Abstract
We compared the inferred transcription regulatory interactions from three high-throughput methods. As these methods utilize different principles, they have few interactions in common, suggesting that they capture distinct facets of the actual transcription regulatory program. In addition, we show that these methods capture disparate biological phenomena, which include long-range interactions between telomeres and transcription factors, downstream effects of interference with ribosome biogenesis and a protein-aggregation response. Through a detailed analysis of the latter we predict components of the system responding to protein-aggregation stress.
Reconstruction of transcriptional regulatory networks
Deciphering the complete transcriptional regulatory program of organisms is an important goal in molecular biology. Identification of the spatial and temporal regulatory interactions between transcription factors (TFs) and their target genes is an important step towards this goal (Text box 1 and Fig 1A). Different high-throughput methods (see Fig S1), are used to infer transcription regulatory interactions in various organisms. However, these methods are based on different principles and it is not clear whether they capture the same or distinct facets, such as combinatorial regulation and back-ups, of the underlying regulatory program. Although numerous studies 1-7 have generated genome-scale transcriptional information, the results from the different studies have not been systematically compared. Therefore, we assembled and compared the genome-scale transcription regulatory networks (TRNs) for yeast, based on data sets from three high-throughput techniques: ChIP-chip, targeted gene disruption and over-expression of transcription factors (see Table S1, Materials and Methods in the online supplementary material). Although there was a significant overlap in TFs between the three reconstructed TRNs (Fig 1B), the number of common regulatory interactions shared by them was <1%. Furthermore, the extent of overlap of inferred regulatory interactions even between pairs of reconstructed TRNs was <5% (Fig 1B), suggesting that the high-throughput methods reveal different aspects of the actual regulatory process (Fig 1B). The level of agreement in regulatory interactions between the reconstructed TRNs did not change even when we restricted the analysis to the TFs shared between the TRNs. Likewise, we did not observe a significant increase in the overlap of interactions when we reconstructed TRNs using different p-values thresholds (see Fig S2 and Table S2). This prompted us to further investigate the nature and significance of regulatory interactions in the three distinct TRNs: TRNCC, TRNGRD and TRNGROE (i.e those generated by the three high-throughput methods – see Glossary). In particular we address the following questions: Are there global and local structural differences amongst the different TRNs? Are the results of the high-throughput methods influenced by disparate biological phenomena? Do they provide novel biological insights apart from the description of the relevant regulatory programs?
Text box 1. Reconstruction of transcription regulatory networks and high-throughput methods.
Although the monumental task of reconstructing regulatory programs for whole organisms is far from complete, recent advances in high-throughput experimental techniques, together with conceptual and representational advances have brought us closer to this objective. Independent experimental approaches enable the genome-scale reconstruction of the transcription regulatory program of an organism either by directly inferring in vivo binding to regulatory sequences or indirectly by identifying the set of genes which are differentially expressed upon over-expression or deletion of the transcription factor (Fig 1A). This regulatory program is best represented as the transcriptional regulatory network (TRN) 15-17, where nodes represent TFs or target genes, and edges represent inferred regulation of a target gene by a TF. As a result the first assemblage of the transcription regulatory network (TRN) for both eukaryotic (Saccharomyces cerevisiae) and prokaryotic (Escherichia coli) model organisms have become available 1, 3, 4, 6, 7, 18. For instance, the high-throughput Chromatin immunoprecipitation-chip (ChIP-chip) has helped in genome-scale reconstruction of the yeast TRN by identifying direct binding events for several transcription factors (TRNCC)3, 4. Similarly, large-scale gene expression analyses involving yeast strains with either deletions or over-expression of individual transcription factors (TFs) have generated independent reconstructions of the yeast TRN 6, 7 TRNGRD (for genetic reconstruction via deletion) and TRNGROE (for genetic reconstruction through over-expression).
The three methods represent major technological landmarks; nevertheless, they have unique pros and cons in terms of experimental design. For example, it is not possible to directly establish the functional relevance of particular DNA-binding events detected in ChIP-chip experiments. The discrimination of direct regulatory interactions from indirect interactions or feed-back mechanisms in genetic methods is also non-trivial (see Fig S1). Some of the technical issues concerning the design of these different experimental approaches have been given in the Fig S1, but here we describe only the comparison of the reconstructed TRNs from these experiments. As a cautionary note, we would like to state that it is not possible to completely discriminate, with the available information, noise (interactions with no biological relevance) from true regulatory interactions in the TRN reconstructions. Hence, there could be still some “noise” in the TRN reconstructions.
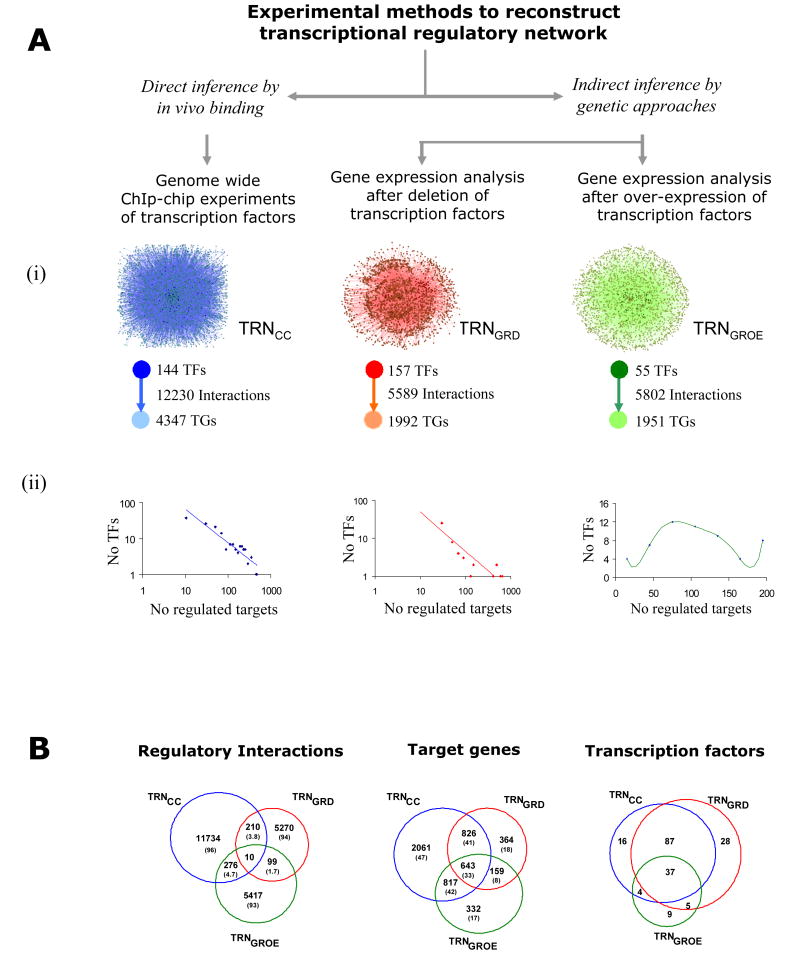
Figure 1.
Comparison of TRNs reconstructed based on data from in vivo binding and genetic studies: (A) Experimental methods and a description of the corresponding high-throughput datasets used in this study. (i) In TRNGRD and TRGGROE, nodes represent TFs or target genes and edges represent differential expression of a target gene upon deletion or over-expression of a particular TF. (ii) The graphs below represent the distribution of the number of TFs and the number of target genes (out-degree distribution) in the TRNCC, TRNGRD and TRNGROE. The out-degree distributions in the TRNCC and TRNGRD are approximated by a power-law equation, suggesting the existence of scale-free structure in these networks. However, TRNGROE shows a centralized out-degree distribution. (B) Venn diagrams showing the extent of overlaps in the regulatory interactions, target genes and transcription factors between the TRNCC, TRNGRD and TRNGROE. Respective percentage values are shown in parentheses. Although the number of shared transcription factors and target genes are high, the number of common regulatory interactions between the three TRNs is strikingly low. There are 200 predicted specific TFs and >6000 TGs in yeast. As the number of TFs and TGs in TRNCC and TRNGRD are comparable to the total number of the predicted TFs, the results are likely to be representative on the whole genome-scale.
Comparison of the local and global structure of the inferred networks
The three distinct TRNs have several interesting similarities and differences in terms of their global and local structure. At the global level, TFs in the TRNCC and TRNGRD have similar distributions in terms of the number of target genes regulated by a given TF (i.e. out-degree distribution), a trend best approximated by a power-law decay2 (Fig 1A). This implies the presence of global regulators or hubs (traditionally defined as the top 20% of TFs with the greatest number of target genes) in the two TRNs. Interestingly, the out-degree distribution in the TRNGROE has a more centralized distribution, rather than a power-law decay, with a peak of 60–120 TGs per TF (Fig 1A). This is suggestive of a downstream homeostatic process, such as increased RNA or protein decay, which channels the effects of over-expression of several functionally distinct TFs via a relatively constant number of responding TGs. Furthermore, the larger average number of inferred target genes per TF in the TRNGROE compared with those in the TRNGRD indicates the propagation of indirect downstream effects due to TF over-expression.
We next identified TFs that are hubs in the TRNCC and TRNGRD, and analyzed the extent of overlap in their inferred regulatory interactions. We found that only seven hubs (Abf1p, Ume6p, Aft1p, Swi4p, Cin5p, Cbf1p and Hsf1p), constituting less than one-quarter of the total number of hubs, are shared between the networks (Fig S3a). Repeating this procedure using different thresholds to define hubs consistently revealed only a few shared hubs between TRNCC and TRNGRD (see Table S3). TRNCC and TRNGRD overlap to a larger extent in terms of number of regulatory interactions (normalized by respective network size), when the yeast TRNs were reconstructed from data accumulated from case-by-case biochemical studies1 or from another comprehensive genetic study8 (Fig S3b and Fig S4). Hence, TRNGRD might be under-representing condition-specific transcriptional responses, as all assays were conducted under standard conditions. Comparing the sub-network for the ubiquitin conjugation system of TRNCC with that of TRNGRD showed that the differences mentioned above were also present at the level of this specific functional sub-system (see SI).
We also discovered differences in the distribution of network motifs between the TRNs via a comprehensive search for different motif types (see SI and Table S4 and Table S5). Multiple-input motifs (MIMs) are most prominent in the TRNCC, suggesting that these in part represent independent back-ups for regulatory interactions, which possibly contribute to the combinatorial robustness of the network9. Furthermore, the relative abundance of MIMs and FFMs (Feed-forward motifs) in TRNGRD and TRNGROE, respectively (Table S1 and SI) implies that: (i) Expression changes in response to TF deletions are less likely than TF over-expression to alter the expression levels of other TFs. (ii) Over-expression of TFs probably tends to affect expression levels of other TFs both directly and indirectly, inducing further gene expression changes. The existence of transcription regulatory events that manifest only under certain conditions, like stress response and cell cycle, could also account for some of the major qualitative differences in the motifs found in the three reconstructed TRNs (See Table S6).
Interference with translation, the telomere effect and a response to protein aggregation confound the different TRNs
We examined the TF hubs in TRNGRD and found that ∼40% of the regulatory interactions in this network were due to the top four of the five major hubs (i.e. Gcr1p, Cst6p, Sfp1p and Mcm1p). None of these four was identified as a hub in TRNCC. These hubs, with the exception of Mcm1p, have regulatory interactions with numerous target genes (∼100) encoding ribosomal components (Fig S5a and Table S7). Furthermore, Sfp1p is a well-characterized major regulator of genes involved in ribosomal biogenesis10, 11. Most ribosomal target genes (∼86%; p<0.01) are inferred to be up-regulated upon deletion of these TFs, implying that the TFs function as direct or indirect transcriptional repressors of ribosomal TGs. Consequently, these TF deletions might alter the stoichiometry of ribosomal components and thereby affect translation. Thus, the major hubs in the TRNGRD seem to have acquired this status predominantly as a result of indirect translational defects. Genetic manipulation of translation has previously been shown to interfere with a large number of unconnected processes, including subsequent transcription12.
We previously noted that a telomere-related effect acts as a confounding factor in TRNCC. TGs in the sub-telomeric regions were inferred to have an unusually large number of binding events (i.e., incoming connections > 13) with functionally diverse TFs. We proposed that this might result from TF-telomere interactions being captured in the ChIP-chip experiments owing to either the telomeres looping back and interacting with chromatin complexes on internal chromosomal sites or due to the interaction of chromosome ends with diverse TFs assembled at the inner nuclear envelope13. We tested this interpretation by comparing the number of incoming regulatory interactions of target genes associated with the subtelomeric regions in the three TRNs. In most cases, the TGs in the sub-telomeric regions show a much greater normalized in-degree (number of distinct TFs regulating a target gene) in the TRNCC than in TRNGRD or TRNGROE (Fig S5b). This further supports the proposal that the ChIP-chip studies captured genuine, potentially long-range, interactions between telomeres and TFs.
A systematic search for high in-degree TGs in TRNGRD or TRNGROE (see SI: Materials and Methods and Fig S6a) identified 42 and 56 such genes, respectively. We classified them into functional categories based on sequence analysis and evidence from the literature (Fig S6b and SI). We found that 16 out of 56 TGs with high in-degree in the TRNGROE were related to stress response pertaining to protein unfolding and oxidative damage (p<0.02; Fig S7 and SI). In particular, we found that three paralogous genes of the DJ-1/ThiJ/PfpI superfamily, [Hsp31, Hsp33 and Hsp34 (Sno4)] have high in-degrees, suggesting that their expression is affected by over-expression of several unrelated TFs. Disruption of DJ1, the human ortholog of these proteins, was implicated with a protein aggregation defect in Parkinson's disease14. Hence, we suggest that over-expression of several TFs and subsequent overproduction of certain proteins causes an increase of aggregated mis-folded polypeptides, in turn triggering a specific stress-response pathway. We hence conjecture that many of the other high in-degree TGs in the TRNGROE are likely to be functionally associated with such a stress response. We also predict that products of these TGs, which include other chaperones, such as Hsp26, Hsp42 and Hsp12, along with the nitrosative stress response protein Yhb1p (all of which have statistically significant high in-degree in the TRNGROE) are likely to cooperate with the DJ-1/ThiJ/PfpI superfamily proteins in a protein-aggregation stress response system. Hence, the analysis of over-expression of TFs could be used as a model to uncover the program that underlies protein misfolding and/or aggregation responses in different cellular systems (see SI for further details).
Concluding remarks
We identified additional effects captured by the high-throughput methods, thereby arguing for the need of post-facto analysis to discriminate functionally relevant regulatory interactions from such effects. The major secondary effects in the three networks, namely the telomere effect (in TRNCC), the ribosomal gene effect (in TRNGRD) and the role of the protein misfolding and/or aggregation response (in TRNGROE), provide leads, some of which were previously unsuspected, to understand disparate biological processes. These observations along with low number of shared regulatory interactions between the networks suggest that the three networks reflect distinct facets such as combinatorial regulation and regulatory backups of the actual transcription regulatory program in yeast. Hence, we envisage that a careful combination of the TRNs reconstructed from more-complete versions of such datasets might enable us to decouple genuine combinatorial regulation from regulatory back-up, and provide an estimate of the resulting robustness. It is therefore important that the results of future high-throughput experiments which aim to reconstruct regulatory programs through biological networks are analyzed with awareness of these secondary effects. Our identification of these effects facilitates two distinct directions of study: (i) a deeper understanding of specific biological phenomena, like protein aggregation response or the telomere effect. (ii) Improved experimental designs to subtract the additional effects and obtain more accurate network reconstructions.
Supplementary Material
Acknowledgments
SB, LMI and LA are funded by the Intramural research program of National Institutes of Health, USA. MMB is funded by the Medical Research Council UK, Darwin College and Schlumberger Ltd. We thank Arthur Wuster and colleagues at the LMB, the editor and the anonymous referees for helpful feedback on previous versions of this manuscript.
Glossary
- TRNCC
The transcriptional network reconstructed from large-scale ChIP-chip experiments. Nodes represent TFs or TGs and edges represent direct binding of the TF in the promoter region of the TG.
- TRNGRD
The transcriptional network reconstructed from analysis of gene expression upon deletion of the relevant TFs. Nodes represent TFs or target genes. A TF is linked to a target gene if it is differentially expressed upon deletion of the TF.
- TRGGROE
The transcriptional network reconstructed from analysis of gene expression upon over-expression of the relevant TFs. Nodes represent TFs or TGs. A TF is linked to a target gene if it is differentially expressed upon over-expression of the TF.
References
- 1.Svetlov VV, Cooper TG. Review: compilation and characteristics of dedicated transcription factors in Saccharomyces cerevisiae. Yeast. 1995;11:1439–1484. doi: 10.1002/yea.320111502. [DOI] [PubMed] [Google Scholar]
- 2.Guelzim N, et al. Topological and causal structure of the yeast transcriptional regulatory network. Nat Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- 3.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 5.Horak CE, et al. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Z, et al. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 7.Chua G, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 9.Balaji S, et al. Uncovering a hidden distributed architecture behind scale-free transcriptional regulatory networks. J Mol Biol. 2006;360:204–212. doi: 10.1016/j.jmb.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Marion RM, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorgensen P, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komili S, et al. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu MM, et al. Estimating the prevalence and regulatory potential of the telomere looping effect in yeast transcription regulation. Cell Cycle. 2006;5:2354–2363. doi: 10.4161/cc.5.20.3386. [DOI] [PubMed] [Google Scholar]
- 14.Wilson MA, et al. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc Natl Acad Sci U S A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babu MM, et al. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 17.Schlitt T, Brazma A. Modelling in molecular biology: describing transcription regulatory networks at different scales. Philos Trans R Soc Lond B Biol Sci. 2006;361:483–494. doi: 10.1098/rstb.2005.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huerta AM, et al. RegulonDB: a database on transcriptional regulation in Escherichia coli. Nucleic Acids Res. 1998;26:55–59. doi: 10.1093/nar/26.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.