Abstract
In Arabidopsis thaliana, the cryptochrome (CRY) blue light photoreceptors and the phytochrome (phy) red/far-red light photoreceptors mediate a variety of light responses. COP1, a RING motif–containing E3 ubiquitin ligase, acts as a key repressor of photomorphogenesis. Production of stomata, which mediate gas and water vapor exchange between plants and their environment, is regulated by light and involves phyB and COP1. Here, we show that, in the loss-of-function mutants of CRY and phyB, stomatal development is inhibited under blue and red light, respectively. In the loss-of-function mutant of phyA, stomata are barely developed under far-red light. Strikingly, in the loss-of-function mutant of either COP1 or YDA, a mitogen-activated protein kinase kinase kinase, mature stomata are developed constitutively and produced in clusters in both light and darkness. CRY, phyA, and phyB act additively to promote stomatal development. COP1 acts genetically downstream of CRY, phyA, and phyB and in parallel with the leucine-rich repeat receptor-like protein TOO MANY MOUTHS but upstream of YDA and the three basic helix-loop-helix proteins SPEECHLESS, MUTE, and FAMA, respectively. These findings suggest that light-controlled stomatal development is likely mediated through a crosstalk between the cryptochrome-phytochrome-COP1 signaling system and the mitogen-activated protein kinase signaling pathway.
INTRODUCTION
The stomatal pores of higher plants act as ports that tightly regulate the uptake of CO2 and the evaporation of water and, thus, not only are critical for photosynthesis but also exert a major influence on global carbon and water cycles (Hetherington and Woodward, 2003). Situated in the epidermis, they are surrounded by a pair of guard cells, which regulate their opening in response to environmental and internal signals, including light, CO2, and phytohormones (Assmann and Wang, 2001; Schroeder et al., 2001). The production of stomata is complex and regulated by environmental factors including light and CO2, as well as by developmental programs (Gray et al., 2000; Lake et al., 2001; Hetherington and Woodward, 2003). In Arabidopsis thaliana, a stomatal lineage arises from an undifferentiated protodermal cell, which divides asymmetrically to give rise to two daughter cells: a larger daughter cell and a meristemoid. The meristemoid undergoes one to three rounds of asymmetric division prior to differentiating into a round, guard mother cell. The guard mother cell divides symmetrically to generate a pair of guard cells that surround a microscopic pore.
Plants have evolved multiple photoreceptor systems to monitor light quality, quantity, and direction. In Arabidopsis, these photoreceptors include the blue/UV-A light-absorbing cryptochromes (CRY: CRY1, and CRY2) and phototropins and the red/far-red light-absorbing phytochromes (phy: phyA, to phyE) (Cashmore et al., 1999; Briggs and Christie, 2002; Quail, 2002; Lin and Shalitin, 2003; Li and Yang, 2007). Cryptochromes and phytochromes act together to regulate photomorphogenic development and photoperiodic flowering (Casal and Mazzella, 1998; Neff and Chory, 1998; Mockler et al., 1999) and entrain the circadian clock (Somers et al., 1998). Also, cryptochromes and phototropins act together to mediate blue light regulation of stomatal opening (Kinoshita et al., 2001; Mao et al., 2005). Very recently, it has been demonstrated that phyB is involved in the regulation of stomatal development (Boccalandro et al., 2009; Casson et al., 2009).
Cryptochomes typically have both an N-terminal photolyase-related domain that shares sequence similarity with photolyase, a family of flavoproteins that catalyze the repair of UV light–damaged DNA, and a distinguishing C-terminal domain that is absent in photolyase and has no strong sequence similarity with known protein domains (Sancar, 1994; Cashmore et al., 1999; Lin and Shalitin, 2003). The C-terminal domain of Arabidopsis CRY1 or CRY2 (CCT1 or CCT2, respectively) is shown to mediate the signaling of CRY1 or CRY2 in response to light activation through its physical interaction with COP1 (Yang et al., 2000; Wang et al., 2001; Yang et al., 2001), a key repressor of photomorphogenesis (Deng et al., 1992). The N-terminal photolyase-related domain of CRY is shown to mediate homodimerization of CRY, which is required for light activation of the photoreceptor activity of CRY (Sang et al., 2005; Yu et al., 2007; Rosenfeldt et al., 2008). It is shown that COP1, together with other COP/DET/FUS proteins, acts constitutively to repress photomorphogenesis (Deng et al., 1991; Chory, 1993; Kwok et al., 1996) and is implicated in the repression of stomatal production (Deng et al., 1992; Deng and Quail, 1992; Wei et al., 1994b). It is known that COP1 acts as an E3 ubiquitin ligase to ubiquitinate the light-signaling transcriptional factors, such as HY5, LAF1, and HFR1, and the key photoperiodic transcriptional activator CONSTANS (CO), promoting degradation of these proteins to repress photomorphogenesis and floral induction, respectively (Osterlund et al., 2000; Seo et al., 2003; Jang et al., 2005, 2008; Yang et al., 2005; Liu et al., 2008). It is proposed that the outcome of the light activation of CRY, through its physical interaction with COP1, is the disruption of the negative regulation of COP1 exerted on its substrates, such as HY5 and CO. In this manner, HY5 and CO are relieved from COP1 and 26S proteasome-dependent proteolysis and are able to perform their role in promoting photomorphogenesis and flowering (Yang et al., 2001; Liu et al., 2008).
In Arabidopsis, the phytochrome family has five members, phyA through phyE, of which phyA and phyB are the best characterized (Smith, 2000; Quail, 2002). Phytochromes are soluble, dimeric chromopeptides with monomers of 120 to 130 kD that possess two photoconvertible forms: Pr (red light-absorbing) and Pfr (far-red light-absorbing). Phytochrome signaling pathway in Arabidopsis is composed of an intricate network of numerous downstream signaling components (Smith, 2000; Quail, 2002), such as COP1, Phytochrome Kinase Substrate1 (Fankhauser et al., 1999), Suppressor of PhyA (SPA1) (Hoecker et al., 1999), and phytochrome interacting factors (PIFs; PIF1, PIF3, PIF4, PIF5, and PIF6) (Castillon et al., 2007), which are basic helix-loop-helix (bHLH) transcriptional factors that act to negatively regulate photomorphogensis. PIFs are the best-characterized components of phytochrome signaling, which are shown to specifically bind to the biologically active Pfr form of phytochromes (Ni et al., 1999; Huq et al., 2004). In the dark, phyB is localized to cytosol. Upon red light illumination, phyB translocates to the nucleus (Nagatani, 2004), where it interacts with PIFs. It has been shown that the phyB-PIF3 interaction can lead to the phosphorylation of PIF3, triggering its degradation through the 26S proteasome–dependent pathway and eventually relieving its negative regulation of photomorphogenesis (Al-Sady et al., 2006).
Recent studies reveal that several negative regulators play a critical role in regulating stomatal development and patterning in Arabidopsis. These include the three ERECTA (ER) family of leucine-rich repeat (LRR) receptor-like kinases (ER, ERL1, and ERL2) (Masle et al., 2005; Shpak et al., 2005), the leucine-rich repeat receptor-like protein TOO MANY MOUTHS (TMM) (Nadeau and Sack, 2002), and their putative ligand EPF1 (Hara et al., 2007), as well as a subtilisin protease, SDD1 (Berger and Altmann, 2000) and the mitogen-activated protein kinase (MAPK) signaling components YDA, MKK4/5, and MPK3/6 (Bergmann et al., 2004; Wang et al., 2007). Loss-of-function mutations in these loci disrupt epidermal patterning with stomata produced adjacent to each other or in clusters. Other recent studies suggest that the SDD-processed ligand, which is presently unknown, is perceived by TMM through potential interaction with ER, ERL1, and ERL2 and transduced to downstream MAPK cascades via YDA to repress stomatal formation (Bergmann et al., 2004; Sack, 2004). Acting in opposition to these stomatal limiting factors are the three closely related bHLH transcriptional factors, SPEECHLESS (SPCH), MUTE, and FAMA, and the two more distantly related bHLHs, ICE1/SCREAM and SCREAM2, which act in concert to consecutively promote initiation of asymmetric divisions, proliferation of transient precursor cells, and differentiation of stomatal guard cells, respectively (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008).
To date, although it is demonstrated that phyB is involved in the regulation of stomatal development (Boccalandro et al., 2009; Casson et al., 2009), whether other photoreceptors are involved in this process and whether the components in the light signaling pathway genetically interact with those in the intrinsic developmental pathway remain unknown. Here, by analyzing the stomatal phenotype of the loss-of-function mutants of CRY, phyB, and phyA in monochromatic blue, red, and far-red lights, we demonstrate that CRY (CRY1 and CRY2) and phyB are the primary photoreceptors mediating blue and red light–induced stomatal development, respectively, and that phyA is likely the sole photoreceptor mediating far-red light–induced stomatal development. We also examined the stomatal phenotype of the loss-of-function mutants of COP1 and YDA in the light and darkness and demonstrate that COP1 and YDA are key negative regulators acting constitutively to repress stomatal development and differentiation in both light and darkness. Furthermore, genetic interaction studies demonstrate that CRY, phyA, and phyB work together to promote light-induced stomatal development and that COP1 acts genetically downstream of CRY, phyA, and phyB and in parallel with TMM, but upstream of YDA, SPCH, MUTE, and FAMA, respectively. The establishment of an overall genetic pathway of light-controlled stomatal development and its genetic interaction with the MAPK signaling pathway advances our understanding of the regulatory mechanisms by which light signals regulate the production of stomata.
RESULTS
CRY1 and CRY2 Mediate Blue Light–Induced Stomatal Development
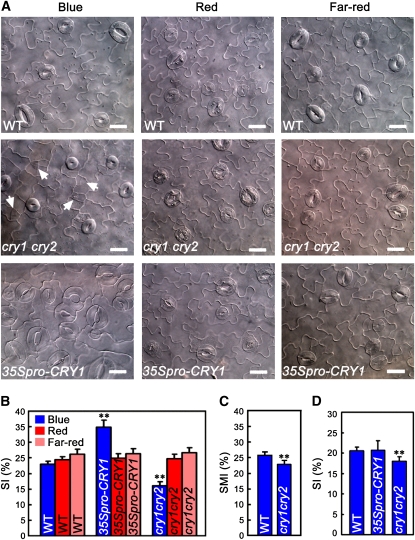
In a previous study, we demonstrated that stomatal opening is reduced in the cry1 cry2 mutant but is enhanced in transgenic plants overexpressing CRY1 (35Spro–CRY1) or CRY2 (35Spro–CRY2) (Mao et al., 2005). When we examined the stomatal phenotype of the cotyledon epidermis of 35Spro–CRY1 seedlings under blue light, we observed dramatic clustered stomata produced in 35Spro–CRY1 cotyledon epidermis (Figure 1A). Measurements of the stomatal index (SI; guard cells per total epidermal cells) demonstrated that the SI of the cotyledon epidermis of 35Spro–CRY1 was significantly greater than that of the wild type (Figure 1B). Under red or far-red light, no clustered stomatal phenotype was observed for 35Spro–CRY1 epidermis, and the SI of 35Spro–CRY1 was similar to that of the wild type (Figures 1A and 1B). This finding suggests that CRY is involved in the blue light regulation of stomatal development.
Figure 1.
Cryptochromes Are Required for Blue Light–Triggered Stomatal Development.
(A) Differential interference contrast (DIC) images of the abaxial cotyledon epidermis of 10-d-old wild-type, cry1 cry2 double mutant, and 35Spro–CRY1 seedlings. Seedlings were grown in blue light (30 μmol·m−2·s−1), red light (50 μmol·m−2·s−1), and far-red light (12 μmol·m−2·s−1). Meristemoids are indicated by arrowheads. Bars = 20 μm.
(B) The SI obtained from the samples in (A). In (B) to (D), the SI and SMI are presented as the percentage of mean ± sd. Asterisks denote significant differences between the indicated genotypes and the wild type (t test, P < 0.01), n = 10.
(C) The SMI obtained from the samples in (A). Asterisks denote significant difference between cry1 cry2 and the wild type (t test, P < 0.01), n = 10.
(D) The SI of the abaxial leaf epidermis of ∼4-week-old wild-type, 35Spro–CRY1, and cry1 cry2 plants grown under blue light (50 μmol·m−2·s−1). Asterisks denote significant differences between cry1 cry2 and the wild type (t test, P < 0.01), n = 5.
To confirm this possibility, we first examined the cotyledon epidermis of cry1 cry2 mutant seedlings under different light spectra (see Supplemental Figure 1 online). We found that stomatal development of the cry1 cry2 mutant was inhibited in a blue light–dependent manner, as shown by the production of many meristemoids and by reduced stomatal size and SI under blue light (Figures 1A and 1B). However, the stomatal and meristemoid index (SMI; guard cells plus meristemoids per total epidermal cells) of the cry1 cry2 mutant was significantly less great than that of the wild type (Figure 1C). We then examined the stomatal phenotype of the true leaf epidermis of 35Spro–CRY1 and cry1 cry2 mutant plants under blue light. For unknown reasons, we observed no stomatal clusters in the true leaf epidermis of 35Spro–CRY1 plants and no difference in the SI between 35Spro–CRY1 and wild-type leaf epidermis either. However, the SI of the cry1 cry2 mutant leaf epidermis was significantly less great than that of the wild type (Figure 1D).
Next, we investigated blue light fluence rate response of stomatal development of cotyledon epidermis of 35Spro–CRY1 and the cry1 cry2 mutant. At the fluence rates of 0.5 and 5 μmol·m−2·s−1 blue light, cry1 cry2 epidermis produced much more meristemoids but fewer and smaller stomata than the wild type, whereas 35Spro–CRY1 epidermis produced fewer meristemoids, but more and larger stomata than the wild type (see Supplemental Figure 2A online). As the fluence rate was increased to 50 μmol·m−2·s−1, the cry1 cry2 mutant still produced meristemoids, whereas the wild type and 35Spro–CRY1 rarely did. Rather, 35Spro–CRY1 produced stomata in clusters. Compared with the wild type, stomatal development of cry1 cry2 mutant epidermis, when expressed as the SI, showed a reduced blue light response, whereas that of 35Spro–CRY1 epidermis showed a hypersensitive blue light response (see Supplemental Figure 2B online). These data indicate that CRY (CRY1 and CRY2) is the primary photoreceptor mediating blue light–induced stomatal development of cotyledon epidermis.
PhyB and phyA Mediate Red and Far-Red Light–Induced Stomatal Development of Cotyledon Epidermis, Respectively
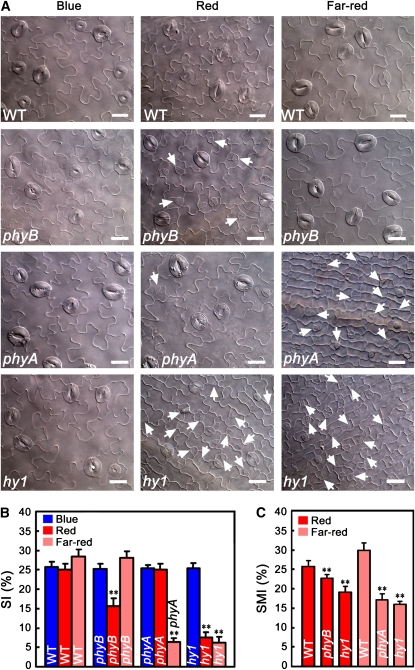
To investigate whether phytochromes are also involved in the regulation of stomatal development, we first examined the stomatal phenotype in cotyledon epidermis of phyB and phyA under blue, red, or far-red light, respectively. We found that stomatal development of phyB mutant was delayed in a red light–dependent manner, as shown by the production of many meristemoids and by reduced stomatal size and SI (Figures 2A and 2B), and that phyA mutant epidermis hardly produced stomata under far-red light (Figure 2A), similar to the stomatal phenotype observed for the wild type in darkness (Figure 3A). Moreover, the SMI of phyB and phyA mutants was significantly less great than that of the wild type under red and far-red light, respectively (Figure 2C). It is known that mutation in the HY1 gene, which encodes a higher-plant heme oxygenase that is required for phytochrome chromophore biosynthesis (Davis et al., 1999), results in a failure in the production of holophytochromes in plants. Next, we examined the stomatal phenotype of hy1 mutant under different light spectra. As expected, hy1 mutant epidermis rarely produced stomata under either red or far-red light (Figure 2A), similar to the stomatal phenotype observed for phyA mutant under far-red light. Consistently, the SMI of hy1 mutant was significantly less than that of the wild type under either red or far-red light (Figure 2C).
Figure 2.
Phytochromes Are Required for Red and Far-Red Light–Triggered Stomatal Development.
(A) DIC images of the abaxial cotyledon epidermis of 10-d-old wild-type, phyB, phyA, and hy1 seedlings grown under blue light (30 μmol·m−2·s−1), red light (50 μmol·m−2·s−1), and far-red light (12 μmol·m−2·s−1). Meristemoids are indicated by arrowheads. Bars = 20 μm.
(B) The SI obtained from the samples in (A). In (B) and (C), the SI and SMI are presented as the percentage of mean ± sd. Asterisks denote significant differences between the indicated genotypes and the wild type (t test, P < 0.01), n = 10.
(C) The SMI obtained from the samples in (A). Asterisks denote significant differences between the indicated genotypes and the wild type (t test, P < 0.01), n = 10.
Figure 3.
COP1 Acts Constitutively to Suppress Stomatal Development and Differentiation and Is Regulated by CRY1, SPA, and DET1.
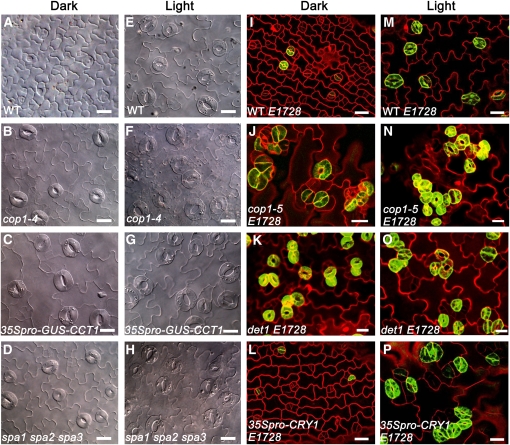
(A) to (D) DIC images of the abaxial cotyledon epidermis of 10-d-old dark-grown wild-type (A), cop1-4 (B), 35Spro–GUS–CCT1 (C), and spa1 spa2 spa3 (D) seedlings.
(E) to (H) DIC images of the abaxial cotyledon epidermis of wild-type (E), cop1-4 (F), 35Spro–GUS–CCT1 (G), and spa1 spa2 spa3 (H) seedlings grown under white light (150 μmol·m−2·s−1) for 10 d.
(I) to (L) Confocal images of the abaxial cotyledon epidermis of 7-d-old dark-grown wild-type expressing E1728 (WT E1728) (I), cop1-5 mutant expressing E1728 (cop1-5 E1728) (J), det1 mutant expressing E1728 (det1 E1728) (K), and 35Spro–CRY1 expressing E1728 (35Spro–CRY1 E1728) (L).
(M) to (P) Confocal images of the abaxial cotyledon epidermis of 7-d-old light-grown WT E1728 (M), cop1-5 E1728 (N), det1 E1728 (O), and 35Spro–CRY1 E1728 (P). Light condition for (M) to (O) is white light (150 μmol·m−2·s−1), and for (P) is blue light (50 μmol·m−2·s−1). In (I) to (P), epidermal cell periphery is highlighted by propidium iodide (PI; red), and mature guard cells are indicated by the E1728 marker (green). Bars = 20 μm.
Next, we investigated red light fluence rate response of stomatal development of cotyledon epidermis of phyB mutant and PHYB-overexpressing (35Spro–PHYB) seedlings. At the fluence rates of 0.5 and 5 μmol·m−2·s−1 red light, the phyB epidermis produced much more meristemoids but fewer and smaller stomata than the wild type, whereas the 35Spro–PHYB epidermis produced much fewer meristemoids but more and larger stomata than the wild type (see Supplemental Figure 3A online). At the fluence rates of 50 and 140 μmol·m−2·s−1 red light, the phyB mutant still produced meristemoids, whereas the wild type and 35Spro–PHYB rarely did. Moreover, the stomatal size of the phyB mutant was smaller than that of the wild type, whereas the stomatal size of 35Spro–PHYB was larger than that of the wild type. Compared with the wild type, stomatal development of phyB mutant epidermis showed a reduced red light response, whereas that of 35Spro–PHYB epidermis showed an enhanced red light response (see Supplemental Figure 3B online). We further examined far-red light fluence rate response of stomatal development of cotyledon epidermis of the phyA mutant. At the fluence rates of 0.4 and 2 μmol·m−2·s−1 far-red light, the wild type epidermis produced meristemoids, but as the fluence rate was increased to 12 and 40 μmol·m−2·s−1, it largely produced mature stomata but rarely produced meristemoids (see Supplemental Figure 4A online). By contrast, under all the fluence rates of far-red light tested, meristemoids and immature stomata were largely arrested in the phyA mutant, and mature stomata were seldom produced. In contrast with the wild type, whose stomatal development showed a clear far-red light response, the stomatal development of the phyA mutant was insensitive to far-red light. Taken together, these data indicate that phyB is the primary photoreceptor mediating red light–induced stomatal development and that phyA is likely the sole photoreceptor mediating far-red light–induced stomatal development.
COP1 Acts Constitutively to Repress Stomatal Development and Differentiation of Cotyledon Epidermis
COP1 is known to be a key negative regulator of photomorphogenesis, floral initiation, and stomatal opening (Deng et al., 1992; Mao et al., 2005; Jang et al., 2008; Liu et al., 2008). To determine whether COP1 is involved in the light regulation of stomatal development, we first analyzed the stomatal phenotype of a weak cop1 mutant allele, cop1-4, in both light and darkness. We found that it constitutively produced stomata that were not attached to each other (Figures 3B and 3F). We then characterized the stomatal phenotype of a lethal cop1 mutant allele, cop1-5, using a mature guard cell–specific green fluorescent protein (GFP) marker, E1728. In the wild-type background, E1728 was expressed in epidermal cells in the light but was rarely expressed in darkness (Figure 3M versus 3I), suggesting that the establishment of the mature guard cell identity is light dependent. Strikingly, the cop1-5 mutant produced clustered stomata in both light and darkness that constitutively expressed E1728 (Figures 3N and 3J), indicating a critical role for COP1 in repressing stomatal development and differentiation.
COP1 is regulated by multiple components of light signaling, including CRY1, CRY2, SPA1, and the CDD (COP10, DDB1, DET1) complex. CRY1 and CRY2 negatively regulate COP1 through physical interaction of their C-terminal domain (CCT1 and CCT2) with COP1 (Wang et al., 2001; Yang et al., 2001), whereas SPA1 and CDD complex positively regulate COP1 through direct interaction with COP1 (Seo et al., 2003; Yanagawa et al., 2004). It is shown that transgenic seedlings constitutively expressing CCT1 or CCT2 fused to β-glucuronidase (GUS) (35Spro–GUS–CCT1 or 35Spro–GUS–CCT2), as well as spa1 spa2 spa3 and det1 mutants show a cop1 mutant-like constitutive photomorphogenic phenotype in darkness (Chory et al., 1989; Yang et al., 2000; Laubinger et al., 2004). Consistent with these studies, 35Spro–GUS–CCT1, spa1 spa2 spa3, and det1 mutants exhibited a constitutive stomatal development phenotype in the dark (Figures 3C, 3D, 3K, and 3O), with det1 constitutively producing stomata in clusters that expressed E1728 in both light and darkness. Unlike 35Spro–GUS–CCT1, 35Spro–CRY1 seedlings are shown to be fully etiolated in the dark, similar to the wild type (Lin et al., 1996). Consistent with this, they did not produce mature stomata in the dark (Figure 3L), similar to the wild type. These results indicate that the light signaling components that modulate the activity of COP1 can also affect stomatal development.
COP1 Acts Constitutively to Regulate Postprotodermal Cell Development
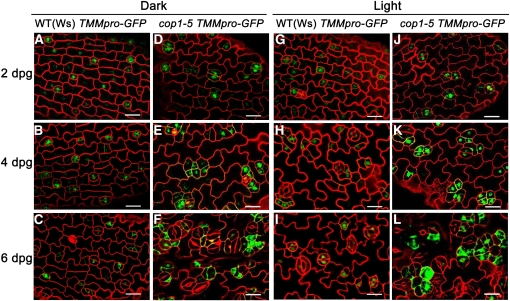
To determine how stomatal clusters are generated in the cop1-5 mutant, we examined the cotyledon epidermal development process of the wild type and cop1-5 mutants using a division-competent cell-specific GFP marker, TMMpro–GFP. At 2 d postgermination (dpg), GFP-positive cells were evenly dispersed in wild-type and cop1-5 epidermis in both light and darkness (Figures 4G, 4J, 4A, and 4D). During the next few days, development of the wild-type epidermis did not proceed further in the dark (Figures 4B and 4C), whereas it did in the light (Figures 4H and 4I). At 4 dpg, more than two adjacent cells established the identity of meristemoids in cop1-5 in both light and darkness (Figures 4E and 4K). The meristemoids later divided either symmetrically to generate guard cells or asymmetrically at random angles, resulting in the production of large stomatal clusters (Figures 4F and 4L). These results demonstrate that mutation in COP1 may constitutively affect postprotodermal cell development through regulating the patterning and/or asymmetric division orientation of meristemoids in both light and darkness.
Figure 4.
Time Sequence of Stomatal Differentiation in the cop1-5 Mutant.
Confocal images of the abaxial epidermis of wild-type and cop1-5 cotyledons. TMMpro–GFP (green) was used to monitor stomatal lineage cells. Red, PI counterstaining. Bars = 20 μm.
(A) to (C) Wild-type epidermis at 2, 4, and 6 dpg in the dark, respectively.
(D) to (F) cop1-5 epidermis at 2, 4, and 6 dpg in the dark, respectively.
(G) to (I) Wild-type epidermis at 2, 4, and 6 dpg in white light (150 μmol·m−2·s−1), respectively.
(J) to (L) cop1-5 epidermis at 2, 4, and 6 dpg in white light (150 μmol·m−2·s−1), respectively.
CRY1, CRY2, phyA, and phyB Act Together to Promote Stomatal Development
To explore the genetic interaction between cryptochromes and phytochromes in regulating stomatal development, we first analyzed the stomatal phenotype of the cotyledon epidermis of the cry1 cry2, phyA phyB, and cry1 phyA phyB mutants under blue plus red plus far-red light. Compared with the wild type, the SI of the cry1 cry2 and phyA phyB mutant epidermis was reduced, with that of the phyA phyB mutant being reduced more pronouncedly (see Supplemental Figures 5A and 5C online), but compared with the phyA phyB double mutant, stomatal development of the cry1 phyA phyB triple mutant was inhibited much more severely, as indicated by the production of many meristemoids and the significantly reduced stomatal size and SI (see Supplemental Figures 5A and 5C online).
We then determined the stomatal phenotype of the true leaf epidermis of these mutants under blue plus red plus far-red light and found that the SI of the leaf epidermis of the cry1 cry2 and phyA phyB mutants was significantly less than that of the wild type, and the SI of the leaf epidermis of the cry1 phyA phyB mutant was significantly less than that of the phyA phyB mutant (see Supplemental Figures 5B and 5D online). These results demonstrate that CRY1, CRY2, phyA, and phyB act additively to enhance stomatal development of both cotyledon and true leaf epidermis.
COP1 Genetically Acts Downstream of CRY, phyA, and phyB to Regulate Stomatal Development
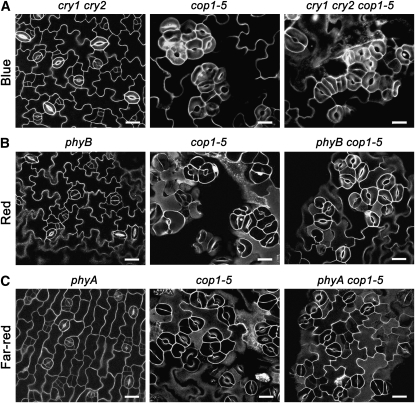
It is shown that COP1 genetically acts downstream of cryptochromes and phytochromes to regulate photomorphogenesis (Ang and Deng, 1994) and downstream of cryptochromes to regulate photoperiodic flowering (Liu et al., 2008). To explore the genetic interaction of COP1 with cryptochromes and phytochromes, we generated cry1 cry2 cop1-5, phyB cop1-5, and phyA cop1-5 mutants and compared the stomatal phenotype of these double or triple mutants with that of their parental single or double mutants under blue, red, and far-red light, respectively. As shown in Figures 5A to 5C, cry1 cry2 cop1-5, phyB cop1-5, and phyA cop1-5 mutants all produced clustered stomata under the corresponding light spectrum, similar to the cop1-5 single mutant, indicating that COP1 is epistatic to CRY1, CRY2, PHYA, and PHYB, respectively. The demonstrations that CRY1, CRY2, PHYA, PHYB, and COP1 are involved in the regulation of stomatal development predict that these genes may express in the stomatal lineage cells. To test this possibility, we investigated the expression pattern of these genes by generating transgenic plants expressing GUS fused to the promoters of CRY1, CRY2, PHYA, PHYB, and COP1, respectively. Histochemical staining for GUS activity indicated that they all are expressed in the stomatal lineage cells as well as in the pavement cells (see Supplemental Figure 6 online).
Figure 5.
COP1 Genetically Acts Downstream of CRY1, CRY2, phyA, and phyB.
(A) Confocal images of the cotyledon epidermis of cry1 cry2, cop1-5, and cry1 cry2 cop1-5 seedlings grown under blue light (30 μmol·m−2·s−1) for 10 d.
(B) Confocal images of the cotyledon epidermis of phyB, cop1-5, and phyB cop1-5 seedlings grown under red light (50 μmol·m−2·s−1) for 10 d.
(C) Confocal images of the cotyledon epidermis of phyA, cop1-5, and phyA cop1-5 seedlings grown under far-red light (12 μmol·m−2·s−1) for 10 d. Cell shapes were visualized by staining by PI. Bars = 20 μm.
CRY, phyA, and phyB Act Antagonistically with TMM to Regulate Stomatal Patterning
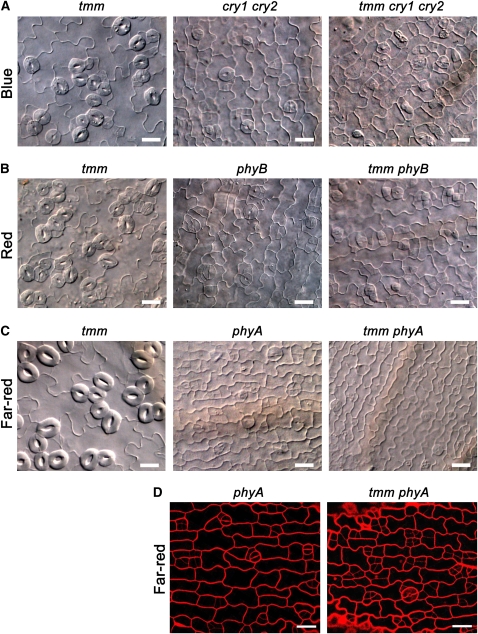
To place cryptochromes and phytochromes within the context of the known genetic pathways for stomatal patterning and differentiation, we constructed various double, triple, and quadruple mutants among cry1 cry2, phyA, and phyB mutants and the putative transmembrane receptor TMM mutant, tmm. Stomatal phenotype analysis of cotyledon epidermis demonstrated that, compared with the tmm single mutant, which produced mature stomata in clusters under blue light, the tmm cry1 cry2 triple mutant predominantly produced arrested meristemoids but still produced adjacent immature stomata and stomatal precursors that were not observed for the cry1 cry2 mutant (Figure 6A). Conversely, the tmm mutant produced mature clustered stomata under red light, whereas the tmm phyB double mutant primarily produced arrested meristemoids but still produced adjacent immature stomata and stomatal precursors that were not observed for the phyB mutant (Figure 6B). Strikingly, while the tmm mutant clearly produced mature clustered stomata under far-red light, the tmm phyA double mutant hardly produced stomata under far-red light and exhibited a phenotype similar to that observed for the phyA single mutant (Figure 6C). Close examination by confocal microscopy indicated that, compared with the phyA single mutant, the tmm phyA double mutant appeared to have executed more entry divisions and/or asymmetric cell divisions, resulting in the production of frequent adjacent meristemoids (Figure 6D).
Figure 6.
Mutations in Cryptochromes or Phytochromes Attenuate the Stomatal Cluster Phenotype of the tmm Mutant.
(A) DIC images of the cotyledon epidermis of tmm, cry1 cry2, and tmm cry1 cry2 seedlings grown in blue light (10 μmol·m−2·s−1) for 10 d.
(B) DIC images of the cotyledon epidermis of tmm, phyB, and tmm phyB seedlings grown in red light (10 μmol·m−2·s−1) for 10 d.
(C) DIC images of the cotyledon epidermis of tmm, phyA, and tmm phyA seedlings grown in far-red light (12 μmol·m−2·s−1) for 10 d.
(D) Confocal images of the cotyledon epidermis of phyA and tmm phyA prepared from (C). PI is presented in red. Bars = 20 μm.
Next, we analyzed the stomatal phenotype of the true leaf epidermis of these mutant plants under blue light plus red light. Compared with the tmm single mutant, no difference was observed for the tmm cry1 cry2 triple mutant in terms of the stomatal cluster size or SI, but a significant reduction in both stomatal cluster size and SI was observed for the tmm phyB double mutant (see Supplemental Figures 7A and 7B online). Moreover, compared with the tmm phyB double mutant, either the number or the size of the clustered stomata was significantly reduced in the tmm cry1 cry2 phyB quadruple mutant. Only occasionally did this quadruple mutant produce small clusters with two stomata adjacent to each other (see Supplemental Figures 7A online). Consistent with these observations, the SI of the tmm cry1 cry2 phyB quadruple mutant was significantly less great than that of the tmm phyB double mutant (see Supplemental Figures 7B online), indicating that CRY acts additively with phyB to attenuate tmm mutant phenotype. Nevertheless, the SI of the tmm cry1 cry2 phyB quadruple mutant is significantly greater than that of the cry1 cry2 phyB triple mutant (see Supplemental Figure 7B online). Taken together, these results demonstrate that CRY, phyA, and phyB act antagonistically with TMM in regulating stomatal patterning.
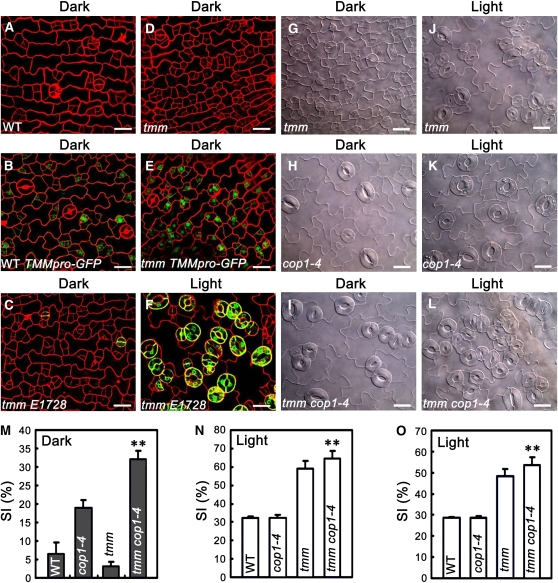
COP1 Acts Together with TMM to Repress Meristemoid Production of Cotyledon Epidermis
To investigate whether COP1 genetically interacts with TMM, we first characterized the stomatal phenotype of tmm mutant in the dark using TMMpro–GFP and E1728 markers. In darkness, tmm mutant cotyledon epidermis produced much more TMMpro–GFP-positive meristemoids than the wild type (Figures 7B and 7E). However, on rare occasions did the tmm mutant epidermal cells express the E1728 marker (Figure 7C), indicating that most of the tmm meristemoids are not able to develop to mature stomata in darkness. Next, we generated a tmm cop1-4 double mutant and analyzed the stomatal phenotype in both light and darkness. In contrast with the cop1-4 single mutant, which did not produce stomatal clusters in either light or darkness (Figures 3F, 3B, 7K, and 7H), the tmm cop1-4 double mutant clearly produced clustered stomata in darkness (Figure 7I) and produced larger stomatal clusters than the tmm single mutant in the light (Figures 7L and 7J). Consistent with these phenotypes, the SI of the tmm cop1-4 double mutant was significantly greater than that of the cop1-4 and tmm single mutants in the dark and in the light, respectively (Figures 7M and 7N). We further analyzed the SI of the true leaf epidermis of these mutants. As shown in Figure 7O, the SI of the tmm cop1-4 double mutant was significantly greater than that of the tmm single mutant. These results, in conjunction with those shown in Figure 4, suggest that TMM and COP1 act in parallel to repress meristemoid production of cotyledon epidermis.
Figure 7.
TMM and COP1 Act Additively to Regulate Stomatal Patterning.
(A) to (E) Confocal images of the cotyledon epidermis of 6-d-old dark-grown wild-type (A), WT TMMpro–GFP (B), tmm E1728 (C), tmm (D), and tmm TMMpro–GFP (E) seedlings.
(F) Confocal images of the cotyledon epidermis of tmm E1728 seedlings grown in white light (150 μmol·m−2·s−1) for 6 d. Red, PI counterstaining; green, GFP fluorescence.
(G) to (I) DIC images of the cotyledon epidermis of 10-d-old dark-grown tmm (G), cop1-4 (H) and tmm cop1-4 (I) seedlings.
(J) to (L) DIC images of the cotyledon epidermis of tmm (J), cop1-4 (K), and tmm cop1-4 (L) seedlings grown in blue (30 μmol·m−2·s−1) plus red (50 μmol·m−2·s−1) plus far-red light (6 μmol·m−2·s−1) for 10 d. Bars = 20 μm.
(M) The SI obtained from (G) to (I). Asterisks denote a significant difference between tmm cop1-4 and cop1-4 mutants (t test, P < 0.01), n = 10.
(N) The SI obtained from (J) to (L). Asterisks denote a significant difference between tmm cop1-4 and tmm mutants (t test, P < 0.01), n = 10.
(O) The SI of the abaxial true leaf epidermis of the various genotypes of adult plants grown in the same light condition as in (J) to (L) for ∼4 weeks. Asterisks denote a significant difference between tmm cop1-4 and tmm mutants (t test, P < 0.01), n = 5.
YDA Genetically Acts Downstream of COP1 to Regulate Stomatal Development and Patterning
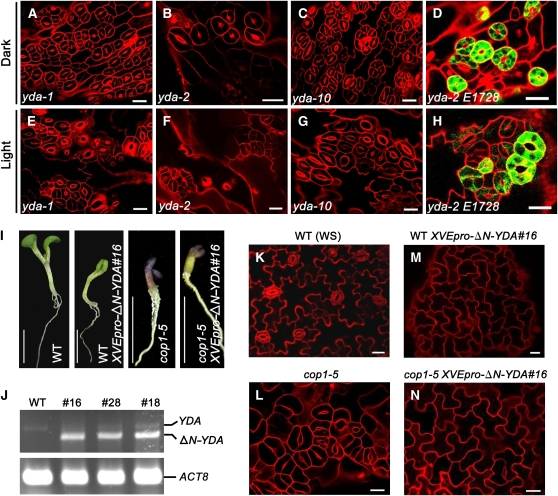
The MAPK signaling component MAPK kinase kinase, YDA, is a critical negative regulator of stomatal development and patterning (Bergmann et al., 2004), which acts upstream of MKK4/5 and MPK3/6 (Wang et al., 2007). It is known that the cotyledon epidermis of the light-grown yda mutants produces large stomatal clusters (Bergmann et al., 2004). To investigate whether YDA acts constitutively to repress stomatal development and patterning, we examined the stomatal phenotype of three mutant alleles of yda, yda-1, yda-2, and yda-10, in the light and darkness, respectively. Strikingly, all these yda mutants constitutively produced large clusters of stomata in both light and darkness. Compared with yda-1 and yda-10 mutants, the yda-2 mutant produced relatively smaller stomatal clusters. To determine whether the stomatal clusters observed for the yda mutants have a mature guard cell identity, we crossed the E1728 marker into the yda-2 mutant background. As shown in Figures 8D and 8H, yda-2 mutant cotyledon epidermis constitutively expressed E1728 in both light and darkness.
Figure 8.
YDA Genetically Acts Downstream of COP1.
(A) to (D) Confocal images of the cotyledon epidermis of 7-d-old dark-grown yda-1 (A), yda-2 (B), yda-10 (C), and yda-2 E1728 (D) seedlings.
(E) to (H) Confocal images of the cotyledon epidermis of yda-1 (E), yda-2 (F), yda-10 (G), and yda-2 E1728 (H) seedlings grown in white light (150 μmol·m−2·s−1) for 7 d. Bars = 20 μm.
(I) Transgenic XVEpro–ΔN–YDA#16 seedlings at 7 dpg under white light (150 μmol·m−2·s−1). Seedlings are in the wild-type or in the cop1-5 mutant background as indicated. Bars = 0.5 cm.
(J) RT-PCR analysis of leaky expression of ΔN–YDA in three independent transgenic lines XVEpro–ΔN–YDA#16, #28, and #18. YDA denotes endogenous YDA expression. ACT8 was used as a loading control.
(K) to (N) Confocal images of the cotyledon epidermis of wild-type (K), cop1-5 (L), WT XVEpro–ΔN–YDA#16 (M), and cop1-5 XVEpro–ΔN–YDA#16 (N) seedlings prepared from (I). Red, PI counterstaining; green, GFP fluorescence. Bars = 20 μm.
The similar constitutive stomatal cluster phenotype observed for yda and cop1-5 mutants prompted us to investigate whether COP1 genetically interacts with YDA. It has been shown that YDA protein lacking the N-terminal fragment (ΔN–YDA) is constitutively active and that the cotyledon epidermis of transgenic seedlings expressing ΔN–YDA produces no stomata (Bergmann et al., 2004). To explore the genetic interaction of COP1 and YDA, we generated a construct expressing ΔN–YDA fused to a chemical-inducible promoter, XVE (Zuo et al., 2000), and transformed it into the heterozygous cop1-5 mutant plants (cop1-5/+). Twenty-nine independent transgenic T1 lines were obtained. In the T2 generation, these cop1-5/+ XVEpro–ΔN–YDA lines were induced with β-estradiol at a variety of concentrations, and they exhibited a hypersensitive response, having a major difficulty in germination and growth, and failing to develop cotyledons. However, of these 29 lines, 12 lines segregated pale-green seedlings with cotyledons that were folded and not fully expanded in T2 generation without β-estradiol induction, similar to the transgenic seedlings expressing ΔN–YDA with the native promoter of YDA reported previously (Figure 2C; Bergmann et al., 2004). The T2 seedlings of one representative line, cop1-5/+ XVEpro–ΔN–YDA#16, are shown in Figure 8I. RT-PCR analysis clearly detected the expression of ΔN–YDA in these lines (Figure 8J), suggesting that the leaky activity of the XVE promoter, which has been reported by others (Degenhardt and Bonham-Smith, 2008; Vlot et al., 2008), is sufficient to drive ΔN–YDA expression and exert biological function. We conducted stomatal phenotype analysis on the seedlings with cotyledons that were folded and not fully expanded from four independent cop1-5/+ XVEpro–ΔN–YDA lines and found that they all failed to produce stomata. The results obtained from the representative line cop1-5/+ XVEpro–ΔN–YDA#16 are shown in Figures 8M and 8N, which demonstrate that expression of ΔN–YDA in either the wild type or the cop1-5 mutant background leads to no production of stomata. These results therefore indicate that YDA is epistatic to COP1.
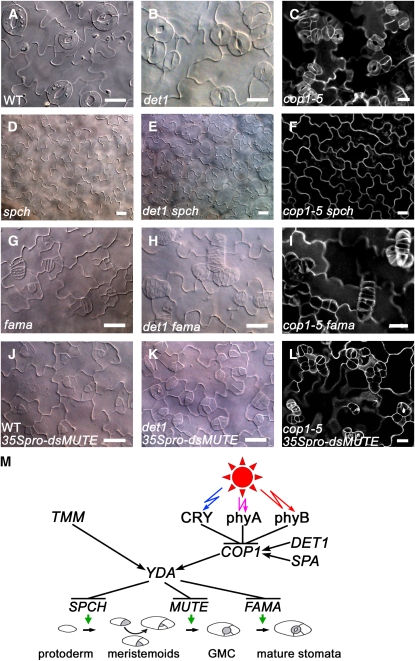
SPCH, MUTE, and FAMA Genetically Act Downstream of COP1 and DET1 to Regulate Stomatal Development and Patterning
It is shown that SPCH genetically acts downstream of YDA (MacAlister et al., 2007). To investigate whether COP1 and DET1 genetically interact with SPCH, MUTE, and FAMA, we first generated det1 spch, cop1-5 spch, det1 fama, and cop1-5 fama double mutants and analyzed the stomatal phenotype. The results demonstrated the cotyledon epidermis of the det1 spch and cop1-5 spch double mutants produced no stomata at all (Figures 9E and 9F), similar to that of the spch single mutant (Figure 9D), and the overall stomatal phenotype of the det1 fama and cop1-5 fama double mutants resembles that of the fama single mutant (Figures 9G to 9I), although the caterpillar-like rows of guard mother cell phenotype observed for the fama mutant seem to be exaggerated in the det1 fama and cop1-5 fama double mutant backgrounds. These results indicate that SPCH and FAMA are epistatic to DET1 and COP1. We then made a construct expressing a double-stranded RNA of MUTE (dsMUTE) fused to the 35S promoter and obtained a transgenic line that constitutively expresses the same 35pro–dsMUTE transgene in the wild type (WT 35pro–dsMUTE), det1 (det1 35pro–dsMUTE), and cop1-5 (cop1-5 35pro–dsMUTE) mutant backgrounds, respectively. As shown in Figures 9J to 9L, the overall stomatal phenotypes of det1 35pro–dsMUTE and cop1-5 35pro–dsMUTE are identical to those of WT 35pro–dsMUTE, although the arrested meristemoids observed for WT 35pro–dsMUTE appear to be aggregated in the det1 35pro–dsMUTE and cop1-5 35pro–dsMUTE. These results indicate that MUTE is also epistatic to DET1 and COP1.
Figure 9.
Genetic Interactions of COP1 and DET1 with SPCH, MUTE, and FAMA.
(A) to (L) Abaxial cotyledon epidermis of 10-d-old white-light (150 μmol·m−2·s−1)-grown wild type (A), det1 (B), cop1-5 (C), spch (D), det1 spch (E), cop1-5 spch (F), fama (G), det1 fama (H), cop1-5 fama (I), WT 35Spro–dsMUTE (J), det1 35Spro–dsMUTE (K), and cop1-5 35Spro–dsMUTE (L). The images in (C), (F), (I), and (L) are PI (white)-outlined photomicrographs, and the others are DIC photomicrographs. Bars = 20 μm.
(M) A genetic model for the light signaling pathway and its interaction with the developmental pathway. COP1 activity is negatively regulated by CRY, phyA, and phyB but is positively regulated by SPA and DET1, presumably through physical interactions (Wang et al., 2001; Yang et al., 2001; Seo et al., 2003; Yanagawa et al., 2004). YDA might be positively regulated by COP1 through yet unknown mechanisms. Arrow, positive regulation; T-bar, negative regulation. GMC, guard mother cell.
DISCUSSION
Previous studies demonstrated that the production of stomata is promoted by light (Lake et al., 2001) and that phyB is involved in this process (Boccalandro et al., 2009; Casson et al., 2009). In this report, we characterized the involvement of CRY, phyA, phyB, and COP1 in the regulation of light-controlled stomatal development and demonstrated that CRY and phyB are the primary photoreceptors mediating blue and red light–induced stomatal development, respectively. We also showed that phyA is the sole photoreceptor mediating far-red light–induced stomatal development and that COP1 is a key repressor of light-promoted stomatal development. Moreover, we established the genetic pathway of cryptochrome- and phytochrome-mediated light-induced stomatal development and its genetic interaction with the developmental pathway. These findings represent a significant progress in our understanding of the genetic basis of light-controlled stomatal development, providing a genetic link between the cryptochrome- and phytochrome-mediated light signaling pathway and the YDA-regulated MAPK signaling pathway.
Cryptochromes and Phytochromes Are Involved in Promoting the Entire Stomatal Development Process
To date, light regulation of stomatal development has not been investigated with the cotyledon epidermis from seedlings grown under monochromatic blue, red, and far-red lights; thus, the role for the photoreceptors that mediate specific light signaling to promote stomatal development during the early developmental stage of seedlings has not been revealed. In this report, by analyzing the stomatal phenotype of the cotyledon epidermis of the various photoreceptor mutants under different monochromatic light, we are able to demonstrate that CRY, phyB, and phyA are responsible for mediating blue, red, and far-red light–induced stomatal development, respectively. To note, based on the decrease in SI, as well as SMI in the blue light–grown cry1 cry2, red light–grown phyB, and far-red light–grown phyA mutants (Figures 1B, 1C, 2B, and 2C), we conclude that the many meristemoids observed for these mutants do not reflect a role for CRY, phyA, and phyB in repressing the production of meristemoids. On the contrary, they reflect arrested stomatal development in these mutants and indicate a role for these photoreceptors in promoting the formation of meristemoids and stomata. Moreover, we showed that stomatal maturation is compromised in the various photoreceptor mutants and the tmm/photoreceptor-deficient double and triple mutants (Figure 6). Therefore, CRY, phyA, and phyB likely function together to positively regulate the entire stomatal development process from the asymmetric cell division throughout the cell division of the guard mother cells.
COP1 Is a Key Negative Regulator of Stomatal Differentiation and Development
Although it is shown that light signals enhance stomatal development of true leaf epidermis (Lake et al., 2001; Casson et al., 2009), the stomatal development of cotyledon epidermis of dark-grown seedlings has not carefully been examined. In this study, using a mature guard cell–specific marker E1728, we demonstrate that, in the wild-type background, stomata are developed to maturity in the light but hardly developed to maturity in the dark, whereas in the cop1-5 mutant background, stomata are constitutively developed to maturity and produced in clusters in both light and darkness. Furthermore, by introducing a meristemoid-specific TMMpro–GFP marker into the cop1-5 mutant background, we demonstrate that COP1 likely constitutively regulates the asymmetric division of meristemoids in both light and darkness. These findings suggest that the establishment of the mature guard cell identity is light dependent and that, in contrast with CRY, phyA, and phyB, COP1 acts to constitutively repress the entire stomatal development process.
COP1 activity is regulated by CRY1 and CRY2 and the multiple components of the COP/DET/FUS proteins, which are negative regulators of photomorphogenic development (Chory et al., 1989; Deng et al., 1991; Misera et al., 1994; Wei et al., 1994a). It has been shown that CRY1 and CRY2 negatively regulate COP1 through physical interaction of CCT1 and CCT2 with COP1 (Wang et al., 2001; Yang et al., 2001). Among the COP/DET/FUS proteins, DET1 was initially shown to be involved in regulating chromatin remodeling and gene expression (Benvenuto et al., 2002; Schroeder et al., 2002). The COP9 signalosome (CSN) is a constitutively nuclear-enriched protein complex consisting of eight distinct subunits that is required for the nuclear accumulation of COP1 (Chamovitz et al., 1996). COP10 is a ubiquitin conjugating enzyme variant (Suzuki et al., 2002), which forms a CDD complex with DDB1 and DET1 and interacts with both CSN and COP1 (Yanagawa et al., 2004). The E3 ligase activity of COP1 also requires SPA1 (Seo et al., 2003), initially identified as a nuclear-localized repressor of far-red light signaling (Hoecker et al., 1998, 1999). Based on these studies, it is reasonable to speculate that the light signaling components that modulate the activity of COP1 would be able to affect stomatal development. Consistent with this speculation, clustered stomata are shown to be produced in the cop10, csn1/cop11, csn4/cop8 (Wei et al., 1994b; Serna and Fenoll, 2000), and det1 mutants, as well as blue light–grown 35Spro–CRY1 seedlings (this report), and the constitutive stomatal development was observed in the spa1 spa2 spa3 mutants and transgenic 35Spro–GUS–CCT1 seedlings (this report).
CRY, phyA, phyB, and COP1 May Act in Parallel with TMM to Regulate Stomatal Patterning
The role for TMM in the control of the orientation of asymmetric divisions that create the minimal one-celled stomatal spacing pattern has been characterized in the light (Nadeau and Sack, 2002). Whether TMM plays a role in regulating stomatal patterning in the dark and in the crytpchrome-, phytochrome-, and COP1-deficient mutants remains unknown. In this study, we demonstrate that the cotyledon epidermis of tmm mutant produces dramatic clustered stomata in the blue, red, and far-red light, respectively. However, the clustered stomatal phenotype observed for tmm mutant is severely compromised in tmm cry1 cry2, tmm phyB, and tmm phyA mutants under blue, red, and far-red light, respectively (Figure 6). From these data, we propose that the multiple photoreceptor-mediated light signals are required for both stomatal maturation and stomatal patterning. This proposition is supported by the demonstrations that stomata are not developed to maturity in tmm mutant in the dark (Figure 7C) and that the clustered stomatal phenotype observed for the true leaf epidermis of tmm mutant is dramatically attenuated in the true leaf epidermis of tmm phyB and tmm cry1 cry2 phyB mutants (see Supplemental Figures 7A and 7B online), being most severely compromised in the latter mutant. By contrast, the cotyledon epidermis of the tmm cop1-4 double mutant makes larger stomatal clusters than the tmm single mutant in the light, and the true leaf epidermis of the tmm cop1-4 double mutant produces more stomata than the tmm single mutant (this report). Therefore, we conclude that CRY, phyA, and phyB act antagonistically with TMM to regulate stomatal patterning and that COP1 and TMM act additively to repress the asymmetric division of meristemoids.
Light-Controlled Stomatal Development May Be Mediated through a Crosstalk between the Cryptochrome-Phytochrome-COP1 Signaling System and the MAPK Signaling Pathway
Our observation that cop1 mutants constitutively produce stomata with a mature guard cell identity in the dark suggests that the downstream transcriptional factors required for promoting stomatal development and differentiation, such as SPCH, MUTE, and FAMA, must be constitutively activated in these mutants. To test this possibility, we investigated whether the expression of SPCH, MUTE, and FAMA is regulated by the light signaling components through quantitative real-time RT-PCR. In the dark, higher expression levels of SPCH, MUTE, and FAMA were detected in the cop1-4 mutant (see Supplemental Figure 8A online). Conversely, the expression of SPCH, MUTE, and FAMA was reduced in the blue light–grown cry1 cry2 mutant, red light–grown phyB mutant, and far-red light–grown phyA mutant, respectively, but was enhanced in the blue light–grown 35Spro–CRY1 and red light–grown 35Spro–PHYB seedlings, respectively (see Supplemental Figures 8B to 8D online). We then generated transgenic plants expressing GUS fused to the native promoters of SPCH, MUTE, and FAMA, respectively. Histochemical staining for GUS activity indicated that they all expressed in cotyledon epidermis of seedlings grown in either continuous light or darkness (see Supplemental Figures 9A and 9B online). Furthermore, it appears likely that dark adaptation of the light-grown seedlings did not make any difference in GUS staining (see Supplemental Figures 9C and 9D online). Thus, the differences in the expression levels of SPCH, MUTE, and FAMA obtained from quantitative RT-PCR might result from the differences in the stomatal lineage cell density in the different genotypes of seedlings. These data therefore indicate that SPCH, MUTE, and FAMA may not primarily be regulated by the light signaling at the transcriptional level and that posttranscriptional regulation mechanisms might be involved. Indeed, SPCH is shown to be regulated by MAPK signaling through phosphorylation (Lampard et al., 2008). It will be worth investigating whether SPCH, MUTE, and FAMA can be posttranscriptionally regulated by light signaling. To note, we do not exclude the possibility that other components, such as ICE1/SCREAM and SCREAM2 (Kanaoka et al., 2008), are regulated by light signaling at the transcriptional level and/or at the posttranscriptional level.
It is intriguing to note that cop1 and yda mutants exhibit a similar constitutive clustered stomatal phenotype in the dark. The demonstrations that mutations in either YDA or COP1 affect postprotodermal cell development (Bergmann et al., 2004; this report), and the constitutive clustered stomatal phenotype observed for the yda mutants appears to be stronger than that observed for the cop1 mutants (Figures 8A to 8H versus Figures 3B, 3F, 3J, and 3N), and that YDA acts genetically downstream of COP1 to regulate stomatal development and patterning strongly indicate that YDA might be regulated by COP1. It is shown that SPCH acts genetically downstream of YDA (MacAlister et al., 2007). Based on previous genetic studies and our results, we establish an overall genetic pathway of light-controlled stomatal development and its interaction with the developmental signaling pathway (Figure 9M), in which COP1 lies downstream of CRY, phyA, and phyB photoreceptors, in parallel with TMM, but upstream of YDA, SPCH, MUTE, and FAMA, respectively. From this genetic model, we propose that COP1- and TMM/ER/ERL-mediated signaling converges to YDA. This proposition is supported by the demonstrations that YDA acts downstream of COP1 and TMM (Bergmann et al., 2004; this report) and that yda mutants exhibit stronger constitutive clustered stomatal phenotype than either cop1 or tmm mutant in the dark (this report). It is intriguing to predict from this genetic model that COP1 plays a pivotal role in integrating the CRY-, phyA-, and phyB-mediated signals to the MAPK cascade consisting of the YDA-MKK4/MKK5-MPK3/MPK6 module (Wang et al., 2007).
How can YDA be regulated by COP1? COP1 is a RING motif–containing E3 ubiquitin ligase and its identified substrates include HY5, LAF1, phyA, HFR1, and CO (Osterlund et al., 2000; Seo et al., 2004; Jang et al., 2005; Yang et al., 2005; Liu et al., 2008). YDA is a MAPK kinase kinase that acts upstream of MKK4/5 and MPK3/6 in the MAPK signaling pathway (Wang et al., 2007). COP1 localizes to the nucleus in the dark, whereas it is relocated to the cytoplasm in the light (von Arnim and Deng, 1994), and this nucleocytoplasmic translocation of COP1 is mediated by CRY1, phyA, and phyB (Osterlund and Deng, 1998). The cellular localization property of YDA is currently unknown. It will be intriguing to investigate whether YDA gene expression and/or the stability and/or the kinase activity and/or the cellular localization property of YDA can be regulated by COP1 through molecular, biochemical, and cytobiological studies. Elucidation of the regulatory mechanisms by which COP1 regulates YDA in future studies will make it possible to establish the molecular link between the cryptochrome-phytochrome signaling pathway and the MAPK signaling pathway in the regulation of stomatal production.
METHODS
Light Source
Monochromatic blue, red, and far-red light was generated from blue diodes (λ max = 469 nm), red diodes (λ max = 680 nm), and far-red diodes (λ max = 730 nm) of either an LED screen (Qiding) or an E-30 LED growth chamber (Percival), respectively. Mixtures of blue light plus red light and blue light plus red light plus far-red light were made by appropriate combinations of these monochromatic lights. Light spectra were analyzed with a HandHeld spectroradiometer (ASD). A Li250 quantum photometer (Li-Cor) was used to measure the fluence rates of blue and red light, and an ILT1400-A radiometric photometer (ILT) was used to measure the fluence rate of far-red light. The light spectra of the monochromatic blue, red, and far-red light and the mixtures of blue light plus red light and blue light plus red light plus far-red light are shown in Supplemental Figure 1 online.
Plant Materials and Growth Conditions
The Arabidopsis thaliana Columbia accession was used as the wild type unless noted otherwise. All mutants were generated in the Columbia background, except for cop1-5 (McNellis et al., 1994), which was generated in the Wassilewskija background, and yda-1 and yda-2 (Lukowitz et al., 2004), which were generated in the Landsberg erecta background. Ethyl methanesulfonate–mutagenized tmm-1 (Nadeau and Sack, 2002), yda-1 and yda-2, T-DNA insertion alleles of cop1-5 (adult-lethal, kanamycin-resistant, and purple-colored seeds and seedlings), fama (Salk_100073, kanamycin-resistant), spch (SAIL_36_B06, basta-resistant), yda-10 (SALK_105078C), det1-1s (adult-lethal, basta-resistant, and purple-colored seeds and seedlings), spa1 (SALK_023840), and spa2 (SALK_083331) were obtained from the ABRC (Ohio State University). spa3 (FLAG_414C10) was generated in the Wassilewskija ecotype background and obtained from the Versailles Genetics and Plant Breeding Laboratory Arabidopsis Resource Centre. cry1-104, cry2-1, phyA-211, phyB-9, cop1-4, cry1 cry2, hy1, 35Spro–CRY1, and 35Spro–GUS–CCT1 have been described previously (Yang et al., 2000; Mao et al., 2005). Seeds were sterilized with 20% bleach, plated on Murashige and Skoog medium (Sigma-Aldrich), and put in a 4°C refrigerator for 3 to 5 d. Germination was then induced in 150 μmol m−2 s−1 fluorescent cool white light (Philips, 36W/840) for 24 h, and finally the seedlings were transferred to the experimental light treatments at 22°C.
Construction of Double, Triple, and Quadruple Mutants
The following double, triple, and quadruple mutants were generated for this study: phyA phyB, cry1 phyA phyB, cry1 cry2 phyB, cry1 cry2 cop1-5, phyA cop1-5, phyB cop1-5, tmm cry1 cry2, tmm phyA, tmm phyB, tmm cry1 cry2 phyB, tmm cop1-4, cop1-5 spch, cop1-5 fama, det1 spch, det1 fama, cop1-5 E1728, det1 E1728, and yda-2 E1728. E1728 is an enhancer trap line (http://enhancertraps.bio.upenn.edu). See Supplemental Table 1 online for a list of parental mutants used for sexual crosses and Supplemental Table 2 online for a list of PCR primer sequences used for genotyping. For details, see Supplemental Methods online.
Construction of Plant Expression Cassettes and Transformation of Plants
The following plant expression vectors were generated for this study: pKYL71–35Spro–PHYB–YFP, pCambia1300–CRY1pro–GUS, pCambia1300–CRY2pro–GUS, pCambia1300–PHYApro–GUS, pCambia1300–PHYBpro–GUS, pCambia1300–COP1pro–GUS, pCambia1300–SPCHpro–GUS, pCambia1300–MUTEpro–GUS, pCambia1300–FAMApro–GUS, pCambia1302–TMMpro–GFP, pHB–35Spro–dsMUTE, and pER8–XVEpro–ΔN–YDA. See Supplemental Table 2 online for a list of PCR primer sequences used for plasmid construction. All plant expression constructs were introduced into the Agrobacterium tumefaciens strain GV3101 and then transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transgenic seeds were screened on half-strength Murashige and Skoog plates containing either 100 mg/mL kanamycin or 50 mg/mL hygromycin. At least 10 independent transgenic lines were characterized for each construct. For details, see Supplemental Methods online.
Microscopy
DIC microscopy was used for phenotypic quantifications and for some qualitative analyses. The cotyledon phenotypes were observed on the 11th day after germination, and the leaves were collected when the leaf margins began to dry up, which indicates full maturity of the leaf. These samples were preserved in 95% ethanol, rehydrated in a graded ethanol series, and placed in the clear solution (glycerol:chloral hydrate: water, 1:8:1) overnight. Samples mounted in the clear solution were visualized using a Leica DM2500 microscope with Nomarsky optics. An Olympus FluoView FV1000 confocal laser scanning microscope was used to capture PI (Sigma-Aldrich) staining and GFP fluorescence images. To counterstain the epidermal cell shapes, tissues were stained with 50 μg/mL PI for a few minutes and rinsed briefly with distilled water before visualization.
Determination of the SI
SI was calculated using the equation SI = number of stomata/(number of stomata + number of pavement cells) × 100%, and SMI was calculated using the equation SMI = (number of stomata + number of meristemoids)/(number of stomata + number of meristemoids + number of pavement cells) × 100%. Only stomata with pores were counted. Counts of stomata and pavement cells were performed using a Leica DM2500 microscope in two square areas of 0.2 mm2 per cotyledon from 10 cotyledons of 10 independent seedlings and five areas per leaf from five leaves of five independent plants, respectively. The SI was calculated for each cotyledon or leaf individually, and the mean and sd were then calculated from these data. For statistical analysis, an unpaired Student's t test was performed.
GUS Staining Assays
The histochemical GUS assays were performed according to standard protocols (Sessions et al., 1999) with minor modifications. Six-day-old seedlings were placed directly into GUS reaction buffer (0.5 mg/mL X-glucuronic acid, 10 mM EDTA, 0.1% Triton X-100, and 2 mM [see Supplemental Figure 6 online] or 10 mM [see Supplemental Figure 9 online] potassium ferri/ferrocyanide in 50 mM phosphate buffer, pH 7.0). After evacuation for 20 min, they were incubated overnight at 37°C. Stained tissues were cleared in 70% ethanol and detected using DIC microscopy, as described above.
RNA Extraction and Quantitative Real-Time PCR
The cotyledons of 6-d-old seedlings were collected and total RNA was extracted using RNArose reagent (Watson). Real-time PCR was performed using the Rotor Gene RG 3000 thermocycler (Corbett Research). For details, see Supplemental Methods online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CRY1 (At4g08920), CRY2 (At1g04400), PHYA (At1g09570), PHYB (At2g18790), COP1 (At2g32950), SPCH (At5g53210), MUTE (At3g06120), FAMA (At3g24140), TMM (At1g80080), YDA (At1g63700), SPA1 (At2g46340), SPA2 (At4g11110), SPA3 (At3g15354), ACT8 (At1g49240), and UBQ10 (At4g05320). T-DNA insertion alleles used were cop1-5 (CS6259), fama (SALK_100073), spch (SAIL_36_B06), yda-10 (SALK_105078C), det1-1s (CS16146), spa1 (SALK_023840), spa2 (SALK_083331), and spa3 (FLAG_414C10).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Light Spectra.
Supplemental Figure 2. The cry1 cry2 Mutant and Transgenic Seedlings Overexpressing CRY1 Exhibit Dramatic Opposing Stomatal Phenotypes in Response to Blue Light.
Supplemental Figure 3. The phyB Mutant and Transgenic Seedlings Overexpressing PHYB Display Opposite Stomatal Phenotypes in Response to Red Light.
Supplemental Figure 4. The phyA Mutant Shows Little Sign of Stomatal Development in Response to Far-Red Light.
Supplemental Figure 5. Cryptochromes and Phytochromes Function Additively to Regulate Stomatal Development and Patterning.
Supplemental Figure 6. Promoter Activities of CRY1, CRY2, PHYA, PHYB, and COP1 Genes in Cotyledon Epidermal Cells.
Supplemental Figure 7. Cryptochromes and Phytochromes Act Additively to Antagonize TMM in the Regulation of Stomatal Patterning.
Supplemental Figure 8. Effects of Light Signaling Components on Relative Expression of SPCH, MUTE, and FAMA Genes.
Supplemental Figure 9. Promoter Activities of SPCH, MUTE, and FAMA Genes in Different Light Conditions.
Supplemental Table 1. Double, Triple, and Quadruple Mutants Constructed and Their Parental Mutants.
Supplemental Table 2. Primers for Mutant Genotyping, Plasmid Construction, RT-PCR, and Quantitative RT-PCR.
Supplemental Methods.
Supplemental References.
Supplementary Material
Acknowledgments
We thank members of the lab for discussions and comments on the manuscript, N.H. Chua for pER8 vector, S. Poethig for E1728 marker, H.W. Xue for a GUS vector, and X.W. Deng for helpful comments. Some stocks were obtained from the ABRC at Ohio State University. This work was supported by the National Natural Science Foundation of China (30830012 to H.-Q.Y.), the Ministry of Science and Technology of China (2006AA10A102), National Special Grant for Transgenic Crops (2009ZX08009-081B to H.-Q.Y.), and the Shanghai Leading Academic Discipline Project (B209).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hong-Quan Yang (hqyang@sjtu.edu.cn).
Online version contains Web-only data.
References
- Al-Sady, B., Ni, W., Kircher, S., Schäfer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., and Deng, X.W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M., and Wang, X.Q. (2001). From milliseconds to millions of years: Guard cells and environmental responses. Curr. Opin. Plant Biol. 4 421–428. [DOI] [PubMed] [Google Scholar]
- Benvenuto, G., Formiggini, F., Laflamme, P., Malakhov, M., and Bowler, C. (2002). The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 12 1529–1534. [DOI] [PubMed] [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bergmann, D.C., Lukowitz, W., and Somerville, C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304 1494–1497. [DOI] [PubMed] [Google Scholar]
- Boccalandro, H.E., Rugnone, M.L., Moreno, J.E., Ploschuk, E.L., Serna, L., Yanovsky, M.J., and Casal, J.J. (2009). Phytochrome B enhances photosynthesis at the expense of water use efficiency in Arabidopsis. Plant Physiol. 150: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7 204–210. [DOI] [PubMed] [Google Scholar]
- Casal, J.J., and Mazzella, M.A. (1998). Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol. 118 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284 760–765. [DOI] [PubMed] [Google Scholar]
- Casson, S.A., Franklin, K.A., Gray, J.E., Grierson, C.S., Whitelam, G.C., and Hetherington, A.M. (2009). Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol. 19: 229–234. [DOI] [PubMed] [Google Scholar]
- Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121. [DOI] [PubMed] [Google Scholar]
- Chory, J. (1993). Out of darkness: Mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 9 167–172. [DOI] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 991–999. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., Kurepa, J., and Vierstra, R.D. (1999). The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc. Natl. Acad. Sci. USA 96 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt, R.F., and Bonham-Smith, P.C. (2008). Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are desperately required for normal development. Plant Physiol. 147 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.W., Caspar, T., and Quail, P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71 791–801. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2 83–95. [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Gray, J.E., Holroyd, G.H., van der Lee, F.M., Bahrami, A.R., Sijmons, P.C., Woodward, F.I., Schuch, W., and Hetherington, A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408 713–716. [DOI] [PubMed] [Google Scholar]
- Hara, K., Kajita, R., Torii, K.U., Bergmann, D.C., and Kakimoto, T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington, A.M., and Woodward, F.I. (2003). The role of stomata in sensing and driving environmental change. Nature 424 901–908. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Xu, Y., and Quail, P.H. (1998). SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jang, I.C., Yang, J.Y., Seo, H.S., and Chua, N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S., Marchal, V., Panigrahi, K.C., Wenkel, S., Soppe, W., Deng, X.W., Valverde, F., and Coupland, G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka, M.M., Pillitteri, L.J., Fujii, H., Yoshida, Y., Bogenschutz, N.L., Takabayashi, J., Zhu, J.K., and Torii, K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M., and Shimazaki, K. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414 656–660. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Misera, S., and Deng, X.W. (1996). A complement of ten essential and pleiotropic arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake, J.A., Quick, W.P., Beerling, D.J., and Woodward, F.I. (2001). Plant development. Signals from mature to new leaves. Nature 411 154. [DOI] [PubMed] [Google Scholar]
- Lampard, G.R., Macalister, C.A., and Bergmann, D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322 1113–1116. [DOI] [PubMed] [Google Scholar]
- Laubinger, S., Fittinghoff, K., and Hoecker, U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q.H., and Yang, H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83 94–101. [DOI] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., and Cashmore, A.R. (1996). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10 893–902. [DOI] [PubMed] [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Liu, L.J., Zhang, Y.C., Li, Q.H., Sang, Y., Mao, J., Lian, H.L., Wang, L., and Yang, H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Roeder, A., Parmenter, D., and Somerville, C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116 109–119. [DOI] [PubMed] [Google Scholar]
- MacAlister, C.A., Ohashi-Ito, K., and Bergmann, D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445 537–540. [DOI] [PubMed] [Google Scholar]
- Mao, J., Zhang, Y.C., Sang, Y., Li, Q.H., and Yang, H.Q. (2005). A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle, J., Gilmore, S.R., and Farquhar, G.D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misera, S., Muller, A.J., Weiland-Heidecker, U., and Jurgens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol. Gen. Genet. 244 242–252. [DOI] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nagatani, A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7 708–711. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400 781–784. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., and Bergmann, D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., and Deng, X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Pillitteri, L.J., Sloan, D.B., Bogenschutz, N.L., and Torii, K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445 501–505. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3 85–93. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt, G., Viana, R.M., Mootz, H.D., von Arnim, A.G., and Batschauer, A. (2008). Chemically induced and light-independent cryptochrome photoreceptor activation. Mol. Plant 1 4–14. [DOI] [PubMed] [Google Scholar]
- Sack, F.D. (2004). Plant sciences. Yoda would be proud: Valves for land plants. Science 304 1461–1462. [DOI] [PubMed] [Google Scholar]
- Sancar, A. (1994). Structure and function of DNA photolyase. Biochemistry 33 2–9. [DOI] [PubMed] [Google Scholar]
- Sang, Y., Li, Q.H., Rubio, V., Zhang, Y.C., Mao, J., Deng, X.W., and Yang, H.Q. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, D.F., Gahrtz, M., Maxwell, B.B., Cook, R.K., Kan, J.M., Alonso, J.M., Ecker, J.R., and Chory, J. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12 1462–1472. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Allen, G.J., Hugouvieux, V., Kwak, J.M., and Waner, D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 627–658. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Serna, L., and Fenoll, C. (2000). Stomatal development in Arabidopsis: How to make a functional pattern. Trends Plant Sci. 5 458–460. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20 259–263. [DOI] [PubMed] [Google Scholar]
- Shpak, E.D., McAbee, J.M., Pillitteri, L.J., and Torii, K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293. [DOI] [PubMed] [Google Scholar]
- Smith, H. (2000). Phytochromes and light signal perception by plants–An emerging synthesis. Nature 407 585–591. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C., Liu, P.P., Cameron, R.K., Park, S.W., Yang, Y., Kumar, D., Zhou, F., Padukkavidana, T., Gustafsson, C., Pichersky, E., and Klessig, D.F. (2008). Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56 445–456. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294 154–158. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ngwenyama, N., Liu, Y., Walker, J.C., and Zhang, S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.W. (1994. a). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, N., Kwok, S.F., von Arnim, A.G., Lee, A., McNellis, T.W., Piekos, B., and Deng, X.W. (1994. b). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., and Deng, X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.Q., Wu, Y.J., Tang, R.H., Liu, D., Liu, Y., and Cashmore, A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103 815–827. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Shalitin, D., Liu, X., Maymon, M., Klejnot, J., Yang, H., Lopez, J., Zhao, X., Bendehakkalu, K.T., and Lin, C. (2007). Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. USA 104 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.