Abstract
Quinolone resistance is rapidly increasing in Neisseria gonorrhoeae and is posing a significant public health threat that requires constant surveillance. A rapid and reliable mutation detection assay has been developed. The assay is based on pre-programmed short DNA sequencing and is designed to detect point mutations in the gyrA gene that are highly related to ciprofloxacin resistance, i.e. in codons 91 and 95. By developing an assay based on pyrosequencing and exploiting the pre-programmed nucleotide dispensation capability of this technology, the sequence comprising the mutations will be analysed and promptly reveal whether the N. gonorrhoeae pathogen carries resistance to ciprofloxacin. A panel of 40 N. gonorrhoeae clinical isolates, of which 27 phenotypically displayed decreased susceptibility or resistance to ciprofloxacin, was used in the present study. All point mutations in the short stretch of the N. gonorrhoeae gyrA gene were easily discriminated, and the genotypic results obtained by pre-programmed sequencing were mainly in agreement with the phenotypically identified decreased susceptibility or resistance to ciprofloxacin. The new method used in the present study has the potential for rapid and reliable identification of known as well as previously unknown drug resistance mutations.
Keywords: DNA sequencing, Ciprofloxacin resistance, Neisseria gonorrhoeae, Pre-programmed DNA sequencing, Pyrosequencing technology
1. Introduction
The ability of pathogenic microorganisms to develop resistance to antimicrobial drug treatments has been recognised for decades [1] and continues to increase. This increasing antimicrobial resistance poses a serious threat to public health; thus, as the effects of antimicrobial therapeutic agents decrease, morbidity, mortality and healthcare expenditure increase [2,3].
Neisseria gonorrhoeae is the aetiological agent of gonorrhoea, one of the most common sexually transmitted diseases globally. Quinolone resistance has increased rapidly in N. gonorrhoeae, mainly as a result of point mutations in the bacterial genes gyrA and parC that code for the target enzymes DNA gyrase and topoisomerase IV, respectively [4]. The level of resistance appears to be correlated with the location and number of mutations in these genes. Treatment failures have been shown to be associated with quinolone resistance due to mutations primarily in the gyrA gene [5–7]. Moreover, resistance to ciprofloxacin in many isolates of N. gonorrhoeae is emerging, and genotyping these mutations is of great importance for resistance surveillance.
DNA sequencing is the gold standard method for genotyping and mutation detection. It produces the highest resolution for nucleic acid-based diagnosis of mutant microorganisms and, moreover, provides de novo mutation information of the region being sequenced. In the present study, we describe a rapid, reliable and cost-effective pre-programmed DNA sequencing approach that uses the pyrosequencing technique [8,9] for point mutation sequencing and analysis of the gyrA gene in a single sequencing reaction. The new approach is time efficient (two times faster than conventional pyrosequencing) and the results and sequence read-outs are significantly improved. Furthermore, we have reduced the cost and labour of amplification by using a new pyrosequencing system (PSQ HS 96A), originally intended for single nucleotide polymorphism (SNP) analysis, that requires significantly lower (10-fold) polymerase chain reaction (PCR) volumes. By modifying the SNP software of the PSQ HS 96A system, we have included a pre-programmed sequencing protocol. We have also utilised a rapid and time-efficient Sepharose bead sample preparation system. Moreover, interpretation of the sequence data of point mutations is simple and user friendly, and de novo mutations are easily detected.
2. Materials and methods
2.1. Bacterial isolates and phenotypic antibiotic susceptibility testing
A total of 40 N. gonorrhoeae clinical isolates cultured from different patients in Sweden between 2001 and 2005 were included to evaluate the pre-programmed gyrA gene pyrosequencing technique (Table 1). Selection of the isolates was based on their minimum inhibitory concentration (MIC) to ciprofloxacin, which was phenotypically determined by the Etest method (see below). Consequently, 12 ciprofloxacin-susceptible (MIC ≤ 0.064 mg/L) N. gonorrhoeae isolates, 9 isolates showing decreased susceptibility to ciprofloxacin and 19 ciprofloxacin-resistant (MIC > 0.25 mg/L) isolates were included (Table 1). Nearly all MIC values within the range detectable by the Etest strip, i.e. from <0.002 to >32 mg/L, were represented by at least one isolate. All isolates were subsequently analysed in a blinded fashion to establish the existence of polymorphisms in the short stretch of the gyrA gene.
Table 1.
Mutations in the sequence encoding amino acid positions 90–96 of the gyrA gene, and the minimum inhibitory concentration (MIC) and phenotypic susceptibility to ciprofloxacin of Neisseria gonorrhoeae isolates (N = 40)
| gyrA gene sequence | No. of isolates | Ciprofloxacin MIC (mg/L) | Phenotypic ciprofloxacin susceptibility |
|---|---|---|---|
| Wild-type | 9 | 0.002–0.047 | Susceptible |
| S91P and D95A alterations | 11 | 0.125–32 | Decreased susceptibility to high resistance |
| S91P and D95G alterations | 10 | 0.125–32 | Decreased susceptibility to high resistance |
| S91P alteration | 8 | 0.047–0.38 | Susceptible |
| D95N alteration | 2 | 0.032–0.064 | Susceptible |
Phenotypic susceptibility to ciprofloxacin was analysed using the Etest method (AB Biodisk, Solna, Sweden) on GC II agar (BBL, Becton Dickinson and Company, Cockeysville, MD) supplemented with 1% haemoglobin and 1% IsoVitaleX enrichment (BBL, Becton Dickinson and Company) as previously described [10].
2.2. DNA extraction and PCR
Isolation of bacterial DNA was performed using magnetic silica particles in a robotized system (MagNA Pure LC; Roche Molecular Biochemicals, Mannheim, Germany) according to the instructions of the manufacturer. In brief, bacterial suspensions (ca. 3 × 108 cells/mL) were prepared in sterile 0.15 M NaCl. A total of 1 mL from each suspension was pelleted and re-suspended in 20 μL of sterile distilled water. Then, 130 μL of Bacteria Lysis Buffer (Roche Diagnostics GmbH, Mannheim, Germany) and 20 μL of Proteinase K solution (Roche Diagnostics GmbH) were added to each sample and the suspensions were incubated at 65 °C for 10 min followed by 95 °C for 10 min. DNA was isolated from 100 μL of these final suspensions with the MagNA Pure LC DNA Isolation Kit III (Roche Diagnostics GmbH) and eluted in 100 μL of elution buffer (Roche Diagnostics GmbH) according to the manufacturer's instructions.
The PCR primers GYRA2-1 and GYRA2-2, used to amplify a stretch of the gyrA gene [11] and optimised for use with the pyrosequencing method, were originally designed using the OLIGO 4.0 software (National Biosciences, Inc., Plymouth, MN) from a known sequence available in Gen-Bank for the gyrA gene (strain MUG116; accession number U08817). The sequencing primer GYRA2-3 [11], used to sequence the amplified gyrA region, was designed in the same way. One of the amplification primers in the primer pair was biotin-labelled for single-strand separation. The primers were synthesised by ThermoHybaid (Ulm, Germany). PCR amplification of the segment of the gyrA gene was performed in a 50 μL mixture containing 1 × PCR gold buffer (Applied Biosystems, Foster City, CA), 2.5 mM MgCl2, 10 pmol of each primer, 0.2 mM mix of deoxynucleoside triphosphates (Sigma Aldrich, St Louis, MO), 1.5 U AmpliTaq Gold™ (Applied Biosystems, Stockholm, Sweden) and 5 μL of the DNA template. The target region was amplified by a PCR protocol using the following temperature profile: initial heating at 95 °C for 10 min; a cycle of denaturation at 94 °C for 1 min, annealing at 57 °C for 30 s and elongation at 72 °C for 2 min, repeated in 35 cycles; and a final elongation at 72 °C for 10 min.
2.3. Sample preparation
The double-stranded DNA amplicons were prepared semi-automatically using a Vacuum Prep Tool and Vacuum Prep Worktable (Biotage, Uppsala, Sweden). In brief, 5 μL of biotinylated PCR product was immobilised onto 2.5 μL streptavidin-coated Sepharose High Performance beads (Amersham Biosciences, Piscataway, NJ) by incubation at room temperature for at least 5 min with agitation at 1400 rpm using an Eppendorf Thermomixer R (Eppendorf AG, Hamburg, Germany). The double-stranded DNA immobilised on Sepharose beads was treated with 70% EtOH for cleaning purposes, 0.2 M NaOH for denaturing of the double-stranded DNA, and TE-Buffer (0.1 M Tris-acetate, pH 7.6), all according to the company's (Biotage) instructions for use of the Vacuum Prep Station. The beads were then suspended in 12 μL annealing buffer (10 mM Tris-acetate pH 7.75, 5 mM Mg-acetate) containing 4 pmol sequencing primer. The single-stranded DNA was hybridised to the sequencing primer at 90 °C for 2 min followed by incubation for 10 min at 60 °C and 5 min at room temperature.
2.4. Pyrosequencing technology
The single-stranded prepared PCR products from the isolates were sequenced using an automated plate-based bench-top PSQ™ HS 96A System (Biotage), at a dispensing pressure of 625 mbar with 4 ms open time and 65 s cycle time. Sequencing was performed in a total volume of 12 μL using the PSQ™ 96 SNP kit (Biotage).
3. Results
The mutation sites in the gyrA gene selected for detection in the present study have been reported previously [12–15] (Table 1). For DNA sequencing, we used only 5 μL of the PCR products (compared with 40–50 μL with conventional pyrosequencing) for single-strand DNA preparation and priming of sequencing primer, which took only 15 min to perform.
To detect ciprofloxacin resistance, the PCR fragments were sequenced using a pre-programmed nucleotide dispensation strategy. The pre-programmed sequencing is significantly more rapid, as the nucleotides are dispensed according to the wild-type sequence of the N. gonorrhoeae DNA and alterations in sequence signal peaks will be determined as mutations. Fig. 1 shows a simulation of different alteration patterns of the sequence encoding amino acids 90–96 of the N. gonorrhoeae gyrA gene. Consequently, the mutations are identified as a loss or addition of sequence signal peaks (single-base mutations occurring in the context of a similar base are observed as alterations in signal peak heights within a homopolymeric region). Fig. 1a represents the wild-type used for comparison with the simulated mutations (Fig. 1b–e).
Fig. 1.
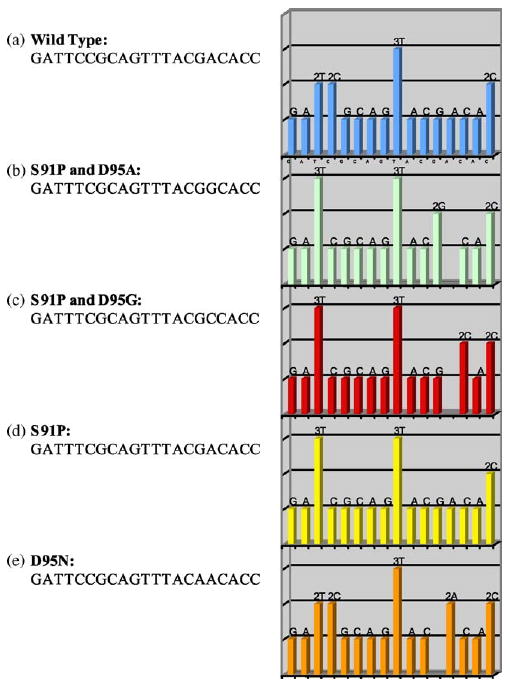
Simulation sequence pattern display of expected mutations related to ciprofloxacin resistance in comparison with the wild-type sequence encoding amino acid positions 90–96 of the Neisseria gonorrhoeae gyrA gene: (a) wild-type sequence; (b–e) expected mutations.
A short stretch of the gyrA gene in 40 N. gonorrhoeae isolates with different MIC values to ciprofloxacin (Table 1) was amplified by PCR. The efficiency of the PCR amplification was evaluated by gel electrophoresis with ethidium bromide staining. All the amplicons were sequenced using the pre-programmed dispensation strategy. The sequence results obtained using the pre-programmed dispensation order generated interpretable sequence information for all isolates. Absent or added signal peaks that differed from the expected sequence pattern, i.e. the pattern of the wild-type gyrA sequence, indicated mutations. The overall time for obtaining the sequence results using the pre-programmed approach was 15 min for a 96-well microplate. Fig. 2 shows representative pyrograms of five amplicons sequenced using the pre-programmed approach. Fig. 2a is the wild-type used for comparison with other samples bearing the mutations that were simulated in Fig. 1 (Fig. 2b–e).
Fig. 2.
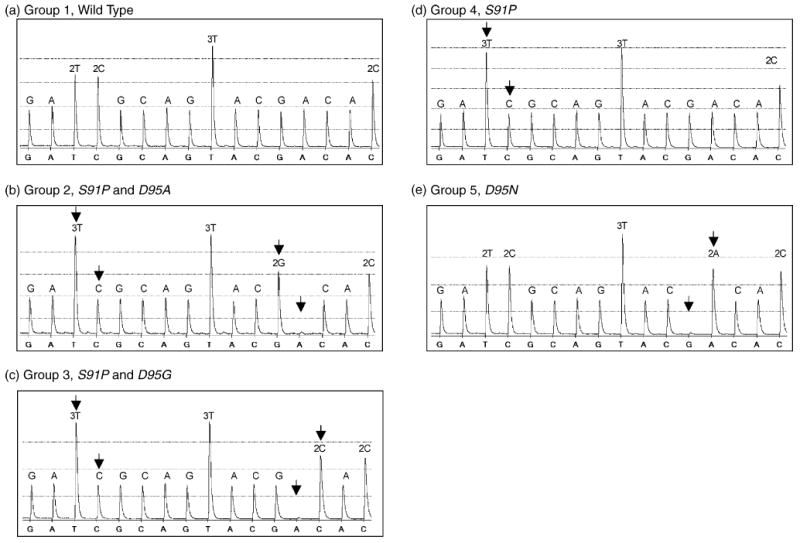
Pyrograms of the sequence encoding amino acid positions 90–96 of the gyrA gene in Neisseria gonorrhoeae isolates obtained by pre-programmed DNA sequencing: (a) wild-type sequence; (b–e) samples harbouring mutations. The arrows show the location of point mutations and alterations in the sequence signal peaks. This figure can be compared with Fig. 1.
Overall, the results of genotypic ciprofloxacin susceptibility testing, i.e. pre-programmed pyrosequencing of the gyrA gene, were highly concordant with the phenotypically identified decreased susceptibility or resistance to ciprofloxacin of the N. gonorrhoeae isolates (Table 1). Notably, all isolates harbouring a gyrA wild-type gene were susceptible (displayed MIC ≤ 0.047 mg/L); the D95N polymorphism alone did not have any significant impact on resistance and was only identified in two susceptible isolates; the S91P alteration alone mainly resulted in a decreased susceptibility or low resistance, but two isolates were also susceptible (MIC = 0.047 mg/L and MIC = 0.064 mg/L, respectively); and the S91P alteration in combination with D95A or D95G alterations mainly resulted in high resistance, but occasional isolates showed only a decreased susceptibility. All isolates displaying a decreased susceptibility or resistance to ciprofloxacin harboured at least one mutation in codons 91 or 95 of the gyrA gene (Table 1).
4. Discussion
Ciprofloxacin resistance caused by point mutations affects the treatment of N. gonorrhoeae infections, mainly owing to reduced binding of this antimicrobial drug to the target sites of DNA gyrase. Therefore, it is important to investigate the molecular mechanisms by which drug resistance is acquired in order to confront drug-resistant gonococcal infections.
Rapid and reliable methods are required for screening and surveillance of these mutations. Pyrosequencing technology has the unique and significant advantage of a pre-programmed nucleotide addition strategy for mutation detection. This is considered an important benefit, since nucleotides are dispensed into the reaction mixture in a programmed order and alterations are discovered easily. In contrast, de novo sequencing uses cyclic nucleotide dispensations, resulting in longer reaction times and, together with product accumulation due to more nucleotide dispensations in the system, causes lower sequence data quality when performing longer reads. The pre-programmed dispensation results in more efficient point mutation detection, as longer and faster reads and more time-effective screening can be achieved. The pre-programmed sequencing is not only suitable for quick screening of known mutations but also for identification of previously unknown mutations.
The sample preparation (single-strand preparation and primer annealing) method used in the present study was relatively quick, allowing for more rapid single-strand separation (ca. 15 min) prior to DNA sequencing. The pyrosequencer (PSQ HS 96A) used for this study has the advantage of using lower volumes of PCR product owing to a more sensitive CCD camera, significantly reducing the cost of performing PCR and sequencing reactions. The PSQ HS 96A system is primarily suitable for SNP analysis. By simple modifications in the SNP software, it is possible to incorporate both the cyclic and pre-programmed approaches in the system.
As the results in Table 1 indicate, all isolates displaying a decreased susceptibility or resistance to ciprofloxacin comprised at least one mutation in codons 91 or 95 of the gyrA gene. The minor discrepancies between the results of genotypic and phenotypic ciprofloxacin susceptibility testing may, in most cases, be due to other additional molecular mechanisms for ciprofloxacin resistance, for instance polymorphisms in the parC gene [6].
We do not suggest that this method can replace cultivation and susceptibility testing for N. gonorrhoeae in patients with suspected gonorrhoea, but it may be useful for laboratories that use only molecular methods for detection of gonococci. Mutations in gyrA were also found in some susceptible strains, but these mutations are probably relevant, since they may represent the first step to complete resistance.
In conclusion, DNA sequencing is the most reliable tool to inspect and analyse the nucleotide sequences of point mutations in clinical samples. With the new approach described in the present study, clinical samples could be screened more efficiently, allowing for attainment of reliable DNA sequencing-based results conveniently and in a shorter period of time. The pre-programmed sequencing is a general approach and can be used for insertion, deletion and mutation detection in other microbial resistance surveillance studies.
Acknowledgments
This study was supported by a grant from the National Institute of Health, 1R21 A1059499-01, PO1-HG000205, and the Scandinavian Society for Antimicrobial Chemotherapy, AFA Health Fund (Sweden) and Tore Nilsons Fond.
References
- 1.Ashley DJ, Brindle MJ. Penicillin resistance in staphylococci isolated in a casualty department. J Clin Pathol. 1960;13:336–8. doi: 10.1136/jcp.13.4.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coast J, Smith RD, Millar MR. Superbugs: should antimicrobial resistance be included as a cost in economic evaluation? Health Econ. 1996;5:217–26. doi: 10.1002/(SICI)1099-1050(199605)5:3<217::AID-HEC200>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Smith RD, Coast J. Antimicrobial resistance: a global response. Bull World Health Organ. 2002;80:126–33. [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka M, Sakuma S, Takahashi K, et al. Analysis of quinolone resistance mechanisms in Neisseria gonorrhoeae isolates in vitro. Sex Transm Infect. 1998;74:59–62. doi: 10.1136/sti.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deguchi T, Saito I, Tanaka M, et al. Fluoroquinolone treatment failure in gonorrhea. Emergence of a Neisseria gonorrhoeae strain with enhanced resistance to fluoroquinolones. Sex Transm Dis. 1997;24:247–50. doi: 10.1097/00007435-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M, Nakayama H, Haraoka M, Nagafuji T, Saika T, Kobayashi I. Analysis of quinolone resistance mechanisms in a sparfloxacin-resistant clinical isolate of Neisseria gonorrhoeae. Sex Transm Dis. 1998;25:489–93. doi: 10.1097/00007435-199810000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Sagiyama K, Haraoka M, Saika T, Kobayashi I, Naito S. Genotypic evolution in a quinolone-resistant Neisseria gonorrhoeae isolate from a patient with clinical failure of levofloxacin treatment. Urol Int. 1999;62:64–8. doi: 10.1159/000030344. [DOI] [PubMed] [Google Scholar]
- 8.Gharizadeh B, Kalantari M, Garcia CA, Johansson B, Nyren P. Typing of human papillomavirus by pyrosequencing. Lab Invest. 2001;81:673–9. doi: 10.1038/labinvest.3780276. [DOI] [PubMed] [Google Scholar]
- 9.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 10.Berglund T, Unemo M, Olcen P, Giesecke J, Fredlund H. One year of Neisseria gonorrhoeae isolates in Sweden: the prevalence study of antibiotic susceptibility shows relation to the geographic area of exposure. Int J STD AIDS. 2002;13:109–14. doi: 10.1258/0956462021924730. [DOI] [PubMed] [Google Scholar]
- 11.Lindback E, Gharizadeh B, Ataker F, et al. DNA gyrase gene in Neisseria gonorrhoeae as indicator for resistance to ciprofloxacin and species verification. Int J STD AIDS. 2005;16:142–7. doi: 10.1258/0956462053057675. [DOI] [PubMed] [Google Scholar]
- 12.Belland RJ, Morrison SG, Ison C, Huang WM. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–80. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 13.Booth SA, Drebot MA, Martin IE, Ng LK. Design of oligonucleotide arrays to detect point mutations: molecular typing of antibiotic resistant strains of Neisseria gonorrhoeae and hantavirus infected deer mice. Mol Cell Probes. 2003;17:77–84. doi: 10.1016/s0890-8508(03)00005-7. [DOI] [PubMed] [Google Scholar]
- 14.Deguchi T, Yasuda M, Asano M, et al. DNA gyrase mutations in quinolone-resistant clinical isolates of Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1995;39:561–3. doi: 10.1128/aac.39.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deguchi T, Yasuda M, Nakano M, et al. Quinolone-resistant Neisseria gonorrhoeae: correlation of alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV with antimicrobial susceptibility profiles. Antimicrob Agents Chemother. 1996;40:1020–3. doi: 10.1128/aac.40.4.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]


