1996; Zinszner et al., 1997; Perrotti et al., 1998; Yang et al., 1998; Andersson et al., 2008; Buratti et al., 2008). Under normal circumstances, both proteins are predominantly localized to the nucleus but shuttle between the nucleus and cytosol (Andersson et al., 2008; Buratti et al., 2008). In fact, it was this functional homology with TDP-43 that led researchers to target FUS early in the process of screening candidate genes in the linked region on chromosome 16 (Kwiatkowski et al., 2009; Vance et al., 2009).
The similarities between FUS and TDP-43 extend to their role in disease. To date, mutations in both genes have predominantly been associated with autosomal dominant forms of classical ALS (Mackenzie et al., 2008b; Kwiatkowski et al., 2009; Vance et al., 2009). Most of the pathogenic mutations in both genes are missense changes, affecting highly conserved sites in the C-terminus. As in ALS with TDP-43 pathology (Arai et al., 2006; Neumann et al., 2006; Davidson et al., 2007), FUS mutations result in abnormal redistribution of the protein from the nucleus to the cytoplasm where it forms insoluble aggregates (Kwiatkowski et al., 2009; Vance et al., 2009). Our findings further extended these similarities by demonstrating that, in FTLD, both proteins form a common range of pathological inclusion bodies, including NCI, NII, DN and glial cytoplasmic inclusions. However, our results also suggest there may be some important differences between pathological forms of FUS and TDP-43. Cells with FUS-positive inclusion bodies retained at least some of the normal nuclear and cytoplasmic distribution of FUS staining, which is in contrast to the dramatic reduction in nuclear staining for TDP-43 in cells bearing TDP-43-positive inclusions (Neumann et al., 2006). Immunoblot analysis of our aFTLD-U cases did not show convincing evidence of abnormal processing of FUS and IHC suggested the inclusions contain the full-length protein. These findings have important implications regarding the pathogenic mechanism of FUS-related neurodegeneration and require more detailed investigation.
The concept that ALS and FTD with FTLD-U pathology are closely related conditions was strongly supported by the identification of TDP-43 as the pathological protein in both groups (Arai et al., 2006; Neumann et al., 2006; Davidson et al., 2007) and is now reinforced by the discovery that FUS plays a pathogenic role in this same spectrum of disease. The consistent involvement of LMN in our aFTLD-U cases, despite an absence of clinical features of ALS, is further support of this overlap. Given initial reports indicating that cases of ALS with FUS mutations have only FUS pathology and no abnormal TDP-43 (Kwiatkowski et al., 2009; Vance et al., 2009); and the finding that our neurological control cases of SALS had pathological TDP-43 but not FUS, it appears that ALS can be caused by abnormalities in these two highly similar but distinct molecular pathways. Comparison of the IHC results of our FTLD-TDP versus aFTLD-U cases, suggests the same dichotomous pathogenicity in FTD.
In summary, we provide evidence that FUS is the pathological protein in a new subtype of FTD, with a unique clinical phenotype and neuropathology (FTLD-FUS). The full spectrum of FTLD-FUS remains to be defined and future studies are needed to examine the possible role of FUS in other types of tau/TDP-43-negative FTLD, including basophilic inclusion body disease, neuronal intermediate filament inclusion disease, hereditary dementia with leukodystrophy and spheroids and FTD-3 caused by CHMP2B mutations. The absence of any identifiable FUS gene abnormality in our aFTLD-U cases is perhaps not surprising given that all appeared to be sporadic. None-the-less, a recent report that mutations in TARDBP may cause both FTD and ALS (Benajiba et al., 2009) indicates that FUS should also be considered a candidate gene in cases of familial FTD, particularly those with confirmed FUS pathology.
Funding
Canadian Institutes of Health Research (grant number 74580 to I.M.); the Pacific Alzheimer Research Foundation (to I.M. and R.R.); National Institute of Health (grant number P50 AG16574 to R.R.); the Deutsche Forschungsgemeinschaft (SFB 596 to M.N.); the Stavros-Niarchos Foundation (to M.N.); the Synapsis Foundation (to M.N.); and the German Brain Bank ‘BrainNet’ (to H.K.).
Acknowledgements
The authors thank Margaret Luk, Mareike Schroff, Mirjam Lutz, Richard Crook, Mariely deJesus and NiCole Finch for their excellent technical assistance.
Glossary
Abbreviations
- aFTLD-U
atypical frontotemporal lobar degeneration with neuronal inclusions composed of an unidentified ubiquitinated protein
- ALS
amyotrophic lateral sclerosis
- FALS
familial amyptrophic lateral sclerosis
- FTD
frontotemporal dementia
- FTLD
frontotemporal lobar degeneration
- FUS
fused in sarcoma
- IHC
immunohistochemistry
- p62
ubiquitin proteasome system associated sequestosome p62
- SALS
sporadic amyotrophic lateral sclerosis
- SDS
sodium dodecyl sulphate
- TDP-43
transactive response DNA-binding protein with Mr 43 kDa.
References
- Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, et al. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, et al. The multifunctional FUS, EWS, and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–11. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLD/FUS and is able to promote D-loop formation. J Biol Chem. 1999;274:34337–42. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–3. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated with cell proliferation. Oncogene. 1999;18:4515–21. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IRA, Neumann M, Lee VMY, Hatanpaa KJ, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 2007a;114:5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007b;171:227–40. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–4. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Davidson Y, Kelley T, Mackenzie IRA, Pickering-Brown S, Du Plessis D, Neary D, et al. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 2007;113:521–33. doi: 10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- Doi H, Okamura K, Bauer PO, Furukawa Y, Shimizu H, Kurosawa M, et al. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in Huntingtin exon 1 with expanded polyglutamine-expressing cells. J Biol Chem. 2008;283:6489–500. doi: 10.1074/jbc.M705306200. [DOI] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005a;15:587–93. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005b;118:5755–65. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, et al. FUS deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–9. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- Holm IE, Englund E, Mackenzie IRA, Johannsen P, Isaacs A. A Reassessment of the Neuropathology of Frontotemporal Dementia Linked to Chromosome 3 (FTD-3) J Neuropathol Exp Neurol. 2007;66:884–91. doi: 10.1097/nen.0b013e3181567f02. [DOI] [PubMed] [Google Scholar]
- Jackson M, Lennox G, Lowe J. Motor neurone disease-inclusion dementia. Neurodegeneration. 1996;5:339–50. doi: 10.1006/neur.1996.0046. [DOI] [PubMed] [Google Scholar]
- Johnson JK, Diehl J, Mendez MF, Neuhaus J, Shapira JS, Forman M, et al. Frontotemporal lobar degeneration. Demographic characteristics of 353 patients. Arch Neurol. 2005;62:925–30. doi: 10.1001/archneur.62.6.925. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Holton JL, Rossor MN, Godbolt AK, Ozawa T, Strand K, et al. Frontotemporal lobar degeneration and ubiquitin immunohistochemistry. Neuropathol Appl Neurobiol. 2004;30:369–73. doi: 10.1111/j.1365-2990.2003.00545.x. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Lin WL, Ahmed Z, Stroh DA, Graff-Radford NR, Dickson DW. Frontotemporal lobar degeneration with ubiquitin-positive, but TDP-43-negative inclusions. Acta Neuropathol. 2008;116:159–67. doi: 10.1007/s00401-008-0397-8. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–4. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG. TLS, EWS, and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, et al. Pathological TDP-43 distinguishes sporadic ALS from ALS with SOD-1 mutations. Ann Neurol. 2007;61:427–34. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Foti D, Woulfe J, Hurwitz TA. Atypical frontotemporal lobar degeneration with ubiquitin-positive, TDP-43-negative neuronal inclusions. Brain. 2008a;131:1282–93. doi: 10.1093/brain/awn061. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Rademakers R. The role of TDP-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr Opin Neurol. 2008b;21:693–700. doi: 10.1097/WCO.0b013e3283168d1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Shi J, Shaw CL, DuPlessis D, Neary D, Snowden JS, et al. Dementia lacking distinctive histology (DLDH) revisited. Acta Neuropathol. 2006;112:539–49. doi: 10.1007/s00401-006-0123-3. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological criteria for fronto-temporal dementia. Arch Neurol. 2001;58:1803–9. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, et al. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–8. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, et al. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–55. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikkarainen M, Hartikainen P, Alafuzoff I. Neuropathologic Features of Frontotemporal Lobar Degeneration With Ubiquitin-Positive Inclusions Visualized With Ubiquitin-Binding Protein p62 Immunohistochemistry. J Neuropathol Exp Neurol. 2008;67:280–98. doi: 10.1097/NEN.0b013e31816a1da2. [DOI] [PubMed] [Google Scholar]
- Prasad DD, Ouchida M, Lee L, Rao VN, Reddy ES. TLD/FUS fusion domain of TDL/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–29. [PubMed] [Google Scholar]
- Roeber S, Mackenzie IR, Kretzschmar HA, Neumann M. TDP-43-negative FTLD-U is a significant new clinico-pathological subtype of FTLD. Acta Neuropathol. 2008;116:147–57. doi: 10.1007/s00401-008-0395-x. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, Dickson D. Update on the neuropathological diagnosis of frontotemporal dementia. J Neuropathol Exp Neurol. 2001;60:1123–6. doi: 10.1093/jnen/60.12.1123. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, de Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761–4. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLD (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110:1741–50. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- Zhukareva V, Shah K, Uryu K, Braak H, Del Tredici K, Sundarraj S, et al. Biochemical Analysis of τ Proteins in Argyrophilic Grain Disease, Alzheimer's Disease, and Pick's Disease. Am J Pathol. 2002;161:1135–41. doi: 10.1016/s0002-9440(10)64390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]