Abstract
The oral cavity of healthy individuals contains hundreds of different bacterial, viral, and fungal species. Many of these can associate to form biofilms, which are resistant to mechanical stress or antibiotic treatment. Most are also commensal species, but they can become pathogenic in responses to changes in the environment or other triggers in the oral cavity, including the quality of an individual's personal hygiene. The complexity of the oral microbiome is being characterized through the newly developed tools of metagenomics. How the microbiome of the oral cavity contributes to health and disease is attracting the interest of a growing number of cell biologists, microbiologists, and immunologists.
“No man is an island, entire of itself”
—John Donne (1572–1631)
Introduction
We have more prokaryotic organisms on or in our bodies than we have eukaryotic cells. In fact, only one out of 10 cells in our bodies is human. These prokaryotic guests perform many biological functions that we could not perform on our own and protect us from invasion by pathogenic microorganisms.
In the early 1990s, scientists were confident that the sequencing of the human genome would be sufficient for understanding the basis for human function and disease, but analysis of the human genome was only an introduction to the genetic composition of our bodies. Humans and their commensal organisms have evolved together over the last two million years and have gradually become dependent on one another (O'Connell et al., 1998; Turnbaugh et al., 2007; Ley et al., 2008). The various commensals include eubacteria, archaebacteria, and fungi, which together comprise the human microbiome (Gill et al., 2006; Turnbaugh et al., 2007). To complicate matters further, the microbial communities within the body respond to different environmental conditions by modifying their species composition and population size. The effects of microbial metabolism in different locations on the human body also correspond to the physiological needs of the sites, which can vary from vitamin K production to the renewal of epithelial cells lining the gut.
While most organisms colonizing our bodies are beneficial to our health, some of them can transition from a commensal relationship to one of pathogenicity, for reasons that are still not understood. According to one view, the disease-causing bacteria are always present in a pathogenic state, but the commensal bacteria, which are more abundant, prevent the dangerous microbes from establishing a foothold (Pennisi, 2005). According to another view, some elusive trigger from the environment, or some temporal cue, stimulates the activity of the bacteria, resulting in infection or disease. Most likely, both possibilities are right. In the case of mucosal biofilms, a confounding issue is the relationship between inflammation and disease, and it is still not clear which comes first: the immune response, or the change in integrity of the mucosal biofilm (Dongari-Bagtzoglou, 2008).
The microbial communities are bound to impact the health of individual humans, and a better understanding of their dynamic complexity may contribute to the next level in medical diagnostic tools. Ideally, this should also lead to more specific treatment, by providing the potential to manipulate the microbiome to optimize personal health. These goals gave rise to the Human Microbiome Project, which aims to identify a core human microbiome, a common set of commensal species that can be defined as a healthy microbiota (Turnbaugh et al., 2007; National Institutes of Health, 2009). This project is being conducted around the world and includes the National Institutes of Health in the United States (Turnbaugh et al., 2007). Technology is now providing advanced sequencing techniques that are more cost effective and faster than ever, allowing for metagenomic analysis of microbial communities found in samples as varied as the human gut and ocean water (National Research Council, 2007). The field of metagenomics allows genomic analysis to be applied to entire communities of microbes, circumventing the need to isolate and culture individual bacterial community members. The new technologies include 454 pyrosequencing, which rapidly and inexpensively obtains genomic sequences without cloning bias, and proteomics (National Research Council, 2007; Keller and Hettich, 2009; Mitreva and Mardis, 2009).
Multiple projects are currently in progress, but the best-described microbiota to date reside in the human gut, which attracted the scientific community's attention due to its high cell density (Turnbaugh et al., 2007) and coevolution with a board range of species, including the causative agent of ulcers, Helicobacter pylori (Backhed et al., 2005).
Other microbiotas of clinical and general interest are now being sequenced, such as the nasal, oral, urogenital, and skin microbiotas (McGuire et al., 2008). At this time researchers are focused mainly on identifying the species, but also beginning to characterize the behavior of individual species and measure interactions between two species in a well-defined laboratory environment. Techniques being developed for studies of the microbiota from the human gut and other tissues are being rapidly adapted to studies of the oral microbiota. The ensuing knowledge should facilitate the understanding of the population diversity and examining the population clonality in oral health and disease.
Commensals in the Oral Cavity
One day in the late summer of 1683, Anthony van Leeuwenhoek decided to examine the film growing in his mouth using his homemade microscope. A new miniature world opened up to him. By September of the same year, Mr. Leeuwenhoek reported his findings to the Royal Society, becoming the first scientist on record to view bacteria (Ford, 2008).
It has been a long journey from these humble beginnings to the study of an oral microbiota consisting of an unknown number of bacteria, archea, and fungi. But not all species that reside in the oral microbiota have been identified yet, and an estimated 750 different species are anticipated (Jenkinson and Lamont, 2005; Paster et al., 2006). Numerous factors impede the identification of this vast number of species. First and foremost, many of the species are not culturable with today's laboratory technologies, and genomic similarities do not allow for organismal determination based on short read lengths. Because of these limitations, researchers have begun by identifying and characterizing the microorganisms with the largest representation within the communities of healthy mouths: Streptococcus, Actinomyces, Veillonella, Fusobacterium, Porphromonas, Prevotella, Treponema, Nisseria, Haemophilis, Eubacteria, Lactobacterium, Capnocytophaga, Eikenella, Leptotrichia, Peptostreptococcus, Staphylococcus, and Propionibacterium (Jenkinson and Lamont, 2005; Wilson, 2005).
Most of these microorganisms exist in our oral cavity in a symbiotic capacity, maintaining relationships with the host that are based on mutual benefits (Los Alamos National Library, 2009). Not only do they not cause harm, but also the commensal populations may keep pathogenic species in check by not allowing them to adhere to mucosal surfaces. The bacteria do not become successful pathogens, causing infection and disease, until they breach the barrier of commensals (Jenkinson and Lamont, 2005).
Biofilms and the Oral Microbiota
One of the greatest challenges facing any pathogen attempting to infect a new host is simply surviving the immune response of the host. Some bacterial invaders overcome the immune response by forming mixed biofilms consisting of commensals and potential pathogens to covertly hide within the host (Costerton, 2007).
In fact, even for commensals, the behavior of microorganisms can be very dynamic, adapting to a wide range of environments and interactions with other microbial species in aggregates called biofilms. The formation of biofilms may occur on a wide variety of surfaces in the oral cavity. Thus, epithelial cells, saliva-coated enamel, dental surfaces, primary colonizing bacteria, and orthodontics all create suitable environments for the establishment of mixed-species biofilms (Jenkinson and Lamont, 2005; Marsh, 2006). With populations in many countries increasing in age and often experiencing poor dental hygiene, biofilms formed by the microbiota of the edentulous are also likely to attract more attention (Sachdeo et al., 2008).
At the beginning of biofilm formation, initial surface attachment occurs by the primary colonizers, resulting in deposition of a microbial monolayer. Next, migration of the early colonizers leads to formation of multilayered microcolonies, which allows the creation of a multicellular matrix (Lemon et al., 2008). The primary colonizers in the oral microbiota for both mucosal and tooth surfaces are usually streptococci, which make up approximately 80% of early biofilms. The wide-ranging species of streptococci also produce the most well-studied oral adhesins, such as antigen I/II, PaG, SspA, amylase-binding proteins, and type 1 fimbriae-associated protein (Rosan and Lamont, 2000; Wilson, 2005). Adhesins produced by other oral bacteria have also been identified (Table 1).
Table 1.
Coaggregates Formed by Oral Bacteria, and Their Adhesins and Receptors
| Species | Adhesin | Receptor | Partner species | References |
|---|---|---|---|---|
| Streptococcus gordonii Streptococcus mitis Streptococcus oralis | Antigen I/II family (SspA SspB) | Bacterial surface proteins, yeast, mannoproteins |
Porphyromonas gingivalis Streptococcus mutans Actinomyces naeslundii Candida albicans |
Rosan and Lamont (2000), Wilson (2005) |
| S. gordonii | CshA, CshB | Bacterial surface proteins, yeast, mannoproteins | S. oralis | Rosan and Lamont (2000), Wilson (2005) |
| S. gordonii | LraI family (ScaA) | A. naeslundii | Rosan and Lamont (2000), Wilson (2005) | |
| S. gordonii | Coaggregation mediating adhesion | Carbohydrate containing lactose or lactose-like moieties | Streptococcus spp. | Rosan and Lamont (2000), Wilson (2005) |
| Streptococcus salivarius | Fibrillar antigen B(VBP) | Veillonella parvula | Rosan and Lamont (2000), Wilson (2005) | |
| Actinomyces naeslundii | Type 2 fimbriae-associated protein | Cell wall polysacchardie contraining Galβ1→3GalNAc and GalNAcβI→3 Gal glycosidic linkage | Streptococcus sanguis Streptococcus spp. | Rosan and Lamont (2000), Wilson (2005), Yoshida et al. (2006) |
| P. gingivalis | Fimbrillin | Surface proteins |
S. gordonii S. oralis A. naeslundii |
Wilson (2005) |
| P. gingivalis | Outer membrane protein | Surface proteins | S. gordonii | Wilson (2005) |
| P. gingivalis | Outer membrane protein | A. naesdlundlii | Wilson (2005) | |
| P. gingivalis | PlaA | Cell wall polysaccharide containing Galβ1 → 3GalNAc and GalNAβI → 3 Gal glycosidic linkage | Streptococcus spp. | Wilson (2005) |
| Prevotella loescheii | Fimbra-associated protein | Actinomyces israelii | Wilson (2005) | |
| Fusobacterium nucleatum | Outer membrane protein | Galactose-containing carbohydrate | P. gingivalis | Wilson (2005) |
| F. nucleatum | Corn cob receptor | Streptococcus cristatus Immunoglobulin A (L-Arginine) | Wilson (2005), Edwards et al. (2006) | |
| Veillonella atypica | Outer membrane protein | Carbohydrate containing lactose or lactose-like moieties | Streptococcus spp. | Wilson (2005) |
| Capnocytopha gingivalis | Outer membrane protein | Cell wall carbohydrate | A. israelii | Wilson (2005) |
| Capnocytophaga ochracea | Outer membrane protein | Cell wall carbohydrate | S. oralis | Wilson (2005) |
| Treponema medium | Outer membrane protein | Fimbriae | P. gingivalis | Wilson (2005) |
| F. nucleatum | RadD |
S. sanguis S. oralis S. gordonii A. naeslundii |
Kaplan et al. (2009) |
This aggregation table summarizes the known adhesins and receptors of oral microorganisms. Blanks indicate that the receptor is still unknown.
Within the oral microbiota, varying environmental conditions contribute to the species composition of biofilms present at each location. Conditional variants include temperature, pH, redox potential, atmospheric conditions, salinity, and water activity from saliva. All these conditions affect any number of biofilms, but saliva flow is specific to the oral cavity. Saliva is used by oral biofilms as a delivery system, bringing nutrients, peptides, and partially dissolved carbohydrates (Kolenbrander et al., 2007). Saliva performs additional functions, including lubrication for digestion, temperature regulation, and host defense (Wilson, 2005). Depending on the location of the biofilm in the oral microbiota, it can be affected differently by the saliva flow. The biofilms closer to origination points are exposed to solutions comprised of electrolytes, immunoglobulins, nitrogenous compounds, and enzymes such as amylase, mucins, and antimicrobials (Scannapieco, 1994; Wilson, 2005), while those at the end are exposed to a richer mixture of fluid, right before being swallowed by the host (Kolenbrander et al., 2007).
The oral microbiota also faces challenges that are not experienced by other microbiotas: the host has the option to maintain good personal hygiene. The cleansing mechanisms, especially deliberate oral hygiene, can alter the natural succession that would otherwise define the climax community. This contributes to the quantity and composition of mixed-species biofilms in the oral cavity. In response to eating, salivating, tooth brushing, tongue movement, flossing, and other agitation, the oral microbial communities have evolved the skills to survive these inhibitory practices.
Heterogeneity of Oral Microbiotas
Although microbiotas are often treated as being homogeneously distributed throughout the host organ, there is in fact a large variety of microbiotas and biofilms in different locations of the oral cavity. The supragingival plaque (dental plaque) encompasses multiple types of biofilms, including biofilms formed on the surface of teeth above the gingival crevice (the highest location were the gum meets the tooth). Other locations for varied oral microbiotas are the subgingival crevice (subgingival plaque), the tongue, mucosal surfaces (buccal cells and the floor of the mouth), and dental prosthetics and fillings (Aas et al., 2005).
Different species of microorganisms find optimal surroundings in each of these microenvironments. Classifying the microorganisms based on their oxygen requirements, the groups include obligate aerobes, obligate anaerobes (including Veillonella and Fusobacterium), facultative anaerobes (including most streptococci and Actinomyces) (Rosan and Lamont, 2000), microaerophiles (species that grow best at low concentrations of O2, from 2% to 10%), and capnophiles (species such as Neisseria that grow best at high CO2 concentrations, from 5% to 10%) (Wilson, 2005).
Bacteria can also participate in synergistic interactions within the biofilms. The bacteria communicate with each other using quorum sensing (QS), which refers to microbial signaling that is mediated by molecules secreted by the bacteria themselves (Fuqua et al., 1994). By controlling communication between the bacteria, QS modulates colony growth and biofilm formation (Irie and Parsek, 2008). Multiple QS systems have been identified in bacteria and fungi, of which the best-known examples are the acyl homoserine lactones, peptide auto inducers, auto inducers-2 (AI-2), and fungal QS systems (Irie and Parsek, 2008). The most prevalent in the oral cavity are the peptide auto inducers. Streptococcus gordonii and Porphyromonas gingivalis both use AI-2, and they continue to grow efficiently in a biofilm even if the AI-2 pathway of one species is inactivated (McNab et al., 2003). Antagonistic relationships in biofilms have also been described, especially when there is competition for resources in mixed-species biofilms (Lepp et al., 2004; Teles et al., 2007).
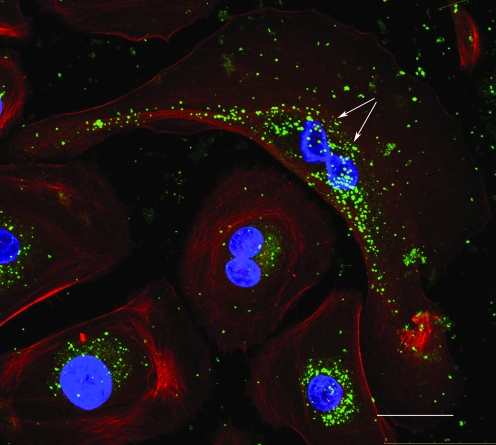
While biofilms are extracellular, the oral microbiota also includes communities of intracellular bacteria that invade the gingival and buccal epithelial cells of the mouth. A prominent member of this community is the opportunistic intracellular species, P. gingivalis, which can behave either as a commensal or pathogen (Rudney et al., 2005; Colombo et al., 2006, 2007; Yilmaz, 2008). Multiple species of intracellular bacteria thought to have pathogenic properties were also identified in healthy epithelium (Yilmaz, 2008). Human beings seem to have evolved a tolerance for these intracellular microbial communities (Fig. 1), which for the most part, still remain uncharacterized.
FIG. 1.
One-day infected primary gingival epithelial cells (actin-red, nuclei-blue) harboring a high number of intracellular Porphyromonas gingivalis bacteria (green) undergo successful mitosis as determined by confocal scanning fluorescence microscopy. White arrows indicate the division of parent nucleus into two daughter nuclei. Scale bar = 10 μm. (This figure is reproduced from Yilmaz, 2008, with permission from Microbiology, Society for General Microbiology.) Color images available online at www.liebertonline.com/dna.
The mere intracellular localization of a bacterial species is not sufficient to predict disease. Thus, one study found high concentrations of P. gingivalis, Tannerella forsythia, and Actinobacillus actinomycetemcomitans in buccal epithelial cells in healthy mouths (Muller et al., 1996; Rudney et al., 2003). Another study demonstrated that the bacterial colonizers on buccal epithelial cells share a similar composition with the supragingival biofilms in the same oral cavity (Rudney et al., 2005).
Each microbiota on the human body is associated with its own difficulties for experimental study. As more information is gleaned, larger numbers of species are discovered, but until now, mainly the species present at high concentrations have been identified or characterized. The challenges facing scientists studying the oral microbiotas include difficulties in reproducing the environment of the oral cavity, ease of sample collection, and a staggering number of species present at low concentrations. For now, researchers mimic dental environments by working with prosthetics from healthy and diseased mouths and common saliva components, but recreating the entire environment of the oral cavity will require reconstitution with different oral mucosal cells along with the saliva flow and pH and temperature ranges of the oral cavity. An unexpected relationship has also been uncovered between placement of orthodontic appliances and increased numbers of the preperiodontal bacteria, A. actimonycetemcomitans, T. forsythia, and various Streptococcus species in adjacent buccal epithelial cells (Leung et al., 2006), revealing the sensitivity of the oral microbiota to subtle changes in the microenvironment.
Characterizing Interactions Between Oral Microorganisms
Until recently, planktonic bacteria have not been studied in settings with more than one species at a time. It was previously known that the commensal Streptococcus gordonii assists in minimizing dental plaque due to its production of hydrogen peroxide, which can kill many oral bacteria. More recently experiments with S. gordonii and Actinomyces naeslundii, two bacterial species that are highly represented in early oral biofilms, showed that A. naeslundii allowed S. gordonii to grow in the absence of arginine and removed hydrogen peroxide from coaggregate cultures, decreasing protein oxidation in S. gordonii. Conversely, hydrogen peroxide produced by S. gordonii inhibited growth of A. naeslundii (Jakubovics et al., 2008). These observations illustrate the complexity of bacterial interactions in multispecies communities that occur widely in nature.
Fusobacterium nucleatum can aggregate with a large range of bacterial species and can bind to host tissues and immunoglobulin A, allowing F. nucleatum to invade epithelial cells and participate in biofilm formation (Edwards et al., 2007). F. nucleatum can also associate with Streptococcus cristatus, and transports noninvasive S. cristatus into oral epithelial cells (Edwards et al., 2006). Conversely, S. cristatus attenuates F. nucleatum–induced cytokine expression in oral epithelial cells (Zhang et al., 2008). The relationship between the two bacterial species can be antagonistic or synergistic, depending perhaps on the composition of the remaining species in the biofilm or other environmental conditions.
Other relationships are purely antagonistic. Streptococcus mutans, leading cause of dental caries, uses QS and releases bacteriocin when introduced to other bacteria (Senadheera and Cvitkovitch, 2008), while Streptococcus, Actinomyces, and Lactobacillus generate an acidic pH, thus inhibiting growth of a variety of bacterial species (Wilson, 2005).
The Oral Microbiota and Disease
Although biofilms are required for health of the oral cavity, biofilms are also known to contain pathogens (Ruby and Barbeau, 2002). Since inflammatory disease of the peridontium is caused by the microflora that are present in the gingival crevice, the mystery is not that disease can be brought on by these pathogenic bacteria, but what changes in their status to bring on the disease.
Periodontal diseases such as chronic gingivitis and periodontitis can result from an increase in the complexity and volume of biofilms located in the gingival crevice. These biofilms are typically comprised mainly of Gram-positive facultative anaerobes (Streptococcus anginosus and A. naeslundii), but in the absence of proper hygiene, the percentage of Gram-negative species (e.g., Porphyromonas spp., Campylobacter spp., T. forsythia, Treponema denticola, and A. actinomycetemcomitans) in the biofilms increases, contributing to periodontal inflammation (Ruby and Barbeau, 2002).
Individuals who do not practice good oral hygiene are also prone to bacterial endocarditis, halitosis, endodontic infection, and actinomycosis (Wilson, 2005). Invasive dental work can also lead to the spread of the bacterial pathogens to the blood stream and subsequently the brain, liver, and lungs (Bahrani-Mougeot et al., 2008; Lockhart et al., 2008; Wilson, 2005). An unhygienic oral cavity thus may increase the probability of gingivitis and periodontal disease, and affects negatively the overall health of an individual.
In particular, there is a strong association between periodontal disease and various forms of lung disease (Raghavendran et al., 2007). Thus, the oral microbiota may act as a reservoir for respiratory pathogens in patients in intensive care units, and intervention ranging from antibiotic treatment, use of antiseptics, and chlorhexidine gluconate rinses results in a large reduction in the number of pneumonia cases observed in the hospitalized patients (DeRiso et al., 1996; Bergmans et al., 2001; Fourrier et al., 2005; Raghavendran et al., 2007).
While many studies of biofilms focus on the role played by bacteria, fungal species can also participate in biofilm structure and function. The presence of Candida albicans is not an automatic inference of illness, since the fungus is present in healthy mouths (Bahrani-Mougeot et al., 2008). However, C. albicans cells can form biofilms on solid surfaces, with characteristic three-dimensional structures that display a high level of antifungal resistance. The requirement for a solid surface is significant for human health because of the large numbers of implanted medical devices that are used in modern medicine (Kumamoto and Vinces, 2005). After initially adhering to the surface, C. albicans biofilms grow on the surface and subsequently produce invasive filaments that penetrate neighboring cells.
Viruses, such as herpesviruses, have also been associated with periodontitis, possibly synergizing with bacteria in causing disease (Slots, 2007; Wu et al., 2007). Conversely, oral bacteria could affect the outcome of viral infection, as shown in the case of P. gingivalis, which upregulates expression of CCR5, a coreceptor for infection by some types of HIV strains (Giacaman et al., 2008). Although not considered a traditional risk factor for AIDS, P. gingivalis from the oral microbiota could thus affect the development of AIDS (Herzberg et al., 2006; Giacaman et al., 2008).
What can we do to minimize disease caused by the oral microbiota? The most obvious recommendation is to improve oral hygiene; however, people with sufficient oral hygiene can still develop chronic infections due to the composition of resident microbiota and changes in the host immune response. Antibiotics can be used for treatment of infections that are already spreading, although they are not usually effective against bacteria embedded in biofilms. In fact, in most biofilms, antibiotics can only slow down the growth of bacteria (Cramton and Gotz, 2004). More worrying is the recent discovery of an ampicillin-resistant F. nucleatum strain in random dental plaques from volunteers (Al-Haroni et al., 2008). The identification of resistance to ampicillin raises the potential for resistance to all β-lactamases, creating the potential for even more virulent oral pathogens. As one cannot overemphasize the importance of better oral hygiene as a safer, more effective barrier to infection, a susceptible host and existing environmental factors could contribute to the complexity of microbial consortia and its resulting negative outcomes.
Future Studies
Given the growing interest in understanding oral microbiotas from different parts of the human body, many technical tools are being developed for facilitating sequencing of microbial communities and data analysis. Multiple databases that include information on the microbiotas from the oral cavity will be available in the near future (Human Oral Microbiome database, 2009; Los Alamos National Library, 2009). The ORALGEN and Human Oral Microbiome Databases offer molecular information on oral pathogens, from viruses to bacteria and fungi. The goal is to provide a one-stop shop where analysis and compilation of oral pathogens can take place simultaneously.
We will likely need to await development of new metagenomics technologies to identify the remaining organisms present in the mixed-species biofilms of the oral cavity. Measurement of microbial diversity in the oral cavity is complicated by changing views on the definition of species. Species are often identified from the overall relatedness of their genomes, but phenotypic traits, such as whether different isolates can cause a particular disease, are also used. For these reasons, many researchers prefer to use the terms operational taxonomic units or phylotype, rather than species, for a cluster of related 16S rRNA gene sequences (Lozupone and Knight, 2008). More work must also be devoted to answer the question of why individual bacteria enter and exit from biofilms, and what triggers their transition from commensal to pathogenic bacteria. Changes in the microbiome that could influence health and disease include the composition of the microbiome (e.g., the presence of different bacteria and ratios of Gram-positive to Gram-negative bacteria), the abundance of particular species, changes in virulence factors produced by the bacteria, and the immune response of the host as a function of genetic susceptibility, environmental factors, or age.
With our populations aging and living longer lives, health management and disease prevention become increasingly important. Improved knowledge of the oral microbiotas and their impact on overall health will contribute greatly to health of society as a whole.
Acknowledgments
This review is supported by NIDCR R01DE16593 and R01DE019444 Grant.
Disclosure Statement
No competing financial interests exist.
References
- Aas A.J. Paster J.B. Stokes L.N. Olsen I. Dewhirst F.E. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Haroni M. Skaug N. Bakken V. Cash P. Proteomic analysis of ampicillin-resistant oral Fusobacterium nucleatum. Oral Microbiol Immunol. 2008;23:36–42. doi: 10.1111/j.1399-302X.2007.00387.x. [DOI] [PubMed] [Google Scholar]
- Backhed F. Ley R.E. Sonnenburg J.L. Peterson D.A. Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bahrani-Mougeot F.K. Paster B.J. Coleman S. Ashar J. Barbuto S. Lockhart P.B. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. 2008;46:2129–2132. doi: 10.1128/JCM.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmans D.C. Bonten M.J. Gaillard C.A. Paling J.C. van der Geest S. van Tiel F.H. Beysens A.J. de Leeuw P.W. Stobberingh E.E. Prevention of ventilator-associated pneumonia by oral decontamination: a prospective, randomized, double-blind, placebo-controlled study. Am J Respir Crit Care Med. 2001;164:382–388. doi: 10.1164/ajrccm.164.3.2005003. [DOI] [PubMed] [Google Scholar]
- Colombo A.V. da Silva C.M. Haffajee A. Colombo A.P. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55:609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- Colombo A.V. da Silva C.M. Haffajee A. Colombo A.P. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodontal Res. 2007;42:236–243. doi: 10.1111/j.1600-0765.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- Costerton J.W. The Biofilm Primer. Springer; Heidelberg: 2007. [Google Scholar]
- Cramton S.E. Gotz F. Biofilm development in Staphylococcus. In Microbial Biofilms. In: Ghannoum M., editor; O'Toole G.A., editor. Chapter 5. ASM Press; Washington, D.C.: 2004. pp. 64–84. [Google Scholar]
- DeRiso A.J., 2nd Ladowski J.S. Dillon T.A. Justice J.W. Peterson A.C. Chlorhexidine gluconate 0.12% oral rinse reduces the incidence of total nosocomial respiratory infection and nonprophylactic systemic antibiotic use in patients undergoing heart surgery. Chest. 1996;109:1556–1561. doi: 10.1378/chest.109.6.1556. [DOI] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A. Mucosal biofilms: challenges and future directions. Expert Rev Anti Infect Ther. 2008;6:141–144. doi: 10.1586/14787210.6.2.141. [DOI] [PubMed] [Google Scholar]
- Edwards A.M. Grossman T.J. Rudney J.D. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun. 2006;74:654–662. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A.M. Grossman T.J. Rudney J.D. Association of a high-molecular weight arginine-binding protein of Fusobacterium nucleatum ATCC 10953 with adhesion to secretory immunoglobulin A and coaggregation with Streptococcus cristatus. Oral Microbiol Immunol. 2007;22:217–224. doi: 10.1111/j.1399-302X.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- Ford B.J. University of California; Berkeley, Berkeley: 2008. [Google Scholar]
- Fourrier F. Dubois D. Pronnier P. Herbecq P. Leroy O. Desmettre T. Pottier-Cau E. Boutigny H. Di Pompéo C. Durocher A. Roussel-Delvallez M PIRAD Study Group. Effect of gingival and dental plaque antiseptic decontamination on nosocomial infections acquired in the intensive care unit: a double-blind placebo-controlled multicenter study. Crit Care Med. 2005;33:1728–1735. doi: 10.1097/01.ccm.0000171537.03493.b0. [DOI] [PubMed] [Google Scholar]
- Fuqua W.C. Winans S.C. Greenberg E.P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacaman R.A. Asrani A.C. Gebhard K.H. Dietrich E.A. Vacharaksa A. Ross K.F. Herzberg M.C. Porphyromonas gingivalis induces CCR5-dependent transfer of infectious HIV-1 from oral keratinocytes to permissive cells. Retrovirology. 2008;5:29. doi: 10.1186/1742-4690-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.R. Pop M. Deboy R.T. Eckburg P.B. Turnbaugh P.J. Samuel B.S. Gordon J.I. Relman D.A. Fraser-Liggett C.M. Nelson K.E. Metagenomics analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg M.C. Weinberg A. Wahl S.M. (C3) The oral epithelial cell and first encounters with HIV-1. Adv Dent Res. 2006;19:158–166. doi: 10.1177/154407370601900128. [DOI] [PubMed] [Google Scholar]
- Human Oral Microbiome Database. Forsyth Institute; Boston, Massachusetts: 2009. [Google Scholar]
- Irie Y. Parsek M.R. Quorum sensing and microbial biofilms. In: Romero T., editor. Bacterial Biofilms. Chapter 4. Springer; Heidelberg: 2008. pp. 67–84. [DOI] [PubMed] [Google Scholar]
- Jakubovics N.S. Gill S.R. Vickerman M.M. Kolenbrander P.E. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H.F. Lamont R.J. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Kaplan C.W. Lux R. Haake S.K. Shi W. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol. 2009;71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. Hettich R. Environmental proteomics: a paradigm shift in characterizing microbial activities at the molecular level. Microbiol Mol Biol Rev. 2009;73:62–70. doi: 10.1128/MMBR.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P.E. Jakubovics N.S. Chalmers N.I. Palmer R.J., Jr. Human oral mulitspecies biofilms: bacterial communities in health and human disease. In: Kjelleberg S., editor. The Biofilm Mode of Life: Mechanisms and Adaptations. Chapter 10. Horizon Bioscience; Norfolk: 2007. pp. 175–194. [Google Scholar]
- Kumamoto C.A. Vinces M.D. Alternative Candida albicans lifestyles: growth on surfaces. Annu Rev Microbiol. 2005;59:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. Earl A.M. Vlamakis H.C. Aguilar C. Kolter R. Biofilm development with an emphasis on Bacillus subtilis. In: Romero T., editor. Bacterial Biofilms. Chapter 1. Springer; Heidelberg: 2008. pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepp P.W. Brinig M.M. Ouverney C.C. Palm K. Armitage G.C. Relman D.A. Methanogenic archaea and human periodontal disease. Proc Natl Acad Sci USA. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N.M. Chen R. Rudney J.D. Oral bacteria in plaque and invading buccal cells of young orthodontic patients. Am J Orthod Dentofacial Orthop. 2006;130:e11–e18. doi: 10.1016/j.ajodo.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Ley R.E. Lozupone C.A. Hamady M. Knight R. Gordon J.I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P.B. Brennan M.T. Sasser H.C. Fox P.C. Paster B.J. Bahrani-Mougeot F.K. Circulation. 2008. Bacteremia associated with toothbrushing and dental extraction; pp. 3118–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los Alamos National Library. ORALGEN. Livermore: 2009. Library LAN ed. [Google Scholar]
- Lozupone C.A. Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P.D. Dental plaque as a biofilm and a microbial community-implications for health and disease. BMC Oral Health. 2006;6(Suppl 1):S14. doi: 10.1186/1472-6831-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.L. Colgrove J. Whitney S.N. Diaz C.M. Bustillos D. Versalovic J. Ethical, legal, and social considerations in conducting the Human Microbiome Project. Genome Res. 2008;18:1861–1864. doi: 10.1101/gr.081653.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R. Ford S.K. El-Sabaeny A. Barbieri B. Cook G.S. Lamont R.J. LuxS-based signaling in Streptococcus gordonii: autooinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M. Mardis E.R. Large-scale sequencing and analytical processing of ESTs. Methods Mol Biol. 2009;533:1–35. doi: 10.1007/978-1-60327-136-3_8. [DOI] [PubMed] [Google Scholar]
- Muller H.P. Zoller L. Eger T. Hoffmann S. Lobinsky D. Natural distribution of oral Actinobacillus actinomycetemcomitans in young men with minimal periodontal disease. J Periodontal Res. 1996;31:373–380. doi: 10.1111/j.1600-0765.1996.tb00506.x. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. Human microbiome project. National Institutes of Health (NIH) Bethesda, MD: 2009. Coordination OoS ed. [Google Scholar]
- National Research Council. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. National Academies Press; Washington D.C.: 2007. [PubMed] [Google Scholar]
- O'Connell J.F. Hawkes K. Blurton Jones N.G. Grandmothering and the evolution of Homo erectus. J Hum Evol. 1998;36:461–485. doi: 10.1006/jhev.1998.0285. [DOI] [PubMed] [Google Scholar]
- Paster B.J. Olsen I. Aas J.A. Dewhirst F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Pennisi E. A mouthful of microbes. Science. 2005;307:1899–1901. doi: 10.1126/science.307.5717.1899. [DOI] [PubMed] [Google Scholar]
- Raghavendran K. Mylotte J.M. Scannapieco F.A. Nursing home-associated pneumonia, hospital-acquired pneumonia and ventilator-associated pneumonia: the contribution of dental biofilms and periodontal inflammation. Periodontol 2000. 2007;44:164–177. doi: 10.1111/j.1600-0757.2006.00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosan B. Lamont R.J. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- Ruby J. Barbeau J. The buccale puzzle: the symbiotic nature of endogenous infections of the oral cavity. Can J Infect Dis. 2002;13:34–41. doi: 10.1155/2002/492656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudney J.D. Chen R. Pan Y. Endpoint quantitative PCR assays for Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J Periodontal Res. 2003;38:465–470. doi: 10.1034/j.1600-0765.2003.00670.x. [DOI] [PubMed] [Google Scholar]
- Rudney J.D. Chen R. Sedgewick G.J. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59–63. doi: 10.1177/154405910508400110. [DOI] [PubMed] [Google Scholar]
- Sachdeo A. Haffajee A.D. Socransky S.S. Biofilms in the edentulous oral cavity. J Prosthodont. 2008;17:348–356. doi: 10.1111/j.1532-849X.2008.00301.x. [DOI] [PubMed] [Google Scholar]
- Scannapieco F.A. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- Senadheera D. Cvitkovitch D.G. Quorum sensing and biofilm formation by Streptococcus mutans. Adv Exp Med Biol. 2008;631:178–188. doi: 10.1007/978-0-387-78885-2_12. [DOI] [PubMed] [Google Scholar]
- Slots J. Herpesviral-bacterial synergy in the pathogenesis of human periodontitis. Curr Opin Infect Dis. 2007;20:278–283. doi: 10.1097/QCO.0b013e3280964da0. [DOI] [PubMed] [Google Scholar]
- Teles R.P. Bogren A. Patel M. Wennstrom J.L. Socransky S.S. Haffajee A.D. A three-year prospective study of adult subjects with gingivitis II: microbiological parameters. J Clin Periodontol. 2007;34:7–17. doi: 10.1111/j.1600-051X.2006.01015.x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J. Ley R.E. Hamady M. Fraser-Liggett C.M. Knight R. Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- Wu Y.M. Yan J. Ojcius D.M. Chen L.L. Gu Z.Y. Pan J.P. Correlation between infections with different genotypes of human cytomegalovirus and Epstein-Barr virus in subgingival samples and periodontal status of patients. J Clin Microbiol. 2007;45:3665–3670. doi: 10.1128/JCM.00374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y. Palmer R.J. Yang J. Kolenbrander P.E. Cisar J.O. Streptococcal receptor polysaccharides: recognition molecules for oral biofilm formation. BMC Oral Health. 2006;6(Suppl 1):S12. doi: 10.1186/1472-6831-6-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. Chen R. Rudney J.D. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J Periodontal Res. 2008;43:408–416. doi: 10.1111/j.1600-0765.2007.01057.x. [DOI] [PubMed] [Google Scholar]