Abstract
Human killer cell immunoglobulin-like receptors (KIRs) are distinguished by expansion of activating KIR2DS, whose ligands and functions remain poorly understood. The oldest, most prevalent KIR2DS is KIR2DS4, which is represented by a variable balance between “full-length” and “deleted” forms. We find that full-length 2DS4 is a human histocompatibility leukocyte antigen (HLA) class I receptor that binds specifically to subsets of C1+ and C2+ HLA-C and to HLA-A*11, whereas deleted 2DS4 is nonfunctional. Activation of 2DS4+ NKL cells was achieved with A*1102 as ligand, which differs from A*1101 by unique substitution of lysine 19 for glutamate, but not with A*1101 or HLA-C. Distinguishing KIR2DS4 from other KIR2DS is the proline–valine motif at positions 71–72, which is shared with KIR3DL2 and was introduced by gene conversion before separation of the human and chimpanzee lineages. Site-directed swap mutagenesis shows that these two residues are largely responsible for the unique HLA class I specificity of KIR2DS4. Determination of the crystallographic structure of KIR2DS4 shows two major differences from KIR2DL: displacement of contact loop L2 and altered bonding potential because of the substitutions at positions 71 and 72. Correlation between the worldwide distributions of functional KIR2DS4 and HLA-A*11 points to the physiological importance of their mutual interaction.
NK cells respond early to infection by killing infected cells and secreting cytokines (Lanier, 1998). Such activation involves integration of signals from a variety of activating and inhibitory receptors, including several that recognize MHC class I molecules (Moretta et al., 1996). Members of the killer cell Ig-like receptor (KIR) family recognize epitopes of HLA-A, -B, and -C. The inhibitory KIRs comprise KIR2DL and KIR3DL, and the activating receptors comprise KIR2DS and KIR3DS.
KIRs with HLA-A, -B, and -C specificity comprise two phylogenetic lineages (Khakoo et al., 2000). In lineage II, KIR3DL1 recognizes the subset of HLA-A and -B allotypes having the Bw4 epitope (Gumperz et al., 1995), and KIR3DL2 recognizes HLA-A3 and -A11 (Döhring et al., 1996; Pende et al., 1996). In lineage III, KIR2DL1 recognizes the subset of HLA-C allotypes having the C2 epitope (HLA-C2) defined by lysine 80, whereas KIR2DL2/3 recognizes the alternative subset having the C1 epitope (HLA-C1) defined by asparagine 80 (HLA-C1; Mandelboim et al., 1996). Unlike the inhibitory KIRs, functions and ligands for the lineage II and III activating KIRs are poorly understood. Few KIR genes are fixed, and activating KIR genes are less common than inhibitory KIR genes (Abi-Rached and Parham, 2005). KIR2DS1 has similar C2 specificity as 2DL1 but much reduced avidity (Biassoni et al., 1997; Stewart et al., 2005; Chewning et al., 2007). Ligands for KIR2DS2, 2DS3, 2DS5, and 3DS1 remain elusive (Kim et al., 1997; Valés-Gómez et al., 1998; Winter et al., 1998; Carr et al., 2007; Della Chiesa et al., 2008; VandenBussche et al., 2009).
KIR2DS4, the most prevalent lineage III–activating KIR, is also the oldest and most divergent, being the only human lineage III KIR with an orthologue in another species: chimpanzee Pt-KIR2DS4 (Khakoo et al., 2000). Before rationalization of the KIR nomenclature (Marsh et al., 2003), KIR2DS4 was alternatively termed p50.3 (Bottino et al., 1996), clone 39 (Wagtmann et al., 1995), NKAT8 (Colonna and Samaridis, 1995; Campbell et al., 1998), and KAR-K1 (Kim et al., 1997). Several early studies failed to detect interactions between 2DS4 and HLA class I (Bottino et al., 1996; Kim et al., 1997; Valés-Gómez et al., 1998; Winter et al., 1998), but two detected weak but potentially significant interactions with HLA-C*03 (Campbell et al., 1998) and HLA-C*04 (Katz et al., 2001). Overall, the weak and ambiguous interactions of activating KIRs with HLA class I led to the physiological relevance of the activating human KIRs being questioned and, in the case of KIR2DS4, to a search for non–MHC class I ligands (Katz et al., 2004).
Epidemiological studies implicate activating KIR genes, often in combination with HLA class I, in a variety of clinical associations (for review see Kulkarni et al., 2008). Although KIR haplotypes differ widely in KIR gene content, they divide into two groups: A, which has mainly inhibitory KIR genes, and B, which has additional activating KIR genes (Uhrberg et al., 1997). All populations examined have both haplotype groups but their relative frequencies vary, and they are likely maintained by balancing selection (Norman et al., 2007). Furthermore, many clinical associations with KIR can be attributed to A and B haplotype differences (Parham, 2005). Overall, the epidemiological studies point to the activating KIRs as having significant influence on the physiology of the human immune response. Of particular importance in this regard is 2DS4, the only activating KIR of A haplotypes. For these compelling reasons, we reinvestigated the HLA class I specificity of KIR2DS4 and its functional implications.
RESULTS
KIR2DS4 recognizes a minority of C1+ and C2+ HLA-C allotypes and HLA-A*11
Previous studies tested the binding of KIR2DS4 to a limited number of HLA class I allotypes (Kim et al., 1997; Valés-Gómez et al., 1998; Winter et al., 1998; Katz et al., 2001). In this study, we examined the binding of soluble 2DS4-Fc fusion protein to 95 HLA class I allotypes (29 HLA-A, 50 HLA-B, and 16 HLA-C). In this analysis, 2DS4-Fc (made from the common 2DS4*001 allotype) was compared with Fc fusion proteins made from the well-characterized inhibitory KIRs that are HLA-C receptors: KIR2DL1, 2, and 3 (Fig. 1 A; Moesta et al., 2008).
Figure 1.
KIR2DS4 binds to HLA-A*11 and subsets of C1+ and C2+ HLA-C allotypes. (A) Titration of KIR2D-Fc fusion proteins made from 2DL1, 2, and 3 and 2DS4 to a panel of beads individually coated with 1 out of 29 HLA-A, 50 HLA-B, 9 C1+ HLA-C, and 7 C2+ HLA-C allotypes. Informative reactions are shown individually; negatively reacting (NR) allotypes are collectively represented by their means. Mean values from three independent experiments are shown. (B) Comparison of the binding of the four KIR2D-Fc fusion proteins at a concentration of 100 µg/ml against the four HLA allotype groups. All binding data for KIR2D-Fc was normalized to the binding obtained with W6/32, an mAb recognizing a conserved epitope of HLA class I. Mean values from three independent experiments are shown.
Significant reactions with 2DS4-Fc were observed for six HLA-C, two HLA-A, and zero HLA-B allotypes (Fig. 1 A). The positively reacting HLA-C allotypes comprised three with the C1 epitope (C*1601, C*0102, and C*1402) and three with the C2 epitope (C*0501, C*0202, and C*0401). Binding of 2DS4-Fc to C*1601 was comparable to that of 2DL3-Fc, but for the other C1+ allotypes (C*0102 and C*1402), 2DS4-Fc bound weakly compared with 2DL3-Fc. The three C2+ HLA-C allotypes to bind 2DS4-Fc are the same ones that react with 2DL2 and 2DL3 (Moesta et al. 2008), and they bind the two KIRs with similar hierarchy: C*0501>*0202>*0401. This analysis is consistent with the previous report of 2DS4 binding to C*0401 (Katz et al., 2001) but not with C*03 recognition (Campbell et al., 1998). The results show that 2DS4 has a specificity for HLA-C that is different from the 2DL1, 2, and 3 specificities, being narrowly focused on a few HLA-C allotypes and having no preference for C1 or C2. Further emphasizing its distinctive specificity, 2DS4-Fc did not bind to any HLA-B allotype, including B*4601 and B*7301, which carry the C1 epitope and are recognized by 2DL2 and 2DL3 (Fig. 1 B; Moesta et al., 2008).
Besides HLA-C reactivity, 2DS4 bound to HLA-A*1101 and A*1102 (Fig. 1). Binding to A*1102 was in the same range as to C*0501 and C*1601, and consistently around fourfold higher than to A*1101, a difference correlating with the single substitution—Glu19 in A*1101 and Lys19 in A*1102—that distinguishes them (Morrell et al., 1999). No binding was detected to any of the other 27 HLA-A allotypes tested, which cover the full range of common HLA-A allotypes. The observed recognition of HLA-A*11 by 2DS4, a lineage III KIR, was unexpected, because previously, KIR recognition of HLA-A*11 had only been attributed to 3DL2, a lineage II KIR (Pende et al., 1996; Hansasuta et al., 2004).
In contrast to 2DS4*001, the other seven 2DS4 alleles (2DS4*003–9) share a deletion of 22 bp in exon 5 that changes the reading frame and causes premature termination. The encoded “KIR1D” protein is soluble and has a single intact Ig-like domain (D1; Hsu et al., 2002). Alleles containing this deletion account for a substantial proportion (up to 89% in Caucasians and ∼30% in Asians) of the total 2DS4 alleles in human populations (Middleton et al., 2007). As the functionality of these truncated forms of 2DS4 is uncertain, we made a fusion protein from 2DS4*003, the common form of KIR1D. The 2DS4*003-Fc construct has the same sequence as 2DS4*001 up to the deletion, at which point the frameshift results in 65 residues of divergent sequence before the predicted stop codon. Fusion to the Fc was performed so that the last residue of 2DS4*003 before the stop codon is immediately followed by the Fc sequence. Although 2DS4*003-Fc reacted well with polyclonal antibodies specific for deleted 2DS4 allotypes, we detected no binding to beads coated with HLA class I or to anti-KIR mAbs (Fig. S1). These results demonstrate that the products of 2DS4 deletion alleles are not HLA class I receptors.
Ligation of KIR2DS4 by antibody or HLA-A*1102 induces NKL cell activation
For functional analysis, KIR2DS4*001 and an inhibitory 2DS4-LT derivative, made by replacing the 2DS4 signaling domain with that of 2DL2, were transduced into G4-NKL cells. G4-NKL is an NKL derivative in which siRNA suppresses LILRB1 expression by >90% (Fig. S2). The use of G4-NKL in functional assays of inhibitory KIR function eliminated the confounding effect of MHC class I–mediated inhibition through LILRB1 observed for NKL (Carr et al., 2005).
To determine the signaling potential of the expressed 2DS4*001 and 2DS4-LT receptors, the FcR-expressing mouse mastocytoma cell line P815 was used as the target cell in redirected cytotoxicity assays (Fig. 2 A, top). The presence of a 2DS4-specific mAb (clone 179315) induced killing of P815 targets by NKL-2DS4 to a greater extent than obtained with the combination of mAbs specific for 2B4 (CD244) and NKG2D, activating receptors that are endogenously expressed by NKL. Lysis further increased when all three antibodies were combined in the assay. In parallel assays, the addition of anti-2DS4 antibody induced inhibitory signals in NKL-2DS4-LT, which countered the activating signals transduced by 2B4 and NKG2D to prevent target-cell lysis. The addition of anti-2DS4 antibody had no effect on either G4-NKL cells or G4-NKL cells expressing 2DL3 (Fig. 2 A, bottom).
Figure 2.
Ligation of KIR2DS4 by HLA-A*1102 or a specific antibody induces NKL cell activation. (A) Redirected cytotoxicity of P815 target cells by G4-NKL and G4-NKL transfectants expressing KIR2DS4, KIR2DL3, and KIR2DS4-LT, a mutant of KIR2DS4 that has a long cytoplasmic tail with inhibitory function. Effector cells were preincubated with combinations of mAbs as indicated (1mg/ml each). Mean values from three independent experiments are shown for E/T ratios from 80:1 to 20:1. (B) Results of cytotoxicity with G4-NKL expressing KIR2DL3, KIR2DS4, KIR3DL2, or no KIR as effector cells and untransfected 221 cells (top) or 221 cells expressing HLA-A*1102 (bottom) as target cells. To reduce the basal level of 221 cell lysis by G4-NKL, mAbs against the activating receptors 2B4 and NKG2D were added to effectors (5 mg/ml; right) for blocking. E/T ratios from 50:1 to 12.5:1 are shown. (C) Killing relative to untransfected G4-NKL cells at a 25:1 E/T ratio against 221 cells or 221 cells expressing HLA-A*1102. Blocking antibodies added to effectors are indicated. Data points shown in B and C are mean values from four independent experiments.
To assess the capacity of 2DS4 to bind HLA class I and activate NK cells, we used a standard cytotoxicity assay and targets made from the class I–deficient B cell line 721.221 (221) expressing A*1102, C*0202, C*0304, C*0401, and C*0501. In these experiments, we detected no consistent functional interactions between 2DS4 and any of the HLA-C allotypes tested (unpublished data). In contrast, lysis of 221 cells by G4-NKL was consistently increased, although to a modest extent, when 221 cells expressed A*1102 and G4-NKL expressed 2DS4. In a control experiment, the lysis of 221 cells expressing A*1102 was decreased when the G4-NKL effectors expressed 3DL2, the inhibitory receptor that recognizes A*1102 (Fig. 2 B, left). These effects were amplified in the presence of antibodies against 2B4 and NKG2D, which served to reduce the basal level of 221 cell lysis by G4-NKL (Fig. 2 B, right). Similar analysis, using 221 cells transfected with A*1101, gave no evidence for a functional interaction with either 3DL2 or 2DS4, consistent with the weaker binding of 2DS4-Fc to this allotype (Fig. 1 B).
In this and a previous work (Moesta et al., 2008), we consistently found that NKL kills 221 cells more effectively than HLA class I–transfected 221 cells (Fig. 2 B, top). A possible explanation for this effect is that leader peptides derived from transfected HLA class I increase the abundance of cell-surface HLA-E, which then engages the inhibitory CD94:NKG2A expressed by NKL (Llano et al., 1998). However, to avoid this mechanism, we had mutated arginine -18 to valine in the leader peptides of the transfected HLA class I. The mutation retains binding to HLA-E but prevents engagement of CD94:NKG2A (Michaëlsson et al., 2002). Furthermore, 221 cell expression of A*0301, which has the same leader peptide as A*1102, did not inhibit NKL killing of 221 cells (Carr et al., 2005). Thus, the effect is unlikely to result from inhibition mediated by CD94:NKG2A or LILRB1 but from another uncharacterized inhibitory HLA class I receptor, which could also explain why antibodies against 2B4 and NKG2D inhibited NKL lysis of A*1102-expressing but not untransfected 221 cells (Fig. 2 B, bottom).
Comparing the effect of KIR-transduced G4-NKL relative to lysis by untransduced G4-NKL showed that the activating interaction between 2DS4 and A*1102 increased lysis by ∼58% in the presence of antibodies against 2B4 and NKG2D (Fig. 2 C). Conversely, the inhibitory interaction of A*1102 with 3DL2 was increased from 28 to 48% inhibition in the presence of mAb. As a control, the lytic capacity of 2DL3-expressing G4-NKL was unaffected by the presence of A*1102 (Fig. 2 C).
KIR2DS4 recognition of HLA-A*11 was conserved while HLA-C recognition diverged
KIR2DS4 is the only human lineage III KIR with an identified orthologue in another species: chimpanzee Pt-KIR2DS4 (Khakoo et al., 2000). The Ig domains of Pt-2DS4 and 2DS4 differ by 10 amino acid substitutions, with none at positions predicted to contact bound HLA class I (Boyington and Sun, 2002). A Pt-2DS4-Fc fusion protein was made and compared with 2DS4-Fc for binding to HLA class I–coated beads (Fig. 3). Both similarities and differences were observed. Pt-2DS4-Fc and 2DS4-Fc shared specificity for HLA-A*11 and the stronger reaction with A*1102 than A*1101. Also in common was a lack of reactivity with HLA-B and reactivity with C*1601. In other respects, Pt-2DS4-Fc and 2DS4-Fc differed in their specificity for HLA-C. Pt-2DS4 did not bind the three C2+ allotypes recognized by 2DS4, and gave only very weak binding to other C2+ allotypes. Of the C1+ allotypes, Pt-2DS4-Fc and 2DS4-Fc both bound strongly to C*1601 and weakly to C*0102, but only Pt-2DS4-Fc bound to C*0801.
Figure 3.
Human and chimpanzee KIR2DS4 bind similarly to HLA-A*11 but have distinctive specificities for HLA-C. Shown is the binding of KIR-Fc fusion proteins (100 µg/ml) made from human 2DS4 and its chimpanzee orthologue, Pt-2DS4, to beads individually coated with 1 out of 95 HLA class I allotypes. Only data for positively reacting allotypes are shown individually. The mean for 27 negatively reacting (NR) HLA-A allotypes is shown. Mean fluorescence values from three independent experiments are shown.
That human 2DS4 and chimpanzee Pt-2DS4 similarly recognize HLA-A*11 and C*1601 indicates that these specificities evolved before the separation of human and chimpanzee ancestors, and were subsequently conserved in both lineages. In other aspects, the HLA-C specificities of human and chimpanzee 2DS4 have diverged.
KIR2DS4 uniquely shares the proline 71–valine 72 (Pro71-Val72) motif with KIR3DL2
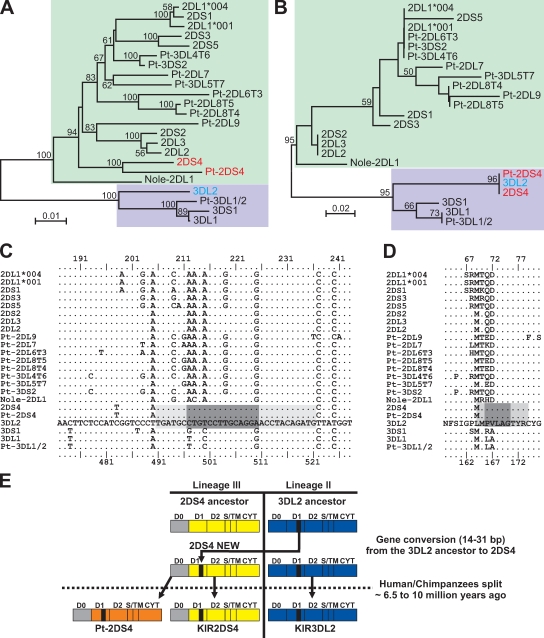
KIR2DS4 and 3DL2 are divergent KIRs from different lineages that both recognize HLA-A*11. Recombination analysis identified a shared 31-bp sequence in exon 4. Although phylogenetic analysis of exon 4 and ∼300 bp of flanking introns placed 2DS4 and 3DL2 on different branches of the tree (Fig. 4 A), analysis of the 31-bp segment grouped 2DS4 and 3DL2 together (Fig. 4 B). Our analysis also showed that the common segment originated with lineage II KIRs, suggesting that 2DS4 acquired this sequence by gene conversion between ancestral 3DL2 and 2DS4 genes. This conversion, involving a segment 14–31 bp in length (Fig. 4 C), replaced the Gln71-Asp72 motif conserved in the D1 domain of other lineage III KIRs with the Pro71-Val72 motif of KIR3DL2 (Fig. 4 D). That both human and chimpanzee 2DS4 have the Pro71-Val72 motif argues for the conversion having occurred before the separation of human and chimpanzee ancestors 6.5–10 million years ago (Benton and Donoghue, 2007).
Figure 4.
Gene conversion with KIR3DL2 was a seminal event in the evolution of KIR2DS4. (A and B) Neighbor-joining tree topology for the genomic segment carrying the exon encoding the D1 domain (A) and for the segment of D1 targeted by the gene conversion between 3DL2 and 2DS4 (B); bootstrap support is indicated where ≥50%. Colored boxes denote KIR genes of lineage II (purple) or III (green). Nole, Nomascus leucogenys (white-cheeked gibbon); Pt, Pan troglodytes (common chimpanzee). (C) Alignment of nucleotide sequences encoding the region of D1 where gene conversion took place. KIR3DL2 is used as the reference sequence, and a dot indicates identity to the reference. Numbering for 2DS4 and other lineage III KIR genes is given at the top, and numbering for 3DL2 and other lineage II KIR genes is at the bottom. The minimal length for the gene conversion is shown by the dark gray shaded box, and the maximal length is shown by the light and dark gray shading. (D) Alignment of amino acid sequences encoded by the nucleotide sequences in C. (E) Model for the gene conversion in which a 3DL2 ancestor was the donor and a 2DS4 ancestor was the recipient. The gene conversion occurred before the speciation event separating humans and chimpanzees 6.5–10 million years ago (Benton and Donoghue, 2007).
Recognition of HLA-A*11 is dependent on the Pro71-Val72 motif in D1
To test the hypothesis that HLA-A*11 recognition by 2DS4 depends on the Pro71-Val72 motif, we made a mutant 2DS4-Fc fusion protein in which the motif was changed to Gln71-Asp72. As predicted, this 2DS4-PV71-72QD-Fc lost A*11 reactivity; in addition, binding to C2+ HLA-C was diminished, and the mutants acquired binding to a range of C1+ HLA-B and -C allotypes not bound by 2DS4-Fc (Fig. 5). These results prove that the motif at residues 71 and 72 determines 2DS4 specificity and that its replacement by gene conversion was responsible for the acquisition of A*11 specificity by 2DS4.
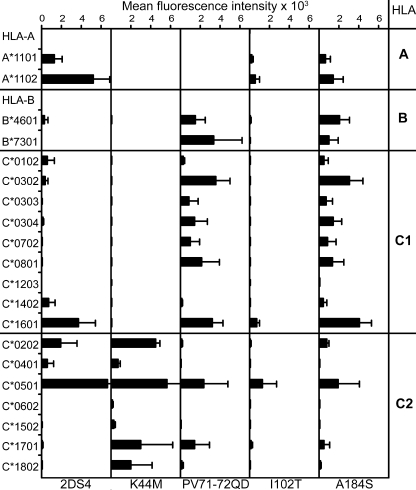
Figure 5.
Mutation at positions 44, 71+72, 102, and 184 alter the specificity and/or avidity of KIR2DS4 for HLA class I. The binding of Fc fusion proteins made from 2DS4 or 2DS4 mutants (K44M, PV71-72QD, I102T, or A184S) to 95 HLA class I allotypes was determined as described in the legend to Fig. 3. Mean values from four independent experiments are shown.
Lysine 44 in D1 was also demonstrated to be essential for 2DS4 recognition of A*11. Mutation to methionine, the residue present in C2-specific 2DL1, abrogated reactivity with A*11, as well as with C1+ HLA-B and -C, while increasing reactivity with C2+ HLA-C. Because Ala184 is unique to 2DS4 among human KIRs and predicted to contact bound HLA class I (Boyington et al., 2000; Fan et al., 2001; Boyington and Sun, 2002), we made a mutant having Ser184, the residue in other KIRs. This mutation acted similarly to replacement of the Pro71-Val72 motif. Increased reactivity with C1 was accompanied by reduced reactivity with C2 and A*11. Of the 13 residues predicted to stabilize the hinge between D1 and D2, only residue 102 is variable: Ile102 in 2DS4 and 2DL1, and Thr102 in Pt-2DS4 and 2DL2/3. Mutating Ile102 to threonine generally reduced binding to C1, C2, and A*11. This was not because of poorly folded fusion protein, because it bound well to conformation-dependent anti-KIR2DS4 and -NKVFS1 antibodies (Fig. S1). The dramatic effect of this mutation points to the functional importance of the polymorphism at position 102. That Pt-2DS4 has Thr102, but binds A*11 and HLA-C more avidly than the 2DS4-Thr102 mutant, shows that additional substitutions between 2DS4 and Pt-2DS4 compensate for the negative effects of Thr102.
The 2DS4 mutants demonstrated that Lys44, Pro71, and Val72 were all necessary for the binding of human 2DS4 to A*11. To complement this analysis of the loss of A*11 reactivity, we aimed to introduce A*11 reactivity into 2DL3, which has Lys44 but lacks the Pro71-Val72 motif. This was accomplished by the single mutation of Glu71 to proline (Fig. S3). The resulting 2DL3-71P-Fc retained the C1 specificity of 2DL3-Fc but bound A*11 more strongly. Acquisition of A*11 specificity was accompanied by some reduction in binding to C1, which varied with allotype. As a control, introducing the same mutation in 2DL1, which has Met44, Gln71, and Asp72, preserved both strength and specificity for C2. These results show that substituting proline for glutamine at position 71 is sufficient for 2DL3 to recognize A*11 (Fig. S3).
The 2DL3 mutant, in which both 71 and 72 were mutated to introduce the Pro71-Val72 motif, lost all interaction with HLA class I despite binding conformation-dependent antibodies (Fig. S3). Thus, the additional substitution at position 72 dramatically perturbed the ligand-binding site of 2DL3. In 2DL1, the effect of the double mutation was less severe, with C2 specificity being retained but with a reduced avidity compared with 2DL1-Fc.
Crystal structure of KIR2DS4
To investigate the impact of the 22–31 substitutions that distinguish the 2DS4 D1 and D2 domains from other lineage III KIRs, we determined the three-dimensional structure of 2DS4 (residues 6–200) by x-ray diffraction to 2.5 Å (Fig. 6 A). The final R factors for the structure are 24.1 and 28.4% for Rcryst and Rfree, respectively (Table I).
Figure 6.
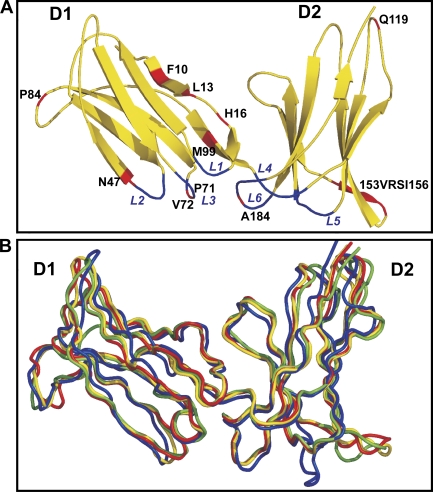
Comparison of the three-dimensional structure of KIR2DS4 to the structures of other lineage III KIRs. (A) An illustration of the structure of the D1 and D2 domains of 2DS4, with the β strands represented by arrows. Binding loops are indicated and shown in blue. 14 out of the 15 unique substitutions in the extracellular domains that distinguish 2DS4 from other human lineage III KIRs are labeled and shown in red; the 15th residue is glutamine at position 1 in D1, which was not included in the construct used to produce the protein for crystallization. (B) Cα trace superposition for 192 equivalent Cα atoms (residues 8–200) of 2DS4 (yellow), 2DL1 (red; 1IM9; Fan et al., 2001), 2DL2 (green; 1EFX; Boyington and Sun, 2002), and 2DS2 (blue; 1M4K; Saulquin et al., 2003).
Table I.
Data collection and refinement statistics for the KIR2DS4 structure
| Parameter | Value | |
| Space group | P212121 | |
| Unit cell dimensions (Å) | 51.232, 62.851, 65.934 | |
| Data collection | Resolution (Å) | 40–2.49 (2.86–2.49) |
| Unique reflections | 7,814 (751) | |
| Completeness (%) | 99.8 (97.9) | |
| Redundancy | 13.6 (10) | |
| Average l/σl | 64 (10) | |
| Rsym (%) | 4.9 (20.7) | |
| Refinement | Resolution (Å) | 40–2.49 |
| Number of reflections | 7,778 (99.69%) | |
| Test reflections | 357 (4.59%) | |
| Rcryst | 24.1% | |
| Rwork | 23.8% | |
| Rfree | 28.4% | |
| Number of atoms | Protein | 1,514 (residues 6–200) |
| Average B factor (Å2) | 45.22 | |
| Water | 53 | |
| Average B factor (Å2) | 42.04 | |
| RMSD from ideal | Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.36 | |
| Ramachandran plot qualitya | Most favored | 85.7% |
| Additionally allowed | 13.7% | |
| Generously allowed | 0.6% | |
| Disallowed | 0% |
SFCheck version 7.2.02, included in the CCP4 package.
Overall structure.
The folding topology of 2DS4 resembles other KIRs (Fan et al., 1997; Maenaka et al., 1999; Snyder et al., 1999; Saulquin et al., 2003). Superimposing 2DS4 onto their structures for 192 equivalent Cα atoms gave a root mean square deviation (RMSD) ranging from 1.04 Å for 2DL1 (in complex with HLA-Cw4) to 2.37 Å for 2DS2 (Fig. 6 B and Table II). The area buried at the interface of D1 and D2 was 940 Å2 (calculated by EMBL-EBI PISA, available at http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), within the range reported for 2DL1 (1,076 Å2; Fan et al., 1997), 2DL2 (919 Å2; Snyder et al., 1999), and 2DL3 (1,050 Å2; Maenaka et al., 1999), as well as the more distantly related LILRB1 (1,245 Å2; Chapman et al., 2000) and FcαR (1,134 Å2; Ding et al., 2003). In 2DS4, the 69° hinge angle between D1 and D2 is comparable to that of 2DL1 when bound to HLA-C*04 (66°), wider than that in 2DL1 alone (55°), and narrower than those in 2DL2 (81°) and 2DL3 (78°). Stabilizing the hinge is a conserved hydrophobic core and an interdomain salt bridge, in which only position 102 varies and influences the HLA specificity of 2DS4, as shown in the previous section. Asp98 and Arg149, which form the interdomain salt bridge in 2DL1, 2DL2, and 2DL3, are present in 2DS4 but are too far apart to bond (7.5 Å), as previously seen in 2DS2 (Saulquin et al., 2003). The interdomain hydrogen bonds involving Thr76-Tyr186 and Asp98-Tyr186 are conserved in 2DS4 and other lineage III KIRs. The 2DS4 hinge is further stabilized by interdomain bonds formed by His16, a residue unique to 2DS4, with the main-chain oxygen of Ala145 (3.3 Å) and the side chain of Glu147 (2.9 Å) in the CC′ loop of D2 (Fig. 7 A). This allows 2DS4 to form a narrow hinge angle, like 2DL1 bound to HLA-C*04, which could account for the avidity of 2DS4 for three HLA-C2 allotypes.
Table II.
RMSD comparing KIR2DS4 to other KIRs
| Domains compared | |||
| 2DS4 compared to | D1/D2 | D1 | D2 |
| Å | Å | Å | |
| KIR2DL1 (1NKR) | 1.91 | 1.67 | 0.76 |
| KIR2DL1 complex (1IM9) | 1.04 | 1.21 | 0.65 |
| KIR2DL2 (2DL2) | 1.53 | 1.45 | 0.98 |
| KIR2DL2 complex (1EFX) | 1.12 | 0.86 | 0.87 |
| KIR2DL3 (1B6U) | 1.71 | 1.53 | 0.98 |
| KIR2DS2 (1M4K) | 2.37 | 0.78 | 2.96 |
Figure 7.
Features distinguishing the KIR2DS4 structure. (A) The 2DS4 unique residue His16 of the D1 domain forms two novel interdomain bonds with the D2 domain. Shown are these bonds (dashed line) formed by His16 with Ala145 (3.3 Å) and the side chain of Glu147 (2.9 Å). The electron-density maps (gray) of the interdomain region are contoured at the 1.2-σ level to indicate the quality of the resolved structure. (B–F) Close-up views of key residues involved in KIR2DL/HLA-C recognition. The D1 and D2 domains of 2DS4 were superimposed on the respective D1 and D2 domains of the structures of 2DL1 bound to HLA-C*04 (Fan et al., 2001) and 2DL2 bound to HLA-C*03 (Boyington et al., 2000). KIRs are colored as in Fig. 6. HLA-C molecules are colored magenta (HLA-C*03) and light blue (HLA-C*04), the GAV peptide of the 2DL2–HLA-C*03 complex is colored in red, and the key water molecule 38 (w38) is shown as a red asterisk. (B) The close proximity (1.6 Å) of Lys44 in 2DL2/3 to Lys80 of HLA-C2 causes steric hindrance, which prevents functional interactions between the two lysine residues. With the backbone displacement of L2 in 2DS4, Lys44 Cα of 2DS4 is separated from that of 2DL2 by 2.5 Å (dashed line), increasing the minimum distance between their Nζ atoms to 4 Å. (C) Lys44 in 2DL2 forms hydrogen bonds with Asn80 and Arg79 of HLA-C*03. The backbone shift widens the distance between Lys44 in 2DS4 and Asn80 to 3.7 Å and eliminates contact with Arg79. Because the benzyl group of Phe45 is rotated 90° clockwise in 2DS4, it is unlikely to make the hydrophobic contacts reported for 2DL2 and HLA-C*03. (D) In both KIR–HLA-C structures, Gln71 and Asp72 in loop 3 form hydrogen and hydrophobic contacts with HLA-C. Gln71 of 2DL2 also forms hydrogen bonds with Ala8 of peptide and water molecule 38. These interactions are abrogated by the replacement of Gln71/Asp72 with Pro71/Val72 in 2DS4, although they do not alter the conformation of loop 3. (E and F) The replacement of Ser184 by alanine is unique to 2DS4. This substitution abolishes the interactions described between Ser184 and Lys146 in the 2DL2–HLA-C*03 structure (E), or between Ser184 and both Lys80 and Lys146 in the 2DL1–HLA-C*04 structure (F).
Comparison of KIR2D receptor ligand binding sites.
Up to 18 residues from six loops at the KIR surface contribute to interaction with HLA-C. Three D1 domain loops (L1, A′B; L2, CC′; and L3, EF) interact with the α1 helix of HLA-C and the bound peptide. The hinge loop (L4) and the BC (L5) and FG (L6) loops of the D2 domain contact the α2 helix of HLA-C. Superimposing 2DS4 on other KIRs, alone or in complex with HLA-C, showed no significant differences in the L1, L4, and L5 contact loops.
Examination of contact loop L2 revealed a backbone displacement of residues 43–45 in 2DS4 away from the KIR–HLA binding groove, as indicated by a 2.5-Å separation of the Lys44 Cα in 2DS4 compared with 2DL2/3. Because of this shift, Lys44, the specificity-determining residue, is unlikely to form the hydrogen bonds with Asn80 and the Nϵ atom of Arg79 that are central to the interaction of 2DL2/3 with HLA-C1. Furthermore, the hydrophobic contacts that Phe45 of 2DL1 and 2DL2 makes with Arg75, Val76, and Arg79 of HLA-C (Boyington et al., 2000; Fan et al., 2001) are unlikely to be achieved by Phe45 of 2DS4, because its benzyl group shows a 90° clockwise rotation away from the HLA heavy chain. The inability of 2DL2/3 to bind HLA-C2 is attributed to steric hindrance and electrostatic repulsion between Lys44 of the KIR and Lys80 of HLA-C2 (Fan et al., 2001). In 2DS4, the backbone displacement is predicted to increase the minimum distance between the Nζ atoms of Lys44 in 2DS4 and Lys80 of HLA-C2 from 1.6 to 4 Å, thus reducing the steric hindrance and permitting some interaction of 2DS4 with HLA-C2. In summary, the backbone displacement can explain both the weaker interactions of 2DS4 with HLA-C1 (compared with 2DL2/3) and its cross-reactivities with HLA-C2 (Fig. 7, B and C).
In loop L6 of 2DS4 and Pt-2DS4, Ser184 is uniquely replaced by alanine. Consequently, the Ser184 Oγ atom, which forms hydrogen and polar interactions with the Nζ atoms of Lys146 and Lys80 in the 2DL1–HLA-C*04 and 2DL2–HLA-C*03 complexes, is absent, and the methyl side chain of Ala cannot form any of these interactions (Fig. 7, E and F). That the presence of Ala184 reduces avidity for 2DS4-Fc is demonstrated by the improved binding of the 2DS4 mutant in which Ala184 was replaced by serine (Fig. 5).
Effect of substitutions in the L3 loop of 2DS4 introduced by gene conversion.
Apart from residue 184, the other residues of the hinge and D2 that contact HLA class I are highly conserved in human lineage III KIRs. In contrast, six out of the seven HLA-contact residues in D1 are polymorphic, consistent with D1 determining HLA specificity. Four polymorphic contact residues are in loop 3: Arg68 interacts with C1, Met70 interacts with C2, and Gln71 and Asp72 interact with both C1 and C2 (Boyington and Sun, 2002). The gene conversion that replaced the Gln71-Asp72 motif with Pro71-Asp72 did not alter the loop configuration but likely affected interactions with HLA class I. Superposition of KIR structures indicates that Pro71 in 2DS4 cannot bond with peptide residue Ala8 and Val76 of HLA-C1, just as Gln71 does in the 2DL2–HLA-C*03 complex (Boyington et al., 2000). The side chain atoms of Pro71 are 5.2 Å away from the Ala8 N atom and 5.3 Å away from water molecule 38, compared with 3 Å for both bonds in the 2DL2–HLA-C*03 complex. In addition the aliphatic side chain of Val72 in 2DS4 cannot form the hydrogen bond observed between Asp72 of 2DL2 and Gln72 of HLA-C*03 (Fig. 7 B). For 2DL3, loss of these bonds to Gln71 and Asp72 is sufficient to abrogate C1 binding, whereas for 2DL1 the binding to C2 is halved. For 2DS4, the replacement of Gln71-Asp72 with Pro71-Val72 by gene conversion diminished interaction with C1 while increasing interactions with C2 and A*11.
Alteration of the KIR–HLA binding interface
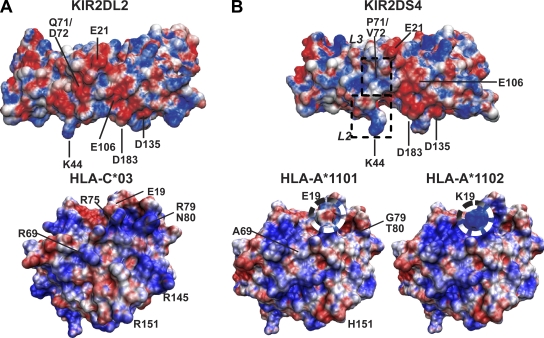
The binding of 2DL to HLA-C is dominated by interactions between complementary charged residues (Boyington et al., 2000; Fan et al., 2001). KIR2DL has an electronegative binding surface that complements an electropositive surface on HLA-C (Fig. 8 A). In 2DS4, the electronegative surface is disrupted by small positive and uncharged patches coming from the L2 and L3 loops and caused by replacement of Asp47 by asparagine and Asp72 by valine (Fig. 8 B, top). These differences likely contribute to the distinctive specificity and avidity of 2DS4 for HLA class I.
Figure 8.
Possible effects of the difference between A*1101 and A*1102 on the interaction with KIR2DS4. Predicted effects of the single substitution between HLA-A*1101 (Glu19) and -A*1102 (Lys19) were modeled using the structure file for HLA-A*1101 (1QVO). Electrostatic potentials were calculated with APBS tools (Baker et al., 2001) for 2DS4, 2DL2 (1EFX), HLA-C*03 (1EFX), HLA-A*1101 (1QVO), and HLA-A*1102 and were used to color the molecular surface drawn on their Cα traces (red, negative [−4]; blue, positive [4]). (A) Characterizing the binding of HLA-C to KIR2DL is an electropositive surface on HLA-C (e.g., HLA-C*03; bottom) that interacts with a complementary electronegative surface on KIR2D (e.g., 2DL2; top). Key acidic residues for KIR2D and basic residues for HLA-C are indicated. (B) In 2DS4 the common electronegative surface is disrupted by small positive and uncharged patches in L2 and L3 (boxes; top). HLA-A*11 has four nonconservative changes in the predicted KIR–HLA-C contact region (A69R, G79R, T80N/K, and H151R), resulting in an uncharged altered electrostatic surface compared with HLA-C. As indicated in the black and white circles, the substitution of Glu19 for Lys19 in A*1102 restores the electropositive surface in this region, which could explain its stronger reaction with 2DS4 than A*1101.
The general inability of KIR2D to recognize HLA-A can be explained by the presence of three or more nonconservative changes in the contact residues. Although A*11 interacts with 2DS4, the absence of basic residues at positions 69, 79, 80, and 151 disrupts the electrostatic surface present in HLA-C (Fig. 8 B, bottom). Consequently, A*11 is unlikely to form the hydrogen bonds and salt bridges that characterize the interaction of HLA-C with KIR2D (Boyington and Sun, 2002). These differences suggest that 2DS4 contacts A*11 in a fashion different from that described for KIR2DL and HLA-C, as also proposed for the interaction of 2DS4 with C*04 (Katz et al. 2001). Consistent with this idea, residue 19 that distinguishes high-avidity A*1102 from low-avidity A*1101 is outside the defined KIR–HLA-C binding site. Comparing the electrostatic surface potentials of the two A*11 subtypes revealed a distinctive electropositive pocket around lysine 19 in HLA-A*1102 that could interact with KIRs and explain the differential avidity of 2DS4 for A*1101 and A*1102.
DISCUSSION
We studied KIR2DS4, the only activating lineage III KIR of the group A haplotypes and the oldest, most divergent member of this lineage, which has uniquely expanded in the evolution of the human species. Previously the ligand specificity of 2DS4 was examined using small numbers of HLA class I variants and assays of limited sensitivity (Kim et al., 1997; Valés-Gómez et al., 1998; Winter et al., 1998; Katz et al., 2001). In this study, we applied a robust, sensitive binding assay to 95 HLA class I allotypes representing the >2,500 HLA-A, -B, and -C variants defined worldwide (available at http://www.ebi.ac.uk/imgt/hla/; Robinson et al., 2003)). Two HLA-A allotypes and six HLA-C allotypes (three carrying the C1 epitope and three carrying the C2 epitope) reliably bound 2DS4 at levels above the low background obtained with this assay. Consistent with the earlier studies, 2DS4 exhibited no reactivity with HLA-B, and the positive reactions were generally weaker than those obtained with the inhibitory HLA-C receptors: 2DL1, 2, and 3.
The HLA-A and -C allotypes recognized by 2DS4 represent a novel specificity not observed for other human KIRs, but combining aspects of the specificities of HLA-C–reactive KIR2DL1, 2, and 3 and A3/11-specific KIR3DL2. The HLA-C specificities of lineage III KIRs correlate with the polymorphism at position 44 in the D1 domain. Methionine 44 gives 2DL1 exquisite C2 specificity, whereas lysine 44 gives 2DL2/3 C1 specificity but also confers cross-reactivity with C2+ C*0501, C*0202, and C*0401. KIR2DS4 has lysine 44, like 2DL2/3, and selectively binds to these same C2+ allotypes as well as to C1+ C*1601, C*0102, and C*1402. Thus, 2DS4 has a unique, selective HLA-C specificity with neither C1 nor C2 bias. No pattern of amino-acid substitution within the HLA-C heavy chain could be correlated with the set of six 2DS4-reactive HLA-C allotypes, pointing to bound peptides as a determining factor.
Distinguishing 2DS4 from other lineage III KIRs is the Pro71-Val72 motif in the D1 domain that is uniquely shared with 3DL2, an inhibitory lineage II KIR with specificity for HLA-A*03 and -A*11. This commonality most likely arose by gene conversion, which transferred the Pro71-Val72 motif from a 3DL2 ancestor to a 2DS4 ancestor. Targeted mutagenesis shows that the functional effects of the gene conversion were to give 2DS4 specificity for A*11 (but not A*03) to reduce overall avidity for C1+ HLA-C and to retain cross-reactivity with C2+ C*0501, C*0202, and C*0401. Chimpanzee Pt-KIR2DS4 shares the Pro71-Val72 motif and similar A*11 specificity with human KIR2DS4. In contrast, their HLA-C specificities differ, with Pt-KIR2DS4 having weaker reactions with C2+ and stronger reactions with C1+ allotypes. The gene conversion occurred before separation of human and chimpanzee ancestors, and in the 6.5–10 million years since then (Benton and Donoghue, 2007), natural selection has conserved A*11 specificity while diversifying MHC-C specificity.
The three dimensional structure of 2DS4 is very similar to those of other lineage III KIRs (Fan et al., 1997; Maenaka et al., 1999; Snyder et al., 1999; Saulquin et al., 2003). Comparison with the structures of 2DL1 bound to C2+ HLA-C*04 (Fan et al., 2001) and 2DL2 bound to C1+ HLA-C*03 (Boyington et al., 2000) revealed two features of the 2DS4 structure that likely influence ligand binding and HLA class I specificity. First, a backbone displacement of the contact loop CC′ (L2) is predicted to perturb interactions between Lys44 and Phe45 of 2DS4 and Arg79 and Asn80 of the C1 epitope. This could explain the overall weaker binding of 2DS4 to C1+ allotypes than 2DL2/3. Conversely, this displacement could favor accommodation of Lys80 in the C2 epitope, thereby increasing avidity for some C2+ allotypes. Second, the Pro71-Val72 motif that gave 2DS4 reactivity with A*11 is part of loop EF (L3), which in KIR2DL contacts bound HLA-C using the Gln71-Asp72 motif. Several observations point to Pro71 being essential for A*11 recognition, whereas Val72 can undergo nonconservative substitutions without causing loss of A*11 binding. Mutating Gln71 to Pro71 in 2DL3 causes a gain of A*11 reactivity in the context of Asp72 that is comparable to that of 2DS4 (Fig. S3). Several orangutan KIRs bind A*11 like 2DS4 but have Pro71 in the context of Thr72 (unpublished data). That Gln71 and Asp72 of 2DL3 are predicted to form strong hydrogen bonds with bound HLA class I ligand can explain why the double mutant with Pro71-Val72 loses HLA class I reactivity. In contrast, Asp72 of 2DL1 makes hydrophobic contact with ligand, but Glu72 does not contribute. Consistent with this difference, mutation at positions 71 and 72 had less effect on ligand-binding specificity and avidity for 2DL1 than 2DL3 (Fig. S3).
In binding and functional assays, A*1102 was always a better 2DS4 ligand than A*1101. This difference comes from substitution of Glu19 in A*1101 for Lys19 in A*1102 (Morrell et al., 1999). Modeling the 2DS4 interaction with A*11 using the 2DL2–HLA-C*03 structure (Boyington et al., 2000) identified two potential hydrophobic contacts between Lys19 of A*1102 and Phe45 of 2DS4 (∼3.7 Å). Similar contacts were not possible with Glu19 of A*1101 (∼5.8 Å), or with A*1102 when modeled into the 2DL1–HLA-C*04 complex (Fig. S5). The interaction of 2DS4 with C*04 was shown to depend on Trp14 (Katz et al., 2001), a residue unique to C*04. Trp14 alters the conformation of the loop delineated by residues 14 and 19, and which packs against the outer side of the α1 helix close to the site of KIR–HLA interaction (Fig. S6). Residues 14 and 19 are close together in the HLA class I structure and can interact with each other (Fig. S6). Residue 19 is located below Arg75, one of the six basic residues in HLA-C to which KIR2DL binds. Of the three basic residues present on the HLA-C α1 helix, only Arg75 is conserved in A*11, with Arg69 and Arg79 being replaced by alanine and glycine, respectively. Substitution of Glu19 for lysine in A*1102 is predicted to form a novel electropositive pocket that increases avidity for KIR2DS4 (Fig. 8). That polymorphisms at positions 14 and 19 in HLA class I both influence avidity for 2DS4 points to this loop of HLA class I playing an essential role in binding 2DS4. Neither position is implicated in peptide interactions, suggesting that their effect is directly on binding to KIR.
KIR2DS4 comprises two lineages based on the presence and absence of a frame-shifting deletion in exon 5 that leads to secretion of a KIR1D protein having only the D1 Ig-like domain (Hsu et al., 2002; Middleton et al., 2007). The common alleles lacking and having the deletion are 2DS4*001 and 2DS4*003, respectively. In contrast to 2DS4*001-Fc, 2DS4*003-Fc bound to none of the 95 HLA-A, -B, and -C allotypes tested. This unequivocal result proves that 2DS4*003 and the other deletion alleles (2DS4*004-009) do not encode HLA class I receptors. Although the possibility that they have another function cannot be ruled out, the low level of their transcripts in NK cells (Hsu et al., 2002; Middleton et al., 2007) is evidence for nonsense-mediated mRNA decay (Rebbapragada and Lykke-Andersen, 2009) and likely loss of function. Either way, the two lineages of 2DS4 alleles clearly specify qualitatively different functions.
That 2DS4*001 and 2DS4*003 are both present at significant frequencies in all human populations shows that these two functionally different forms have been maintained since the origin of the modern human species. Such balance is a consistent feature of HLA (Solberg et al., 2008) and KIR (Yawata et al., 2006; Norman et al., 2007) variation, and is unlikely the product of chance, but of balancing selection. Depending on circumstances, competitive advantage is conferred to individuals who are homozygous for either 2DS4*001 or 2DS4*003, or heterozygotes. The relative frequency of full-length and deleted 2DS4 varies with population, with the full-length form predominating in Asians with frequencies of up to 80%, compared with 29–45% in Caucasians (Yawata et al., 2006). Population comparisons revealed a significant correlation (P < 0.001) between the presence of 2DS4*001 and A*11 (Fig. S4): A*1101 is prevalent in East and Southeast Asia and the aboriginal populations of Papua New Guinea and Australia (allele frequency, 20–64%) and less common (3–10%) in Caucasians, whereas A*1102 is restricted to East and Southeast Asia and exhibits the highest frequency (5–12%) in aboriginal Taiwanese (Middleton et al., 2003).
HLA-A*11 initiates strong cytotoxic T cell responses to EBV infections (Gavioli et al., 1993; Rickinson and Moss, 1997), and complexes of HLA-A*11 with EBV peptides preferentially bind to 3DL2 (Hansasuta et al., 2004). As 2DS4 has similar A*11 specificity to 3DL2, it too may focus on binding A*11 complexes with EBV peptides. Latent herpes viruses have dynamic evolutionary relationships with NK cell–mediated host immunity (Gumá et al., 2006), and thus, the combination of A*11 and 2DS4*001 could help control EBV infection. In Southeast Asia, EBV infection is associated with nasopharyngeal cancer (Thompson and Kurzrock, 2004), and evidence for pressure on the NK cell response is the emergence and abundance of the unusual HLA-B*46 allotype that carries the C1 epitope and is a strong NK cell educator (Yawata et al., 2008).
That the functional A*11-binding form of 2DS4 is significantly correlated with the presence of A*11 is evidence for the functional significance of this interaction and the action of natural selection to preserve it. Further, supporting this thesis are the results of functional experiments showing that interaction between A*1102 and 2DS4*001 activates NKL cell–mediated killing of 221 cells. In contrast, we could not detect a similar functional effect of the interactions with A*1101 or the 2DS4*001-binding C*0202, C*0401, and C*0501 (C*0101, C*1402, and C*1601 were not tested) even though some bound more strongly than A*1102, and C*04 was previously shown to be a functional ligand for 2DS4*001 (Katz et al., 2001). Several factors likely contribute to the difference. Comparison of human and chimpanzee 2DS4 shows that A*11 reactivity has been preserved for 6.5–10 million years, whereas HLA-C reactivity has changed. Consistent with this thesis, many chimpanzee Patr-A allotypes are related to HLA-A*11 (Adams and Parham, 2001; Robinson et al., 2005). Thus, the functional interaction of 2DS4 with A*11 could be more optimized than those with HLA-C. As noted by others (Katz et al., 2001), the interactions and functional response of KIR2DS4 and KIR3DL2 are weaker than those of the inhibitory KIRs and particularly sensitive to assay conditions. In this regard, the bound peptides that allow binding of 2DS4 to HLA-C may not be identical to the ones that trigger a functional response (Stewart et al., 2005). Lastly, we cannot rule out the possibility that NKL cells differ from physiological NK cells in being selectively less sensitive to engagement of 2DS4 by HLA-C and A*1101 ligands than to engagement of A*1102 ligand.
The level of KIR2D-Fc binding to beads coated with HLA class I varies widely, but reproducibly, depending on the combination of KIR and HLA variants. Such variation probably depends on the HLA class I allotype and on the peptides bound to the allotype. Because of the considerable washing and dilution in the binding assay, it is likely that only high avidity interactions are detected, and thus, differences in binding level, as observed for 2DS4 interaction with A*1101 and A*1102, will in part reflect differences in the peptide repertoires that the two allotypes bind.
KIR3DL2 is a conserved, framework gene that defines the telomeric end of the human KIR locus. In all group A and some group B haplotypes, KIR2DS4 is the neighboring gene to 3DL2. In this study, we have demonstrated that these two adjoining KIR genes encode A*11-reactive receptors with an activating and inhibitory function, respectively. Such properties raise the possibility that 2DS4 and 3DL2 play a balancing role in the NK cell response to EBV infection in individuals carrying A*11. Such pairs of activating and inhibitory receptors with similar ligand specificity have been described for several other families of immunoreceptors (Arase and Lanier, 2004; Barclay and Hatherley, 2008).
MATERIALS AND METHODS
KIR-Fc fusion proteins and binding assays to HLA class I.
These methods were previously described (Moesta et al., 2008). DNA constructs encoding KIR2DS4*001, Pt-KIR2DS4, 2DL1*003, 2DL2*001, and 2DL3*001 were cloned into pcr2.1 and mutated using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). Using a recombination PCR strategy, exons encoding the Ig domains and stems of KIRs were fused in frame with exons encoding the Fc domains of human IgG1. For the deletion variant KIR2DS4*003, the last residue of KIR2DS4*003 before the premature stop codon was placed next to the Fc portion. Chimeric products were inserted into the transfer vector pACgp67, and soluble fusion proteins were produced as described. Purified fusion proteins were assessed against a panel of 29 HLA-A, 50 HLA-B, and 16 HLA-C allotypes using commercially available LABScreen single-antigen bead sets (One Lambda), according to the manufacturer's directions. KIR-Fc fusion proteins at various concentrations ranging from 400 to 0.1 µg/ml were incubated with LABScreen microbeads and visualized on a Luminex 100 reader (Luminex Corp.; provided by J.-H. Lee, One Lambda Inc., Canoga Park, CA). The results were expressed as relative fluorescence ratios and normalized against the binding obtained with the positive control mAb W6/32, which recognizes a common epitope of HLA class I allotypes.
The quality of KIR-Fc fusion proteins was assessed by flow cytometry using KIR conformation-dependent mAbs (Fig. S1). 100 µg/ml KIR-Fc was incubated with 20 µl anti–human IgG-coated beads (Bangs Laboratories) for 30 min at 4°C, and washed and stained for 30 min at 4°C in 100 µl PBS containing 1–2 µl of a PE-conjugated specific antibody for KIR2DS4 (CD158i-PE; Beckman Coulter), KIR2DL2/3 (DX27-PE; BD), KIR2DL1 (EB6-PE; Beckman Coulter), and the PE-conjugated mouse anti–human KIR antibody (clone NKVFS1; AbD Serotec), which is specific for human lineage III KIR. Flow cytometry was performed using a FACScan instrument (BD).
Generation of the G4-NKL derivative of NKL cells.
The NKL cell line (Robertson et al., 1996), which expresses no endogenous cell-surface KIRs, was maintained as previously described (Moesta et al., 2008). To eliminate the confounding effect of MHC class I–mediated inhibition through LILRB1 (Carr et al., 2005), we generated a derivative of the NKL cell line, G4-NKL, in which the endogenous expression of LILRB1 is suppressed by >90% by siRNA. Total RNA from NKL was extracted using TriReagent (Sigma-Aldrich), and cDNA was synthesized using Superscript III (Invitrogen). LILRB1 was amplified from NKL cDNA using specific oligonucleotide primers (LILRB1F, 5′-GCACCGAGGGCTCATCCATCCA-3′; LILRB1r, 5′-AGTTCCCAGAGTCTCCTGGGG-3′). The two LILRB1 alleles identified (available from GenBank/EMBL/DDBJ under accession nos. AF009220 and NM_006669) were targeted for knockdown using siRNA. The sense and antisense primers used for the LILRB1 knockdown (LILR-KILRs, 5′-GATCCGCGTGACTTCCTTCAGCTCGTTCAAGAGACGAGCTGAAGGAAGTCACGTTTTTTGCTAGCG-3′ and its reverse complement) are unique to exon 5 of LILRB1 that encodes the third Ig domain (D3). The two primers were annealed together, ligated into the pSIREN-RetroQ vector (Takara Bio Inc.), and transfected into φ-NX cells, as previously described (Moesta et al., 2008). The supernatant containing recombinant amphotrophic retrovirus was used to transfect growing NKL cells, giving stable transfectants that express the LILRB1-directed siRNA. Transfected NKL cells were maintained in 2 µg/ml puromycin, screened, and regularly monitored for LILRB1 expression using the antibody CD85j-PE (BD; Fig. S2).
Generation of NK effector and target cell lines.
G4-NKL cells, which express no endogenous cell-surface KIRs, were used as effector cells and transduced to express selected KIRs. Full-length coding regions of 2DL1*003, 2DL3*001, 2DS4*001, and 3DL2*001 were amplified by PCR and cloned into the pBMN retroviral vector. An inhibitory long-tailed derivative of 2DS4*001 (KIR2DS4-LT) was made by replacing the signaling domain with that of 2DL2*001. Recombinant retrovirus was generated by transfection into φ-NX cells, from which supernatants were used to infect G4-NKL. Transduced cells were sorted for equivalent KIR expression levels using a cell sorter (FACStar; BD), as previously described (Moesta et al., 2008).
The 221 cell line and 221 transfectants expressing selected HLA class I allotypes were used as target cells. Transfected HLA class I constructs encoded mutated leader peptides that were nonpermissive for HLA-E expression, as previously described (Michaëlsson et al., 2002; Carr et al., 2007). Transfectants were maintained in 1 mg/ml G418 (Invitrogen) and sorted for equivalent levels of HLA class I expression using an FITC-conjugated W6/32 anti–human HLA class I mAb (eBioscience).
Cytotoxicity assay.
The effect of KIR2DS4 ligation on G4-NKL–mediated cytotoxic activity was determined in redirected cytotoxicity assays using the FcγR-expressing mouse mastocytoma cell line P815 as the target. Effectors were first incubated for 30 min with 1 mg/ml of the KIR2DS4-specific mAb clone 179315 (R&D Systems) or with a combination of anti–human NKG2D and anti–human 2B4 (CD244) mAbs (BD). Effector cells were added to targets at E/T ratios ranging from 1:1 to 100:1. Killing of transfected and untransfected 221 cells by G4-NKL transductants was assayed as previously described (Carr et al., 2005). In brief, effector cells incubated for 60 min with or without the combination of anti–human NKG2D and 5 mg/ml anti–human 2B4 were added to 51Cr-loaded target cells for 4 h at 37°C at ratios ranging from 100:1 to 1:1. After incubation, supernatants were harvested and 51Cr was quantified using a β-scintillation counter (Wallac). Specific lysis was calculated using the following formula: (specific release − spontaneous release)/(total release − spontaneous release) × 100. Experiments were conducted in triplicate for each condition, and each experiment was independently replicated at least three times. Anti-KIR3DL2 antibody DX31 was provided by L. Lanier (University of California, San Francisco, San Francisco, CA).
Recombination and phylogenetic analysis.
To identify potential recombination events between KIR3DL2 and KIR2DS4, we used a dataset of primate KIR genes and the Recombination Detection Program (Martin et al., 2005). To reconstruct the evolution of the lineage III KIR D1 sequences, a genomic segment containing the exon encoding the D1 domain, as well as ∼300 bp of intron on each side of the exon, was used. Nucleotide sequences were aligned with MAFFT (Katoh et al., 2002), and the resulting alignments were corrected manually. Neighbor-joining phylogenetic analyses were performed with MEGA4 (Kumar et al., 2004) using the Tamura-Nei method with 500 replicates. The same approach was used for the analysis of the 31-bp segment involved in the gene conversion between KIR3DL2 and KIR2DS4 ancestors.
Crystallization, data collection, refinement, and data deposition.
The cDNA encoding residues 3–200 of KIR2DS4*001 was cloned with a HIS-Tag at the C terminus in frame with the pET-28c vector (EMD). The sequence was verified by DNA sequencing and transformed into BL21-CodonPlus(DE3)-RIL–competent cells (Agilent Technologies). Cells carrying the expression plasmid were grown from a single colony and induced at logarithmic phase with 0.5 mM IPTG to produce protein. Cells were harvested by centrifugation, resuspended, and lysed by freezing and thawing. The insoluble inclusion bodies were harvested by using a FrenchPress (Thermo Fisher Scientific), washed extensively to remove contaminating proteins, and dissolved in 8 M urea, 20 mM Tris (pH 8), 0.125 mM EDTA, and 0.125 mM dithiothreitol. 300 mg of solubilized KIR2DS4 receptor inclusion bodies were refolded at 4°C for 24 h in 1 liter of buffer containing 300 mM Tris HCl (pH 8.5), 400 mM l-arginine, 2.5 mM EDTA, 0.5 mM of oxidized gluthathione, and 5 mM of reduced glutathione. The refolded protein was concentrated and dialyzed at 4°C, and afterward purified by ion metal affinity chromatography using Ni-NTA resin (QIAGEN) and then by a Superdex 75 (GE Healthcare) liquid chromatography gel filtration column.
Crystals were grown in 1–2- µl hanging drops at 10°C containing a 1:1 mixture of protein solution (5 mg/ml) and reservoir solution of 0.22 M Mg-formate/100 mM BIS-Tris (pH 5.5), soaked briefly in reservoir solution containing 30% ethylene glycol, and flash cooled with liquid nitrogen. Data were collected from a single crystal to 2.5 Å at beamline 11-1 of the Stanford Synchotron Radiation Laboratory on 7 January 2008, with the cryocooling temperature set at 100 K, wavelength 0.979 Å, one degree oscillation, using a MAR325 detector. Diffraction data, completed to 2.5 Å, were indexed and integrated using HKL2000 (Otwinowski and Minor, 1997), and merged and scaled with SCALA of CCP4 (Collaborative Computational Project No. 4 Software for Macromolecular X-Ray Crystallography, available at http://www.ccp4.ac.uk/). X-ray diffraction data were collected to 2.5 Å resolution. The crystals belong to the space group P212121 with unit cell 51.23 Å, 62.85 Å, 65.93 Å, 90°, 90°, 90°. Molecular replacement was performed using PHASER (McCoy et al., 2005), with chain D available from the PDB database under accession no. 1IM9. Refinement was performed using PHENIX (Adams et al., 2002), with subsequent inspection of maps and manual rebuilding performed with COOT (Emsley and Cowtan, 2004). The final Rwork of the model was 23.84%, with an Rfree of 28.42%. The hinge angle of KIR2DS4 was calculated using DynDom (Hayward and Berendsen, 1998). The atomic coordinates of the KIR2DS4 structure are available from the PDB database under accession no. 3H8N.
Online supplemental material.
Fig. S1 shows the binding of four monoclonal anti-KIR antibodies to 17 KIR-Fc fusion proteins. Fig. S2 shows that the G4-NKL derivative of the NKL cell line exhibits a reduced amount (<10%) of LILRB1 at the cell surface, an effect induced by stable transfection of LILRB1-directed siRNA. Fig. S3 shows how mutations at positions 71, 72, and 84 in the D1 domain of KIR2DL1 and KIR2DL3 change the specificity and avidity of these KIRs for HLA class I. Fig. S4 shows how the frequency of HLA-A*11 in human populations correlates positively with the frequency of the full-length and functional form of KIR2DS4 (2DS4*001). Fig. S5 shows a structural model that can explain why KIR2DS4 binds more strongly to HLA-A*1102 than to HLA-A*1101. Fig. S6 shows how residues 14 and 19 of HLA class I interact with each other and affect the conformation of the loop formed by residues 14–19. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091010/DC1.
Acknowledgments
We thank J.-H. Lee for the Luminex reader and L. Lanier for the DX31 antibody.
This work was supported by National Institutes of Health (NIH) grants AI24258, AI22039, and PO1 11412 to P. Parham, and by a Dr. Mildred Scheel Foundation for Cancer Research fellowship to T. Graef. Some of the research was performed at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the Office of Basic Energy Sciences, United States Department of Energy. The SSRL Structural Molecular Biology Program is supported by the Office of Biological and Environmental Research, United States Department of Energy; the Biomedical Technology Program, NIH National Center for Research Resources; and the National Institute of General Medical Sciences.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- KIR
- killer cell Ig-like receptor
- RMSD
- root mean square deviation
References
- Abi-Rached L., Parham P. 2005. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J. Exp. Med. 201:1319–1332 10.1084/jem.20042558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams E.J., Parham P. 2001. Species-specific evolution of MHC class I genes in the higher primates. Immunol. Rev. 183:41–64 10.1034/j.1600-065x.2001.1830104.x [DOI] [PubMed] [Google Scholar]
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- Arase H., Lanier L.L. 2004. Specific recognition of virus-infected cells by paired NK receptors. Rev. Med. Virol. 14:83–93 10.1002/rmv.422 [DOI] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 98:10037–10041 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay A.N., Hatherley D. 2008. The counterbalance theory for evolution and function of paired receptors. Immunity. 29:675–678 10.1016/j.immuni.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M.J., Donoghue P.C. 2007. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24:26–53 10.1093/molbev/msl150 [DOI] [PubMed] [Google Scholar]
- Biassoni R., Pessino A., Malaspina A., Cantoni C., Bottino C., Sivori S., Moretta L., Moretta A. 1997. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur. J. Immunol. 27:3095–3099 10.1002/eji.1830271203 [DOI] [PubMed] [Google Scholar]
- Bottino C., Sivori S., Vitale M., Cantoni C., Falco M., Pende D., Morelli L., Augugliaro R., Semenzato G., Biassoni R., et al. 1996. A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur. J. Immunol. 26:1816–1824 10.1002/eji.1830260823 [DOI] [PubMed] [Google Scholar]
- Boyington J.C., Sun P.D. 2002. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol. Immunol. 38:1007–1021 10.1016/S0161-5890(02)00030-5 [DOI] [PubMed] [Google Scholar]
- Boyington J.C., Motyka S.A., Schuck P., Brooks A.G., Sun P.D. 2000. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 405:537–543 10.1038/35014520 [DOI] [PubMed] [Google Scholar]
- Campbell K.S., Cella M., Carretero M., López-Botet M., Colonna M. 1998. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur. J. Immunol. 28:599–609 [DOI] [PubMed] [Google Scholar]
- Carr W.H., Pando M.J., Parham P. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175:5222–5229 [DOI] [PubMed] [Google Scholar]
- Carr W.H., Rosen D.B., Arase H., Nixon D.F., Michaelsson J., Lanier L.L. 2007. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 178:647–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T.L., Heikema A.P., West A.P., Jr., Bjorkman P.J. 2000. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2). Immunity. 13:727–736 10.1016/S1074-7613(00)00071-6 [DOI] [PubMed] [Google Scholar]
- Chewning J.H., Gudme C.N., Hsu K.C., Selvakumar A., Dupont B. 2007. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J. Immunol. 179:854–868 [DOI] [PubMed] [Google Scholar]
- Colonna M., Samaridis J. 1995. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 268:405–408 10.1126/science.7716543 [DOI] [PubMed] [Google Scholar]
- Della Chiesa M., Romeo E., Falco M., Balsamo M., Augugliaro R., Moretta L., Bottino C., Moretta A., Vitale M. 2008. Evidence that the KIR2DS5 gene codes for a surface receptor triggering natural killer cell function. Eur. J. Immunol. 38:2284–2289 10.1002/eji.200838434 [DOI] [PubMed] [Google Scholar]
- Ding Y., Xu G., Yang M., Yao M., Gao G.F., Wang L., Zhang W., Rao Z. 2003. Crystal structure of the ectodomain of human FcalphaRI. J. Biol. Chem. 278:27966–27970 10.1074/jbc.C300223200 [DOI] [PubMed] [Google Scholar]
- Döhring C., Scheidegger D., Samaridis J., Cella M., Colonna M. 1996. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 156:3098–3101 [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fan Q.R., Mosyak L., Winter C.C., Wagtmann N., Long E.O., Wiley D.C. 1997. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature. 389:96–100 10.1038/38028 [DOI] [PubMed] [Google Scholar]
- Fan Q.R., Long E.O., Wiley D.C. 2001. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2:452–460 [DOI] [PubMed] [Google Scholar]
- Gavioli R., Kurilla M.G., de Campos-Lima P.O., Wallace L.E., Dolcetti R., Murray R.J., Rickinson A.B., Masucci M.G. 1993. Multiple HLA A11-restricted cytotoxic T-lymphocyte epitopes of different immunogenicities in the Epstein-Barr virus-encoded nuclear antigen 4. J. Virol. 67:1572–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumá M., Angulo A., López-Botet M. 2006. NK cell receptors involved in the response to human cytomegalovirus infection. Curr. Top. Microbiol. Immunol. 298:207–223 10.1007/3-540-27743-9_11 [DOI] [PubMed] [Google Scholar]
- Gumperz J.E., Litwin V., Phillips J.H., Lanier L.L., Parham P. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 181:1133–1144 10.1084/jem.181.3.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansasuta P., Dong T., Thananchai H., Weekes M., Willberg C., Aldemir H., Rowland-Jones S., Braud V.M. 2004. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 34:1673–1679 10.1002/eji.200425089 [DOI] [PubMed] [Google Scholar]
- Hayward S., Berendsen H.J. 1998. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 30:144–154 [DOI] [PubMed] [Google Scholar]
- Hsu K.C., Liu X.R., Selvakumar A., Mickelson E., O'Reilly R.J., Dupont B. 2002. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J. Immunol. 169:5118–5129 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz G., Markel G., Mizrahi S., Arnon T.I., Mandelboim O. 2001. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J. Immunol. 166:7260–7267 [DOI] [PubMed] [Google Scholar]
- Katz G., Gazit R., Arnon T.I., Gonen-Gross T., Tarcic G., Markel G., Gruda R., Achdout H., Drize O., Merims S., Mandelboim O. 2004. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J. Immunol. 173:1819–1825 [DOI] [PubMed] [Google Scholar]
- Khakoo S.I., Rajalingam R., Shum B.P., Weidenbach K., Flodin L., Muir D.G., Canavez F., Cooper S.L., Valiante N.M., Lanier L.L., Parham P. 2000. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 12:687–698 10.1016/S1074-7613(00)80219-8 [DOI] [PubMed] [Google Scholar]
- Kim J., Chwae Y.J., Kim M.Y., Choi I.H., Park J.H., Kim S.J. 1997. Molecular basis of HLA-C recognition by p58 natural killer cell inhibitory receptors. J. Immunol. 159:3875–3882 [PubMed] [Google Scholar]
- Kulkarni S., Martin M.P., Carrington M. 2008. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 20:343–352 10.1016/j.smim.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lanier L.L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:359–393 10.1146/annurev.immunol.16.1.359 [DOI] [PubMed] [Google Scholar]
- Llano M., Lee N., Navarro F., García P., Albar J.P., Geraghty D.E., López-Botet M. 1998. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur. J. Immunol. 28:2854–2863 [DOI] [PubMed] [Google Scholar]
- Maenaka K., Juji T., Stuart D.I., Jones E.Y. 1999. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure. 7:391–398 10.1016/S0969-2126(99)80052-5 [DOI] [PubMed] [Google Scholar]
- Mandelboim O., Reyburn H.T., Valés-Gómez M., Pazmany L., Colonna M., Borsellino G., Strominger J.L. 1996. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 184:913–922 10.1084/jem.184.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S.G., Parham P., Dupont B., Geraghty D.E., Trowsdale J., Middleton D., Vilches C., Carrington M., Witt C., Guethlein L.A., et al. 2003. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 55:220–226 10.1007/s00251-003-0571-z [DOI] [PubMed] [Google Scholar]
- Martin D.P., Williamson C., Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 21:260–262 10.1093/bioinformatics/bth490 [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Storoni L.C., Read R.J. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458–464 10.1107/S0907444905001617 [DOI] [PubMed] [Google Scholar]
- Michaëlsson J., Teixeira de Matos C., Achour A., Lanier L.L., Kärre K., Söderström K. 2002. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J. Exp. Med. 196:1403–1414 10.1084/jem.20020797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D., Menchaca L., Rood H., Komerofsky R. 2003. New allele frequency database: http://www.allelefrequencies.netTissue Antigens. 61:403–407 [DOI] [PubMed] [Google Scholar]
- Middleton D., Gonzalez A., Gilmore P.M. 2007. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum. Immunol. 68:128–134 10.1016/j.humimm.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Moesta A.K., Norman P.J., Yawata M., Yawata N., Gleimer M., Parham P. 2008. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 180:3969–3979 [DOI] [PubMed] [Google Scholar]
- Moretta A., Bottino C., Vitale M., Pende D., Biassoni R., Mingari M.C., Moretta L. 1996. Receptors for HLA class-I molecules in human natural killer cells. Annu. Rev. Immunol. 14:619–648 10.1146/annurev.immunol.14.1.619 [DOI] [PubMed] [Google Scholar]
- Morrell G., Whalley J., Stewart A., Day S., Lewis L., Makar Y., Fuggle S.V., Ross J., Dunn P.P. 1999. Identification of an HLA-A11 serological variant and its characterization by sequencing based typing. Tissue Antigens. 53:591–594 10.1034/j.1399-0039.1999.530612.x [DOI] [PubMed] [Google Scholar]
- Norman P.J., Abi-Rached L., Gendzekhadze K., Korbel D., Gleimer M., Rowley D., Bruno D., Carrington C.V., Chandanayingyong D., Chang Y.H., et al. 2007. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 39:1092–1099 10.1038/ng2111 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Parham P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5:201–214 10.1038/nri1570 [DOI] [PubMed] [Google Scholar]
- Pende D., Biassoni R., Cantoni C., Verdiani S., Falco M., di Donato C., Accame L., Bottino C., Moretta A., Moretta L. 1996. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J. Exp. Med. 184:505–518 10.1084/jem.184.2.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I., Lykke-Andersen J. 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 21:394–402 10.1016/j.ceb.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Rickinson A.B., Moss D.J. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405–431 10.1146/annurev.immunol.15.1.405 [DOI] [PubMed] [Google Scholar]
- Robertson M.J., Cochran K.J., Cameron C., Le J.M., Tantravahi R., Ritz J. 1996. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 24:406–415 [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Parham P., de Groot N., Bontrop R., Kennedy L.J., Stoehr P., Marsh S.G. 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31:311–314 10.1093/nar/gkg070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Stoehr P., Marsh S.G. 2005. IPD—the Immuno Polymorphism Database. Nucleic Acids Res. 33:D523–D526 10.1093/nar/gki032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulquin X., Gastinel L.N., Vivier E. 2003. Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j). J. Exp. Med. 197:933–938 10.1084/jem.20021624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder G.A., Brooks A.G., Sun P.D. 1999. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc. Natl. Acad. Sci. USA. 96:3864–3869 10.1073/pnas.96.7.3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O.D., Mack S.J., Lancaster A.K., Single R.M., Tsai Y., Sanchez-Mazas A., Thomson G. 2008. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69:443–464 10.1016/j.humimm.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.A., Laugier-Anfossi F., Vély F., Saulquin X., Riedmuller J., Tisserant A., Gauthier L., Romagné F., Ferracci G., Arosa F.A., et al. 2005. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA. 102:13224–13229 10.1073/pnas.0503594102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.P., Kurzrock R. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10:803–821 10.1158/1078-0432.CCR-0670-3 [DOI] [PubMed] [Google Scholar]
- Uhrberg M., Valiante N.M., Shum B.P., Shilling H.G., Lienert-Weidenbach K., Corliss B., Tyan D., Lanier L.L., Parham P. 1997. Human diversity in killer cell inhibitory receptor genes. Immunity. 7:753–763 10.1016/S1074-7613(00)80394-5 [DOI] [PubMed] [Google Scholar]
- Valés-Gómez M., Reyburn H.T., Erskine R.A., Strominger J. 1998. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc. Natl. Acad. Sci. USA. 95:14326–14331 10.1073/pnas.95.24.14326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBussche C.J., Mulrooney T.J., Frazier W.R., Dakshanamurthy S., Hurley C.K. 2009. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 10:162–173 10.1038/gene.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagtmann N., Biassoni R., Cantoni C., Verdiani S., Malnati M.S., Vitale M., Bottino C., Moretta L., Moretta A., Long E.O. 1995. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 2:439–449 10.1016/1074-7613(95)90025-X [DOI] [PubMed] [Google Scholar]
- Winter C.C., Gumperz J.E., Parham P., Long E.O., Wagtmann N. 1998. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 161:571–577 [PubMed] [Google Scholar]
- Yawata M., Yawata N., Draghi M., Little A.M., Partheniou F., Parham P. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203:633–645 10.1084/jem.20051884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata M., Yawata N., Draghi M., Partheniou F., Little A.M., Parham P. 2008. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 112:2369–2380 10.1182/blood-2008-03-143727 [DOI] [PMC free article] [PubMed] [Google Scholar]