Abstract
Oncogenic mutations in the small GTPase Ras are highly prevalent in cancer, but an understanding of the vulnerabilities of these cancers is lacking. We undertook a genome-wide RNAi screen to identify synthetic lethal interactions with the KRAS oncogene. We discovered a diverse set of proteins whose depletion selectively impaired the viability of Ras mutant cells. Among these we observed a strong enrichment for genes with mitotic functions. We describe a pathway involving the mitotic kinase PLK1, the anaphase promoting complex/cyclosome and the proteasome that, when inhibited, results in prometaphase accumulation and the subsequent death of Ras mutant cells. Gene expression analysis indicates that reduced expression of genes in this pathway correlates with increased survival of patients bearing tumors with a Ras transcriptional signature. Our results suggest a previously underappreciated role for Ras in mitotic progression and demonstrate a pharmacologically tractable pathway for the potential treatment of cancers harboring Ras mutations.
INTRODUCTION
A major challenge in cancer therapeutics is the identification of cellular drug targets whose inhibition leads to the selective killing of cancer cells while sparing normal cells. Recent advances in mammalian RNA interference (RNAi) technologies have made it possible to systematically interrogate the human genome for genes whose loss of function constitute synthetic lethality either with the oncogenic state or with particular oncogenic mutations (Ngo et al., 2006; Schlabach et al., 2008; Silva et al., 2008). We have developed barcoded, retroviral/lentiviral-based short hairpin RNA (shRNA) libraries targeting the entire human genome to enable genome-wide loss-of-function analysis through stable gene knockdown (Silva et al., 2005). Our design also allowed us to develop a multiplex screening platform that enables the highly parallel screening of >10,000 shRNAs in a pool-based format using microarray deconvolution (Schlabach et al., 2008; Silva et al., 2008). These technological breakthroughs have made it possible to rapidly interrogate the genome for functional vulnerability of cancer cells and here we apply these to the Ras oncogene.
The Ras family of small GTPases are frequently mutated in human cancers [Reviewed in (Karnoub and Weinberg, 2008)]. Ras is a membrane-bound signaling molecule that cycles between the inactive, GDP-bound state and the active, GTP-bound state. Growth factor receptor signaling promotes GTP loading and activation of Ras, which in turn activates an array of downstream pathways to promote cell proliferation and survival. Among the major Ras effector pathways is the MAP kinase pathway, the PI3-kinase (PI3K) pathway, RalGDS proteins, phospholipase-Cε and Rac. Each of these has been implicated in mediation of Ras oncogenesis. Ras GAPs (GTPase activating proteins) inactivate Ras by stimulating GTP hydrolysis. Oncogenic mutations in Ras are invariably point mutations that either interfere with Ras GAP binding or directly disrupt Ras GTPase activity, locking Ras in a constitutively active, GTP-bound state. Oncogenic mutations have been found in all three members of the Ras gene family with KRAS being the most frequently mutated. KRAS mutations are found at high frequencies in pancreatic, thyroid, colon, lung and liver cancers and in myelodyspastic syndrome and are correlated with poor prognosis (Karnoub and Weinberg, 2008).
Despite its prominent status as a cancer drug target, therapeutics aimed at disrupting the Ras pathway have proven challenging thus far. Inhibitors of farnesyl transferase, the enzyme that prenylates Ras for its membrane localization, have met with only limited success (Karnoub and Weinberg, 2008). Chemical screens in isogenic Ras mutant and wild type cell lines have identified compounds that exhibit preferential toxicity towards Ras mutant cells (Torrance et al., 2001; Dolma et al., 2003). However, the translation of these chemical screens into clinical practice has been impeded by the challenge in identifying the protein targets of these chemical entities and subsequent drug development. Inhibitors targeting various Ras effecter pathways could also prove efficacious in treating tumors with Ras mutations, as it was recently shown that a combined application of MEK and PI3K/mTOR inhibitors can reduce tumor burden in a mouse model of Ras-driven lung cancer (Engelman et al., 2008). However, the prevalence of de novo and acquired drug resistance to other targeted therapies suggests that combinations of multiple therapeutic agents will be required to effectively inhibit malignant progression.
In principle, tumors can be attacked by either reversing the effects of oncoproteins through inhibition (i.e. exploiting oncogene addiction), or by attacking tumor-specific vulnerabilities caused by the oncogenic state, often by inhibiting proteins that are not oncoproteins themselves (i.e. exploiting non-oncogene addiction) (Solimini et al., 2008;Luo et al., 2009). The inappropriate rewiring of cellular signaling through oncogene activation should result in vulnerabilities that could be exploited for cancer therapies in theory. Since these vulnerabilities are not obvious and cannot be predicted, the most direct approach to their discovery is through genetic exploration. The systematic identification of genes and pathways necessary for the Ras-driven oncogenic state would provide additional drug targets for therapeutic exploration, shed new light on Ras’ mechanisms of action and potentially provide new biomarkers for patient stratification. To this end, we screened our shRNA library for genes whose inhibition constitutes synthetic lethality with the KRAS oncogene. We identified a functionally diverse set of genes including a number of mitotic proteins and demonstrate that pharmacological inhibitors targeting these mitotic proteins can selectively impair the viability of Ras mutant cells. These findings point to a previously underappreciated role of Ras in mitotic progression and demonstrate that mitotic stress induced by the Ras oncogene might be exploited for therapeutic purposes.
RESULTS
Genome-wide RNAi synthetic lethal screen against the KRAS oncogene
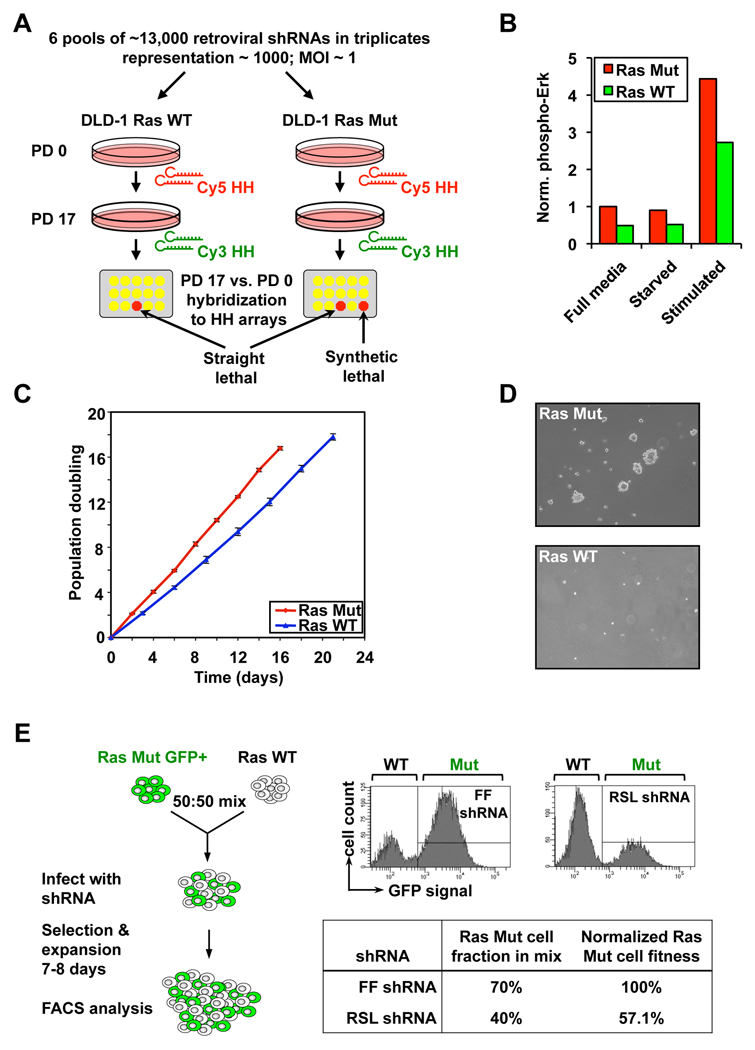
We chose the colorectal cancer cell line DLD-1 for the primary screen (Figure 1A). These cells carry an endogenous activating K-Ras G13D point mutation required for maintaining their oncogenic state (Shirasawa et al., 1993; Torrance et al., 2001). An isogenic clone of DLD-1 cells with the KRASG13D allele disrupted shows decreased MAP kinase signaling, reduced proliferation on adherent surfaces and is no longer able to sustain anchorage-independent growth in vitro (Figure 1 B-D) or tumor growth in vivo. Thus, they clearly exhibit addiction to the KRAS oncogene and their malignant phenotype critically depends on mutant K-Ras function.
Figure 1. Scheme of the Ras synthetic lethal screen.
A. Schematic of the primary screen. Change in a particular shRNA’s abundance in the pool over time is tracked by competitive hybridization between the initial (PD 0) and final samples (PD 17). A low Cy3/Cy5 ratio indicates the dropout of an anti-proliferative shRNA from the pool. Synthetic lethal shRNAs are selectively depleted from the Ras Mut cells. PD, population doubling; HH, half-hairpin.
B. MAPK pathway activity in DLD-1 cells. Phosphorylation on p42/p44 Erk kinases in cells that were either in full media, serum starved overnight, or serum-starved and then stimulated with full media.
C. Growth curve of DLD-1 cells in culture.
D. Anchorage independence colony formation of DLD-1 cells in soft-agarose, assessed 2 weeks after seeding.
E. Schematic of the competition assay used to validate shRNAs from the primary screen. Ras Mut cells expressing GFP and Ras WT cells were mixed and co-infected with the same retroviral shRNA. The Mut to WT cell ratio at the end of the experiment is measured by FACS. The percentage of Ras Mut cells in the mixture infected with a candidate RSL shRNA was normalized against that of a control shRNA targeting firefly luciferase (FF).
We screened the parental KRAS WT/G13D (Ras Mut) DLD-1 cells and the isogenic KRAS WT/− (Ras WT) DLD-1 control cells with our library of 74,905 retroviral shRNAs targeting 32,293 unique human transcripts (including 19,542 RefSeq transcripts). The library was screened in 6 pools of ∼13,000 shRNAs per pool using a protocol described previously (Figure 1A) (Schlabach et al., 2008). We analyzed the change in relative abundance of each shRNA over time by microarray hybridization to identify those that are anti-proliferative and are thus depleted from the population. We compared the lethality signature of the Ras Mut and WT cells to identify those shRNAs showing selective depletion in the Ras Mut but not Ras WT cells. Such shRNAs are potential Ras synthetic lethal (RSL) candidates. Relaxed statistical criteria identified 1,741 RSL shRNAs targeting 1,613 genes, whereas a more stringent cutoff identified a subset 379 RSL shRNAs targeting 368 genes (Table S1 and Table S2).
We devised a multicolor competition assay (MCA) to test the reproducibility of candidate RSL shRNAs from the primary screen (Smogorzewska et al., 2007). Individual shRNAs were packaged into retroviruses and infected into a 50:50 mixture of GFP+ DLD-1 Ras Mut cells and GFP− (colorless) DLD-1 Ras WT cells. The relative ratio of Ras Mut versus WT cells at ∼7 days post shRNA infection was analyzed by FACS and compared to that of the same cells infected with a negative control shRNA targeting luciferase (Figure 1E). Using this assay we tested 320 candidate RSL shRNAs from the primary screen and found 83 shRNA (26%) targeting 77 genes to preferentially decrease the fitness of Ras Mut cells compared to Ras WT cells (Table S3). To rule out cell-line specific effects, we performed MCAs in a second isogenic pair of colorectal cancer cell lines: HCT116 KRAS WT/G13D and HCT116 KRAS WT/−, which were derived in the same manner as the DLD-1 isogenic pair (Shirasawa et al., 1993; Torrance et al., 2001). Many shRNAs that scored in the DLD-1 cells also showed synthetic lethality in the HCT116 cells (50 of 68 tested, 73.5%, Table S3), indicating the majority of candidate RSL shRNAs are likely to interact genetically with KRAS.
Functional diversity of candidate RSL genes
We recovered shRNAs against KRAS itself from the screen. In accordance with the phenotype of the Ras WT isogenic controls, shRNA-mediated knockdown of K-Ras expression (both Mut and WT protein) in both DLD-1 and HCT116 cells resulted only in modest decrease in growth on adhesive surfaces but severely impaired colony formation in soft-agarose (Figure S1), thus confirming the inhibition of K-Ras to be sufficient for suppressing the malignant phenotype of these cells. Both the MAP kinase and PI 3-kinase pathways have been implicated in Ras-driven oncogenesis, However, we recovered few genes in these pathways and the MEK inhibitor U0126 and PI3K inhibitor LY294002 did not cause selective toxicity towards Ras Mut cells compared to Ras WT cells, indicating that inhibiting MEK or PI3K alone in these cells are not sufficient to constitute synthetic lethality (Figure S1) (Haigis et al., 2008).
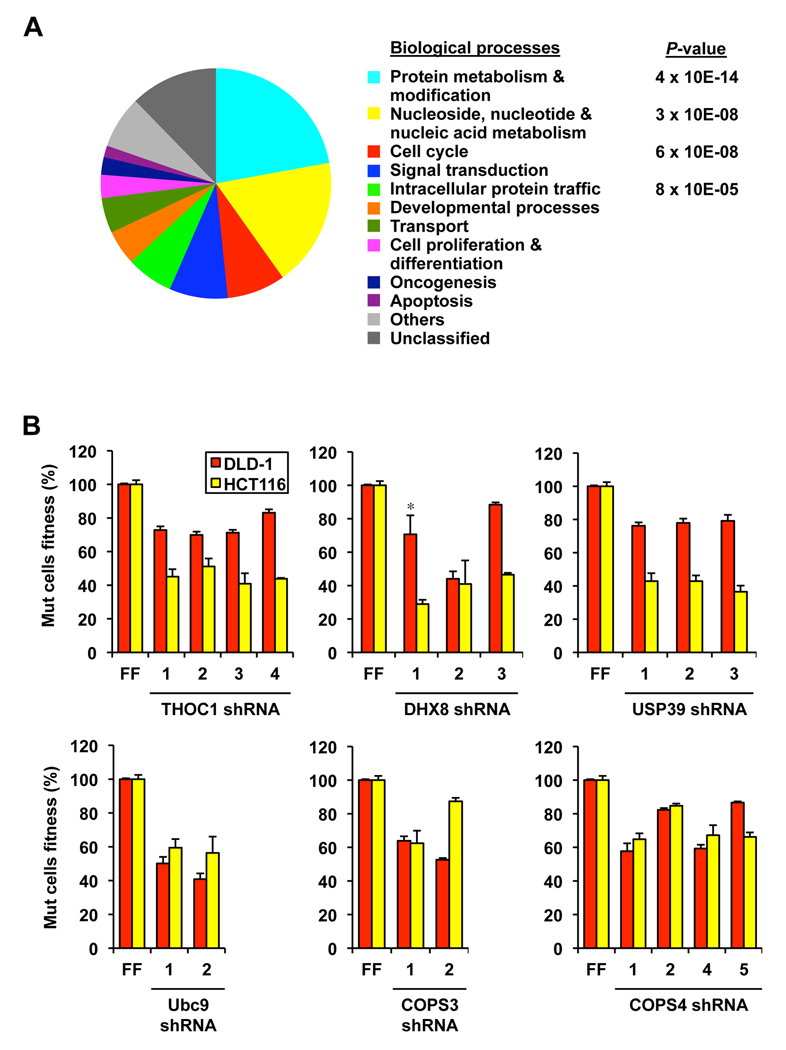
The list of candidate RSL genes is functionally diverse (Figure 2A and Table S3). Using PANTHER (Thomas et al., 2003), we identified several biological processes including protein modification, nucleic acid metabolism, cell cycle and signal transduction to be enriched in our screen. We also identified a number of pathways upon which Ras Mut cells are more dependent. For example, we identified shRNAs against genes in ribosomal biogenesis and translation control (BXDC2, FBL, NOL5A, EIF3S8, EIF3S4, GSPT1, HNRNPC and METAP1), in protein neddylation (COPS3, COPS4, COPS8, NEDD8, NAE1/APPBP1) and sumoylation pathways (SAE1, UBA2 and UBE2I) and in RNA splicing (FIP1L1, NXF1, USP39, DHX8 and THOC1). While many of these are essential genes under circumstances of complete depletion, we have found that partially reducing their activity leads to enhanced growth defects in Ras Mut cells. To rule out off-target effects, we tested multiple shRNAs against several of these genes, and in the majority of cases we were able to identify additional shRNAs that give the same phenotype (Figure 2B).
Figure 2. Functional diversity of candidate RSL genes.
A. Functional classification of candidate RSL genes in Supplemental Table 3 based on biological processes as annotated in PANTHER. The P-value denotes selective enrichment for genes in the corresponding biological process.
B. Validation of candidate RSL genes with multiple shRNAs using the competition assay. FF, negative control shRNA targeting firefly luciferase (for each shRNA p< 0.01 compared to the respective FF control, except cases marked with * which have p<0.05). Error bars indicate standard deviations in all figures unless otherwise indicated.
These findings suggest that Ras requires additional support from many genes to maintain the oncogenic state. For example, we identified the RNA processing/export factor THOC1, a member of the TREX mRNA transport complex, to be synthetically lethal with Ras (Figure 2B). THOC1 was recently shown to be selectively required for the Ras-driven proliferation and transformation of murine fibroblasts (Li et al., 2007). We also identified several subunits of the COP9 signalosome (Cope and Deshaies, 2003), which regulates the activity of SCF ubiquitin ligases as RSLs. Depleting either COPS3 or COPS4 impaired the fitness of Ras Mut cells relative to Ras WT cells in the competition assay (Figure 2). Furthermore, DLD-1 Ras Mut cells with stable depletion of COPS4 exhibit both impaired growth on adherent surfaces and in soft agar (Figure S2).
Ras mutant cells are sensitive to mitotic perturbations
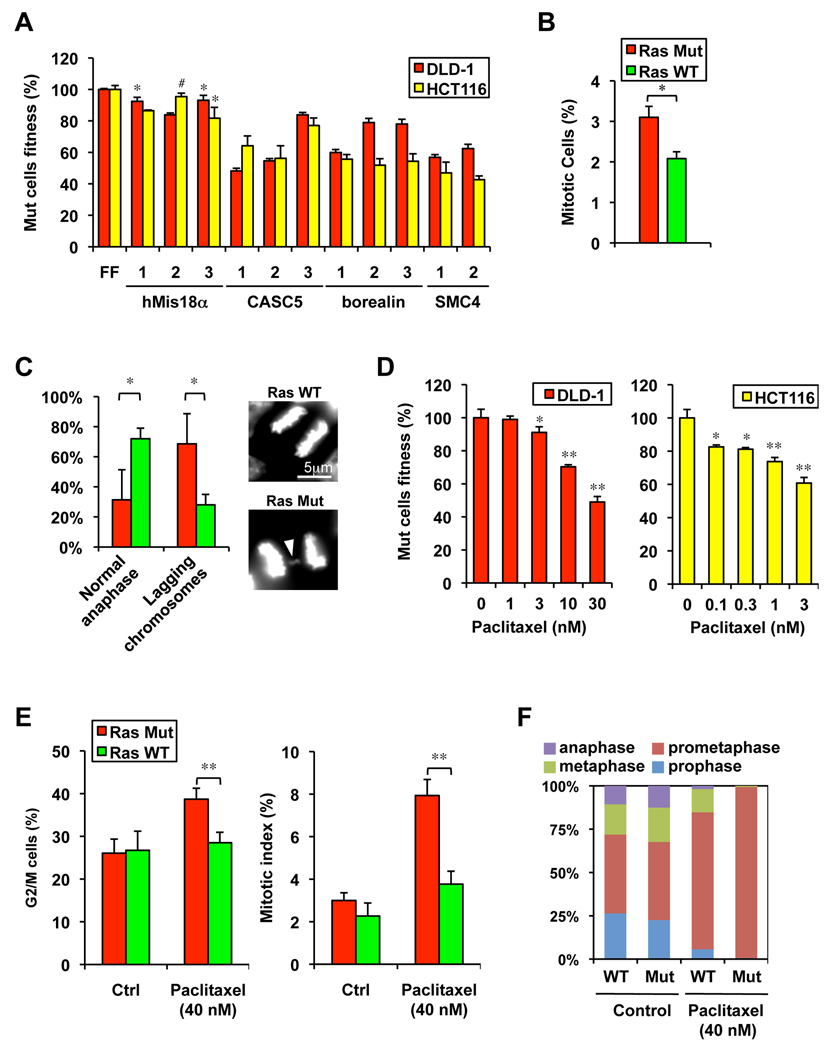
Strikingly, we identified a number of genes involved in the regulation of mitosis as RSL genes. Among these are cyclin A2 (CCNA2), hMis18α and hMis18 β (C21ORF45 and OIP5), borealin (CDCA8), KNL-1 (CASC5), MCAK (KIF2C), subunits of the APC/C complex (ANAPC1, ANAPC4, CDC16 and CDC27), SMC4 and the mitotic kinase PLK1 (Table S1 and Table S3). Their depletion phenotypes are likely to be on-target effects as multiple shRNAs gave similar results (Figure 3A and Figure S3A).
Figure 3. Hypersensitivity of Ras Mut cells to mitotic stress.
A. Examples of mitotic genes with multiple shRNAs showing synthetic lethality with mutant Ras (for each shRNA p< 0.01 compared to the respective FF control, except * p<0.05 and # not significant).
B. Mitotic index in asynchronous DLD-1 cells growing in log-phase as measured by phospho-H3 Ser10 staining (* p<0.05).
C. Ras Mut DLD-1 cells show a higher frequency of abnormal anaphases (lagging chromosomes) 40 minutes after released from the metaphase block by monastrol (* p<0.05). An example of Ras Mut cell with a lagging chromosome (arrowhead) is shown.
D. The microtubule stabilizer paclitaxel selectively decrease the fitness of Ras Mut cells in a dose dependent fashion. The competition assay was carried out in the presence of paclitaxel for 5 days (* p<0.05, ** p<0.01 compared to untreated samples).
E. Paclitaxel preferentially induces the G2 and mitotic accumulation of Ras Mut DLD-1 cells as assessed by FACS using DNA and phospho-H3 Ser10 staining, respectively (** p<0.01).
F. Paclitaxel causes strong prometaphase arrest in mitotic DLD-1 Ra Mut cells (shown are mean values of independent triplicates).
The identification of many mitotic genes as RSL candidates suggest that Ras Mut cells might experience heightened mitotic stress. Indeed, despite having a modestly faster doubling time (Figure 1C) and a similar percentage of G2 cells relative to Ras WT cells (26.1±3.3 vs. 26.7±4.5), DLD-1 Ras Mut cells show a 50% higher mitotic index, indicative of slower mitotic progression (Figure 3B). Furthermore, when released from a mitotic block induced by the Eg5 kinesin inhibitor monastrol (Mayer et al., 1999), a significantly higher fraction of Ras Mut cells exhibit lagging chromosome in anaphase (Figure 3C). To further explore the consequences of mitotic stress pharmacologically we examined the sensitivity of the Ras Mut and WT cells towards two inhibitors of mitotic spindle function, nocodazole and paclitaxel (Peterson and Mitchison, 2002). Whereas both the Ras Mut and WT cells show comparable sensitivity to the microtubule depolymerizer nocodazole (Figure S3B), both DLD-1 and HCT116 Ras Mut cells show increased sensitivity to the microtubule stabilizer paclitaxel relative to their respective WT counterparts (Figure 3D and Figure S3C). Cell cycle analysis reveals that, at the synthetic lethal concentration, paclitaxel causes a strong G2/M arrest in Ras Mut cells but not in Ras WT cells that is attributed to a striking pro-metaphase block in the Ras Mut cells (Figure 3E and F). Together these findings indicate that the Ras oncogene causes increased mitotic stress and renders the cell hypersensitive to perturbation of the mitotic machinery.
Ras mutant cells are hypersensitive to inhibition of PLK1 function
Polo-like kinase 1 (PLK1) plays a key role in mitosis (Barr et al., 2004; Petronczki et al., 2008). Its activity is often deregulated in cancer cells and inhibitors against PLK1 have been developed as potential cancer therapeutics (Strebhardt and Ullrich, 2006). We discovered that multiple shRNAs against PLK1 show increased toxicity towards Ras Mut cells compared to Ras WT cells in both DLD-1 and HCT116 isogenic pairs (Figure 4A, Figure S4A). Furthermore, siRNAs against PLK1 also yielded enhanced toxicity towards Ras Mut cells (Figure S4B).
Figure 4. Hypersensitivity of Ras Mut cells to PLK1 inhibition.
A. PLK1 depletion by shRNA leads to enhanced toxicity in Ras Mut cells (for each shRNA p< 0.01 compares to respective FF control, except * p<0.05).
B. The PLK1 inhibitor BI-2536 selectively decreases the fitness of Ras Mut cells in a dose-dependent fashion. The competition assay was carried out in the presence of BI-2536 for 5 days (* p<0.05, ** p<0.01 compared to untreated samples).
C. Effect of synthetic lethal concentrations of BI-2536 on cell cycle distribution of Ras Mut and Ras WT DLD-1 cells after 26 hours of treatment. Cell cycle profiles shown are representative of 3 experiments.
D. BI-2536 causes strong prometaphase arrest in mitotic DLD-1 Ras Mut cells (shown are mean values of independent triplicates).
E. BI-2536 does not differentially affect mitotic entry in DLD-1 Ras Mut and WT cells. Cells synchronized at G2/M by RO-3306 were released into nocodazole (100 ng/ml) together with indicated concentrations of BI-2536 for 1 hour. Mitotic index was measured as the percentage of cells staining positive for phospho-H3 Ser10. NR, no release.
F. BI-2536 differentially affects mitotic progression in DLD-1 Ras Mut and WT cells. Mitotic cells collected by nocodazole shake-off were released into indicated concentrations of BI-2536 for 2 hours. Mitotic index was measured as above (* p<0.05, ** p<0.01 compared to samples without BI-2536 treatment). NR, no release.
G. Effect of BI-2536 (25 nM) on cell cycle distribution of DLD-1 Ras Mut and WT cells over a 48-hour period (* p<0.05, ** p<0.01).
To further confirm this, we tested the effect of BI-2536, a highly selective small molecule inhibitor of PLK1 (Steegmaier et al., 2007). We observed increased sensitivity of Ras Mut cells towards BI-2536 in both DLD-1 and HCT116 isogenic pairs (Figure 4B, Figure S4C), with the strongest effect found at 25 nM in DLD-1 cells. We next analyzed cell cycle distribution of DLD-1 cells after treatment with either 25 nM or 50 nM of BI-2536 for 24h. Whereas the cell cycle profile of Ras WT cells is only modestly affected, Ras Mut cells show a profound G2/M accumulation in the presence of BI-2536 (Figure 4C). Microscopy analysis revealed the G2/M accumulation in Ras Mut cells is due to a strong block in prometaphase: whereas a substantial number of metaphase and anaphase cells could still be found among Ras WT cells in the presence of BI-2536, few such cells were found among Ras Mut cells (Figure 4D and Figure S4D).
PLK1 functions at multiple stages during mitosis (Petronczki et al., 2008). To investigate whether PLK1 inhibition delays mitotic entry in DLD-1 Ras Mut cells, we synchronized cells at the G2/M boundary using the CDK1 inhibitor RO-3306 (Vassilev et al., 2006) and released them with or without the presence of BI-2536 into nocodazole to trap mitotic cells. Mitotic entry was faster for Ras Mut cells but was unaffected by BI-2536 in either Ras WT or Ras Mut cells (Figure 4E and Figure S4E). We next released mitotic cells synchronized in nocodazole and tested their ability to complete mitosis in the presence of BI-2536. Whereas BI-2536 had minimal effect on Ras WT cells in this respect, it caused a profound delay in mitotic exit in Ras Mut cells (Figure 4F and Figure S4F). This finding further supports the notion that Ras Mut cells are more dependent upon PLK1 for mitotic progression. The mitotic arrest of Ras Mut cells in BI-2536, however, was not sustained over time. Prolonged treatment with BI-2536 for 48h results in an elevated sub-G1 population in Ras Mut cells, indicative of cell death (Figure 4G and Figure S4G).
PLK1 is transcriptionally upregulated in late S and in G2 phase, whereas its catalytic activation requires phosphorylation at Thr210 by the kinase Aurora-A at the G2/M transition (Seki et al., 2008; Macurek et al., 2008). A simple explanation of our results would be if Ras Mut cells had lower PLK1 protein levels or activity during mitosis. However, the levels of both total and activated PLK1 protein are actually slightly elevated in Ras Mut cells during mitosis, particularly at G2/M (Figure S4H). Together our findings suggest that activated Ras adversely affects mitotic progression and renders cells more dependent on PLK1 activity for proper mitosis progression.
Ras mutant cells are hypersensitive to APC/C and proteasome inhibition
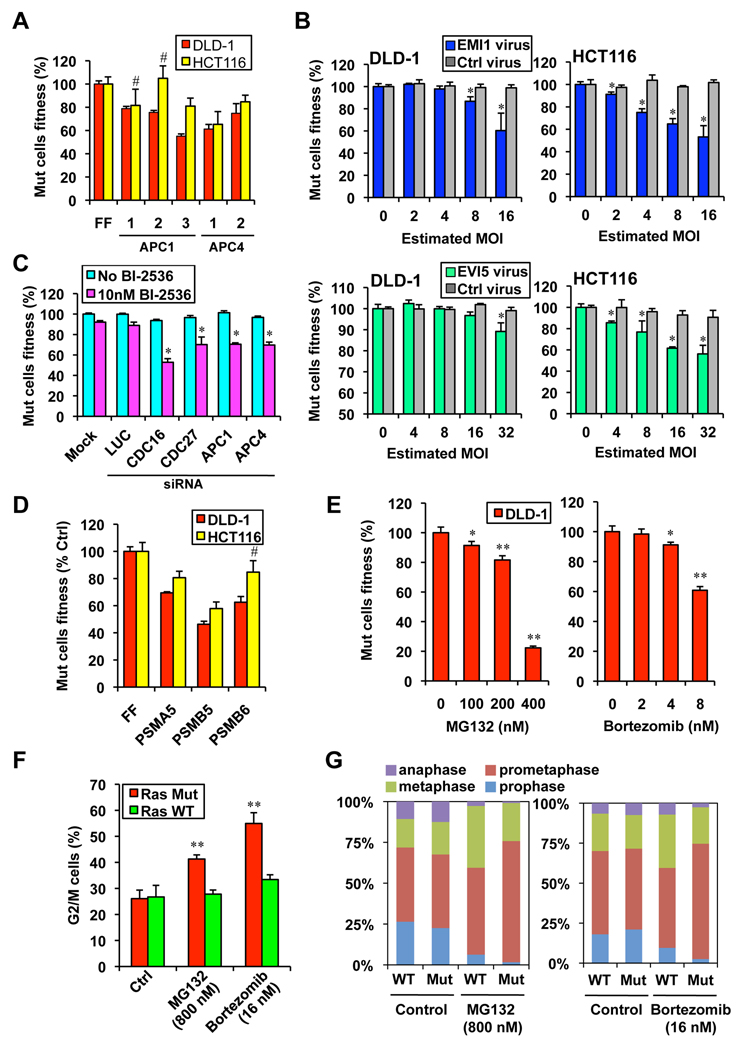
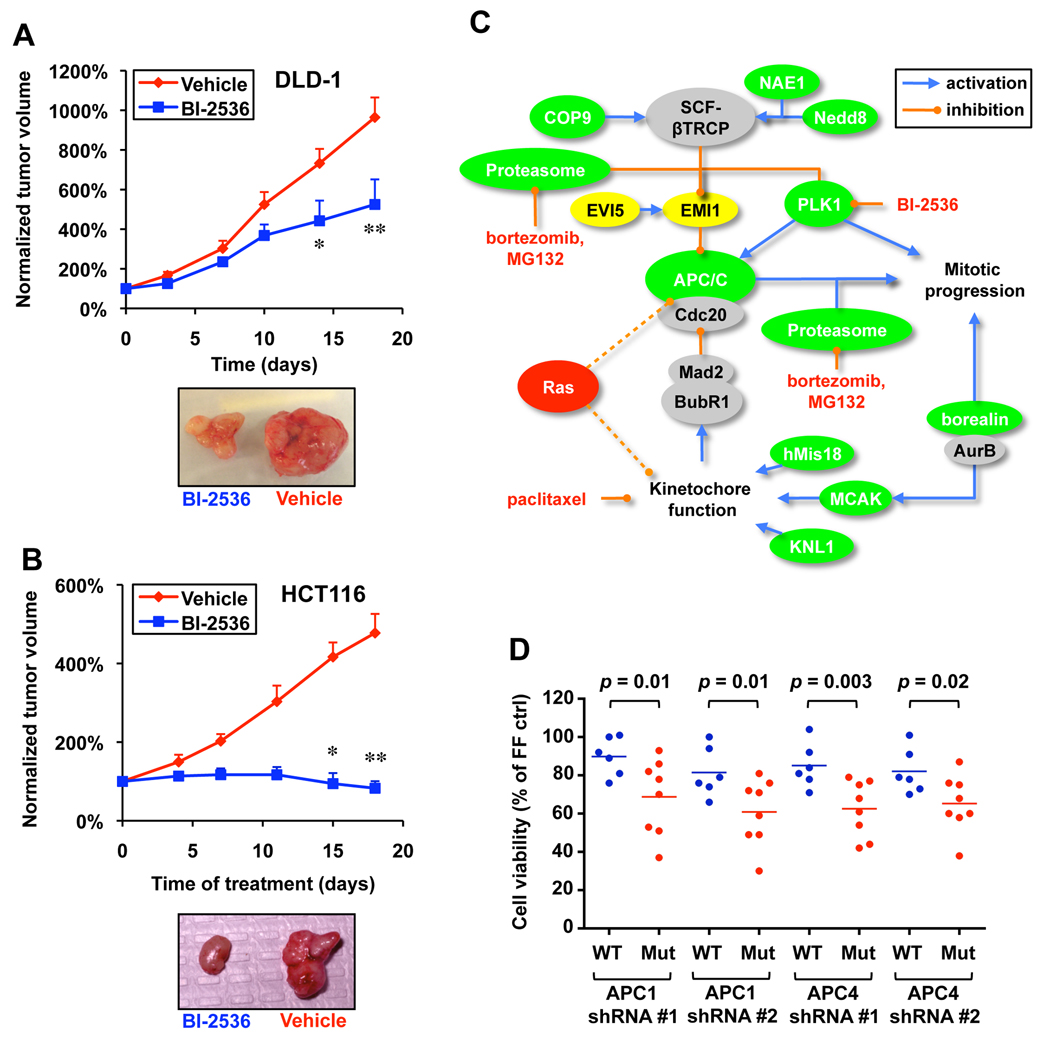
Mitotic progression is controlled by the activity of the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase that promotes the orderly degradation of key mitotic proteins (Peters, 2006). Several APC/C subunits including APC1/ANAPC1, APC4/ANAPC4, Cdc16 and Cdc27 scored in our screen (Table S1 and Table S3, Figure 5A), suggesting that Ras Mut cells are more dependent on APC/C activity for mitotic progression. The activity of APC/C is inhibited by EMI1 prior to mitosis, until PLK1 phosphorylates EMI1 and targets it for degradation via the SCF-βTRCP E3 ligase (Reimann et al., 2001; Hansen et al., 2004). The binding partner of EMI1, EVI5, on the other hand, blocks PLK1 phosphorylation of EMI1 and thereby antagonizes APC/C activation (Eldridge et al., 2006). Thus if Ras Mut cells are more dependent on APC/C activity, they might also be more sensitive to EMI1 or EVI5 overexpression. This is indeed the case. When we lentivirally over-expressed exogenous EMI1 and EVI5 in these cells, the viability of Ras Mut cells was specifically impaired (Figure 5B). Our results suggest that either APC activation might be reduced in Ras Mut cells or that these cells show a higher dependence on normal APC/C activity for survival. Consistent with these models, sub-phenotypic siRNA knockdown of APC subunits strongly synergizes with sub-phenotypic low concentrations of BI-2536 to confer synthetic lethality in DLD-1 Ras Mut cells (Figure 5C).
Figure 5. Ras Mut cells are hypersensitive to APC/C and proteasome inhibition.
A. Multiple shRNAs against APC1 and APC4 confer synthetic lethality in Ras Mut cells as measured by the competition assay (for each shRNA p< 0.05 compared to respective FF control, except # not significant).
B. Over expression of the APC/C inhibitor EMI1 and its binding protein EVI5 selectively impair the viability of Ras Mut cells days (* p<0.05 compared to uninfected samples).
C. siRNA mediated knockdown of APC/C subunits synergizes with low concentration of BI-2536 to selectively impair the viability of DLD-1 Ras Mut cells as measured by the competition assay. Cells were transfected with pools of 4 siRNAs against each gene, 2 days post transfection cells were treated with 10 nM of BI-2536 for 3 days before analysis by FACS (* p<0.05 compared to untransfected samples in the sample treatment group).
D. shRNAs targeting various proteasome subunits exhibit synthetic lethality in Ras Mut cells (for each shRNA p< 0.05 compared to respective FF control).
E. The proteasome inhibitors MG132 and bortezomib selectively decreased the fitness of DLD-1 Ras Mut cells in a dose dependent fashion over 4 days (* p<0.05,** p<0.01 compared to untreated samples).
F. MG132 and bortezomib preferentially induce the accumulation of Ras Mut DLD-1 cells as assessed by FACS using staining (** p<0.01 compared to samples without drug treatment).
G. MG132 and bortezomib cause strong prometaphase arrest in mitotic DLD-1 Ras Mut cells (shown are mean values of independent triplicates).
Two steps in the central pathway identified in our screen require proteolysis. Both activation of the APC/C through EMI1 ubiquitination and APC/C targeted ubiquitination of mitotic proteins ultimately require proteasome activity for degradation. Importantly, our screen also identified shRNAs against several proteasome subunits including PSMA5, PSMB5 and PSMB6 (Figure 5D). Furthermore, two structurally distinct small molecule inhibitors of the proteasome, MG132 and bortezomib (Velcade), both exhibit synthetic lethality with Ras Mut cells (Figure 5E and Figure S5A). Consistent with the model that the hypersensitivity of Ras Mut cells to proteasome inhibition is in part due to mitotic defects, Ras Mut cells are more sensitive to MG132- and bortezomib-induced G2/M arrest (Figure 5F and Figure S5B), again due to a profound prometaphase block (Figure 5G). Interestingly, we found that DLD-1 Ras Mut cells have higher levels of cyclin B1, a key mitotic cyclin that must be degraded by the APC/C during metaphase-anaphase transition (Figure S5C). Together these results suggest that the Ras oncogene causes a heightened dependency on the APC and renders cells sensitive to further inhibition of this complex.
Mitotic inhibitors attenuate tumor xenograft growth in vivo
To assess whether targeting the mitotic machinery could inhibit the growth of DLD-1 and HCT116 cells in vivo using mouse xenograft models, we treated nude mice bearing subcutaneous DLD-1 or HCT116 tumors with the PLK1 inhibitor BI-2536. In both cases we found tumor growth to be significantly attenuated in animals treated with BI-2536 (Figure 6A and 6B). In agreement with their in vitro sensitivity, we find HCT116 tumors to be more sensitive to BI-2536 compared to DLD-1 tumors.
Figure 6. A model of mitotic regulation by Ras.
A. BI-2536 attenuates DLD-1 tumor growth in vivo. Intravenous injection of BI-2536 started 1 week after subcutaneous injection of DLD1 cells. Tumor volume at each time point was normalized to the initial tumor volume (* p<0.05 and ** p<0.01, 2-way ANOVA, error bars indicate S.E.M.). Representative images of tumors after treatment are shown.
B. BI-2536 attenuates HCT116 tumor growth in vivo. Treatment of HCT116 xenografts was similar to that described for DLD-1 tumors (* p<0.05 and ** p<0.01, 2-way ANOVA, error bars indicate S.E.M.).
C. A model in which oncogenic Ras introduces mitotic stress that can be exacerbated to produce lethality by interfering with kinetochore and APC/C function. Genes shaded green are RSL genes, while yellow genes cause Ras-specific lethality when overproduced. Dotted lines illustrate hypothetical connections between Ras and aspects of mitotic regulation leading to mitotic stress.
D. Sensitivity of a panel of NSCLCs to APC/C shRNAs. NSCLCs with endogenous Ras mutations (H23, H358, H647, H1299, H1734, H2030, H2122 and H2228) and without Ras mutations (H522, H838, H1437, H1650, H1838 and H1975) were infected with shRNAs against APC1 or APC4. Cell viability was compared to control FF shRNA infected samples 6 days post infection.
The mitotic machinery as an Achilles’ heel for Ras mutant cancer cells
Taken together, our results suggest that cancer cells with mutant Ras experience elevated mitotic stress and are more dependent on key mitotic proteins such as PLK1, the APC complex, the COP9 signelosome and the proteasome for proper mitotic progression (Figure 6C), and we showed that targeting selected mitotic proteins could exacerbate this mitotic stress to selectively kill Ras mutant cancer cells. We have shown that small molecule inhibitors that disrupt mitosis, including paclitaxel, BI-2536, bortezomb (Velcade) and MG132, all constitute synthetic lethality with Ras mutant cells. Strikingly, transient treatment of DLD-1 cells for 24 hours with these drugs, which approximates the length of one cell cycle for these cells, is sufficient to selectively impair the viability of Ras Mut cells (Figure S6). This indicates that many Ras Mut cells might not recover from their drug-induced mitotic arrest and fail to complete normal mitosis following the inhibitor removal.
To assess whether this mitotic stress might also be associated with other oncogenes, we tested potential synthetic lethal interactions between paclitaxel, BI-2536, bortezomib or MG132 and the PIK3CA oncogene. DLD-1 and HCT116 cells also harbor oncogenic mutations in PIK3CA encoding the p110α catalytic subunit of PI3K. Isogenic DLD-1 and HCT116 cell lines in which the PIK3A oncogene has been deleted by homologous recombination have been generated (Samuels et al., 2005). In contrast to Ras Mut DLD-1 cells, PI3K Mut DLD-1 cells are more resistant to these inhibitors compared to PIK3CA WT DLD-1 cells. Furthermore, HCT116 PI3K Mut and WT cells showed very little difference when treated with these inhibitors (Figure S7). These results indicate that the increased mitotic stress we observed is specific for oncogeneic Ras.
Although the detailed mechanisms by which the Ras oncogene affects the activity of various mitotic machineries remain to be elucidated, our data suggest that either impaired APC function or an enhanced requirement for APC function might be a critical oncogenic stress associated with Ras mutation. We tested this proposition using a panel of non-small cell lung cancer (NSCLC) cell lines with or without Ras mutations by assessing their sensitivity to shRNA-mediated knockdown of either APC1 or APC4. Since these cell lines are not isogenic and harbor many additional, different mutations, they are expected to display a range of sensitivities to APC/C knockdown. However, as a group the NSCLC cells with Ras mutations are generally more sensitive to shRNAs against APC1 and APC4, thereby further supporting the notion that Ras mutation is associated with mitotic stress (Figure 6D).
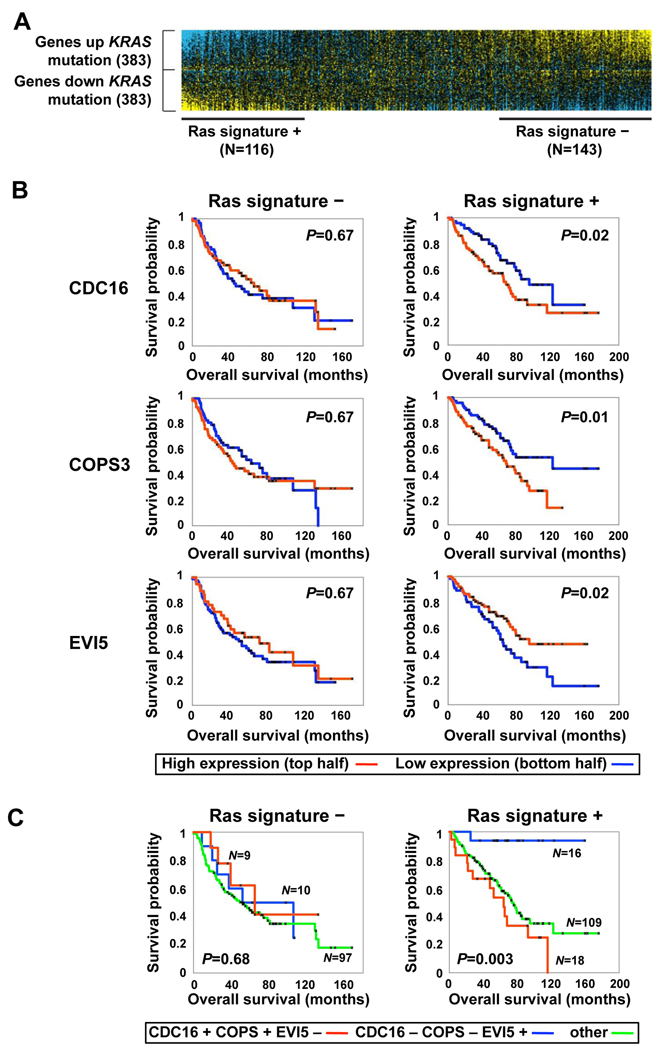
If our in vitro and mouse xenograft analysis are relevant to Ras-driven tumors, patients bearing tumors with activated Ras could benefit from decreased APC activity. Support for this hypothesis comes from analysis of lung cancer tumor samples. We analyzed whether the expression of any of our core mitotic RSL candidate genes and other genes directly associated with APC function (all APC subunits, EMI1, EVI5, Cdc20, Cdh1 and UbcH10) correlated with patient prognosis in a large cohort of lung cancer samples (Shedden et al., 2008). As the mutation status of the Ras genes in these tumors is currently unknown but their transcriptional profiles are known, we first derived a Ras expression signature from a separate, smaller set of lung tumors whose Ras mutation status was known (Bhattacharjee et al., 2001). Importantly, we were able to validate the predictive power of our Ras signature in two additional cohorts of lung tumor samples (Figure S8). We applied this Ras signature to our large cohort to stratify them as having positive, negative or neutral Ras signatures (Figure 7A). We defined 143 tumors as having a strong Ras mutant signature (Ras signature +) and 116 as having a WT-Ras signature (Ras signature -). We then asked if the expression levels of any of the 24 APC/C and COPS9-associated core genes were associated with prognosis in a manner that was also dependent on the tumor’s Ras signature status. Three genes, COPS3, CDC16 and EVI5, showed such a correlation. Strikingly, lower expression of COPS3 and CDC16, and higher expression of EVI5 – all consistent with potentially decreased APC/C activity – are each associated with enhanced survival for patients bearing tumors with a positive Ras signature but have no prognostic value in patients bearing tumors with a negative Ras signature (Figure 7B). The correlation pattern of these genes remains significant after correction for multiple hypothesis testing (p = 0.02). When the tumors were simultaneously investigated for all three signatures, those with lower COPS3 and CDC16 together with higher EVI5 expression levels were associated with a striking enhancement of survival for patients bearing tumors that exhibit a positive Ras signature (Figure 7C). These findings are consistent with the hypothesis that APC activity could present a limiting factor in Ras mutant cancer cells and present an attractive drug target for cancers with Ras mutation.
Figure 7. Candidate RSL genes associated with prognosis in human lung adenocarcinomas showing activation of the Ras pathway.
A. A gene expression signature for lung cancers with activated Ras pathway. A previously derived gene expression signature (766 genes) of KRAS mutant versus wild-type lung tumors was used as a probe in an additional set of expression profiles from 442 human lung adenocarcinomas. Both tumors with significant (P<0.01) similarity or dissimilarity to the KRAS signature (“Ras signature +” and “Ras signature –” tumors, respectively) were considered for subsequent survival analyses.
B. RSL pathway genes that correlate with prognosis among “Ras signature+” tumors. The expression of two RSL genes, COPS3 and CDC16, are inversely correlated with better survival whereas expression of EVI5 correlates with better survival (P<0.05, both log-rank and Cox, log-rank P-values shown). Within each of the two tumor subsets considered (“Ras signature +” and “Ras signature −”), tumors with expression levels for the given gene greater than the median (red line) were compared to the rest of the tumors in the subset (blue line), using Kaplan-Meier analysis.
C. Prognosis in “Ras signature +” and “Ras signature −” tumors using combined information from COPS3CDC16, and EVI5. For each of the three genes, “+” and “−” is relative to their median expression across the subset of tumors. Within each of the two tumor subsets, three tumor groups were compared by Kaplan-Meier analysis: tumors with high CDC16, high COPS3, and low EVI5 (“CDC16+ COPS3+ EVI−”); tumors with low CDC16, low COPS3, and high EVI5 (“CDC16− COPS3− EVI+”); and tumors not falling into these two groups (“other”). Log-rank P-values indicate significant differences among any of the three groups.
DISCUSSION
Using our pool-based shRNA platform in a genome-wide screen, we identified candidate Ras synthetic lethal (RSL) genes whose depletion constitute synthetic lethality with the KRAS oncogene. The vast majority of these are implicated in Ras function for the first time. Our study suggests that a broad genetic network spanning multiple cellular functions is required to support the Ras oncogenic state. Many genes in this network could be exploited as potential therapeutic targets, as demonstrated by the synthetic lethal effect of their depletion by RNAi.
Mitotic stress as a hallmark of the Ras oncogenic state
We identified multiple mitotic genes whose inhibition results in synthetic lethality with mutant Ras. These findings underscore a previously underappreciated role for Ras in regulating mitotic progression and suggest that the Ras oncogene renders cells more dependent on key mitotic proteins for survival, possibly by compromising the fidelity of mitosis itself and generating mitotic stress. It has been shown that mutant Ras can induce chromosome instability (Denko et al., 1994). How Ras affects mitotic progression is presently unclear although our data suggest a role in regulation of prometaphase events, possibly chromosome congression. Several reports have implicated the Ras/MAPK pathway in mitosis. Previously, we showed yeast Ras genetically interacts with the kinetochore DASH/Dam1-Duo1 complex and thus might influence spindle attachment and resolution of syntelic attachments (Li et al., 2005). In mitotic Xenopus oocyte extracts, MAPK activity is required for mitotic entry and the maintenance of the mitotic state, whereas its inactivation is required for mitotic exit (Wang et al., 2007; Guadagno and Ferrell, 1998). In Xenopus oocytes arrested at metaphase of meiosis II, a signaling pathway involving Mos-MAPK activates Erp1/Emi2 to inhibit APC/C activation (Inoue et al., 2007; Nishiyama et al., 2007). Activated MAP kinase localizes to kinetochores (Zecevic et al., 1998) and a hyperactive MAPK pathway can promote spindle checkpoint bypass (Eves et al., 2006). In addition, the Ras effector, RASSF1A, has been proposed to negatively regulate the APC in mammals (Whitehurst et al., 2008). We observe that Ras activation causes a delay in mitotic progression, lengthening it by nearly 50%. In addition, we find that transient treatment with monastrol arrests Ras mutant cells that upon release display a massive increase in lagging chromosomes during anaphase. Our data suggest that Ras mutants are experiencing mitotic stress and exacerbating this stress in particular ways such that interference with KNL-1, PLK1 or MCAK, or addition of paclitaxel leads to stress overload and cell death. In support of our findings, depletion of survivin and TPX2 has deleterious effects on Ras mutant cells (Sarthy et al., 2007; Morgan-Lappe et al., 2007). Importantly, not all mitotic perturbations selectively kill Ras mutants, as nocodazole treatment displayed no synthetic lethality. It will be important to precisely dissect these mitotic defects to determine how they might best be exploited to kill Ras mutant cells.
New drug targets for tumors with Ras mutations
A key goal in cancer research is the discovery of new drug targets that will selectively impair the viability of cancer cells. Our approach enables the rapid identification of functional vulnerabilities in cancer cells for therapeutic exploitation. Based on our genetic analysis, we have identified at least 7 such enzymatic targets in the mitotic pathway alone, NAE1 (the E1 for Nedd8), the COP9 signalosome (a deneddylase), PLK1, MCAK, the APC, the proteasome and possibly the SCF. Our studies strongly suggest that inhibiting each of these enzymatic complexes could have efficacy in selectively killing cancer cells. Of these targets, all have some relationship to control of the APC, which, along with NAE1 and the COP9 signalosome, presents itself as a novel target in cancer cells. In support of this, the overproduction of two inhibitors of the APC, EMI1 and EVI5 are also more toxic to Ras mutant cells. Furthermore, we showed that NSCLCs with Ras mutations tend to be more sensitive to APC knockdown. Thus, inhibiting APC function could prove efficacious in treating Ras-driven tumors of diverse genetic backgrounds. In addition to mitotic regulators, our screen has identified a functionally diverse set of RSL candidates with enzymatic activities and thus are potential drug candidates. Examples are the helicase DHX8, the kinases JAK1, ERK5/MAPK7, ROIK1, the glutaminyl t-RNA synthase QARS, the histone H3-K9 methryltransferase SUV39H2, the E2 SUMO conjugating enzyme UBA2/SAE2, and the E1 and E2 ubiquitin conjugating enzymes UBE1 and UBC9/UBE2I, respectively. Further validation efforts will be needed to assess the addiction of Ras mutant cells to these genes and determine their utility as drug targets.
Illumination of the mitotic stress phenotype of Ras mutants led us to the identification of a number of small molecule inhibitors potentially useful for treating tumors with Ras mutations. Two of these, the microtubule stabilizer paclitaxel and the proteasome inhibitor bortezomib (Velcade), have already been approved for the treatment of certain types of cancers. Velcade has been approved for use in multiple myeloma (MM). Perhaps not coincidentally, MM has a high frequency of Ras mutation, 30–50% in different studies and up to 80% in patients experiencing a relapse (Liu et al., 1996; Bezieau et al., 2001). In addition, a significant number of patients that do not have activating mutations in Ras, have an activated Ras pathway through IL-6 signaling (Ogata et al., 1997; Rowley and Van Ness, 2002). Thus, the efficacy of Velcade in treating MM might be in part due to their high prevalence of Ras mutations and acute activation of Ras, and it would be important to determine if patients with Ras mutations respond better to Velcade, thereby allowing Ras status to be used as a marker for patient stratification in future clinical trials. The third agent, the PLK1 inhibitor BI-2536, is currently undergoing phase I clinical trials (Mross et al., 2008). Although their efficacy with respect to Ras mutational status remains to be tested clinically, finding three existing molecules as targets based on this simple genetic screen bodes well for genetic dissection of cancers in the future.
Additional uses of the RSL genes in cancer analysis
In addition to the discovery of novel cancer drug targets and potential patient stratification markers for clinical trials such as Ras for proteasome inhibitors, this screen has provided markers that provide prognostic value for patient survival. Expression levels of three genes identified in our study, COPS3, CDC16 and EVI5 show good correlations with patient survival in the direction that reducing APC/C activity promotes survival in Ras-driven lung cancers but has no predictive value in lung tumors lacking the Ras transcriptional signature. In support of this, we found that although our Ras synthetic lethal screen was carried out in colon cancer cells, the genetic dependency of the Ras oncogene on APC appears to be conserved in NSCLCs as well. These findings lend strong support to the idea that the activity of the RSL genes will affect tumorigenesis and survival in humans. It will be interesting to see whether these transcriptional patterns correlate with survival in other Ras-dependent tumor types. A priori, there is no reason that these genes in particular will correlate with survival in other cancer types but it is possible that other members of this RSL gene cohort will display a similar ability in different tumor contexts as each of them can become rate limiting for this process. Furthermore, it is possible that other synthetic lethal cohorts for other oncogenes or cancer types will also provide predictive value for other tumors types.
Our study illustrates the rapidity in which synthetic lethality analysis can be translated into better therapeutic strategies. We find that therapeutic strategies aimed at suppressing the Ras oncogenic pathway directly (e.g. RNAi against Ras), inhibiting the stress support pathways protecting the cancer cells from oncogenic stress (e.g. RNAi and inhibitors against the proteasome and the APC), or enhancing the stress phenotype of cancer cells (e.g. paclitaxel) could all selectively impair the viability of Ras mutant cancer cells. Complementary to the physical mapping of cancer genomes, functional approaches such as these can identify genetic dependencies of cancer cells regardless of the mutational status of the gene of interest. Indeed, a key point from this study is that among the many genes identified and verified in this screen as potential drug targets, only Ras itself is a known oncogene. This suggests that there is a much larger set of non-oncogen`es that can serve as drug targets in treating cancer. We have recently proposed the concept of “non-oncogene addiction” (NOA) to describe the extensive dependency of cancer cells on the function of diverse networks of genes – many of which are neither mutated in cancer or oncogenic – for their growth and survival (Solimini et al., 2008; Luo et al., 2009). Our study provides a glimpse of the landscape of NOA and suggests this is an area that is likely to shed new light on the mechanisms of tumorigenesis and presents new opportunities for cancer therapeutics that cannot be discovered by examination of alterations in tumors.
Experimental Procedures
shRNA library screen and micoarray hybridization
The pool-based shRNA screen using half-hairpin (HH) barcode deconvolution was carried out as described in (Schlabach et al., 2008) at a representation of 1000, screened as 6 pools of ∼13,000 shRNAs per pool in independent triplicates. For each pair of corresponding PD0 and PD17 samples, shRNA HH barcodes were PCR-recovered from genomic samples and competitively hybridized to a microarray containing the corresponding probes. Custom microarrays with HH barcode probe sequences were from Roche Nimblegen. For additional information, see the supplemental information.
Cell culture, molecular biology and reagents
Sequence information for additional shRNAs and siRNAs used for validation is in Supplemental Table 4. The DLD-1 and HCT116 isogenic cells were maintained in McCoy’s 5A media supplemented with 10% FBS and antibiotics. NSCLC lines were maintained in RPMI-1640 media with 10% FBS and antibiotics. For proliferation assays, cells were seeded in either 24-well or 96-well plates and cell number was measured using CellTiterGLO (Promega). For adherent colony assays, 1000 cells were seeded in each well of a 6-well plate and colonies were counted 10 days later by Coomassie staining. For anchorage independent colony assays, 1000 cells were seeded in each well of a 6-well plate in media containing 0.35% low-melting point agarose and colonies were counted 3 weeks later by crystal violet staining.
Analysis of human lung tumor profiles
A gene signature of KRAS mutant versus wild-type tumors was defined, using a published dataset of 84 lung adenocarcinomas (Bhattacharjee et al., 2001) for which the KRAS mutation status of each tumor was known (genes with P<0.01, two-sided t-test were selected). This KRAS signature was validated by both gene signature similarity scores and by gene set enrichment analysis using two independent cohorts of lung cancer where the KRAS mutation status was known for the tumors (Ding et al., 2008; Beer et al., 2002), (Figure S8). This KRAS gene signature was then applied to analyze a gene expression profile dataset of 442 human lung adenocarcinomas (Shedden et al., 2008). Each tumor was scored for manifestation of the Ras pathway as follows: The Shedden tumor profiles were generated among four laboratories, and so within each laboratory subset, expression values for each gene were normalized to standard deviations from the mean. Within each Shedden tumor profile, the average of the genes high (“up”) in the Bhattacharjee KRAS signature were compared with the average of the genes low (“down”) in the signature: tumors with higher expression of the “up” genes as compared to the “down” genes (P<0.01, t-test) were classified as showing Ras pathway manifestation (“Ras signature+”); tumors with higher expression of the “down” genes (P<0.01) were classified as not showing Ras pathway manifestation (“Ras signature–”); tumors that were intermediate between the above two groups were not used in subsequent analyses. Within the Shedden “Ras signature+” tumors (N=143), tumors with expression levels greater than the median for the given gene were compared with the rest of the tumors in the subset, using Kaplan-Meier analysis for time to patient death. In addition, univariate Cox analysis evaluated the expression of genes as a continuous variable for correlation with outcome. The same Kaplan-Meier and Cox analyses were also carried out using the Shedden “Ras signature-” tumors (N=116). To correct for multiple hypothesis testing, we carried out simulation tests to estimate the probability of obtaining 3 out of 24 APC and COPS9 signalosome associated genes that both had a p-value of <0.05 and whose relative expression levels changed in the predicted direction in KRAS mutant tumors. Based on these criteria, the expected number of chance significant hits was 0.57 genes (based on 1000 simulations, where both survival p-value and direction of change was randomly generated for each of the 24 genes), and the probability of getting 3 or more genes due to multiple testing was p = 0.02.
Cell cycle analysis
For cell cycle analysis, cells were analyzed using a BD LSR II FACS analyzer. Cells were synchronization by double thymidine (Steegmaier et al., 2007) and were released into media with the indicated drug, RO-3306 (10µM) (Vassilev et al., 2006), BI-2536 (100 nM) (Steegmaier et al., 2007) and nocodazole (100 ng/ml). For one-step arrest in G2/M or in prometaphase, cells were treated overnight with either RO-3306 (10 µM) or monastrol (100 µM), respectively.
Mouse xenograft tumor models
DLD-1 cells (5 million cells per site) or HCT116 cells (2 million cells per site) in suspension were mixed with equal volumes of matrigel (BD bioscience) and injected subcutaneously into both flanks of 6 week-old female nude mice (Charles River laboratory). Drug treatment began after one week of cell injection. BI-2536 was given by i.v. under a twice per week schedule. The dose of BI-2536 used for DLD-1 xenografts was 50 mg/kg for the first 2 weeks and then increased to 75 mg/kg for the last week. The dose of BI-2536 used for HCT116 xenografts was 50 mg/kg. Measurement of tumor size was performed twice a week, and tumor volume was estimated using the formula: volume = Length x Width2 x 0.5. The volume of each tumor at serial time points was normalized with the initial tumor volume and comparison of tumor volumes between the drug treatment group and vehicle control group was carried out using 2-way ANOVA with Bonferroni post-tests.
Supplementary Material
ACKNOWLEDGEMENT
We thank Drs. Bert Vogelstein and Victor Velculescu for the DLD-1 and HCT116 cells and Dr. Nathanael Gray and SAI Advantium for help with the synthesis of BI-2536. We thank Matthew Buttarazzi, Nicholas Kwiatkowski, Dror Berel and Dr Chandra Miduturu for technical assistance and Drs. Greg Hannon, Nicole Solimini, Tim Mitchison and Randal King for helpful discussions. J.L. is supported by the AACR Prevent Cancer Foundation AstraZeneca Fellowship in Translational Lung Cancer Research. M.J.E. is the Philip O’Bryan Montgomery Jr., MD fellow of the Damon Runyon Cancer Research Foundation (DRG-1996-08). C.J.C. and T.F.W. are supported in part by NIH grant P30 CA125123. TFW is supported by the Susan G. Komen for the Cure Foundation (KG090355). This work is supported by grants from NIH and DOD to S.J.E. and S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include 8 figures, 4 tables and Supplemental Experimental Procedures and can be found with this article online.
REFERENCE
- Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bezieau S, Devilder MC, Avet-Loiseau H, Mellerin MP, Puthier D, Pennarun E, Rapp MJ, Harousseau JL, Moisan JP, Bataille R. High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Hum Mutat. 2001;18:212–224. doi: 10.1002/humu.1177. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci U S A. 1994;91:5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson PK. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell. 2006;124:367–380. doi: 10.1016/j.cell.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras and PIK3CA murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR. Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol Cell. 2006;23:561–574. doi: 10.1016/j.molcel.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadagno TM, Ferrell JEJ. Requirement for MAPK activation for normal mitotic progression in Xenopus egg extracts. Science. 1998;282:1312–1315. doi: 10.1126/science.282.5392.1312. [DOI] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the APC by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol Biol Cell. 2004;15:5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Ohe M, Kanemori Y, Nobui T, Sagata N. A direct link of the Mos-MAPK pathway to Erp1/Emi2 in meiotic arrest of Xenopus laevis eggs. Nature. 2007;446:1100–1104. doi: 10.1038/nature05688. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JM, Li Y, Elledge SJ. Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the ras/PKA pathway and a novel subunit required for Ask1 association. Mol Cell Biol. 2005;25:767–778. doi: 10.1128/MCB.25.2.767-778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lin AW, Zhang X, Wang Y, Goodrich DW. Cancer cells and normal cells differ in their requirements for Thoc1. Cancer Res. 2007;67:6657–6664. doi: 10.1158/0008-5472.CAN-06-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Leong T, Quam L, Billadeau D, Kay NE, Greipp P, Kyle RA, Oken MM, Van Ness B. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood. 1996;88:2699–2706. [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, Vernetti L, Schurdak M, Wang J, Fesik SW. Identification of Ras-related nuclear protein, targeting protein for xenopus KLP 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 2007;67:4390–4398. doi: 10.1158/0008-5472.CAN-06-4132. [DOI] [PubMed] [Google Scholar]
- Mross K, Frost A, Steinbild S, Hedbom S, Rentschler J, Kaiser R, Rouyrre N, Trommeshauser D, Hoesl CE, Munzert G. Phase I dose escalation and pharmacokinetic study of BI 2536 in patients with advanced solid tumors. J Clin Oncol. 2008;26:5511–5517. doi: 10.1200/JCO.2008.16.1547. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Ohsumi K, Kishimoto T. Phosphorylation of Erp1 by p90rsk is required for cytostatic factor arrest in Xenopus laevis eggs. Nature. 2007;446:1096–1099. doi: 10.1038/nature05696. [DOI] [PubMed] [Google Scholar]
- Ogata A, Chauhan D, Teoh G, Treon SP, Urashima M, Schlossman RL, Anderson KC. IL-6 triggers cell growth via the Ras-dependent mitogen-activated protein kinase cascade. J Immunol. 1997;159:2212–2221. [PubMed] [Google Scholar]
- Peters JM. The APC/C: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Peterson JR, Mitchison TJ. Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton. Chem Biol. 2002;9:1275–1285. doi: 10.1016/s1074-5521(02)00284-3. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the APC. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Rowley M, Van Ness B. Activation of N-ras and K-ras induced by IL-6 in a myeloma cell line: implications for disease progression and therapeutic response. Oncogene. 2002;21:8769–8775. doi: 10.1038/sj.onc.1205387. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Sarthy AV, Morgan-Lappe SE, Zakula D, Vernetti L, Schurdak M, Packer JC, Anderson MG, Shirasawa S, Sasazuki T, Fesik SW. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol Cancer Ther. 2007;6:269–276. doi: 10.1158/1535-7163.MCT-06-0560. [DOI] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ERr, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening. Nat Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, He G, Nelman-Gonzalez M, Ashorn CL, Gallick GE, Stukenberg PT, Kirschner MW, Kuang J. Regulation of Cdc25C by ERK-MAP kinases during G2/M. Cell. 2007;128:1119–1132. doi: 10.1016/j.cell.2006.11.053. [DOI] [PubMed] [Google Scholar]
- Whitehurst AW, Ram R, Shivakumar L, Gao B, Minna JD, White MA. The RASSF1A tumor suppressor restrains APC/C activity during the G1/S phase transition to promote cell cycle progression. Mol Cell Biol. 2008;28:3190–3197. doi: 10.1128/MCB.02291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic M, Catling AD, Eblen ST, Renzi L, Hittle JC, Yen TJ, Gorbsky GJ, Weber MJ. Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.