Abstract
DNA pooling can provide an economic and efficient way to detect susceptibility loci to complex diseases. We carried out a genome screen with 400 microsatellite markers spaced at approximately 10 cm in two DNA pools consisting of 119 schizophrenia (SZ) patients and 119 controls recruited from a homogenous population in the Chang Le area of the Shandong peninsula of China. Association of D6S289, a dinucleotide repeat polymorphism in the JARID2 gene with SZ, was found and confirmed by individual genotyping (X2 = 17.89; P = .047). In order to refine the signal, we genotyped 14 single nucleotide polymorphisms (SNPs) covering JARID2 and the neighboring gene, DNTBP1, in an extended sample of 309 cases and 309 controls from Shandong peninsula (including the samples from the pools). However, rs2235258 and rs9654600 in JARID2 showed association in allelic, genotypic and haplotypic tests with SZ patients from Chang Le area. This was not replicates in the extended sample, we conclude that JARID2 could be a susceptibility gene for SZ.
1. Introduction
Schizophrenia, SZ (MIM 181500) is a severe psychiatric disorder which affects ~1% of the general population and has a strong genetic component [1]. However, the model of transmission is complex and is most likely polygenic [2]. Over the last two decades, linkage and association studies have identified a number of chromosomal regions and several promising candidate genes [3, 4]. Those for which the evidence is the strongest are neuregulin 1 (NRG1), Dysbindin (DTNBP1), Disrupted-in-Schizophrenia 1 (DISC1), and regulator of G-protein signalling 4 (RGS4). In addition, D-amino oxidase activator (DAOA) has been implicated in multiple studies of schizophrenia and bipolar disorder (BP) [5]. Support for the involvement of these genes in the pathophysiology of psychotic disorders comes from expression and neurobiological studies as well as from linkage and association studies [6–9]. However, in spite of what might appear an impressive weight of evidence, in no case there has been consistent replication for the same markers and haplotypes across studies, and specific risk or protective nucleotide variants have not been consistently and unambiguously identified. While such consistency might emerge as these genes are subjected to more detailed and systematic analysis in sufficiently large samples, the strength and consistency of the genetic association evidence in support of these candidates is not currently as strong as that reported for other complex disorders such as type 1 and 2 diabetes, rheumatoid arthritis, and macular degeneration [10–16]. Much hope is now pinned on the results of genome-wide association (GWA) studies, as it is now possible to conduct such studies with hundreds of thousands of SNPs. However, such studies are expensive and require very large sample sizes to compensate for the necessary correction for multiple testing, and are, therefore, impossible to conduct by smaller research teams. Small laboratories can, however, rely on DNA pooling studies which cut dramatically the cost [17].
Although the sensitivity and specificity of pooled DNA analyses are imperfect, previous research suggests that such analysis can offer an economic alternative to individual genotyping, although clearly, not all associated loci will be detected. Association studies using microsatellite markers distributed across the human genome every 100 to 150 kb were reported to have distinct advantages over SNP typing because of the much longer extent of linkage disequilibrium (LD) of microsatellites and their higher informativeness [18].
When an appropriate isolated population of relatively recent origin can be studied, it might be possible to use fewer microsatellite markers to screen the entire genome [19]. Based on this theory, we investigated the immigration history of the population residing in Shandong peninsula of China. In particular, we identified the population of the Chang Le area as having the elements of an isolate. This area is located in the middle east of Shandong peninsula and the population there expanded from a limited number of ancestors who emigrated from Shan Xi Hong Dong district in 1388. We reasoned that the 600 year-long history of this population is short enough to preserve long LD, so that some it might be captured by only 400 microsatellite markers spaced at about 10 cm throughout human genome. We admit that there is no sufficient evidence that this is a homogenous population and that our coverage of the genome is likely to be insufficient. Ideally, we would have preferred to cover the genome with 3 000–6 000 markers, but this was beyond our resources.
2. Materials and Methods
2.1. Subjects
A number of 119 unrelated SZ cases and 119 controls from Chang Le County in Shandong peninsula of China were recruited for the first stage of the DNA pooling genome screen. According to the historical records (Ming Taizu Record), the population of Shandong peninsula in the fourteenth century was reduced dramatically due to continuous war, natural disasters (flood, plague, and grasshoppers) and most of the villages were devastated. Many people left the area. In contrast, no war or natural disasters took place in Shan Xi province at that time. The two provinces are separated by 1000-kilometer distance and high mountains, serving as a natural barrier. To balance the population distribution in China, the Emperor of ming dynasty (Y.Z. Zhu) initiated two massive immigration waves from Shan Xi to Shandong peninsula from 1388 to 1389 in order to repopulate that region. It appears that the present-day populations of Shandong peninsula are mostly offspring expanded from different groups of limited number of ancestors that settled there 600 years ago. So, we expect a high degree of homogeneity in the genetic structure of this population, compared with the outbred populations formed in the recent 50 years in many other parts of China, especially in the big cities. The average age of the cases was 33.98 ± 11.99, and the male/female ratio was 1.05(61/58); the average age of the controls was 22.14 ± 5.59, and the male/female ratio was 1.05(61/58), the difference in average age of the cases and controls is significant.
Other two groups of SZ cases and controls from Jinan and Zichuan area were recruited giving a total of 309 cases and 309 controls for the second stage of the study. All SZ cases were from inpatient departments of local psychiatric hospitals. The average age was 27.52 ± 10.84, the male/female ratio was 1.73(196/113). All controls were from the corresponding local blood transfusion centers that were geographically matched to the three psychiatric hospitals. Controls were not screened for mental illness. The average age of controls was 34.21 ± 22.22 and the male/female ratio was 1.83(200/109). All cases and controls are Han Chinese. Diagnoses were made according to DSM- IV criteria [20] by at least two experienced psychiatrists, on the basis of empirical diagnostic interviews and review of medical records. Written consent was obtained from all subjects and this study was approved by the Ethics Committee of the Institute of Basic Medicine of Shandong Academy of Medical Sciences.
2.2. DNA Pools Construction
Genomic DNA was extracted from peripheral venous blood by a modified phenol–chloroform method. The concentration was measured twice on a DU-530 Life science UV/V spectrophotometer (BECKMAN). Samples showing > 10% difference were measured again and we used the mean of the three measurements. DNA samples were diluted to 20 ng/μL working concentrations, and two DNA pools from SZ cases and controls were constructed, respectively, by adding up 10 μL of each sample.
2.3. Genotyping
We used microsatellite markers from the ABI PRISM Linkage Mapping Set MD-10 (Applied Biosystems, Foster City, Calif, USA) comprising 400 markers with an average spacing of 10 cm. Polymerase Chain Reaction (PCR) in a 15 μL volume was performed with 20 ng of genomic DNA, 1.5 mM MgCI2, 10x reaction buffer, 0.2 mM of each dNTP, 0.3 μM of each primer, and 1 U of Taq polymerase. Amplification was performed on a Gene Amp 9700 thermocycler (Applied Biosystems) with the following parameters: 95°C for 12 minutes initial denaturation, followed by 94°C 15 seconds, 55°C 15 seconds, 72°C 30 seconds, 10 cycles; then 89°C 15 seconds, 55°C 15 seconds, 72°C 30 seconds, 20 cycles, then 72°C 10 minutes, and 4°C hold. Electrophoresis was carried out on ABI PRISM 3100-Avant Genetic Analyzer, by mixing up to 9 PCR products simultaneously, which had no overlap of products of the same size range labelled with the same dye. Genotyping was performed with GeneMapper 3.5 software (Applied Biosystems).
SNPs for fine mapping were selected from the HapMap database (www.hapmap.org), ensuring that they are polymorphic in the Chinese population (CHB). LD blocks were determined with the criteria R2 > 0.80 and minimum MAF 2%, using HAPLOVIEW ver.3.2 software [21]. Haplotype tag SNPs (htSNPs) were defined as those capturing 90% of the haplotype diversity within each LD block with no exclusion criterion of minimum minor allele frequency. Fourteen SNPs were selected around DTNPB1 and JARID2 (Table 2). Thirteen of those were genotyped using the Sequenom Mass ARRAY system as per the manufacturer's instructions with either hME or iplex chemistries. SNP rs4715984 was genotyped with allele-specific PCR using the Amplifluor system [22, 23], as it would not fit on a Sequenom panel. All assays used to type the full association sample were optimized initially by genotyping DNA from the 30 CEPH parent–offspring trios used in the international HapMap project. All genotypes were called blind to sample identity and affected status.
Table 2.
Association Analysis of SNPs within JARID2 and DTNBP1 from a homogenous population in Shandong peninsula of China. 177 individuals with nonmissing phenotypes, 89 cases, 88 controls and 0 missing, 87 males, 90 females, 12 of 177 individuals removed for low genotyping (MIND >0.1), Total genotyping rate in remaining individuals is 0.933333.
| SNP | A1 | F_A | F_U | A2 | CHISQ | P | OR |
|---|---|---|---|---|---|---|---|
| jarid2rs2235258 | 3 | 0.3851 | 0.25 | 1 | 6.881 | .00871 | 1.879 |
| jarid2rs9654600 | 1 | 0.2644 | 0.1731 | 4 | 3.979 | .04607 | 1.717 |
| jarid2rs1474587 | 4 | 0.08046 | 0.05769 | 2 | 0.6576 | .4174 | 1.429 |
| jarid2rs13198512 | 4 | 0.1322 | 0.2051 | 2 | 3.151 | .07587 | 0.5902 |
| dtnbp1rs742106 | 3 | 0.3908 | 0.3846 | 1 | 0.01327 | .9083 | 1.026 |
| dtnbp1rs16876575 | 4 | 0.1092 | 0.141 | 2 | 0.7659 | .3815 | 0.7466 |
| dtnbp1rs9296977 | 2 | 0.09195 | 0.0641 | 4 | 0.8791 | .3484 | 1.478 |
| dtnbp1rs4712253 | 4 | 0.3563 | 0.3718 | 2 | 0.0851 | .705 | 0.9353 |
| dtnbp1rs4715984 | 1 | 0.02299 | 0.03205 | 2 | 0.2547 | .6138 | 0.7106 |
| dtnbp1rs9358063 | 4 | 0.1092 | 0.08974 | 2 | 0.3458 | .5565 | 1.243 |
| dtnbp1rs2619533 | 1 | 0.08621 | 0.05769 | 4 | 0.9917 | .3193 | 1.541 |
| dtnbp1rs4715986 | 4 | 0.2701 | 0.2436 | 1 | 0.3026 | .5822 | 1.149 |
| dtnbp1rs2619542 | 2 | 0.2759 | 0.2436 | 4 | 0.4446 | .5049 | 1.183 |
| dtnbp1rs9476886 | 4 | 0.3391 | 0.3205 | 2 | 0.1282 | .7203 | 1.088 |
2.4. Statistical Analysis
The predicted allele frequencies of microsatellites between cases and controls in DNA pools were first estimated by dividing each allele peak height obtained by the GeneMapper v3.5 software with the sum of all peak heights of that marker. This ratio was converted into allele counts and the statistical significance was calculated with CLUMP [24] which can analyze multiallelic markers. The analysis of individual genotyping of D6S289 was also performed by CLUMP.
Tests of allelic, haplotype association, and Hardy Weinberg equilibrium were performed with HAPLOVIEW3.32 [21]; Genotypic and allelic association analyses were performed with PLINK [25]. Haplotype association of the microsatellite marker D6S289 and SNPs was performed with UNPHASED [26].
3. Results
All 400 microsatellites were genotyped, some after repeated PCR and/or electrophoresis. A P value of <.05 produced by CLUMP was set as a cut-off point for follow-up with individual genotyping. Markers meeting this standard were repeated in both PCR and electrophoresis, and the number of replications varied from 2 to 5 times depending on the quality of the genotypes achieved. A total of 126 loci were repeated and only three markers remained significant: D4S403, D6S276, and D6S289.
D6S289 is located on chromosome 6p23 within the linkage locus for schizophrenia (SCZD3). It lies within JARID2 and immediately adjacent to DTNBP1, a strong candidate gene for SZ. Therefore, we chose it as a priority for individual genotyping, which confirmed the predicted association: X2 = 17.89, P = .047. A total of eleven alleles were observed, and three alleles (209, 211 and 213) were more common in cases than in controls (Table 1).
Table 1.
Allele frequencies of genotyping of D6S289 in 119 SZ cases and 119 controls from a homogenous population in Shandong peninsula of China. Normal chi-square (T1) = 17.89, P = .047). This was reached 4657 times in 100 000 simulations by clump analysis.
| Alleles (bp) | 207 | 209 | 211 | 213 | 215 | 217 | 219 | 221 | 223 | 225 | 227 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | 24 | 20 | 20 | 18 | 62 | 64 | 22 | 6 | 1 | 0 | 1 |
| Control | 39 | 17 | 7 | 14 | 65 | 61 | 19 | 8 | 5 | 3 | 0 |
SNP genotyping was performed on the samples included in the pool, and in additional cases and controls, giving a total of 309 cases and 309 controls. Genotypes of the 14 SNPs located in JARID2 and DTNBP1 in the same 90 CEU DNA samples used in the HapMap Project were 100% concordant with HapMap data. However 13 of the 14 SNPs were in Hardy Weinberg equilibrium, with rs4715984 showing HW P value of .001. No significant association with illness was observed for any single marker by allele or genotype in the whole cohort of 309 cases and 309 controls (data not shown).
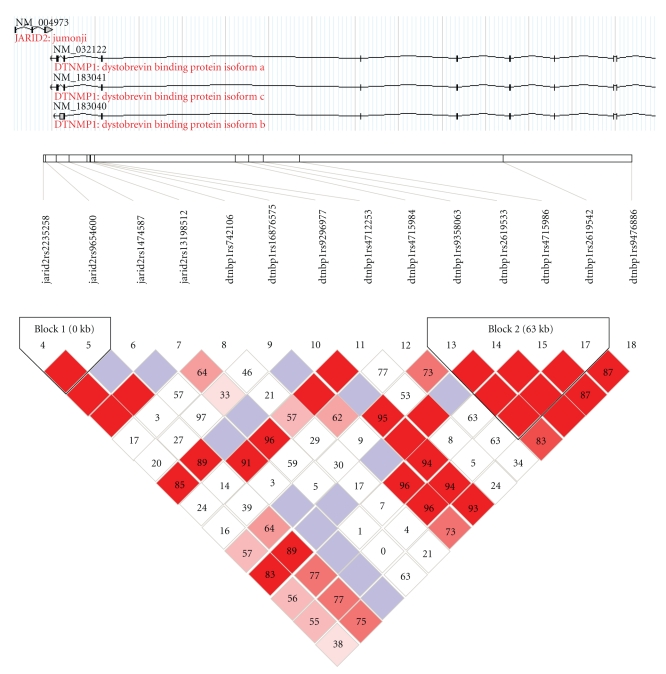
Then we tested separately the association in the three geographical samples. In the sample from Chang Le (89 cases and 88 controls available from the 119 pooling cohort), two SNPs, rs2235258 and rs9654600 (both 3’ of JARID2) showed allelic (Table 2), genotypic (rs2235258, X2 = 7.029, P = .02976; rs9654600, X2 = 4.006, P = .04533 DOM) and haplotypic association; Three of the eleven observed haplotypes between D6S289 with rs2235258 or rs9654600 were more common in cases than in controls: 211 bp-A-T (6% in cases, 0% in controls, X2 = 10.56, P = .0011); 211 bp-G-T (10.3% in cases, 3% in controls, X2 = 9.42, P = .0021); 213 bp-G-A (9.5% in cases, 1.8% in controls, X2 = 5.52, P = .018); haplotype 207 bp-A-T was more common in controls (11.9% in cases, 23.1% in controls, X2 = 7.9, P = .0049). No association was detected in samples from Ji Nan and Zi Cuan area. The SNPs genotyped across the two genes are in a high LD with each other (Figure 1).
Figure 1.
Linkage disequilibrium in JARID2 and DTNBP1 in a homogenous population in Shandong peninsula of China. The AT haplotype in JARID2 (block1) more in cases than in controls (Frequency = 0.667, Case control ratios = 108.0 : 68.0, 125.6 : 48.4; X2 = 4.622, P = .0316), allelic and genotypic association also observed. This samples was from an isolated population with about 600 years history of immigration in the middle east of Shandong peninsular of China.
4. Discussion
In this paper, we first performed a DNA pooling study with 119 SZ cases and 119 healthy controls from an isolated population in China. We covered the entire genome with 400 microsatellite markers, assuming that in an isolated population the LD would cover larger distances and some association signals can be detected with this space coverage. The extent of linkage disequilibrium (LD) in general populations is unlikely to extend for more than 1 Mb [27]. Therefore, genome screens to map disease loci by case control design usually require at least 3 000–6 000 highly polymorphic, evenly spaced microsatellite markers. However, when an appropriate isolated population of relatively recent origin is available for association mapping of disease, a few hundred markers could be sufficient to scan the whole genome, if it can be assumed that linkage disequilibrium extends to ≥ 10 Mb [19]. Based on this view, we investigated the immigration history of the population in Shandong peninsula, and chose samples from three geographical areas.
From the pooling study, we identified a positive signal on chromosome 6, near DTNPB1, and confirmed the association with individual genotyping of the microsatellite marker, X2 = 17.89, P = .047. We then saturated the region with 14 SNPs and typed them individually in an extended sample of 309 cases and 309 controls (including 89 cases and 88 controls in the pools). SNPs genotyping showed association with two of them in the original sample included in the pool. Allelic association was observed with rs2235258 and rs9654600 of P = .0087 and P = .046, respectively. The association was not replicated in the additional samples from other regions, LD between the microsatellite marker and SNPs in the region extended up to 8 Mb in the relatively isolated population of Chang Le, although we admit that we do not know the corresponding extent of LD in the other Chinese samples, as they were not genotyped for the microsatellite. The maintenance of homogeneity in genetic structure in the Chang Le population is mainly due to the economy, which has long been dominated by agriculture. According to the “Ming Tai Zu Record,” this population expanded from a limited number of ancestors who were resettled from the Shanxi province by Emperor YZ Zhu during the ming dynasty (AD1388). No further immigration happened in the following 600 years. Relatively isolated populations have produced some promising findings in psychiatric genetics in the past [28], such as for the genes NRG1 [29] and DAOA [30].
One weakness of the current study is that only 89 cases from Chang Le are available for the SNP genotyping. Such a small sample size could lead to a false positive finding (type 1 error). However, the possibility of type 1 error is reduced because our analysis was carried out on an isolated population, reducing genetic heterogeneity. In addition, the association findings were based on an analysis of a gene targeted by our hypothesis-free genome wide linkage study, a region previously linked to schizophrenia by many groups.
The association was not present in additional samples from the same peninsula. This could indicate that the genetic structure of the population in Shandong peninsula is not uniform, and the relatively homogenous population might be only restricted to a certain range of geographic area (100–150 km according to this study). Our findings suggest that the power to detect association is not merely related to enlargement of the sample size, but more importantly, to the genetic structure in the samples. Therefore, using a heterogeneous population increases the chance of producing a false negative result (type II error).
JARID2 (jumonji, AT rich interactive domain 2) is an ortholog of the mouse jumonji gene, which encodes a nuclear protein essential for mouse embryogenesis, including neural tube formation. Over expression of mouse jumonji negatively regulates cell proliferation. The jumonji proteins contain a DNA-binding domain, called an AT-rich interaction domain (ARID), and share regions of similarity with human retinoblastoma-binding protein-2 and the human SMCX protein. The first report of JARID2 as a candidate gene for 6p23-linked SZ was from the USA [31] and our study suggests that JARID2, rather than DTNBP1, is a candidate gene for 6p22.3-linked SZ. A significant difference in allele distribution for a tetra nucleotide repeat polymorphism in the JARID2 gene that affects an AT-rich domain was found in that study. In our study, for D6S289, a dinucleotide repeat polymorphism of 11 alleles in JARID2, alleles of 209, 211, and 213 bp were more frequent in SZ cases than controls from Chang Le area of Shandong peninsula. This association was also present in two SNPs within the gene, rs2235258 and rs9654600.
Straub et al. described the results of family-based association analysis on chromosome 6p and first reported genetic variation in the DTNBP1 gene, encoding dysbindin, which was associated with schizophrenia and related phenotypes [32]. The association has been well replicated in several populations, including Caucasian, Chinese (Shanghai, Sichuan) and Japanese samples [33–41]. Furthermore, variation in DTNBP1 conferring susceptibility to schizophrenia through reduced expression has also been reported [6, 42]. However, no disease-causing alleles have been identified so far and some replication studies failed to provide positive results [43–48].
Including the current one, there have been a total of four studies on DTNBP1 in Chinese SZ samples: three of these samples were recruited from the southern part of Yangtze River, which is the biggest natural barrier in China and divides the Chinese population into two parts from an anthropological point of views. In Sichuan samples, a single haplotype, which included rs2619538 and P1583; and another rare haplotype, composed of P1320 and P1757, were significantly associated with SZ [37]. In samples from Shanghai, only a haplotypic (TGTCA) but no allelic or genotypic association was observed [49]. No association was reported with SZ in families from Taiwan either [45]. Therefore, the association of DTNBP1 with SZ in the Chinese population is far from convincing. The evidence implicating DTNBP1 as a susceptibility gene by so many investigators could not be regarded as invalid, and the extremely close proximity of JARID2 and DTNBP1 (within 1553 bp), makes them very likely to be regulated coordinately. DTNBP1 could be a downstream target of the JARID2 transcription factor, or both are coordinately regulated by the same cis or trans-acting factor [31]. The design of the SNPs genotyping was originally for testing the DTNBP1 association in this Chinese SZ samples, therefore, the JARID2 5’ end was not analyzed in this study.
5. Web Resources
The URLs for data presented herein are as follows:
(1) Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia; SCZD, schizophrenia3; SCZD3);
(2) Entrez Gene, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene (for JARID2 and DTNBP1);
(3) UCSC, http://genome.ucsc.edu/ (for SNPs primer design);
(4) GDB, http://www.gdb.org/ (for d6s289 primer synthesis);
(5) HapMap, http://www.hapmap.org/ (for the comparison of SNP genotype in CEPH samples and the selection of tag SNPs);
(6) Haploview 3.32, http://www.broad.mit.edu/personal/jcbarret/haploview/ (for the tests of allelic, haplotypic association and Hardy Weinberg equilibrium);
(7) Clump, http://www.smd.qmul.ac.uk/statgen/dcurtis/software.html (For the analysis of pooled and individual genotyping of microsatellite markers);
(8) PLINK, http://pngu.mgh.harvard.edu/purcell/plink/ (for the analysis of genotypic and allelic association of SNPs);
(9) UNPHASED, http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/ (for analysis of the association between d6s289 and all SNPs).
Acknowledgments
The authors would like to thank Dr. Lyudmila Georgieva; Dr. Dilihan Gumus; Dr. Tim Peirce, Dr. Hywel Williams, and Sarah Dwyer for their technical assistance. The authors would like to thank Dr. Jan P Zhu and Dr. Zheng Weng for assistance in collecting the blood samples. The authors would like also to thank all the patients and controls participated in this research. This work was supported by grants from the Natural Science Foundation of Shandong Province, China (Grants IDZ2004C10; Z2008C11), the National Natural Science Foundation of China (Grant no. 30440042; 30770780), the Medical Research Council (UK) and the National Institute of the Neuroscience of Mental Disorders (Grant no. MH066392).
References
- 1.Gottesman II. Schizophrenia Genesis: The Origins of Madness. New York, NY, USA: W. H. Freeman; 1991. [Google Scholar]
- 2.Owen MJ. Genomic approaches to schizophrenia. Clinical Therapeutics. 2005;27(supplement 1):S2–S7. doi: 10.1016/j.clinthera.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 3.Kirov G, O'Donovan MC, Owen MJ. Finding schizophrenia genes. The Journal of Clinical Investigation. 2005;115(6):1440–1448. doi: 10.1172/JCI24759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen MJ, Craddock N, O'Donovan MC. Schizophrenia: genes at last? Trends in Genetics. 2005;21(9):518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biological Psychiatry. 2006;60(2):106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Bray NJ, Preece A, Williams NM, et al. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Human Molecular Genetics. 2005;14(14):1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- 7.Hall D, Gogos JA, Karayiorgou M. The contribution of three strong candidate schizophrenia susceptibility genes in demographically distinct populations. Genes, Brain and Behavior. 2004;3(4):240–248. doi: 10.1111/j.1601-183X.2004.00078.x. [DOI] [PubMed] [Google Scholar]
- 8.Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(17):6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4) Biological Psychiatry. 2006;60(6):534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 10.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marx J. Gene offers insight into macular degeneration. Science. 2006;314(5798):p. 405. doi: 10.1126/science.314.5798.405a. [DOI] [PubMed] [Google Scholar]
- 12.Pennisi E. New gene found for inherited macular degeneration. Science. 1998;281(5373):p. 31. doi: 10.1126/science.281.5373.31. [DOI] [PubMed] [Google Scholar]
- 13.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 14.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nature Genetics. 2007;39(12):1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314(5801):992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 16.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirov G, Nikolov I, Georgieva L, Moskvina V, Owen MJ, O'Donovan M. Pooled DNA genotyping on affymetrix SNP genotyping arrays. BMC Genomics. 2006;7(1, article 27) doi: 10.1186/1471-2164-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamiya G, Shinya M, Imanishi T, et al. Whole genome association study of rheumatoid arthritis using 27 039 microsatellites. Human Molecular Genetics. 2005;14(16):2305–2321. doi: 10.1093/hmg/ddi234. [DOI] [PubMed] [Google Scholar]
- 19.Hästbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nature Genetics. 1992;2(3):204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 20.APA. Diagnostic and Statistical Manual of Mental Disorders DSM-IV-TR. 4th edition. Washington, DC, USA: American Psychiatric Association; 1994. [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Hawkins JR, Khripin Y, Valdes AM, Weaver TA. Miniaturized sealed-tube allele-specific PCR. Human Mutation. 2002;19(5):543–553. doi: 10.1002/humu.10060. [DOI] [PubMed] [Google Scholar]
- 23.Myakishev MV, Khripin Y, Hu S, Hamer DH. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Research. 2001;11(1):163–169. doi: 10.1101/gr.157901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Annals of Human Genetics. 1995;59(1):97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genetic Epidemiology. 2003;25(2):115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 27.Dawson E, Abecasis GR, Bumpstead S, et al. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002;418(6897):544–548. doi: 10.1038/nature00864. [DOI] [PubMed] [Google Scholar]
- 28.Venken T, Del-Favero J. Chasing genes for mood disorders and schizophrenia in genetically isolated populations. Human Mutation. 2007;28(12):1156–1170. doi: 10.1002/humu.20582. [DOI] [PubMed] [Google Scholar]
- 29.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. American Journal of Human Genetics. 2002;71(4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chumakov I, Blumenfeld M, Guerassimenko O, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(21):13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedrosa E, Ye K, Nolan KA, et al. Positive association of schizophrenia to JARID2 gene. American Journal of Medical Genetics B. 2007;144(1):45–51. doi: 10.1002/ajmg.b.30386. [DOI] [PubMed] [Google Scholar]
- 32.Straub RE, Jiang Y, MacLean CJ, et al. Genetic variation in the 6p22.3 Gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. American Journal of Human Genetics. 2002;71(2):337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeRosse P, Funke B, Burdick KE, et al. Dysbindin genotype and negative symptoms in schizophrenia. American Journal of Psychiatry. 2006;163(3):532–534. doi: 10.1176/appi.ajp.163.3.532. [DOI] [PubMed] [Google Scholar]
- 34.Funke B, Finn CT, Plocik AM, et al. Association of the DTNBP1 locus with schizophrenia in a U.S. population. American Journal of Human Genetics. 2004;75(5):891–898. doi: 10.1086/425279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirov G, Ivanov D, Williams NM, et al. Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biological Psychiatry. 2004;55(10):971–975. doi: 10.1016/j.biopsych.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 36.Kohn Y, Danilovich E, Filon D, et al. Linkage disequlibrium in the DTNBP1 (dysbindin) gene region and on chromosome 1p36 among psychotic patients from a genetic isolate in Israel: findings from identity by descent haplotype sharing analysis. American Journal of Medical Genetics B. 2004;128(1):65–70. doi: 10.1002/ajmg.b.30044. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Zhang F, Liu X, et al. Identifying potential risk haplotypes for schizophrenia at the DTNBP1 locus in Han Chinese and Scottish populations. Molecular Psychiatry. 2005;10(11):1037–1044. doi: 10.1038/sj.mp.4001718. [DOI] [PubMed] [Google Scholar]
- 38.Numakawa T, Yagasaki Y, Ishimoto T, et al. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Human Molecular Genetics. 2004;13(21):2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- 39.Schwab SG, Knapp M, Mondabon S, et al. Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. American Journal of Human Genetics. 2003;72(1):185–190. doi: 10.1086/345463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Den Bogaert A, Schumacher J, Schulze TG, et al. The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. American Journal of Human Genetics. 2003;73(6):1438–1443. doi: 10.1086/379928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams NM, Preece A, Morris DW, et al. Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1) Archives of General Psychiatry. 2004;61(4):336–344. doi: 10.1001/archpsyc.61.4.336. [DOI] [PubMed] [Google Scholar]
- 42.Weickert CS, Straub RE, McClintock BW, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Archives of General Psychiatry. 2004;61(6):544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 43.Datta SR, McQuillin A, Puri V, et al. Failure to confirm allelic and haplotypic association between markers at the chromosome 6p22.3 dystrobrevin-binding protein 1 (DTNBP1) locus and schizophrenia. Behavioral and Brain Functions. 2007;3(1, article 50) doi: 10.1186/1744-9081-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo E-J, Lee K-Y, Jeong SH, Ahn Y-M, Koo YJ, Kim YS. The dysbindin gene (DTNBP1) and schizophrenia: no support for an association in the Korean population. Neuroscience Letters. 2006;407(2):101–106. doi: 10.1016/j.neulet.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Liu C-M, Liu Y-L, Fann CS-J, et al. No association evidence between schizophrenia and dystrobrevin-binding protein 1 (DTNBP1) in Taiwanese families. Schizophrenia Research. 2007;93(1–3):391–398. doi: 10.1016/j.schres.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Morris DW, McGhee KA, Schwaiger S, et al. No evidence for association of the dysbindin gene [DTNBP1] with schizophrenia in an Irish population-based study. Schizophrenia Research. 2003;60(2-3):167–172. doi: 10.1016/s0920-9964(02)00527-3. [DOI] [PubMed] [Google Scholar]
- 47.Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM, Sklar P. Analysis of high-resolution HapMap of DTNBP1 (dysbindin) suggests no consistency between reported common variant associations and schizophrenia. American Journal of Human Genetics. 2006;79(5):903–909. doi: 10.1086/508942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turunen JA, Peltonen JO, Pietiläinen OPH, et al. The role of DTNBP1, NRG1, and AKT1 in the genetics of schizophrenia in Finland. Schizophrenia Research. 2007;91(1–3):27–36. doi: 10.1016/j.schres.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 49.Tang JX, Zhou J, Fan JB, et al. Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Molecular Psychiatry. 2003;8(8):717–718. doi: 10.1038/sj.mp.4001287. [DOI] [PubMed] [Google Scholar]