Abstract
Runx proteins are essential for hematopoiesis and play an important role in T cell development by regulating key target genes, such as CD4 and CD8 as well as lymphokine genes, during the specialization of naive CD4 T cells into distinct T helper subsets. In regulatory T (T reg) cells, the signature transcription factor Foxp3 interacts with and modulates the function of several other DNA binding proteins, including Runx family members, at the protein level. We show that Runx proteins also regulate the initiation and the maintenance of Foxp3 gene expression in CD4 T cells. Full-length Runx promoted the de novo expression of Foxp3 during inducible T reg cell differentiation, whereas the isolated dominant-negative Runt DNA binding domain antagonized de novo Foxp3 expression. Foxp3 expression in natural T reg cells remained dependent on Runx proteins and correlated with the binding of Runx/core-binding factor β to regulatory elements within the Foxp3 locus. Our data show that Runx and Foxp3 are components of a feed-forward loop in which Runx proteins contribute to the expression of Foxp3 and cooperate with Foxp3 proteins to regulate the expression of downstream target genes.
Runx family transcription factors are essential for embryonic development and have key roles in hematopoiesis. They bind DNA through the conserved Runt domain and can activate or repress transcription by cooperating with other sequence-specific transcription factors, coactivators, and corepressors (Durst and Hiebert, 2004; Taniuchi and Littman, 2004). Runx1 (AML1) and Runx3 (AML2) are intimately linked to T cell fate choice and function by regulating target genes that specify CD4/CD8 development in the thymus and the postthymic differentiation of Th1, Th2, and Th17 cells. Runx1 and Runx3 sequentially interact with the Cd4 silencer to block CD4 expression in double-negative and CD8 single-positive thymocytes (Taniuchi et al., 2002). Runx3 facilitates CD8 T cell lineage choice by repressing Zbtb7b, the gene encoding the CD4 lineage signature transcription factor Th-POK (Egawa and Littman, 2008; Setoguchi et al., 2008) and also plays a role in mature cytotoxic T cells (Cruz-Guilloty et al., 2009). Runx1 is predominant in naive CD4 T cells and transiently down-regulated in response to activation. Runx1 is reexpressed under Th2 culture conditions (Naoe et al., 2007), whereas Th1 cells express Runx3 (Djuretic et al., 2007; Naoe et al., 2007), which cooperates with T-bet to silence Il4 and activates Ifng expression (Djuretic et al., 2007). Runx1 promotes Th17 cell differentiation by up-regulating Rorc, the gene encoding the Th17 signature transcription factor Ror-γt, and II17 (Zhang et al., 2008). In T reg cells, Runx1 and the T reg signature transcription factor Foxp3 cooperate at the protein level to regulate downstream target genes (Ono et al., 2007), but the transcriptional regulation of T reg cell determinants by Runx proteins remains to be investigated.
These data prompted us to explore the role of Runx proteins in the regulation of Foxp3 in conventional CD4 T cells and in natural T reg (nT reg) cells. We find that the overexpression of Runt, the isolated Runx DNA binding domain with dominant-negative activity, interfered with de novo expression of Foxp3 in conventional CD4 T cells induced by TGF-β and other signals that drive induced T reg (iT reg) cell differentiation. Interestingly, Runt also interfered with Foxp3 expression by established nT reg cells. The conditional ablation of core-binding factor β (Cbfb), the gene encoding the shared β subunit of Runx protein complexes, resulted in a progressive decline of Foxp3 expression by nT reg cells. Consistent with a role for Runx complexes in the regulation Foxp3, Cbfb binds to regulatory elements within the Foxp3 locus in T reg cells but not in conventional CD4 T cells. Runx proteins therefore contribute to the induction and the maintenance of Foxp3 expression. Because Foxp3 cooperates with Runx proteins in the regulation of target genes (Ono et al., 2007), our finding that Runx itself regulates Foxp3 expression suggests that Runx and Foxp3 are components of a feed-forward loop, in which Runx proteins act first as regulators of Foxp3 expression and subsequently as interaction partners of Foxp3.
RESULTS AND DISCUSSION
Runx3 promotes de novo Foxp3 expression
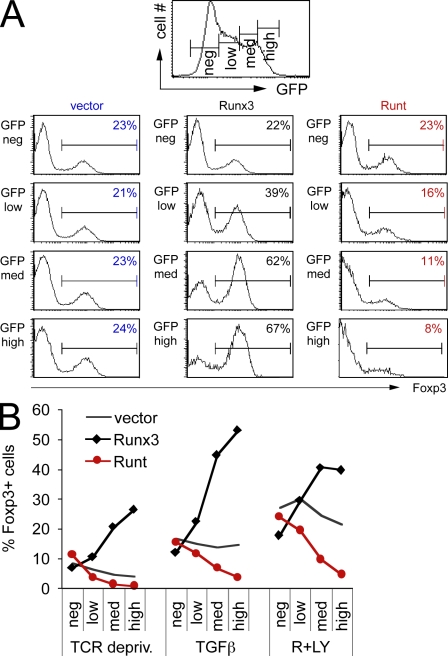
Naive CD4 T cells (CD4+ CD25− CD62Lhigh) were isolated and depleted of preexisting T reg cells by flow cytometry. After overnight activation with beads coated with anti-CD3/CD28, the cells were transduced with Runx3-IRES-GFP (Runx3), Runx3 DNA binding domain–IRES-GFP (Runt), or control IRES-GFP (vector) retroviruses. 0.3 ng/ml TGF-β was added 24 h after retroviral transduction, and Foxp3 expression was assessed 2 d later by intracellular staining. Full-length Runx3 synergized with TGF-β in promoting de novo Foxp3 expression in a dose-dependent fashion, as judged by gating on cells with increasing levels of Runx3-IRES-GFP expression (Fig. 1 A). Only modest levels of expression were achieved with the closely related Runx1, which did not consistently promote Foxp3 induction (Fig. S1 and not depicted). Because the overexpression of transcription factors does not necessarily indicate physiological significance, we tested the impact of the isolated Runt DNA binding domain. Runt acts as a dominant negative for endogenous Runx proteins, presumably by competing for access to Runx DNA binding sites (Durst and Hiebert, 2004; Telfer et al., 2004). Runt expression antagonized the induction of Foxp3 in conventional CD4 T cells in a dose-dependent manner (Fig. 1 A). De novo Foxp3 expression in newly activated CD4 T cells can be induced not only by TGF-β but also by the premature termination of TCR signaling and by inhibitors of phosphatidyl inositol 3 kinase (PI3K) and the mammalian target of rapamycin (mTOR; Gao et al., 2007; Bruno and Merkenschlager, 2008; Haxhinasto et al., 2008; Sauer et al., 2008), a signaling network downstream of the TCR. Full-length Runx3 synergized with TCR signal deprivation and the PI3K inhibitor LY294002 and the mTOR inhibitor rapamycin in the induction of Foxp3. In contrast, the isolated Runt DNA binding domain counteracted Foxp3 induction (Fig. 1 B). IL2 is involved in iT reg differentiation and Il2 is a known Runx target gene (Djuretic et al., 2007; Ono et al., 2007), but the dominant-negative effect of Runt on Foxp3 induction persisted in the presence of exogenous IL2 (Fig. S2), indicating that Runt blocks Foxp3 induction independently of IL2 expression. Collectively, these data point to an involvement of Runx proteins in the induction of de novo Foxp3 expression.
Figure 1.
Foxp3 induction is promoted by full-length Runx3 and antagonized by the expression of dominant-negative Runt domain proteins. (A) Naive CD4 T cells were activated for 18 h with anti-CD3/anti-CD28 beads and retrovirally transduced with Runx3-IRES-GFP (Runx3), Runt-IRES-GFP (Runt), or control-IRES-GFP (vector). After 24 h, 0.3 ng/ml TGF-β was added and Foxp3 expression was assessed 48 h later by intracellular staining after gating on the level of GFP expression (negative [neg], low, medium [med], or high). Data are representative of four independent experiments. (B) Naive CD4 T cells were activated and retrovirally transduced as in A. After 24 h, the anti-CD3/anti-CD28 beads were removed (TCR depriv.) and TGF-β or rapamycin and LY294002 (R+LY) were added as indicated. Foxp3 expression was assessed 48 h later by intracellular staining after gating on the level of GFP expression (negative, low, medium, or high). Data are representative of four independent experiments.
Dominant-negative Runt domain antagonizes established Foxp3 expression in nT reg cells
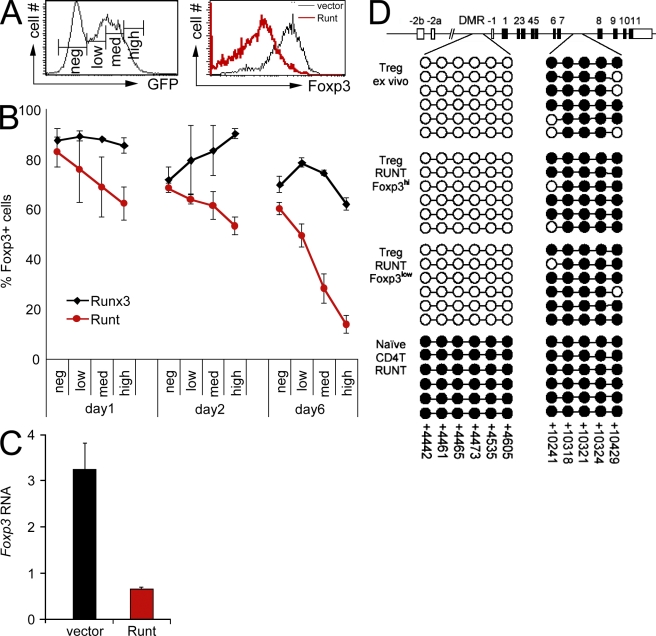
We next analyzed the impact of antagonizing Runx function in established nT reg cells. CD4+ CD25+ LN T cells were sorted, activated, and transduced with Runt-IRES-GFP (Runt) or IRES-GFP (control vector). 4 d later, GFPhigh cells were examined for Foxp3 expression by intracellular staining. Runt-transduced cells showed reduced expression of Foxp3 with a broad profile that ranged from negative to high, whereas control cells remained uniformly Foxp3 positive (Fig. 2 A). Analysis at different times after retroviral infection showed that Foxp3 expression in Runt-transduced nT reg cells declined gradually and in a dose-dependent fashion (Fig. 2 B). Real time RT-PCR established that the reduced expression of Foxp3 protein reflected substantially reduced Foxp3 RNA levels in Runt-transduced GFPhigh nT reg cells (Fig. 2 C).
Figure 2.
Runx proteins control the maintenance of Foxp3 expression in established T reg cells. (A) CD4+ CD25+ LN T cells were sorted, activated for 18 h with anti-CD3/anti-CD28 beads in the presence of IL-2, and retrovirally transduced. Histograms show GFP expression (left) and Foxp3 expression (right) in GFPhigh cells 4 d after transduction with vector-IRES-GFP (black line, right histogram) or Runt-IRES-GFP (red line, right histogram). Data are representative of five independent experiments. (B) CD4+ CD25+ LN T cells were sorted, activated, and retrovirally transduced with Runx3-IRES-GFP (black) or Runt-IRES-GFP (red), and Foxp3 expression was assessed on the indicated days by intracellular staining and gating on the level of GFP (negative, low, medium, or high as defined in A). Data are mean ± SE of two independent experiments. (C) CD4+ CD25+ LN T cells were sorted, activated, and retrovirally transduced with IRES-GFP (vector) or Runt-IRES-GFP (Runt). 5 d later, GFPhigh cells were sorted and analyzed for Foxp3 RNA expression by real time RT-PCR. Data are mean ± SD of five independent experiments. (D) Runt-transduced T reg cells with down-regulated Foxp3 expression have a demethylated Foxp3 DMR, which is consistent with a history of Foxp3 expression. CD4+ CD25+ LN T cells were sorted, activated, and transduced with Runt-IRES-GFP (Runt). 5 d later, GFPhigh Foxp3high and Foxp3low cells were sorted and analyzed for the methylation status of the indicated regions in the Foxp3 locus by bisulfite sequencing. A schematic of the locus with coding (filled rectangles) and noncoding exons (open rectangles) is shown on top. Each chain of circles represents one sequenced allele with methylated (filled circles) and unmethylated CpG dinucleotides (open circles). The positions of CpG dinucleotides are indicated (Floess et al., 2007).
To explore the developmental history of Foxp3low “exT reg” cells that arose from the enforced expression of the dominant-negative Runt domain in nT reg cells, we exploited the finding that a differentially methylated region (DMR) upstream of Foxp3 exon −1 is unmethylated in nT reg cells but methylated in conventional T cells (Floess et al., 2007; Kim and Leonard, 2007). GFPhigh Foxp3high and Foxp3low cells were isolated by cell sorting 5 d after transduction of nT reg cells with Runt-IRES-GFP. Genomic DNA was analyzed for the CpG methylation status of the Foxp3 DMR by bisulfite sequencing (CpG dinucleotides in intron 7 that are methylated in both conventional and nT reg cells served as a control). We found that the DMR was unmethylated in Runt-transduced Foxp3high and Foxp3low exT reg cells and in freshly isolated control nT reg cells. This indicated that Foxp3low cells arose from nT reg cells with an unmethylated DMR and that the Runt-mediated down-regulation of Foxp3 did not result in the remethylation of the DMR within the time frame of our analysis. The DMR remained fully methylated in Runt-transduced conventional CD4 T cells, excluding the formal possibility that enforced Runt expression itself might cause DMR demethylation (Fig. 2 D). We conclude from these data that the dominant-negative Runt domain interferes with the maintenance of Foxp3 expression in established nT reg cells and, therefore, that Runx proteins are likely regulators of Foxp3 expression in nT reg cells.
Genetic evidence for a role of Runx proteins in the maintenance of Foxp3 expression in nT reg cells
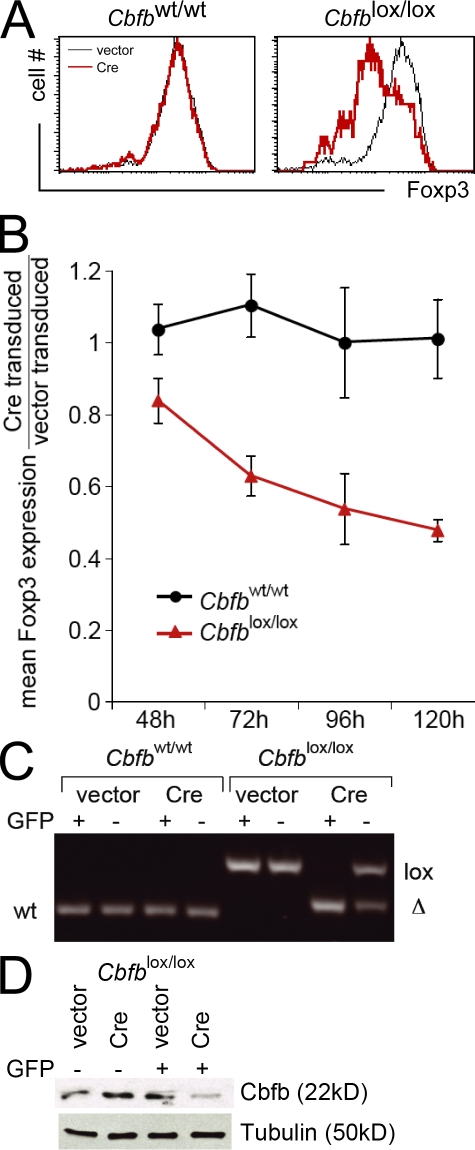
The activity of Runx proteins depends on the non-DNA binding subunit Cbfb (Durst and Hiebert, 2004), and we used the conditional deletion of Cbfb to probe the role of Runx proteins in the maintenance of Foxp3 expression in nT reg cells. CD4+ CD25+ LN T cells were sorted from control Cbfbwt/wt and Cbfblox/lox mice (Naoe et al., 2007), activated, and retrovirally transduced with vector-IRES-GFP or Cre-IRES-GFP. The impact of Cre-mediated Cbfb deletion was monitored by intracellular Foxp3 staining 4 d later and showed a selective decline in Cbfblox/lox cells transduced with Cre-IRES-GFP (red). Foxp3 expression remained unaffected in vector-transduced Cbfblox/lox cells (Fig. 3 A, black) and in Cbfbwt/wt cells transduced with either vector-IRES-GFP (Fig. 3 A, black) or Cre-IRES-GFP (Fig. 3 A, red). This effect was quantified as the ratio of Foxp3 expression in GFP+ Cre-transduced versus vector-transduced Cbfblox/lox (Fig. 3 B, red) and Cbfbwt/wt (Fig. 3 B, black) cells between 2 and 5 d after retroviral transduction and showed a progressive decline in Cre-transduced Cbfblox/lox nT reg cells (Fig. 3 B, red) but not in Cbfbwt/wt nT reg cells (Fig. 3 B, black; mean ± SD; n = 3). The effective deletion of the floxed Cbfb locus was monitored by genomic PCR (Fig. 3 C) and Cbfb protein expression by Western blotting (Fig. 3 D). These genetic data and the results obtained with the dominant-negative Runt domain make a strong case that Runx complexes are required for the maintenance of Foxp3 expression in nT reg cells.
Figure 3.
Genetic evidence for a role of Runx proteins in the maintenance of Foxp3 expression in nT reg cells. (A) CD4+ CD25+ LN T cells were sorted from control Cbfbwt/wt and Cbfblox/lox mice, activated for 18 h with anti-CD3/anti-CD28 beads in the presence of IL-2, and retrovirally transduced with vector-IRES-GFP (vector) or Cre-IRES-GFP (Cre). 4 d later, Foxp3 expression had declined selectively in Cbfblox/lox cells transduced with Cre-IRES-GFP (red). Foxp3 expression remained unaffected in vector-transduced Cbfblox/lox cells (black) and in Cbfbwt/wt cells transduced with either vector-IRES-GFP (black) or Cre-IRES-GFP (red). Data are representative of three independent experiments. (B) Time course of Foxp3 expression by GFP+ Cre-IRES-GFP–transduced relative to vector-IRES-GFP–transduced Cbfblox/lox nT reg cells (red) and Cbfbwt/wt nT reg cells (black; mean ± SD of three independent experiments). (C) GFP-positive and -negative nT reg cells were sorted 4 d after transduction with vector-IRES-GFP (vector) or Cre-IRES-GFP (Cre), and Cbfb loci were analyzed by genomic PCR. The position of bands corresponding to the wild-type (wt), floxed (lox), and deleted (Δ) locus are indicated. One of three independent experiments is shown. (D) GFP-positive and -negative Cbfblox/lox cells were sorted 4 d after transduction with vector-IRES-GFP (vector) or Cre-IRES-GFP (Cre), and Cbfb protein expression was analyzed by Western blotting. Tubulin was used as a loading control. One of three independent experiments is shown.
The Foxp3 locus as a target of Runx/Cbfb
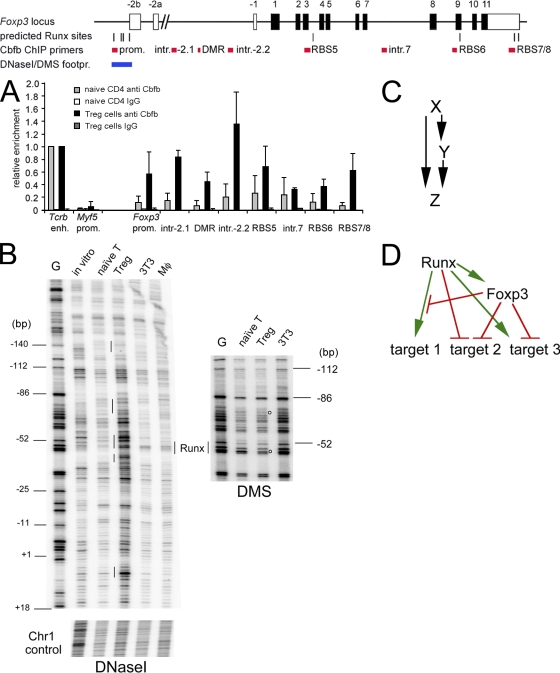
The experiments described in the previous sections link Runx proteins with the control of Foxp3 expression. To investigate whether this link is likely to be direct, we performed chromatin immunoprecipitation (ChIP) experiments using an antibody to Cbfb (Naoe et al., 2007), the shared subunit of all Runx protein complexes (Durst and Hiebert, 2004). We detected strong binding at the TCR-β chain enhancer, a known target of Runx/Cbfb binding (Naoe et al., 2007) in naive CD4 T cells and in ex vivo–isolated nT reg cells (Fig. 4 A). No binding was observed at the promoter of a negative control locus, Myf5 (Fig. 4 A). Interestingly, the Foxp3 promoter, which contains four predicted Runx binding sites (RBSs), was associated with Runx/Cbfb selectively in T reg cells but not in naive CD4 T cells (Fig. 4 A). T reg cell–specific Runx/Cbfb binding was also found at the predicted RBSs 5 and 7/8, whereas other potential Runx sites and a control site in intron 7 showed less binding (Fig. 4 A). Finally, Cbfb ChIP showed enrichment around the DMR upstream of Foxp3 exon −1 (primers intron 2.1, DMR, and intron 2.2). Further analysis by DNaseI footprinting showed that, compared with naive CD4 T cells, ex vivo–isolated nT reg cells had increased DNaseI accessibility of the Foxp3 promoter, which was visible as stronger bands in T reg samples (Fig. 4 B). This increased accessibility was selective because a control region on chromosome 1 was equally accessible in T reg cells and naive CD4 T cells. DMS footprinting showed protected (i.e., weaker) bands in T reg cells compared with naive CD4 T cells, which is indicative of the presence of DNA binding proteins in T reg cells (Fig. 4 B). One of the DMS-protected bands is within a predicted RBS in the Foxp3 promoter, 52 bp upstream of the Foxp3 transcription start site. These data establish the Foxp3 locus as a direct target for Runx/Cbfb in T reg cells.
Figure 4.
Runx and Foxp3 as components of a feed-forward loop. (A) Cbfb ChIP in naive CD4 T cells (gray bars) and ex vivo–isolated T reg cells (black bars). The Tcrb enhancer (enh.) and Myf5 promoter (prom.) are used as positive and negative controls, respectively. Predicted RBSs are indicated, and sites 2, 3, 4, and 5 are conserved between mouse and human. Regions within the Foxp3 locus examined for Cbfb binding are indicated in red. IgG is used instead of anti-Cbfb as a ChIP control. The region examined by footprint analysis is in blue. PCR signals are normalized to input and shown relative to the positive control site Tcrb enh. (mean ± SD; n = 3 independent experiments). (B) DNaseI (left) and DMS footprinting (right) of the Foxp3 promoter region. G, G sequencing reaction; in vitro, naked DNA; naive T, naive CD4 T cells; T reg, ex vivo–isolated T reg cells; 3T3, NIH 3T3 cells; Mφ, macrophages. Base positions are shown relative to the Foxp3 transcription start site. Note that T reg cells show increased DNaseI accessibility of the Foxp3 promoter (stronger bands in the T reg sample) but not of a control region on chromosome 1. Vertical lines indicate local DNaseI sensitivity in T reg cells. Open circles indicate DMS-protected (weaker) bands in T reg cells, which may indicate the position of DNA binding proteins. A predicted Runx site is indicated. Data are representative of an extensive analysis of both forward and reverse strands with primers for the proximal and distal Foxp3 promoter region using independent template preparations for DNaseI and DMS treatment. (C) Scheme of a feed-forward loop where X regulates Y and X and Y together regulate Z. (D) Model describing a relationship between Runx and Foxp3 where Runx acts on Foxp3 expression and Foxp3 either blocks Runx activity, converts Runx from an activator into a repressor, or cooperates with Runx to activate or repress downstream target genes.
Runx and Foxp3 as regulatory circuit components
Because Foxp3 protein cooperates with Runx proteins in the control of target genes (Ono et al., 2007), our demonstration that Foxp3 is itself regulated by Runx proteins points to a regulatory circuit in which Runx proteins are both inducers and interaction partners of Foxp3. Genetic feed-forward loops are significant network motifs in both prokaryotes and eukaryotes (Mangan and Alon, 2003) and involve two transcription factors: X regulates the expression of Y and both factors together regulate one or more downstream targets Z (Fig. 4 C). In the case described here, X is a Runx family transcription factor and Y is Foxp3. Runx and Foxp3 interact at regulatory DNA sequences or independently of DNA to control the expression of target genes (Fig. 4 D) like Il2 and Ifng (Ono et al., 2007). Feed-forward regulation is a recurring theme in the biology of Foxp3, which interacts not only with Runx but also with other transcription factors including Ror-γt (Ichiyama et al., 2008; Zhou et al., 2008) and NFAT (Wu et al., 2006). Runx1 and Foxp3 both interact with Ror-γt, indicating the potential for a complex regulatory network (Ichiyama et al., 2008; Zhou et al., 2008), and NFAT, like Runx, contributes to the regulation of Foxp3 expression (Mantel et al., 2006; Wu et al., 2006; Tone et al., 2008). Foxp3 replaces the NFAT partner AP1 (Wu et al., 2006) and, together, NFAT and Foxp3 regulate target genes both positively (Ctla4 and Il2ra) and negatively (Il2 and Ifng; Mantel et al., 2006). Depending on the sign of regulation between their elements, feed-forward circuits have the potential to fine-tune gene expression levels and to determine response parameters like on/off kinetics (Mangan and Alon, 2003). The identification of Foxp3 as a recurring element of feed-forward loops that involve transcription factors controlling T cell function suggests a distinct perspective, namely that Foxp3 subverts T cell regulatory circuits to establish T reg cell–specific gene activation and repression programmes.
Concluding remarks
Our experiments show that Runx proteins play an important role in the transcriptional regulation of Foxp3 during the induction of Foxp3 in conventional CD4 T cells and the maintenance of Foxp3 expression in established T reg cells. Our experiments do not speak to the relative contribution of Runx family members. However, Runx1 is predominant in nT reg cells as well as in naive CD4 T cells (Fig. S3). Runx1 is down-regulated after activation (Djuretic et al., 2007), but inducers of iT reg differentiation, such as TGF-β or PI3K/mTOR inhibitors, result in the reexpression of Runx1 as well as moderate Runx3 expression (Fig. S3). Runx proteins are known to physically interact with Foxp3 protein in established T reg cells (Ono et al., 2007), providing one mechanism by which Runt may affect nT reg cells. However, our data that Runx3 promotes iT reg cell differentiation by increasing the frequency of conventional CD4 T cells that initiate the expression of Foxp3 clearly show an impact of Runx proteins on Foxp3 expression by previously Foxp3 negative cells. This function is novel and distinct from the known role of Runx proteins as interactors of Foxp3 protein in nT reg cells (Ono et al., 2007). Together with the T reg cell–specific binding of Runx/Cbfb at the Foxp3 locus, these data argue that Runx proteins are bona fide regulators of Foxp3 expression.
MATERIALS AND METHODS
Mouse strains, cell sorting, culture, and viral infections.
Animal work was performed according to the Animals (Scientific Procedures) Act under the authority of project licences PPL70/5936 and PPL70/6845 issued by the Home Office, UK. Naive CD4+ CD25− CD62Lhigh LN T cells or CD4+ CD25+ T reg cells were sorted by flow cytometry and activated at 106/ml either with 200 ng/ml of plate-bound anti–TCR-β (H57) and 2 µg/ml of anti-CD28 (BD) or with anti-CD3/CD28 beads (Invitrogen) as previously described (Sauer et al., 2008). Exogenous IL2 was added where indicated. Activated T cells were retrovirally transduced with control mouse stem cell virus–IRES-GFP or Runx vectors (Telfer et al., 2004) by spin infection (90 min, 2,500 rpm, 35°C, no polybrene). Where indicated, activated cells were deprived of TCR signals by removing anti-CD3/CD28 beads or by moving the cells to fresh wells not coated with anti-TCR. 10 µg/ml LY294002, 25 nM rapamycin, or TGF-β was added as indicated. Cells were fixed and permeabilized (eBioscience) and stained with CD4 (Invitrogen) and Foxp3 as advised (eBioscience). Retrovirally transduced cells were fixed for 24 h to improve the retention of GFP. T reg cells from Cbfblox/lox mice (Naoe et al., 2007) were isolated by flow cytometry, activated, and infected with Cre-IRES-GFP or control IRES-GFP retrovirus. Deletion of Cbfb exon 5 was monitored by genomic PCR (Naoe et al., 2007) and Foxp3 expression was evaluated by intracellular staining.
Immunoblotting.
Immunoblotting was done as previously described (Sauer et al., 2008) using rabbit anti-Runx1/3 (Hayashi et al., 2000; gift from M. Satake, Tohoku University, Sendai, Japan) and donkey anti–rabbit IgG-HPR (GE Healthcare). Lamin (Santa Cruz Biotechnology, Inc.) or tubulin (Sigma-Aldrich) antibodies were used as loading controls.
Real time RT-PCR.
Total RNA was isolated using RNA-Bee (Tel-Test, Inc.) and reverse transcribed. PCR reactions included 2× SYBR PCR Master Mix (QIAGEN), 300 nM of primers, and 2 µl of complementary DNA as a template in a 50-µl reaction volume. Cycle conditions were 94° C for 8 min, 40 cycles of 94° C for 30 s, 55° C for 30 s, and 72° C for 1 min, followed by plate read. All primers amplified specific complementary DNAs with at least 95% efficiency. Data were normalized to the geometrical mean of two housekeeping genes, using the CT method as outlined in the Applied Biosystems protocol for RT-PCR (Sauer et al., 2008). Primer sequences were the following: Ywhaz, forward 5′-CGTTGTAGGAGCCCGTAGGTCAT-3′ and reverse 5′-TCTGGTTGCGAAGCATTGGG-3′; Ube, forward 5′-AGGAGGCTGATGAAGGAGCTTGA-3′ and reverse 5′-TGGTTTGAATGGATACTCTGCTGGA-3′; Runx1, forward 5′-GCCTCCTTGAACCACTCCAC-3′ and reverse 5′-GTTCTGCAGAGAGGCTGGTC-3′; Runx3, forward 5′-CGACCGCTTTGGAGACCTGC-3′ and reverse 5′-GCGTAGGGAAGGAGCGGTCA-3′; Cbfb, forward 5′-GGACCAGAGGAGCAAGTTCG-3′ and reverse 5′-CTGGAGAGACAGATTGGTTC-3′; and Pim1, forward 5′-CGAGTGTACAGTCCTCCAGAG-3′ and reverse 5′-GGTGCTGACACTCTGAAGAG-3′.
ChIP.
Cells were fixed with 1% PFA for 10 min at room temperature and the reaction was stopped by 125 mM glycine. Cells were washed twice in PBS at 4°C and lysed in 1% SDS, 10 mM EDTA, pH 8.0, 50 mM Tris-HCl, pH 8.1, and protease inhibitors (Roche), and chromatin was sonicated to a fragment size of 400–1,000 bp, cleared by centrifugation, diluted in 140 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% Na deoxycholate, and protease inhibitors, and incubated with blocked Protein A Dynabeads (Invitrogen) for 2 h at 4°C. Chromatin from 2 × 106 cell equivalents was incubated with Dynabeads saturated with anti-Cbfb or rabbit anti–mouse rabbit IgG at 4°C overnight. Immune complexes were washed twice in RIPA buffer (140 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% Na deoxycholate, and protease inhibitors), twice in high-salt RIPA (500 mM NaCl), once in LiCl RIPA (250 mM LiCl), and once in TE, pH 8.0, and eluted at 68°C overnight in 20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl, 50 µg/ml proteinase K, and 5 µg/ml RNase A. DNA was analyzed by SYBR green (QIAGEN) real-time PCR using the following primer sequences: Tcrb enhancer, forward 5′-AGAATGGCCACCTGCCATAG-3′ and reverse 5′-GGTGATAGCTAGAGGCTGAG-3′; Myf5 promoter, forward 5′-GGAGATCCGTGCGTTAAGAATCC-3′ and reverse 5′-CGGTAGCAAGACATTAAAGTTCCGTA-3′; Foxp3 promoter, forward 5′-TGAGGTTTGGAGCAGAAGGAAG-3′ and reverse 5′-TGTGAGACGTTGGAGGATCGC-3′; Foxp3 intron 1a, forward 5′-CAATTGCCTTCCAAAACCCTTCC-3′ and reverse 5′-ACCACCATTTCAGGCTGATACTGC-3′; Foxp3 DMR, forward 5′-ATGGACGTCACCTACCACATCCG-3′ and reverse 5′-CCCACAGGTTTCGTTCCGAGA-3′; Foxp3 intron 1b, forward 5′-GCCCTTCCCTCTTCTACATCCTCAT-3′ and reverse 5′-AGGAATGCAGTATGAACTCTGAGGGTT-3′; Foxp3 RBS5, forward 5′-TGATCAGCCTCCCACCACCTT-3′ and reverse 5′-CCGTGACTCTTAAGACCCTTCTATACC-3′; Foxp3 RBS6, forward 5′-AGACACAGCTTACGGCGGGT-3′ and reverse 5′-ACCCCCTACCCCACCTTTGTCT-3′; Foxp3 RBS7/8, forward 5′-ACTGACCCCAGTTCCCTACCCA-3′ and reverse 5′-CCAGCTGTCTGCTTATATGTTCCC-3′; and Foxp3 intron 9, forward 5′-AGTGGCATGCACCTGTAGTCCTGG-3′ and reverse 5′-TCCCCAACCGTGTTTCTTTGTG-3′.
In vivo footprinting.
Dimethylsulfate (DMS) and DNase I treatment of cells and naked DNA and ligation-mediated PCR were performed as described previously (Lefevre et al., 2005). 1 µg DNA was used as input for the ligation-mediated PCR. The following primers were positioned relative to the Foxp3 transcription start site: promoter reverse primer 1 (+116 to +92), biotin 5′-ATACCTCTCTGCCACTTTC-3′; primer 2 (+71 to +49), 5′-AGAGCAGGGACACTCGCTCACCT-3′; and primer 3 (+62 to +35), 5′-ACACTCGCTCACCTTGGTGAAGTGGACT-3′. The following control primers corresponded to a gene-free region on chromosome 1: primer 1, biotin 5′-ATGCCTCATCCTCTAGGGAAGATAT-3′; primer 2, 5′-TCTAGGGAAGATATGGCCTTGAA-3′; and primer 3, 5′-GGGAAGATATGGCCTTGAACATAGATG-3′.
DNA methylation analysis.
DNA methylation was assessed by bisulphite sequencing. 1.5 µg of genomic DNA was bisulfite converted using EZ-DNA Methylation kit (Zymo Research). Approximately 25 ng of bisulfite-converted DNA was amplified using gene-specific PCR primers (MethPrimer software; www.urogene.org/methprimer). Products were gel purified and cloned into pCRII vector (Invitrogen). Plasmid DNA from individual clones was mini-prepped and sequenced. Primer sequences were the following: DMR, forward 5′-TTTGTGGGGTAGATTATTTGTT-3′ and reverse 5′-AACCAACCAACTTCCTACACTA-3′; and intron 7, forward 5′-AGAGGTTGAAGGAGGAGTATTT-3′ and reverse 5′-ACTATCTATCCAATTCCCCAAC-3′.
Online supplemental material.
Fig. S1 provides information on the overexpression of Runx1 and Runx3 achieved by retroviral gene transfer. Fig. S2 shows that overexpression of Runt blocks Foxp3 induction in the presence as well as in the absence of exogenous IL-2. Fig. S3 summarizes the expression of Runx1 and Runx3 proteins in freshly isolated and cultured T cells and relates Runx expression to inducible Foxp3 expression.
Acknowledgments
We thank Dr. Masanobu Satake for Runx1/3 antibody, Dr. Nancy Speck for Runx1 antibody, Drs. Victor J. Dzau and Richard Pestell for plasmids, Dr. Masahiro Ono for discussion, and Eric O’Connor, Eugene Ng, and Phil Hexley for cell sorting.
This work was supported by the Medical Research Council, UK.
The authors have no financial conflict of interest.
Footnotes
Abbreviations used:
- Cbfb
- core-binding factor β
- ChIP
- chromatin immunoprecipitation
- DMR
- differentially methylated region
- iT reg
- induced T reg
- mTOR
- mammalian target of rapamycin
- nT reg
- natural T reg
- PI3K
- phosphatidyl inositol 3 kinase
- RBS
- Runx binding site
References
- Bruno L., Merkenschlager M. 2008. Directing T cell differentiation and function with small molecule inhibitors. Cell Cycle. 7:2296–2298 [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F., Pipkin M.E., Djuretic I.M., Levanon D., Lotem J., Lichtenheld M.G., Groner Y., Rao A. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206:51–59 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuretic I.M., Levanon D., Negreanu V., Groner Y., Rao A., Ansel K.M. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153 10.1038/ni1424 [DOI] [PubMed] [Google Scholar]
- Durst K.L., Hiebert S.W. 2004. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 23:4220–4224 10.1038/sj.onc.1207122 [DOI] [PubMed] [Google Scholar]
- Egawa T., Littman D.R. 2008. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9:1131–1139 10.1038/ni.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E., et al. 2007. Epigenetic control of the Foxp3 locus in regulatory T cells. PLoS Biol. 5:e38 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Lu Y., El Essawy B., Oukka M., Kuchroo V.K., Strom T.B. 2007. Contrasting effects of cyclosporine and rapamycin in de novo generation of alloantigen-specific regulatory T cells. Am. J. Transplant. 7:1722–1732 10.1111/j.1600-6143.2007.01842.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxhinasto S., Mathis D., Benoist C. 2008. The AKT–mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205:565–574 10.1084/jem.20071477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Natsume W., Watanabe T., Abe N., Iwai N., Okada H., Ito Y., Asano M., Iwakura Y., Habu S., et al. 2000. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J. Immunol. 165:6816–6824 [DOI] [PubMed] [Google Scholar]
- Ichiyama K., Yoshida H., Wakabayashi Y., Chinen T., Saeki K., Nakaya M., Takaesu G., Hori S., Yoshimura A., Kobayashi T. 2008. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J. Biol. Chem. 283:17003–17008 10.1074/jbc.M801286200 [DOI] [PubMed] [Google Scholar]
- Kim H.P., Leonard W.J. 2007. CREB/ATF-dependent T cell receptor–induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 204:1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P., Lacroix C., Tagoh H., Hoogenkamp M., Melnik S., Ingram R., Bonifer C. 2005. Differentiation-dependent alterations in histone methylation and chromatin architecture at the inducible chicken lysozyme gene. J. Biol. Chem. 280:27552–27560 10.1074/jbc.M502422200 [DOI] [PubMed] [Google Scholar]
- Mangan S., Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 100:11980–11985 10.1073/pnas.2133841100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel P.Y., Ouaked N., Rückert B., Karagiannidis C., Welz R., Blaser K., Schmidt-Weber C.B. 2006. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 176:3593–3602 [DOI] [PubMed] [Google Scholar]
- Naoe Y., Setoguchi R., Akiyama K., Muroi S., Kuroda M., Hatam F., Littman D.R., Taniuchi I. 2007. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med. 204:1749–1755 10.1084/jem.20062456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Yaguchi H., Ohkura N., Kitabayashi I., Nagamura Y., Nomura T., Miyachi Y., Tsukada T., Sakaguchi S. 2007. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 446:685–689 10.1038/nature05673 [DOI] [PubMed] [Google Scholar]
- Sauer S., Bruno L., Hertweck A., Finlay D., Leleu M., Spivakov M., Knight Z.A., Cobb B.S., Cantrell D., O’Connor E., et al. 2008. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA. 105:7797–7802 10.1073/pnas.0800928105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi R., Tachibana M., Naoe Y., Muroi S., Akiyama K., Tezuka C., Okuda T., Taniuchi I. 2008. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 319:822–825 10.1126/science.1151844 [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Littman D.R. 2004. Epigenetic gene silencing by Runx proteins. Oncogene. 23:4341–4345 10.1038/sj.onc.1207671 [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., Littman D.R. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633 10.1016/S0092-8674(02)01111-X [DOI] [PubMed] [Google Scholar]
- Telfer J.C., Hedblom E.E., Anderson M.K., Laurent M.N., Rothenberg E.V. 2004. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J. Immunol. 172:4359–4370 [DOI] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]
- Zhang F., Meng G., Strober W. 2008. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9:1297–1306 10.1038/ni.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Lopes J.E., Chong M.M., Ivanov I.I., Min R., Victora G.D., Shen Y., Du J., Rubtsov Y.P., Rudensky A.Y., et al. 2008. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 453:236–240 10.1038/nature06878 [DOI] [PMC free article] [PubMed] [Google Scholar]