Abstract
Diffuse large B cell lymphoma (DLBCL) is the most common type of lymphoma in humans. The aggressive activated B cell–like (ABC) subtype of DLBCL is characterized by constitutive NF-κB activity and requires signals from CARD11, BCL10, and the paracaspase MALT1 for survival. CARD11, BCL10, and MALT1 are scaffold proteins that normally associate upon antigen receptor ligation. Signal-induced CARD11–BCL10–MALT1 (CBM) complexes couple upstream events to IκB kinase (IKK)/NF-κB activation. MALT1 also possesses a recently recognized proteolytic activity that cleaves and inactivates the negative NF-κB regulator A20 and BCL10 upon antigen receptor ligation. Yet, the relevance of MALT1 proteolytic activity for malignant cell growth is unknown. Here, we demonstrate preassembled CBM complexes and constitutive proteolysis of the two known MALT1 substrates in ABC-DLBCL, but not in germinal center B cell–like (GCB) DLBCL. ABC-DLBCL cell treatment with a MALT1 protease inhibitor blocks A20 and BCL10 cleavage, reduces NF-κB activity, and decreases the expression of NF-κB targets genes. Finally, MALT1 paracaspase inhibition results in death and growth retardation selectively in ABC-DLBCL cells. Thus, our results indicate a growth-promoting role for MALT1 paracaspase activity in ABC-DLBCL and suggest that a pharmacological MALT1 protease inhibition could be a promising approach for lymphoma treatment.
Diffuse large B cell lymphoma (DLBCL) accounts for 30–40% of all human lymphoid malignancies (Anderson et al., 1998; Coiffier, 2001). Gene expression profiling has identified distinct DLBCLs subtypes that differ in their oncogenic mechanisms. The two main categories are activated B cell–like (ABC) DLBCL, and germinal center B cell–like (GCB) DLBCL (Alizadeh et al., 2000; Rosenwald et al., 2002, 2003; Savage et al., 2003; Wright et al., 2003). The ABC subtype shows the most aggressive clinical behavior and a gene expression signature that corresponds to the profile of B lymphocytes stimulated through their antigen receptor (B cell receptor). Its key feature is the activation of the NF-κB signaling pathway (Alizadeh et al., 2000; Davis et al., 2001).
NF-κB proteins are ubiquitously expressed transcription factors (Hayden and Ghosh, 2008). A large variety of physiological stimuli can rapidly activate NF-κB through a canonical IκB kinase (IKK)–dependent pathway to induce expression of target genes that include potent survival factors, cell cycle regulators, and growth-promoting cytokines. ABC-DLBCL cells exhibit uncontrolled and stimulus-independent activation of the IKK–NF-κB pathway (Davis et al., 2001; Lam et al., 2005) and small molecule IKK inhibitors are selectively toxic to ABC but not to GCB-type DLBCL cells in vitro (Davis et al., 2001; Lam et al., 2005). These findings marked the NF-κB pathway as an attractive target for ABC-DLBCL therapy. Yet, as the IKK–NF-κB axis is ubiquitously expressed and contributes to cellular homeostasis in multiple tissues, a general IKK–NF-κB inhibition is associated with significant toxicities in vivo, and thus not ideal for lymphoma treatment (Baud and Karin, 2009).
Lymphocyte-specific IKK and NF-κB activation in response to normal antigen receptor signaling requires the immune cell–restricted CARD11–BCL10–MALT1 (CBM) complex consisting of the adapter proteins CARD11 (also called CARMA1) and BCL10 and the paracaspase MALT1 (Ruland and Mak, 2003; Thome, 2004). Antigen receptor ligation induces a rapid assembly of the CBM complex at the plasma membrane by recruiting the constitutively interacting proteins BCL10 and MALT1 to CARD11. The CBM complex is then responsible for a further recruitment of IKKs and other factors into distinct microdomains to trigger context-specific IKK activation for NF-κB–mediated lymphocyte survival and proliferation (Thome, 2004; Rawlings et al., 2006).
Oncogenic gain of function mutations of CARD11 that enforce NF-κB activation are recurrently detected in ABC-DLBCL biopsies (Lenz et al., 2008; Compagno et al., 2009; Kato et al., 2009). Additional genetic events are thought to induce aberrant CBM activity in other cases, as CARD11, BCL10, and MALT1 are all required for the survival and expansion of ABC-DLBCL cells (Ngo et al., 2006). Thus, a selective interference with CBM signaling could be a promising strategy for ABC-DLBCL treatment (Ngo et al., 2006) that would not inhibit the IKK–NF-κB pathway in nonlymphoid cells.
As CARD11 and BCL10 are scaffold proteins without enzymatic function, they are not ideal drug targets. However, the paracaspase MALT1 contains a caspase-like domain (Uren et al., 2000) that possesses inducible arginine-specific cysteine protease activity, in addition to protein–protein interaction motifs (Coornaert et al., 2008; Rebeaud et al., 2008). The MALT1 paracaspase activity is physiologically activated upon antigen receptor stimulation or after PMA and ionomycin (PMA + Iono) treatment and requires CARD11- and BCL10-mediated MALT1 oligomerization (Coornaert et al., 2008; Rebeaud et al., 2008). Two MALT1 substrates have been identified. Activated MALT1 cleaves its binding partner BCL10 and the ubiquitin-editing enzyme A20, an inhibitor of the IKK–NF-κB pathway (Beyaert et al., 2000) that emerges as an important tumor suppressor in the B cell lineage (Compagno et al., 2009; Kato et al., 2009; Malynn and Ma, 2009; Schmitz et al., 2009). MALT1 protease activity is responsible for optimal NF-κB activation and IL-2 production in antigen receptor–stimulated T cells, suggesting that this activity is key for full physiological lymphocyte activation (Coornaert et al., 2008). Whether MALT1 protease activity plays a functional role in malignant lymphocyte growth is currently unknown.
Here, we demonstrate that ABC-DLBCL cells, but not GCB-DLBCL cells, possess constitutively assembled CBM complexes and that A20 and BCL10 are continuously and selectively processed in ABC-DLBCL cells. Lymphoma cell treatment with a MALT1 protease inhibitor blocks A20 and BCL10 cleavage, reduces NF-κB activity and decreases the expression of the NF-κB targets BCL-XL, IL-6, and IL-10. Finally, MALT1 paracaspase inhibition results in ABC-DLBCL cell death and growth retardation. These results indicate a growth-promoting role for MALT1 paracaspase activity, specifically in ABC-DLBCL cells and highlight MALT1 protease activity as a potential target for pharmacological treatment of ABC-DLBCL.
RESULTS AND DISCUSSION
Constitutive and selective CBM complex assembly and MALT1 protease activity in ABC-DLBCL cells
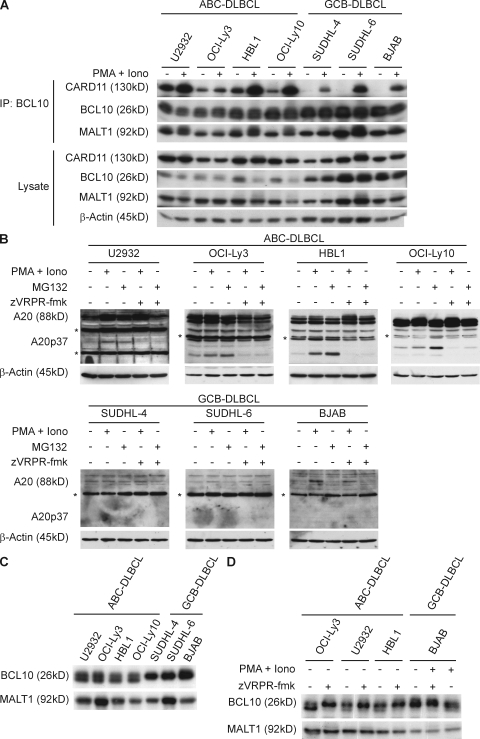
The MALT1 protease is physiologically activated upon antigen receptor triggering and subsequent assembly of the CBM complex (Coornaert et al., 2008; Rebeaud et al., 2008). As ABC-DLBCL cells require constitutive signals from the CBM complex for survival and expansion (Ngo et al., 2006), we first investigated whether CBM complexes might be preassembled in ABC-DLBCL. To this end, we used well-characterized ABC- or GCB-DLBCL cell lines (Davis et al., 2001; Ngo et al., 2006; Lenz et al., 2008) and immunoprecipitated BCL10 in the absence of stimulation and upon stimulation with PMA + Iono. We then tested the association of BCL10 with MALT1 or CARD11 by immunoblotting. BCL10 and MALT1 bind constitutively to each other in all DLBCL cell lines tested (Fig. 1 A). However, stimulus-independent trimolecular association of BCL10/MALT1 with CARD11 was only detected in the ABC-DLBCL cell lines OCI-Ly3, HBL1, U2932, and OCI-Ly10, but not in the GCB-DLBCL lines SUDHL-4, SUDHL-6, and BJAB (Fig. 1 A), although PMA + Iono treatment could induce CBM complexes in all cell lines. These results indicate constitutive CBM complex formation selectively in ABC-DLBCL.
Figure 1.
Constitutive CBM complex assembly and MALT1 protease activity in ABC-DLBCL. (A) ABC-DLBCL or GCB-DLBCL cell lines were left untreated or stimulated with PMA + Iono for 20 min. Lysates were immunoprecipitated with anti-BCL10 and immunocomplexes were analyzed for the presence for CARD11, BCL10, or MALT1 by immunoblotting (top). Total cellular contents of CARD11, BCL10, MALT1, or β-actin were analyzed in lysates without immunoprecipitation (bottom). (B–D) Immunoblot analysis for the presence of A20 (B) or BCL10 (C and D) cleavage products. (B) A20 processing. Individual cell lines were either left untreated, stimulated for 2 h with PMA + Iono or incubated for 2 h in the presence of MG132; zVRPR-fmk was added 30 min before stimulation where indicated. Filled arrow, full length A20; open arrow, A20p37 cleavage product; *, nonspecific band. β-Actin blotting served as a loading control. (C) BCL10 processing. Lysates from nonstimulated ABC-DLBCL or GCB-DLBCL cell lines were immunoblotted with anti-BCL10. (D) DLBCL cells were left untreated or treated with zVRPR-fmk. Filled arrow, full length BCL10; open arrow, BCL10Δ5 fragment. The MALT1 immunoblot indicates equal loading. Data are one representative of three (A and D) or four (B and C) independent experiments.
To study whether the preassembled CBM complexes in ABC-DLBCL cells might be associated with constitutive MALT1 protease activity, we next analyzed the processing of the two known MALT1 substrates A20 and BCL10 (Fig. 1, B–D). MALT1 can cleave A20 after arginine 439, generating the short-lived A20-p37 fragment that is rapidly degraded by the proteasome (Coornaert et al., 2008). A20 is an NF-κB target gene, and as such it is part of a negative feedback loop (Chen et al., 2006; Hayden and Ghosh, 2008). In contrast to the GCB-DLBCL cells, the ABC-DLBCL cells exhibit high expression levels of A20 (Fig. 1 B). Intriguingly, the A20-p37 cleavage product, detected by a monoclonal antibody to the C terminus of A20, is constitutively present in all ABC-DLBCL cell lines. Moreover, proteasome inhibition with MG132 to block A20-p37 degradation (Coornaert et al., 2008) further augmented A20-p37 concentrations in these cells. Likewise, lymphoma cell treatment with PMA + Iono to exogenously induce MALT1 protease activity (Coornaert et al., 2008; Rebeaud et al., 2008) also increased A20-p37 concentrations above constitutive levels. To test whether MALT1 is responsible for A20 cleavage in DLBCL cells, we selectively blocked MALT1 protease function without affecting MALT1 protein expression. To this end, we used the recently developed antagonistic tetrapeptide zVRPR-fmk, which is an irreversible inhibitor of MALT1 protease activity in vitro and in cultured cells (Rebeaud et al., 2008). Treatment of ABC-DLBCL cells with zVRPR-fmk strongly inhibited both constitutive as well as PMA + Iono induced A20 processing, indicating that the MALT1 paracaspase is constitutively active in these cells and responsible for A20 cleavage.
Next, we studied the proteolysis of BCL10, which is present in both the ABC and GCB DLBCL cell lines (Fig. 1, A and C). BCL10 cleavage by MALT1 generates the BCL10Δ5 fragment that lacks the five carboxy-terminal amino acids from BCL10 and migrates slightly faster in gels (Rebeaud et al., 2008). The BCL10Δ5 fragment was constitutively present in all ABC-DLBCL lines, but not in GCB-DLBCL cells (Fig. 1 C), although PMA + Iono treatment induced BCL10 cleavage in the GCB-DLBCL line BJAB (Fig. 1 D). The MALT1 inhibitor zVRPR-fmk blocked both the constitutive processing of BCL10 in the ABC-DLBCL cells (Fig. 1 D), as well as PMA + Iono–induced BCL10 processing in the GCB-DLBCL line, again indicating that MALT1 is responsible for BCL10 cleavage. Together, these results indicate that the MALT1 paracaspase is preactivated and uncoupled from antigenic stimulation in ABC-DLBCL, but not in GCB-DLBCL cells.
MALT1 paracaspase inhibition reduces NF-κB activity in ABC-DLBCL cells
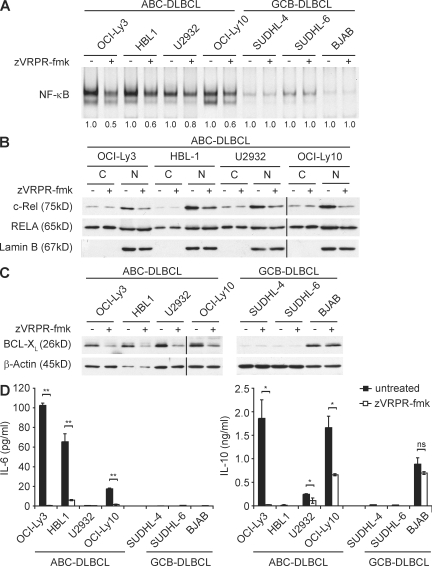
MALT1-mediated proteolysis is required for full NF-κB activation and optimal production of the NF-κB–dependent factor IL-2 in antigen receptor–stimulated T cells (Coornaert et al., 2008; Rebeaud et al., 2008). In line with previously published data (Davis et al., 2001), the ABC-DLBCL lines OCI-Ly3, HBL1, U2932, and OCI-Ly10 exhibit constitutive NF-κB activity that is much higher than in GCB-DLBCL lines (Fig. 2 A). Intriguingly, cell treatment with the MALT1 antagonistic peptide zVRPR-fmk specifically reduced NF-κB DNA-binding in ABC-DLBCL cells, indicating that the high constitutive NF-κB activity is in part controlled by MALT1 protease function. To further characterize the effects of MALT1 protease inhibition on NF-κB signaling in ABC-DLBCL, we separated nuclear from cytoplasmic extracts before and after zVRPR-fmk treatment. Consistent with high NF-κB DNA binding activity, all ABC-DLBCL cell lines exhibit profound levels of the c-Rel and RELA subunits in the nucleus (Fig. 2 B). Cell treatment with the MALT1 inhibitor reduced nuclear c-Rel levels in all ABC-DLBCL lines substantially, although it only marginally affected RELA. These results further indicate that MALT1 protease activity is required for the full constitutive NF-κB signal in ABC-DLBCL cells. These data are in line with the known selective requirement of MALT1 in the control of c-Rel in B cells (Ferch et al., 2007).
Figure 2.
Inhibition of MALT1 protease activity interferes with NF-κB DNA binding in ABC-DLBCL. (A) Gel-mobility-shift assay of nonstimulated ABC-DLBCL or GCB-DLBCL cells left untreated or incubated with zVRPR-fmk for 48 h. Nuclear extracts were analyzed by electrophoretic mobility shift assay for NF-κB DNA-binding activity. NF-κB signals were quantified by densitometry and are expressed below the respective lanes as values relative to untreated cells. (B) Immunoblot analysis of the abundance of c-Rel and RELA in cytoplasmatic (C) and nuclear (N) extracts of ABC-DLBCL cells left untreated or incubated with zVRPR-fmk for 48 h. Western blotting for the nuclear marker protein Lamin B confirms successful separation into cytoplasmic and nuclear extracts and equal nuclear protein loading. (C) Immunoblot analysis of BCL-XL expression in untreated or zVRPR-fmk–treated (48 h) ABC-DLBCL or GCB-DLBCL cell lines. β-Actin blot serves as a loading control. (D) Concentrations of secreted IL-6 and IL-10 were determined in the supernatants of untreated or zVRPR-fmk–treated (48 h) ABC-DLBCL and GCB-DLBCL cell lines by ELISA. Data from one out of three (A, C, and D) or four (B) independent experiments with similar results are shown.
To study the consequences of the MALT1 protease inhibition on gene expression, we next investigated the expression of the NF-κB-controlled antiapoptotic factor BCL-XL in DLBCL cells as a marker of NF-κB activity (Chen et al., 2000). Treatment with the MALT1 inhibitor zVRPR-fmk substantially reduced BCL-XL protein levels in all ABC-DLBCL cells (Fig. 2 C). In contrast, the GCB-DLBCL cell lines were either lacking constitutive BCL-XL expression (SUDHL-4 and SUDHL-6) or, as in the case of BJAB, BCL-XL expression was not sensitive to MALT1 inhibition.
A recent study has shown that the NF-κB activity triggers the production of the cytokines IL-6 and IL-10 in ABC-DLBCL, which contribute to malignant cell growth in an autocrine manner (Lam et al., 2008). We therefore investigated whether the secretion of these factors might be MALT1 protease dependent. The ABC-DLBCL lines OCI-Ly3, HBL1, and OCI-Ly10 produce substantial amounts of IL-6, which was significantly inhibited by zVRPR-fmk (Fig. 2 D). Moreover, the ABC-DLBCL lines OCI-Ly3, U2932, and OCI-Ly10 secreted IL-10, which was also sensitive to MALT1 protease inhibition (Fig. 2 D). The IL-10 production by the GCB-DLBCL line BJAB was, in contrast, not significantly reduced by MALT1 protease inhibition. Collectively, we can conclude that MALT1 paracaspase inhibition also impairs expression of NF-κB targets such as BCL-XL, IL-6, and IL-10 specifically in ABC-DLBCL cells.
MALT1 protease activity is required for survival and proliferation of ABC-DLBCL cells
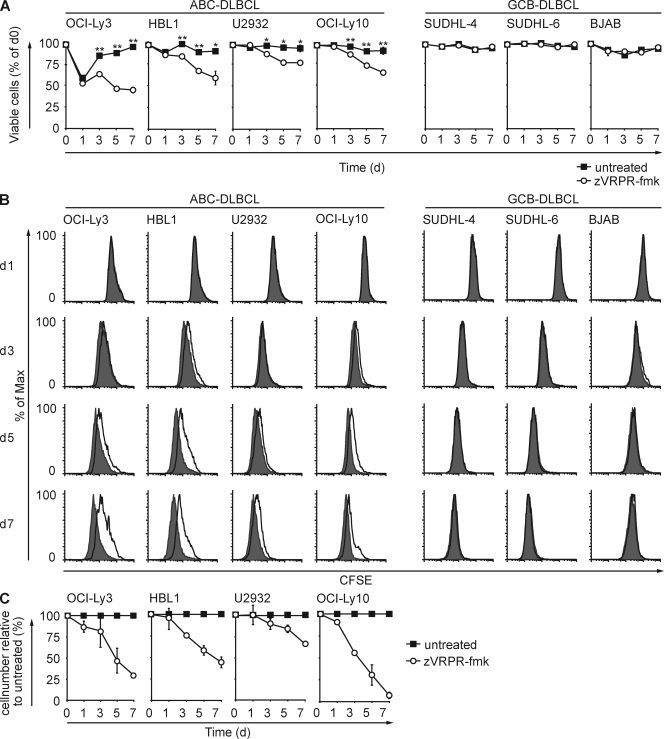
Knockdown experiments with siRNAs have shown that the presence of the MALT1 protein is required for ABC-DLBCL cell growth (Ngo et al., 2006). Even a prolonged incubation of GCB-DLBCL cells with zVRPR-fmk did not affect the survival of these tumor cells (Fig. 3 A) excluding general cytotoxic effects. Yet, the frequency of viable ABC-DLBCL cells declined significantly and continuously over time in all four ABC-DLBCL lines compared with untreated controls.
Figure 3.
MALT1 protease inhibition selectively affects survival and proliferation of ABC-DLBCL cells. (A) Cell viability. ABC-DLBCL or GCB-DLBCL cell lines either left untreated or cultured in the presence of zVRPR-fmk for the indicated times were stained with Annexin V and propidium iodide, and viability was subsequently determined by flow cytometry. Depicted is the mean percentage of viable cells ± SD normalized to day 0 from one representative experiment out of three. *, P < 0.05; **, P < 0.01. (B) Cell proliferation. DLBCL cell lines (5 × 104) labeled with CFSE and cultured in the absence (shaded histograms) or presence of zVRPR-fmk (open histograms) for the indicated times; cell division was tracked by flow-cytometric analysis of CFSE fluorescence in 104 viable cells. Data are representative of three independent experiments. (C) Total cell numbers. ABC-DLBCL cell were cultured with our without zVRPR-fmk, and total viable cell numbers were counted at the indicated time points. Depicted is the percentage of absolute viable cell numbers ± SD after zVRPR-fmk treatment relative to untreated cells. Shown is the mean ± SD from three independent experiments.
To determine the consequences of zVRPR-fmk treatment on DLBCL cell proliferation, the cells were labeled with CFSE and division was tracked by CFSE dilution measurement in the viable population (Fig. 3 B). Similar to cell viability, the GCB-DLBCL cell division rate was unaffected by MALT1 protease inhibition. In contrast, zVRPR-fmk treatment significantly impaired the proliferation of all ABC-DLBCL cell lines. Thus, the MALT1 protease inhibitor zVRPR-fmk reduces both survival and division selectively in ABC-DLBCL cells but has no apparent cytotoxic effects on GCB-DLBCL cells.
To quantify the effects of MALT1 paracaspase inhibition on overall ABC-DLBCL cell growth, we finally compared the absolute cell numbers in the presence or absence of the MALT1 antagonistic peptide (Fig. 3 C). Intriguingly, after 1 wk of treatment, the combined effects on cell survival and proliferation reduced the total ABC-DLBCL cell numbers between ∼35% (U2932) and 95% (OCILy10; Fig. 3 C). Together, these data reveal a vital role for MALT1 protease activity in ABC-DLBCL cell growth, which can be blocked by pharmacological means.
Concluding remarks
A hallmark of the aggressive ABC subtype of DLBCL is the constitutive activation of the NF-κB pathway that occurs through various genetic events (Alizadeh et al., 2000; Lenz et al., 2008; Compagno et al., 2009; Kato et al., 2009). Although the respective upstream mechanism differs between individual cases, the CBM complex is required for ABC-DLBCL cell survival and expansion (Ngo et al., 2006; Lenz et al., 2008; Compagno et al., 2009). As markers for constitutive MALT1 paracaspase activity, we observed selective and stimulus-independent proteolysis of A20 and BCL10 in ABC-DLBCL. The functional role of processed BCL10 in lymphomagenesis is still unclear. Yet, MALT1-mediated cleavage of the tumor suppressor A20 might be functionally relevant, as A20 can limit malignant B cell growth (Kato et al., 2009; Malynn and Ma, 2009). All ABC-DLBCL lines that we used (OCI-Ly3, HBL1, U2932, and OCI-Ly10) are heterozygous at the TNFAIP3 gene locus, which encodes for A20, and thus contain one WT allele (Compagno et al., 2009). Consistently, all lines express full-length A20 that is prone to proteolysis. Although MALT1 activity does not deplete the total cellular A20 content, it cleaves the A20 molecules in the direct vicinity of activated CBM and IKK complexes (Coornaert et al., 2008). We hypothesize that these A20 molecules are responsible for NF-κB inhibition and growth control. Thus, paracaspase-mediated cleavage of A20 could be one mechanism of A20 inactivation in addition to the genomic mutations that are detected in other subsets of B cell lymphoma (Compagno et al., 2009; Kato et al., 2009; Schmitz et al., 2009).
Although additional functionally relevant MALT1 substrates may exist, our report represents the first non–small interfering RNA–based approach that targets a component of the CBM complex to study effects on malignant cell growth. We demonstrate that MALT1 paracaspase inhibition down-regulates NF-κB signaling and results in growth inhibition specifically in CBM/IKK-dependent ABC-DLBCL cells. MALT1 is the only known paracaspase in mammals, and proteases are ideal targets for the development of small molecule inhibitors with several clinically successful examples (Turk, 2006). MALT1-deficient mice exhibit mainly immunological defects, but are otherwise viable and healthy (Ruefli-Brasse et al., 2003; Ruland et al., 2003). Thus, the development of MALT1-specific small molecule protease inhibitors could lead to novel targeted therapeutics for lymphomas with aberrant MALT1 activity, which are expected to have limited toxic side effects in vivo.
MATERIALS AND METHODS
Cell culture and reagents.
All cell lines were cultured as previously described (Davis et al., 2001). In brief, OCI-Ly3 and OCI-Ly10 were maintained in Iscove's modified essential medium with 20% heparinized human plasma, penicillin, streptomycin, and β-mercaptoethanol. HBL1, U2932, SUDHL-4, SUDHL-6, and BJAB cell lines were maintained in RPMI 1640 medium supplemented with 10% FCS, l-glutamine, penicillin, streptomycin, and β-mercaptoethanol. The cell lines OCI-Ly3, HBL1, U2932, and OCI-Ly10 have recently been shown to be hemizygous for TNFAIP3 (Compagno et al., 2009). Moreover, several cell lines harbor additional mutations in NF-κB pathway components (OCI-Ly3, CARD11; OCI-Ly10, RANK; U2932, TAK1; Compagno et al., 2009). zVRPR-fmk (Alexis Biochemicals) was solved in ddH2O at a concentration of 75 µM throughout all experiments. MG132 and PMA + Iono (Sigma-Aldrich) were reconstituted in DMSO (final DMSO concentration 0.1% or 0.01%, respectively).
Survival and proliferation assays.
DLBCL cells were seeded in a density of 106 cells/ml in a 96-well plate and cultured in the absence or presence of zVRPR-fmk (75 µM), as previously described (Rebeaud et al., 2008). Cell viability was quantified by flow cytometry after Annexin V and propidium iodide (eBioscience) staining. Cell proliferation was determined by flow cytometry after CFSE (Sigma-Aldrich) staining. Data were analyzed with FlowJo software (Tree Star, Inc.) according to standard protocols.
Immunoblot.
Standard Western blot extracts were generated using Triton X-100 buffer (1% Triton X-100, 50 mM TRIS, pH 8.0, 150 mM NaCl, 10 mM NaF, 1 mM NaVO4 and protease inhibitors (Calbiochem)). For fractionation into cytoplasmic and nuclear extracts, lymphocytes were first lysed using Buffer A (0.2% Nonidet P40, 10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and protease inhibitors). Protein extracts of pelleted nuclei were generated using RIPA buffer (0.5 M Tris-HCl, pH 7.4, 1.5 M NaCl, 2.5% deoxycholic acid, 10% Nonidet P40, 10 mM EDTA, 10 mM NaF, 1 mM NaVO4, and protease inhibitors).
Coimmunoprecipitation.
DLBCL cells were lysed in coimmunoprecipitation buffer (0.2% Nonidet P40, 150 mM NaCl, 50 mM Hepes, pH 7.5, 1 mM glycerol, 10 mM NaF, 8 mM β-glycero-phosphate, 1 mM Na3VO4, and protease inhibitors). Immunoprecipitation was performed overnight at 4°C, and after incubation of sepharose beads (GE Healthcare) precipitates were boiled and analyzed by immunoblot.
Gel-mobility-shift assay.
Nuclear extracts from 5 × 106 DLBCL cells were prepared using standard methods (Ferch et al., 2007). Concentrations were adjusted according to Bradford assay and nuclear protein extracts (2.5 µg) were incubated in 20-µl binding buffer (100 mM Tris, pH 7.5, 500 mM KCl, 10 mM dithiothreitol and 1 µg poly (dI-dC)) with IRDye700-labeled, double-stranded oligonucleotide probes (NF-κB, 5′-AGTTGAGGGGACTTTCCCAGGC–3′). Upon electrophoretic separation on a 5% polyacrylamide gel, EMSAs were analyzed using Odyssey Infrared Imaging System (Licor Biosciences) and Software for densitometric quantification.
ELISA.
ELISAs were performed using human IL-6 and IL-10 ELISA sets (BD) according to manufacturer’s protocol.
Statistics.
P-values were determined by applying Student’s two-tailed t test for independent samples, assuming equal variances on all experimental datasets using the Microsoft excel t test calculator.
Acknowledgments
This work was supported by a grant of Deutsche Krebshilfe to D.K and SFB grants from Deutsche Forschungsgemeinschaft and by a Max-Eder-Program grant from Deutsche Krebshilfe to J. Ruland.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ABC
- activated B cell–like
- CBM
- CARD11–BCL10–MALT1
- DLBCL
- diffuse large B cell lymphoma
- GCB
- germinal center B cell–like
- IKK
- IκB kinase
- PMA + Iono
- PMA and ionomycin
References
- Alizadeh A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X., et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- Anderson J.R., Armitage J.O., Weisenburger D.D. 1998. Epidemiology of the non-Hodgkin’s lymphomas: distributions of the major subtypes differ by geographic locations. Non-Hodgkin’s Lymphoma Classification Project. Ann. Oncol. 9:717–720 10.1023/A:1008265532487 [DOI] [PubMed] [Google Scholar]
- Baud V., Karin M. 2009. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 8:33–40 10.1038/nrd2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaert R., Heyninck K., Van Huffel S. 2000. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem. Pharmacol. 60:1143–1151 10.1016/S0006-2952(00)00404-4 [DOI] [PubMed] [Google Scholar]
- Chen C., Edelstein L.C., Gélinas C. 2000. The Rel/NF-kappaB family directly activates expression of the apoptosis inhibitor Bcl-x(L). Mol. Cell. Biol. 20:2687–2695 10.1128/MCB.20.8.2687-2695.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Bhoj V., Seth R.B. 2006. Ubiquitin, TAK1 and IKK: is there a connection? Cell Death Differ. 13:687–692 10.1038/sj.cdd.4401869 [DOI] [PubMed] [Google Scholar]
- Coiffier B. 2001. Diffuse large cell lymphoma. Curr. Opin. Oncol. 13:325–334 10.1097/00001622-200109000-00003 [DOI] [PubMed] [Google Scholar]
- Compagno M., Lim W.K., Grunn A., Nandula S.V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A., et al. 2009. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B cell lymphoma. Nature. 459:717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z.J., Marynen P., Beyaert R. 2008. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat. Immunol. 9:263–271 10.1038/ni1561 [DOI] [PubMed] [Google Scholar]
- Davis R.E., Brown K.D., Siebenlist U., Staudt L.M. 2001. Constitutive nuclear factor κB activity is required for survival of activated B cell–like diffuse large B cell lymphoma cells. J. Exp. Med. 194:1861–1874 10.1084/jem.194.12.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferch U., zum Büschenfelde C.M., Gewies A., Wegener E., Rauser S., Peschel C., Krappmann D., Ruland J. 2007. MALT1 directs B cell receptor-induced canonical nuclear factor-kappaB signaling selectively to the c-Rel subunit. Nat. Immunol. 8:984–991 10.1038/ni1493 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell. 132:344–362 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Kato M., Sanada M., Kato I., Sato Y., Takita J., Takeuchi K., Niwa A., Chen Y., Nakazaki K., Nomoto J., et al. 2009. Frequent inactivation of A20 in B cell lymphomas. Nature. 459:712–716 [DOI] [PubMed] [Google Scholar]
- Lam L.T., Davis R.E., Pierce J., Hepperle M., Xu Y., Hottelet M., Nong Y., Wen D., Adams J., Dang L., Staudt L.M. 2005. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin. Cancer Res. 11:28–40 [PubMed] [Google Scholar]
- Lam L.T., Wright G., Davis R.E., Lenz G., Farinha P., Dang L., Chan J.W., Rosenwald A., Gascoyne R.D., Staudt L.M. 2008. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-kappaB pathways in subtypes of diffuse large B-cell lymphoma. Blood. 111:3701–3713 10.1182/blood-2007-09-111948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G., Davis R.E., Ngo V.N., Lam L., George T.C., Wright G.W., Dave S.S., Zhao H., Xu W., Rosenwald A., et al. 2008. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 319:1676–1679 10.1126/science.1153629 [DOI] [PubMed] [Google Scholar]
- Malynn B.A., Ma A. 2009. A20 takes on tumors: tumor suppression by an ubiquitin-editing enzyme. J. Exp. Med. 206:977–980 10.1084/jem.20090765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo V.N., Davis R.E., Lamy L., Yu X., Zhao H., Lenz G., Lam L.T., Dave S., Yang L., Powell J., Staudt L.M. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 441:106–110 10.1038/nature04687 [DOI] [PubMed] [Google Scholar]
- Rawlings D.J., Sommer K., Moreno-García M.E. 2006. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat. Rev. Immunol. 6:799–812 10.1038/nri1944 [DOI] [PubMed] [Google Scholar]
- Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E.M., Rufer N., et al. 2008. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 9:272–281 10.1038/ni1568 [DOI] [PubMed] [Google Scholar]
- Rosenwald A., Wright G., Chan W.C., Connors J.M., Campo E., Fisher R.I., Gascoyne R.D., Muller-Hermelink H.K., Smeland E.B., Giltnane J.M., et al. ; Lymphoma/Leukemia Molecular Profiling Project 2002. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346:1937–1947 10.1056/NEJMoa012914 [DOI] [PubMed] [Google Scholar]
- Rosenwald A., Wright G., Leroy K., Yu X., Gaulard P., Gascoyne R.D., Chan W.C., Zhao T., Haioun C., Greiner T.C., et al. 2003. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 198:851–862 10.1084/jem.20031074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruefli-Brasse A.A., French D.M., Dixit V.M. 2003. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 302:1581–1584 10.1126/science.1090769 [DOI] [PubMed] [Google Scholar]
- Ruland J., Mak T.W. 2003. Transducing signals from antigen receptors to nuclear factor kappaB. Immunol. Rev. 193:93–100 10.1034/j.1600-065X.2003.00049.x [DOI] [PubMed] [Google Scholar]
- Ruland J., Duncan G.S., Wakeham A., Mak T.W. 2003. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 19:749–758 10.1016/S1074-7613(03)00293-0 [DOI] [PubMed] [Google Scholar]
- Savage K.J., Monti S., Kutok J.L., Cattoretti G., Neuberg D., De Leval L., Kurtin P., Dal Cin P., Ladd C., Feuerhake F., et al. 2003. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 102:3871–3879 10.1182/blood-2003-06-1841 [DOI] [PubMed] [Google Scholar]
- Schmitz R., Hansmann M.L., Bohle V., Martin-Subero J.I., Hartmann S., Mechtersheimer G., Klapper W., Vater I., Giefing M., Gesk S., et al. 2009. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 206:981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M. 2004. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat. Rev. Immunol. 4:348–359 10.1038/nri1352 [DOI] [PubMed] [Google Scholar]
- Turk B. 2006. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 5:785–799 10.1038/nrd2092 [DOI] [PubMed] [Google Scholar]
- Uren A.G., O’Rourke K., Aravind L.A., Pisabarro M.T., Seshagiri S., Koonin E.V., Dixit V.M. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 6:961–967 [DOI] [PubMed] [Google Scholar]
- Wright G., Tan B., Rosenwald A., Hurt E.H., Wiestner A., Staudt L.M. 2003. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA. 100:9991–9996 10.1073/pnas.1732008100 [DOI] [PMC free article] [PubMed] [Google Scholar]