Abstract
AGAMOUS-Like15 (AGL15) is a MADS domain transcriptional regulator that promotes somatic embryogenesis by binding DNA and regulating gene expression. Chromatin immunoprecipitation (ChIP) analysis previously identified DNA fragments with which AGL15 associates in vivo, and a low-throughput approach revealed a role for AGL15 in gibberellic acid catabolism that is relevant to embryogenesis. However, higher throughput methods are needed to identify targets of AGL15. Here, we mapped AGL15 in vivo binding sites using a ChIP-chip approach and the Affymetrix tiling arrays for Arabidopsis thaliana and found that ∼2000 sites represented in three biological replicates of the experiment are annotated to nearby genes. These results were combined with high-throughput measurement of gene expression in response to AGL15 accumulation to discriminate responsive direct targets from those further downstream in the network. LEAFY COTYLEDON2, FUSCA3, and ABA INSENSITIVE3, which encode B3 domain transcription factors that are key regulators of embryogenesis, were identified and verified as direct target genes of AGL15. Genes identified as targets of the B3 genes are also targets of AGL15, and we found that INDOLEACETIC ACID-INDUCED PROTEIN30 is involved in promotion of somatic embryo development. The data presented here and elsewhere suggest that much cross-regulation occurs in gene regulatory networks underpinning embryogenesis.
INTRODUCTION
Although Arabidopsis thaliana is a powerful molecular genetic model, studies on embryogenesis, in particular the early stages of embryogenesis, are impaired by the small size of Arabidopsis embryos as well as the fact that they are embedded within maternal tissues. Somatic embryogenesis has been used as a more accessible model for zygotic embryogenesis, but it is itself poorly understood (Vogel, 2005; Rose and Nolan, 2006). One approach to understanding embryogenesis has been isolation of mutants defective in this process. However, many of these embryo-defective mutants are in fact deficient in gene products essential for life, rather than for embryogenesis per se (McElver et al., 2001; Tzafrir et al., 2003). The minority of embryo-defective genes encode key regulators of embryogenesis that are expressed primarily or specifically during embryo development and when ectopically expressed are sufficient to drive embryogenic programs in somatic cells. These include the LEAFY COTYLEDON (LEC) genes that have also been shown to be necessary for somatic embryogenesis (Lotan et al., 1998; Stone et al., 2001; Gazzarrini et al., 2004; Gaj et al., 2005; Wang et al., 2007).
Other genes have been identified that, when ectopically expressed, are able to promote somatic embryogenesis, including WUSCHEL (Gallois et al., 2004), BABY BOOM (Boutilier et al., 2002), SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (Hecht et al., 2001), LEAFY COTYLEDON 1-LIKE (Kwong et al., 2003) PGA37/MYB118 and MYB115 (Wang et al., 2009), and AGAMOUS-LIKE15 (AGL15; Harding et al., 2003; Thakare et al., 2008). Loss-of-function mutants in most of these genes do not have defects during zygotic embryo development, perhaps indicating the presence of redundant factors, as is common with ∼90% of gene functions protected by redundancy (Pickett and Meeks-Wagner, 1995; Meinke et al., 2003). Loss-of-function mutants that show ectopic embryo development, but again, often without obvious zygotic phenotypes, have also been isolated. A number of genes involved or potentially involved in chromatin remodeling when defective in expression produce somatic embryos on postgerminative tissues. These include pkl (Ogas et al., 1997), hdac6/9 RNA interference (Tanaka et al., 2008), swn clf (Chanvivattana et al., 2004), and val1 val2 (Suzuki et al., 2007) mutants.
AGL15 is a member of the MIKC subfamily of MADS domain transcription factors that accumulates primarily, although not exclusively, during embryogenesis (Heck et al., 1995; Rounsley et al., 1995). AGL15-specific antibodies detected accumulation of immunoreactive protein in nuclei of cells developing as embryo tissue in all situations tested in angiosperms (Perry et al., 1996, 1999). Ectopic expression of AGL15 promotes development of somatic embryos from zygotic embryo explants and then maintains development in this mode for extended periods (over 12 years to date; Harding et al., 2003). A 35Spro:AGL15 transgene also promotes somatic embryo development from the shoot apical region of seedlings that had completed germination in liquid media containing 2,4-D (Harding et al., 2003; Thakare et al., 2008). Furthermore, a putative ortholog of AGL15 from soybean (Glycine max) enhanced somatic embryo development in this species (Thakare et al., 2008). Although loss-of-function alleles of agl15 do not have any obvious impairment in zygotic embryo development, agl15, in some systems as a double mutant with a knockout allele in the closest family member agl18, showed decreased ability to produce somatic embryos (Thakare et al., 2008).
Previous work used chromatin immunoprecipitation (ChIP) as a step to identify genes directly regulated by AGL15 (Wang et al., 2002, 2004; Tang and Perry, 2003; Zhu and Perry, 2005; Hill et al., 2008; Nakaminami et al., 2009). One gene expressed in response to AGL15 was found to encode a gibberellin 2-oxidase (Arabidopsis GA2ox6), and expression of this gene was demonstrated to in part explain AGL15's promotion of somatic embryo development (Wang et al., 2004). However, a knockdown allele of GA2ox6 was unable to completely block promotion of somatic embryo development in response to 35Spro:AGL15, suggesting that either a full knockout is needed or other genes regulated by AGL15 also impact somatic embryogenesis. To better understand AGL15 and how this gene is able to promote somatic embryo development, we used high-throughput methods to map in vivo binding sites for AGL15 and to determine the response of genes to AGL15 accumulation. Together, these approaches allow one to distinguish putative direct target genes from indirect targets. Interestingly, several key regulators of embryogenesis were identified as being directly expressed in response to AGL15, and a number of other targets may be shared among these regulators.
RESULTS
Genome-Wide Identification of in Vivo AGL15 Binding Sites
In order to globally map in vivo binding sites of AGL15, we combined ChIP with Affymetrix GeneChip Arabidopsis Tiling 1.0R arrays in a ChIP-chip approach. Three independent biological replicates of the ChIP experiment using anti-AGL15–specific antiserum or preimmune serum as a control were performed. Partek GS (Genomics Suite) software and a ChIP-chip workflow was used to analyze results (Downey, 2006) as described in Methods. Overlap in binding sites was detected between biological replicates and reported as overlap on all three replicates of the experiment or on at least two of the three replicates. Approximately 2000 DNA fragments were present as bound by AGL15 in all three replicates of ChIP-chip (2028 sites) and were assigned to nearby genes (see Supplemental Data Set 1 online). Of these, 1706 were uniquely assigned to a particular Arabidopsis Genome Initiative (AGI) locus. The rest were assigned to two or more contiguous loci because they were located between genes or spanned multiple genes.
CisGenome was also used to analyze the data (Ji and Wong, 2005; Ji et al., 2008). Approximately 73% of sites (1485 in total) identified by Partek were also identified by CisGenome when all three replicates were assessed at the same time. Cisgenome identified 3708 peaks (false discovery rate < 0.01) and assigned the peaks to 3360 genes (see Supplemental Data Set 2 online). Sixty-nine percent of these were also present on at least two of the three replicates, while 44% were present on all three replicates as analyzed using Partek GS.
A number of in vivo sites for AGL15 were previously identified as well as a number of DNA fragments not bound by AGL15 (Wang et al., 2002, 2004; Tang and Perry, 2003; Zhu and Perry, 2005; Hill et al., 2008; K. Hill, W. Tang, Y. Zheng, C. Zhu, and S.E. Perry, unpublished data). Of 10 previously identified targets, six were present on all three replicates as summarized in Supplemental Table 1 online, while regulatory regions corresponding to two others were on two of three replicates. The two that were not present were classified as weakly bound sites in previous work and both encoded unknown proteins (Tang and Perry, 2003; W. Tang and S.E. Perry, unpublished data). The genomic regions identified by ChIP-chip overlapped with those previously identified by ChIP followed by sequencing. Conversely, of the eight DNA fragments previously characterized as not being occupied by AGL15, none were present on all three arrays (see Supplemental Table 1 online).
Enrichment tests were used to determine if previously unknown targets of AGL15 identified by ChIP-chip were truly occupied in vivo by AGL15. Oligonucleotides corresponding to the region bound by AGL15 and oligonucleotides corresponding to a region not bound by AGL15 were used in multiplex PCR. Assuming equal efficiency of PCR, after PCR, the target and control band should be present in approximately equal amounts in the input (total) DNA. After enrichment of in vivo associated sites by ChIP, the target DNA fragment should be present at higher frequency than control, and the multiplex PCR should reflect this. All of the more than a dozen targets identified by ChIP-chip were verified by enrichment tests (see Supplemental Table 2 online; Figure 1A). Verified targets included regulatory regions corresponding to other MADS box genes (FLOWERING LOCUS C and SHORT VEGETATIVE PHASE), to genes involved in gibberellic acid (GA) metabolism or perception (GIBBERELLIN 2-OXIDASE2 and GIBBERELLIC ACID INSENSITIVE) and to genes that function as central regulators of embryogenesis (LEC2, FUSCA3 [FUS3], and ABA INSENSITIVE3 [ABI3]; Figure 1).
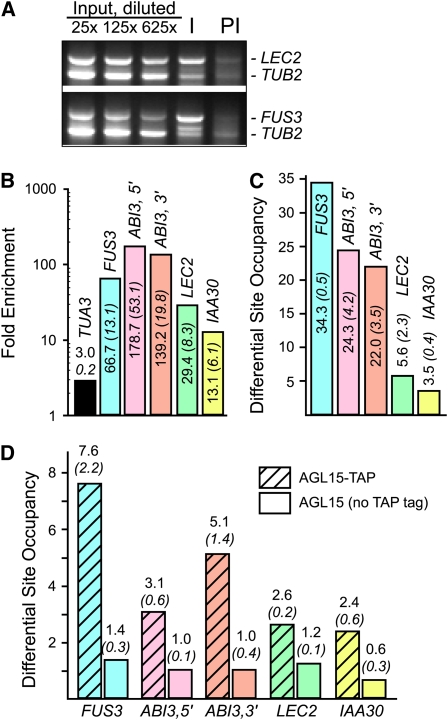
Figure 1.
Verification of in Vivo Association of AGL15 with Select DNA Fragments.
(A) Multiplex enrichment tests using oligonucleotide primers that will amplify the suspected target (top band, LEC2, FUS3 regulatory regions) and primers that will amplify a nonbound control (TUB2) on input DNA, DNA recovered by ChIP with anti-AGL15 serum (I), and preimmune control (PI) show specific enrichment of the target relative to the control in the immune (I) precipitation.
(B) Fold enrichment calculations from qPCR on three independent ChIP experiments. Please note the log10 scale.
(C) DSO calculations from qPCR on three independent ChIP experiments. Recovery of target by coimmunoprecipitation with anti-AGL15 antiserum was compared with recovery of a nonbound control (TUA3) in the same immune precipitation.
(D) Coprecipitation of select DNA fragments using IgG-Sepharose to isolate complexes via the protein A domain in a TAP-tag added to the C-terminal end of AGL15. Nontagged tissue served as a control. DSO calculations from three independent experiments comparing recovery of target to nonbound control (TUA3) in the same immune precipitation is shown. For (B) to (D), means (standard error of the mean) are shown.
[See online article for color version of this figure.]
We used quantitative PCR (qPCR) to quantitate association of DNA fragments with AGL15 using three biological replicates of the ChIP experiment that were independent from those used for ChIP-chip. When data were analyzed to calculate the fold-change between the ChIP (anti-AGL15 immune serum) and preimmune control, all of the targets tested were indeed present at higher amounts in the immune than in the preimmune precipitation (Figure 1B). Differential site occupancy (DSO) was also calculated by comparing the amplicon of the target in the immune precipitation to that of a control that was not expected to be bound by AGL15 (TUA3; Figure 1C) in the same immune precipitation. All of the targets tested were also enriched compared with the nonbound control. Regulatory regions corresponding to LEC2 and INDOLEACETIC ACID-INDUCED PROTEIN30 (IAA30) were found on only two of the three biological replicates of the ChIP-chip experiment, and they also showed lower fold enrichment and DSO than the other targets that were present on all three ChIP-chip arrays.
To further support our conclusion that select DNA fragments are targets of AGL15, we precipitated AGL15-DNA complexes independently of the AGL15-specific antiserum. Tissue expressing a form of AGL15 with a TAP tag (Puig et al., 2001) consisting of a calmodulin binding peptide and IgG binding regions of protein A and IgG-Sepharose beads were used to isolate AGL15-TAP-DNA complexes. Untagged tissue served as a control. As shown in Figure 1D, DSO was higher for the targets when tissue accumulating AGL15-TAP was used compared with the nontagged AGL15 control.
Genome-Wide Location of in Vivo Binding Sites and Functional Classification of Putative AGL15 Targets
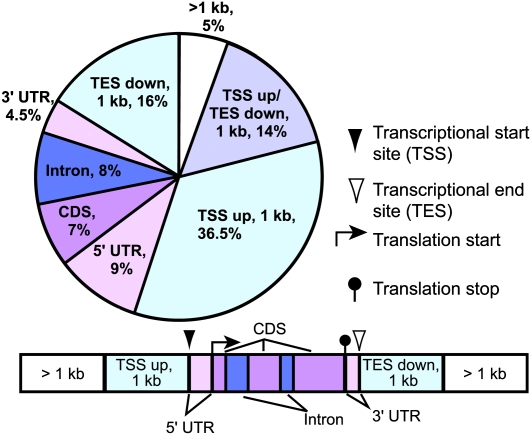
Regions identified as bound by AGL15 on all three replicates of the ChIP-chip experiment by Partek GS or by CisGenome were mapped to the genome using CisGenome, with very similar results (Ji et al., 2008). As shown in Figure 2, the majority (∼72%) of the sites identified by CisGenome were intergenic and most of these were within 1 kb of the transcription unit. More were found 5′ of the transcription start site than downstream of the transcription end site. Some sites were associated with both of these categories, likely due to the relatively compact genome of Arabidopsis. Of sites categorized as intragenic, more were associated with the 5′ untranslated region (UTR) than the 3′ UTR. Of the regions bound, 57.6% had at least one CArG motif of form C[A/T]8G or C[A/T]7GG that is preferentially bound by AGL15 (Tang and Perry, 2003) or the canonical form (also bound by AGL15) of CC[A/T]6GG, where the subscript represents the size of the A/T stretch. When CArGs of form CC[A/T]4NNGG were also considered, 64.2% of the fragments contained at least one binding site. We compared these results to five separate matched control regions generated by CisGenome. The average frequency of C[A/T]8G, C[A/T]7GG, and CC[A/T]6GG for the five matched controls was 43.6%, and inclusion of CC[A/T]4NNGG increased this to 48.6%. When comparing the regions identified as bound by AGL15 to each of the matched control regions, the AGL15-bound fragments show an overrepresentation of potential CArG motifs using a χ2 test (P < 0.0001).
Figure 2.
The Location of Binding Sites for AGL15 Relative to Nearby Gene(s).
UTR refers to the untranslated regions of transcripts, 5′ and 3′ of the gene. CDS refers to the regions coding for protein. TSS and TES refer to the transcriptional start site and transcriptional end site, and regions within 1000 bp of these sites are reported. Some sites are located further away than 1 kb of the transcriptional start site or transcriptional end site, and these are reported as >1 kb.
[See online article for color version of this figure.]
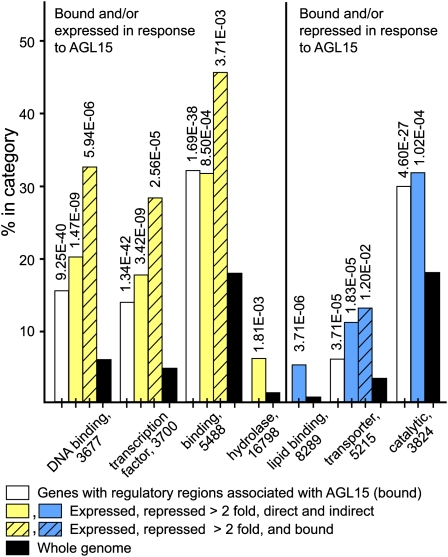
Of the sites identified by both programs and assigned to AGI loci, 13.8% correspond to putative regulatory regions of genes encoding proteins involved in transcription factor activity, a significant overrepresentation compared with the fraction of total genes encoding known proteins with transcription factor activity (4.5%; analysis performed with GO Term Enrichment tool AmiGO on The Arabidopsis Information Resource's website with GO database release 2009-07-03; Carbon et al., 2009). This category and select other categories showing significant overrepresentation when compared with the whole genome are summarized in Figure 3 (cf. white to black bars).
Figure 3.
Functional Categorization of AGL15 Directly Bound and/or Responsive Genes Using AmiGO GO Term Enrichment.
Genes with regulatory regions identified as bound by AGL15 in all three replicates of the ChIP-chip experiment and by both Partek GS and CisGenome were characterized for GO Molecular Function and compared with the whole genome (white bars compared with the black bars within each category). Genes responsive to AGL15 (changing by at least twofold for one comparison and a consistent or no change for the other comparison) that may be direct or indirect targets were also characterized in this manner (shaded bars, light for expressed, dark for repressed) as were genes directly bound and responsive (shaded and hatched bars). Only GO terms with significant difference between the bound and/or responsive gene sets and the whole genome are shown and only a subset for some comparisons. P values are indicated above the bars and GO term and accession number category below the bars.
[See online article for color version of this figure.]
Gene Expression Changes in Response to AGL15 Accumulation
Work with other transcriptional regulators indicates that binding may occur without consequences for gene expression (Wyrick and Young, 2002; Lee et al., 2007; Oh et al., 2009). Thus, it is important to assess which genes respond to alterations in AGL15 accumulation.
To assess gene expression, we used a shoot apical meristem somatic embryo (SAM SE) system where seeds are allowed to complete germination in liquid media containing 2,4-D and at some frequency will produce somatic embryos at the shoot apex by 3 weeks in culture (Mordhorst et al., 1998). Accumulation of AGL15 is positively correlated with production of SAM SEs (Thakare et al., 2008). Because an agl15 agl18 double mutant more consistently showed reduction in somatic embryogenesis, and because AGL18 functions redundantly with AGL15 in other developmental processes (Adamczyk et al., 2007), we used the double mutant for our experiments. Affymetrix ATH1 arrays were hybridized with probes generated from the agl15 agl18 double mutant, Columbia (Col), wild type, and 35Spro:AGL15 (all Col ecotype) from 10-d-old cultures, before any obvious embryo development was apparent.
Approximately 4000 genes were reported as having significant changes (P < 0.01) between the three populations. When sorted for those showing at least a twofold difference between Col wild type and either the double mutant or 35Spro:AGL15, and a consistent change for the other comparison, the list of repressed genes (i.e., 35Spro:AGL15/Col ≤ 0.5 and agl15 agl18/Col ≥ 1; or 35Spro:AGL15/Col ≤ 1 and agl15 agl18/Col ≥ 2) numbered 244 genes. The list of induced genes (agl15 agl18/Col ≤ 0.5 or 35Spro:AGL15/Col ≥ 2 and no or consistent change for the other comparison) was 205 (see Supplemental Data Set 3 online). GO molecular function terms that are significantly overrepresented compared with the whole genome are shown in Figure 3 (cf. light- and dark-shaded bars to black bars). The most obvious deviations from whole-genome categorization were for DNA binding/transcription factor activity for genes upregulated in response to AGL15 and transporter activity for genes repressed in response to AGL15.
Identification of Putative Direct and Indirect Targets of AGL15
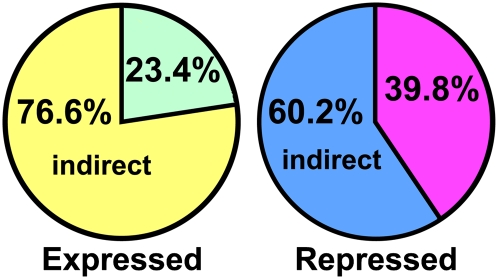
Combining the ChIP-chip and expression array results allows discrimination of genes that are directly bound by AGL15 and responsive from genes that change in transcript amounts but are likely indirect targets. As shown in Figure 4, of the 244 genes repressed at least twofold, 97 were also associated with DNA fragments directly bound by AGL15 (40%). Forty-eight of the 205 (23%) expressed targets appeared to be directly regulated by AGL15. Lists of genes directly expressed and directly repressed are shown in Tables 1 and 2, respectively. When these lists were examined for overrepresentation of GO terms, directly expressed genes were overrepresented in the DNA binding/transcription factor activity categories, and directly repressed genes were overrepresented in the transporter activity category (Figure 3, cf. hatched, shaded bars with the black bars). The distribution of locations of the regions bound for the directly responsive genes was similar to that found for all sites bound (Figure 2) except that all of the regions bound for the directly responsive genes that were intergenic were within 1 kb of the transcription start site and/or end site.
Figure 4.
Fraction of the Expressed and Repressed Genes That Are Also Bound by AGL15.
The expressed and repressed genes that are also bound by AGL15 may represent direct targets compared with responsive but not bound that are likely to represent indirect targets.
[See online article for color version of this figure.]
Table 1.
Genes Directly Bound and Expressed in Response to AGL15
| AGI | Description | agl15agl18/Col, wild type | 35Spro:AGL15/Col, wild type |
|---|---|---|---|
| At5g13790* | Floral homeotic protein AGL-15 (AGL15) | 0.26 | 15.20 |
| At5g64340* | Expressed protein | 0.29 | 1.12 |
| At1g19220 | Transcription factor B3 family protein/auxin-responsive factor AUX/IAA-related | 0.31 | 1.05 |
| At5g48560 | Basic helix-loop-helix family protein | 0.32 | 1.04 |
| At1g04030 | Expressed protein | 0.33 | 1.07 |
| At3g21550 | Expressed protein | 0.34 | 1.40 |
| At3g04320* | Trypsin and protease inhibitor family protein/Kunitz family protein | 0.37 | 1.02 |
| At5g42630* | Myb family transcription factor (KAN4) | 0.40 | 1.01 |
| At4g02800 | Expressed protein | 0.41 | 1.00 |
| At4g37110 | Expressed protein | 0.43 | 1.00 |
| At4g31800 | WRKY family transcription factor | 0.43 | 1.10 |
| At5g51850* | Expressed protein | 0.47 | 1.55 |
| At3g10740 | Glycosyl hydrolase family protein 51 | 0.47 | 1.20 |
| At5g25580* | Expressed protein | 0.49 | 1.02 |
| At5g48800 | Phototropic-responsive NPH3 family protein | 0.50 | 1.28 |
| At2g46590 | Dof zinc finger protein DAG2/Dof affecting germination 2 (DAG2) | 0.50 | 1.18 |
| At2g46710* | Rac GTPase activating protein, putative | 0.51 | 1.09 |
| At1g23000* | Heavy metal–associated domain-containing protein | 0.51 | 1.57 |
| At3g18035 | Histone H1/H5 family protein | 0.51 | 1.27 |
| At1g05070 | Expressed protein | 0.51 | 1.13 |
| At5g46910 | Transcription factor jumonji (jmj) family protein | 0.51 | 1.34 |
| At1g45180 | Zinc finger (C3HC4-type RING finger) family protein | 0.51 | 1.02 |
| At1g69010 | Basic helix-loop-helix family protein | 0.52 | 1.01 |
| At3g04730* | Auxin-responsive protein/indole acetic acid–induced protein 16 (IAA16) | 0.52 | 1.21 |
| At4g34160 | Cyclin δ-3 (CYCD3) | 0.53 | 1.21 |
| At5g25210* | Expressed protein | 0.53 | 1.50 |
| At1g11000 | Seven transmembrane MLO family protein/MLO-like protein 4 (MLO4) | 0.53 | 1.03 |
| At4g30090* | Expressed protein | 0.53 | 1.21 |
| At5g65910* | BSD domain-containing protein | 0.53 | 1.02 |
| At1g53860 | Remorin family protein | 0.54 | 1.23 |
| At1g12780* | UDP-glucose 4-epimerase/UDP-galactose 4-epimerase | 0.54 | 1.20 |
| At5g50180* | Protein kinase, putative | 0.54 | 1.09 |
| At4g28400 | Protein phosphatase 2C, putative/PP2C, putative | 0.55 | 1.15 |
| At4g02210 | Expressed protein | 0.55 | 1.03 |
| At5g25180* | Cytochrome P450 71B14, putative (CYP71B14) | 0.56 | 1.99 |
| At1g29090 | Peptidase C1A papain family protein | 0.57 | 2.62 |
| At1g25550* | Myb family transcription factor | 0.66 | 2.23 |
| At4g38620 | Myb family transcription factor (MYB4) | 0.72 | 1.95 |
| At5g66300 | No apical meristem (NAM) family protein | 0.76 | 2.01 |
| At1g70210 | Cyclin δ-1 (CYCD1) | 0.77 | 2.07 |
| At1g49320 | BURP domain-containing protein | 0.78 | 2.36 |
| At1g68670* | Myb family transcription factor | 0.92 | 2.19 |
| At4g03210* | Xyloglucan:xyloglucosyl transferase, putative/xyloglucan endotransglycosylase | 0.93 | 1.98 |
| At4g36700 | Cupin family protein | 0.95 | 4.45 |
| At4g35070* | Expressed protein | 0.97 | 2.10 |
| At4g24670 | Alliinase family protein, TAR2 | 0.98 | 2.71 |
| At3g54820 | Aquaporin, putative | 0.99 | 2.31 |
|
At1g75190 |
Expressed protein |
1.00 |
2.41 |
Bolded text AGI indicates that the gene was identified as associated with AGL15 directly bound fragments in all three biological replicates of the experiment and performing the analysis with Partek GS. Italicized text indicates that the site was identified on two of the three replicates and performing the analysis with Partek GS. An asterisk indicates that the site was also identified by CisGenome. Upright text indicates that the site was identified by CisGenome, but not Partek.
Table 2.
Genes Directly Bound and Repressed in Response to AGL15
| AGI | Description | agl15agl18/Col wild type | 35Spro:AGL15/Col wild type |
|---|---|---|---|
| At5g53870* | Plastocyanin-like domain-containing protein | 3.73 | 0.21 |
| At2g03020* | Heat shock protein-related | 3.06 | 0.68 |
| At1g80450* | VQ motif-containing protein | 3.03 | 0.92 |
| At2g23130 | Arabinogalactan-protein (AGP17) | 2.93 | 0.71 |
| At5g19260 | Expressed protein | 2.85 | 0.58 |
| At1g35230 | Arabinogalactan-protein (AGP5) | 2.78 | 0.98 |
| At2g42360 | Zinc finger (C3HC4-type RING finger) family protein | 2.78 | 0.95 |
| At3g48700 | Expressed protein | 2.74 | 0.73 |
| At2g32210 | Expressed protein | 2.69 | 0.77 |
| At3g60690* | Auxin-responsive family protein | 2.58 | 0.85 |
| At1g09750 | Chloroplast nucleoid DNA binding protein-related | 2.56 | 0.69 |
| At2g47560 | Zinc finger (C3HC4-type RING finger) family protein | 2.54 | 0.84 |
| At5g11730* | Expressed protein | 2.44 | 0.93 |
| At1g77640* | AP2 domain-containing transcription factor, putative | 2.43 | 0.90 |
| At3g60900 | Fasciclin-like arabinogalactan-protein (FLA10) | 2.43 | 0.74 |
| At1g07750 | Cupin family protein | 2.37 | 0.88 |
| At1g33250* | Fringe-related protein | 2.36 | 0.96 |
| At4g12310* | Cytochrome P450, putative | 2.36 | 0.44 |
| At4g12730 | Fasciclin-like arabinogalactan-protein (FLA2) | 2.36 | 0.40 |
| At4g13340 | Leucine-rich repeat family protein/extensin family protein | 2.35 | 0.64 |
| At3g01930 | Nodulin family protein | 2.31 | 0.49 |
| At2g25620 | Protein phosphatase 2C, putative/PP2C, putative | 2.30 | 0.84 |
| At4g29140 | MATE efflux protein-related | 2.29 | 0.83 |
| At4g30430* | Senescence-associated family protein | 2.16 | 0.91 |
| At1g80530 | Nodulin family protein | 2.16 | 0.88 |
| At2g43780 | Expressed protein | 2.12 | 0.97 |
| At5g39760* | Zinc finger homeobox protein-related/ZF-HD homeobox protein-related | 2.11 | 0.76 |
| At1g21670* | Expressed protein | 2.10 | 0.60 |
| At2g04780 | Fasciclin-like arabinogalactan-protein (FLA7) | 2.05 | 0.93 |
| At1g28390* | Protein kinase family protein | 2.05 | 0.90 |
| At3g07270* | GTP cyclohydrolase I | 2.02 | 0.99 |
| At2g39710* | Aspartyl protease family protein | 2.01 | 0.41 |
| At5g13100 | Expressed protein | 2.00 | 0.78 |
| At1g07090* | Expressed protein | 2.00 | 0.36 |
| At5g19820 | PBS lyase HEAT-like repeat-containing protein | 1.99 | 0.78 |
| At5g14120* | Nodulin family protein | 1.98 | 0.93 |
| At2g28120* | Nodulin family protein | 1.98 | 0.58 |
| At2g16660 | Nodulin family protein | 1.96 | 0.77 |
| At1g24530* | Transducin family protein/WD-40 repeat family protein | 1.95 | 0.96 |
| At3g51860* | Cation exchanger, putative (CAX3) | 1.94 | 0.51 |
| At4g38250 | Amino acid transporter family protein | 1.84 | 0.53 |
| At2g35860 | β-Ig-H3 domain-containing protein/fasciclin domain-containing protein | 1.79 | 0.40 |
| At4g37450* | Arabinogalactan-protein (AGP18) | 1.78 | 0.44 |
| At3g60490 | AP2 domain–containing transcription factor TINY, putative | 1.66 | 0.47 |
| At3g05360* | Disease resistance family protein/LRR family protein | 1.65 | 0.52 |
| At4g15490 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.65 | 0.36 |
| At5g05600 | Oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.63 | 0.47 |
| At5g41400* | Zinc finger (C3HC4-type RING finger) family protein | 1.62 | 0.42 |
| At2g36830* | Major intrinsic family protein / MIP family protein | 1.59 | 0.53 |
| At3g06470* | GNS1/SUR4 membrane family protein | 1.56 | 0.47 |
| At5g45800* | Leucine-rich repeat transmembrane protein kinase, putative | 1.55 | 0.49 |
| At1g52880 | No apical meristem (NAM) family protein | 1.55 | 0.49 |
| At5g01840 | Ovate family protein | 1.54 | 0.50 |
| At5g40170* | Disease resistance family protein | 1.54 | 0.47 |
| At3g63160 | Expressed protein | 1.52 | 0.49 |
| At1g17620 | Expressed protein | 1.50 | 0.52 |
| At1g69870* | Proton-dependent oligopeptide transport (POT) family protein | 1.47 | 0.30 |
| At5g16970* | NADP-dependent oxidoreductase, putative (P1) | 1.45 | 0.55 |
| At5g61520 | Hexose transporter, putative | 1.44 | 0.41 |
| At1g71880 | Sucrose transporter/sucrose-proton symporter (SUC1) | 1.44 | 0.48 |
| At3g07390 | Auxin-responsive protein/auxin-induced protein (AIR12) | 1.42 | 0.52 |
| At5g16010 | 3-Oxo-5-α-steroid 4-dehydrogenase family protein/steroid 5-α-reductase | 1.40 | 0.34 |
| At4g37310 | Cytochrome P450, putative | 1.39 | 0.26 |
| At4g22470* | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.38 | 0.22 |
| At2g33050* | Leucine-rich repeat family protein | 1.36 | 0.51 |
| At1g11700* | Expressed protein | 1.35 | 0.54 |
| At5g48930* | Transferase family protein | 1.35 | 0.40 |
| At3g22120 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.34 | 0.30 |
| At2g39010 | Aquaporin, putative | 1.33 | 0.25 |
| At3g15840 | Expressed protein | 1.31 | 0.51 |
| At2g42610 | Expressed protein | 1.27 | 0.35 |
| At3g50340* | Expressed protein | 1.27 | 0.53 |
| At1g70230* | Expressed protein | 1.23 | 0.42 |
| At5g05340 | Peroxidase, putative | 1.22 | 0.37 |
| At1g31290 | PAZ domain-containing protein/piwi domain-containing protein | 1.21 | 0.41 |
| At1g27480* | Lecithin:cholesterol acyltransferase family protein/LACT family protein | 1.21 | 0.53 |
| At3g05180* | GDSL-motif lipase/hydrolase family protein | 1.20 | 0.33 |
| At3g48690* | Expressed protein | 1.18 | 0.21 |
| At2g33860* | Auxin-responsive factor (ARF3)/ETTIN protein (ETT) | 1.18 | 0.54 |
| At4g37320* | Cytochrome P450 family protein | 1.17 | 0.50 |
| At3g06810* | Acyl-CoA dehydrogenase-related, IBR3 | 1.16 | 0.42 |
| At5g06870* | Polygalacturonase inhibiting protein 2 (PGIP2) | 1.15 | 0.37 |
| At5g07010* | Sulfotransferase family protein | 1.15 | 0.50 |
| At5g55930 | Oligopeptide transporter OPT family protein | 1.15 | 0.39 |
| At5g46330 | Leucine-rich repeat transmembrane protein kinase, putative | 1.14 | 0.48 |
| At5g65380* | Ripening-responsive protein, putative | 1.13 | 0.46 |
| At2g41800* | Expressed protein | 1.11 | 0.54 |
| At1g07180* | Pyridine nucleotide-disulphide oxidoreductase family protein | 1.10 | 0.54 |
| At1g35350 | EXS family protein/ERD1/XPR1/SYG1 family protein | 1.06 | 0.55 |
| At2g37040 | Phe ammonia-lyase 1 (PAL1) | 1.06 | 0.48 |
| At4g21680 | Proton-dependent oligopeptide transport (POT) family protein | 1.06 | 0.51 |
| At5g21105* | l-ascorbate oxidase, putative | 1.05 | 0.54 |
| At1g71400 | Disease resistance family protein/LRR family protein | 1.05 | 0.50 |
| At5g46050 | Proton-dependent oligopeptide transport (POT) family protein | 1.05 | 0.33 |
| At5g50570 | Squamosa promoter-binding protein, putative | 1.05 | 0.51 |
| At2g39210* | Nodulin family protein | 1.04 | 0.46 |
| At1g54570 |
Esterase/lipase/thioesterase family protein |
1.03 |
0.41 |
Bold and italicized terms and asterisks are as in Table 1.
Key Regulators of Embryogenesis Are Direct Targets of AGL15
Because previous work demonstrated that AGL15 accumulation correlates with production of somatic embryos (Perry et al., 1999; Harding et al., 2003; Thakare et al., 2008), we were particularly interested in genes that also lead to expression of embryo programs when ectopically expressed, such as LEC1, LEC2, and FUS3. While regulatory regions corresponding to LEC1 were not identified as being bound by AGL15, regions corresponding to LEC2 and FUS3 were identified as being bound on the tiling array. In addition, ABI3 was potentially a direct target of AGL15 based on the ChIP-chip results. Because a network of interaction between LEC2, FUS3, and ABI3 has been proposed (To et al., 2006), we also investigated whether ABI3 may be a directly regulated target of AGL15.
The region of FUS3 identified as being bound on all three replicates of the ChIP-chip experiment encompassed ∼653 bp, including the 5′ UTR and intergenic region. Two CArG motifs of form C[A/T]7GG preferentially bound by AGL15 (Tang and Perry, 2003) were within this region. Two regions that could correspond to regulatory regions of ABI3 were also present as overlapping sites on all three biological replicates of the ChIP-chip experiment. These sites corresponded to regions in the intergenic region 5′ of ABI3 and in the 3′ UTR and intergenic region of ABI3, both of which contained sites recognized by AGL15. For LEC2, only two of three arrays showed an overlap in a region bound by AGL15, and this region encompassed ∼226 bp that included the last intron. Putative sites for binding of MADS factors were present, including a canonical CC[A/T]6GG site. In vivo association of AGL15 with these DNA fragments was confirmed (Figure 1).
Does association of AGL15 with regulatory regions of LEC2, FUS3, and ABI3 lead to changes in transcript accumulation? Signal from probes corresponding to LEC2 was absent in the expression microarray experiment. However, signal from probes corresponding to FUS3 was present on two of the three arrays hybridized with probe generated from 35Spro:AGL15 tissue and showed a ratio of 2.2 when compared with wild-type tissue where AGL15 accumulates only from the endogenous gene, and where signal corresponding to FUS3 was absent or marginal. Signal corresponding to ABI3 transcript was present in the wild type and 35Spro:AGL15 but variable on the arrays hybridized with probe derived from loss-of-function tissue. Because only a subset of cells at the shoot apical region that produce the embryos may be expressing these factors and because transcriptional regulators tend to be expressed at relatively low levels, expression microarrays may not be sensitive enough to detect these transcripts and changes in response to AGL15. Therefore, we tested transcript accumulation of FUS3, LEC2, and ABI3 using qRT-PCR.
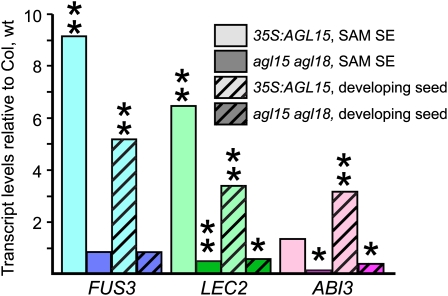
As shown in Figure 5, FUS3 and LEC2 showed increased transcript abundance in 35Spro:AGL15 compared with the wild type in 10-d-old SAM SE culture tissue. While LEC2 transcript was significantly decreased in the agl15 agl18 double mutant compared with the wild type, FUS3 did not show any reproducible change. The single agl15 or agl18 mutant did not show a consistent pattern in transcript abundance when compared with the wild type. ABI3 did not show an increase in response to ectopic or increased AGL15 accumulation in SAM SE culture, but the agl15 agl18 double mutant had significantly less ABI3 transcript than did the wild type (Figure 5). The single agl15 and agl18 mutants also had consistently decreased transcript abundance for ABI3, although whether the difference was significant (P < 0.05) depended on the experiment.
Figure 5.
Genes Encoding Embryo B3 Domain Proteins Respond to AGL15/18 Accumulation.
qRT-PCR to assess transcript abundance in wild-type and gain- and loss-of-function SAM SE and developing seed tissue was performed and compared with Col wild type. Similar results were obtained with at least two additional biological replicates of the experiments. **, Significant at P < 0.01; *, significant at P < 0.05, as determined using a Student's t test.
[See online article for color version of this figure.]
Although we tested gene expression in SAM SE culture at an early stage before any obvious embryos were apparent, it is still possible and likely that early stages of embryogenesis were occurring, and the presence or absence of key embryo regulators may simply correlate with embryo development rather than with AGL15 accumulation directly. Therefore, we investigated the abundance of LEC2, FUS3, and ABI3 transcripts in developing staged Arabidopsis seeds, where wild-type, loss-of-function, and gain-of-function genotypes for AGL15 proceed though morphogenesis at the same rate and where there are no apparent embryo defects (Fernandez et al., 2000; Lehti-Shiu et al., 2005). We used biological replicates of staged 7 to 8 d postanthesis (dpa) or 9 to 10 dpa seed. Results for developing seeds were in reasonable agreement with those found for SAM SE culture. FUS3, LEC2, and ABI3 transcripts are reproducibly increased in 35Spro:AGL15 seed. As in SAM SE culture, FUS3 transcript is not reduced in response to loss of function of agl15 and/or agl18. However, both LEC2 and ABI3 show reduced levels of transcript in the double mutant compared with the wild type, and for ABI3, there is a significant reduction in both of the single mutants as well.
In summary, in both developing staged seed and SAM SE culture, AGL15 is sufficient to increase LEC2 and FUS3 transcript abundance, but only LEC2 shows a decrease in response to loss of agl15/18. By contrast, ABI3 does not show an increase in transcript in both contexts but does show decreased transcript in loss of function of agl15 and/or agl18. Because AGL15 associates in vivo with regulatory regions of these genes, AGL15 (and AGL18) may directly contribute to the regulation of expression of these genes.
Potential for Extensive Cross-Regulation of Embryogenesis
Several genes have been identified as potentially being direct targets of LEC2 and FUS3. One such target is a gene involved in GA biosynthesis, Arabidopsis GA3ox2, that is repressed in response to LEC2 and FUS3, resulting in lower GA/abscisic acid (ABA) ratios and embryo identity of organs (Curaba et al., 2004; Gazzarrini et al., 2004; Lumba and McCourt, 2005). Previous work demonstrated that AGL15 directly upregulated a gene whose product is involved in GA catabolism (GA2ox6), also leading to a reduction in biologically active GA. Regulatory regions 5′ to the transcriptional start site of GA2ox6 were found to be bound by AGL15 in the ChIP-chip experiment (two of three arrays) (Wang et al., 2004). GA2ox6 was also found to be differentially expressed in response to AGL15 on the ATH1 arrays, with more than a fourfold increase in transcript levels in 35Spro:AGL15 compared with the wild type. However, the agl15 agl18 mutant did not show a significant change in transcript abundance for GA2ox6 compared with the wild type. Conversely, regulatory regions of the gene encoding the biosynthetic enzyme GA3ox2 were directly bound by AGL15 (three of three arrays), and agl15 agl18 showed a significant increase in transcript (2.3-fold, P < 0.01), indicating that this gene is repressed by AGL15 and/or AGL18. No significant change in transcript abundance was found for 35Spro:AGL15 compared with the wild type.
Another gene encoding a transcriptional regulator that was previously identified as a putative direct target of LEC2 encodes IAA30, a noncanonical Aux/IAA that lacks domain II that leads to degradation of Aux/IAA proteins in response to auxin signaling (Sato and Yamamoto, 2008). DNA fragments corresponding to regulatory regions 5′ of sequences encoding IAA30 were bound by AGL15 on two of three ChIP-chip replicates, and binding has been verified by qPCR on independent ChIP populations (Figures 1B to 1D). Expression of IAA30 is upregulated in response to AGL15; transcript accumulation was 2.2-fold higher in 35Spro:AGL15 than in Col wild-type tissue on the expression arrays. Subsequent experiments using real-time qRT-PCR confirmed significant increases in IAA30 transcript accumulation for 35Spro:AGL15 compared with the wild type, both in SAM SE cultures and in 9 to 10 dpa developing seed in at least three independent experiments for each tissue type. Loss of function was variable and did not show any consistent pattern.
IAA30 Promotes Somatic Embryogenesis
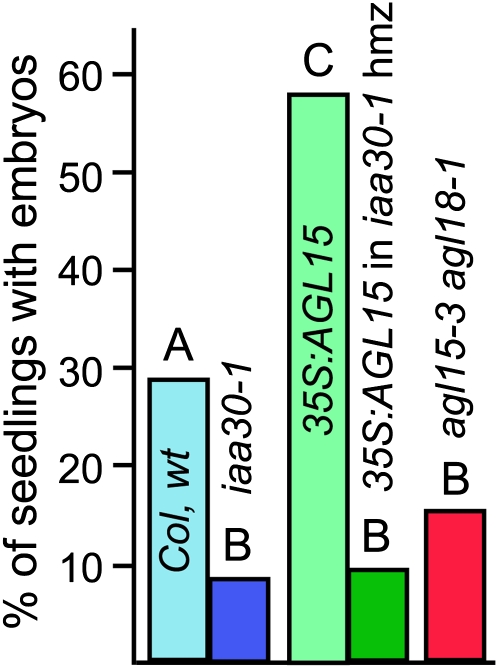
Because IAA30 was identified as a target of LEC2 as well as AGL15, and because both LEC2 and AGL15 promote somatic embryogenesis when constitutively expressed, we tested whether IAA30 was potentially involved in the promotion of somatic embryo development from the shoot apical region of seedlings in liquid media containing 2,4-D. A confirmed knockout line of iaa30-1 was obtained from the ABRC (SALK_065384C; Alonso et al., 2003), and lack of transcript accumulation was verified (see Supplemental Figure 1 online). iaa30-1 was found to produce significantly less SAM SE than Col wild type (8.5% compared with 29.0%; numbers are averages from three independent experiments with independently generated seed lots; Figure 6). A second allele was more recently obtained (JIC SM line, ABRC stock number CS112165; Sundaresan et al., 1995), and it also showed a reduction in the percentage of seedlings with SAM SE (5.3% of iaa30-2) compared with 19.2% of Col wild type and 1.4% of iaa30-1. Both iaa30 alleles are significantly different in terms of the number of seedlings with SAM SE from the wild type at P < 0.01 but are not different from each other. When the iaa30-1 knockout was crossed into 35Spro:AGL15, and the iaa30 homozygous knockout reestablished, 9.3% of seedlings had SAM SE development compared with 58.7% of 35Spro:AGL15 IAA30 seedlings (Figure 6). Therefore, AGL15 promotes SE in part by directly inducing expression of IAA30.
Figure 6.
SAM SE Development Is Reduced by Loss of Function of IAA30.
Means from three independent experiments with independently generated seed lots are shown. Different letters indicate significance at P < 0.0001 as determined using a Student's t test.
[See online article for color version of this figure.]
DISCUSSION
Numbers and Types of Genes Directly Regulated by AGL15
Although ChIP has been used quite extensively to verify suspected protein/DNA interactions, relatively few studies have been conducted to map in vivo associations between a protein and DNA on a genome-wide scale. Benhamed et al. (2008) generated a custom chip representing ∼20,000 Arabidopsis promoters and found that the histone acetyltransferase GCN5 associated with 40% of the promoters and that the bromodomain was required for association with 11% of promoters. Turck et al. (2007) used an Arabidopsis tiling array representing chromosome 4 to map sites with which TFL2/LHP1 associated and found interactions with hundreds of small domains on this chromosome. Lee et al. (2007) used a 60-nucleotide oligomer microarray to map in vivo association of HY5 and found >3000 sites that may represent HY5 binding targets. By contrast, TGA2 showed a fairly limited number of binding sites (Thibaud-Nissen et al., 2006). More recently, sites for PIL5/PIF1 were mapped globally and with fairly stringent cutoffs, 748 in vivo binding sites were identified (Oh et al., 2009). ChIP-sequence and ChIP-chip were used to map binding sites for another Arabidopsis MADS domain protein, SEP3/AGL9, and >4000 sites were identified as being bound by this protein (Kaufmann et al., 2009).
We identified ∼2000 sites annotated to genes with which AGL15 associates in vivo in all three replicates of the ChIP-chip experiment. Similar to results reported for HY5, the majority of AGL15 binding sites were located within 1 kb upstream of the transcription start site, although some were located in other parts of the gene/genome, and for LEC2, intronic binding of AGL15 may result in changes in LEC2 expression. The majority of DNA fragments identified as being associated with AGL15 in vivo have potential binding sites for MADS domain proteins, but some lack a perfect copy of this cis-element. In these cases, AGL15 may still associate directly with DNA via a variation of the CArG motif. Sequences other than those with an uninterrupted core of six to eight A or T nucleotides were isolated in binding site selection assays, indicating that binding can occur to these types of sites in vitro (Tang and Perry, 2003). Other MADS domain proteins can bind CArG motifs with a shorter A/T core (Huang et al., 1993). Alternatively, AGL15 may associate with some DNA fragments via other proteins.
Based on work with other DNA binding factors (e.g., Lee et al., 2007; Kaufmann et al., 2009; Oh et al., 2009), the large number of binding sites for AGL15 is not surprising. However, the minority of nearby genes show a significant and consistent response to AGL15 accumulation. This too appears to be a trend rather than an exception. For HY5, only 5.6% of bound sites showed a twofold change in expression when comparing the wild type to hy5, using a cutoff of P < 0.05 (Lee et al., 2007) and the minority responded for PIL5/PIF1 (Oh et al., 2009). As reviewed by Wyrick and Young (2002), binding without obvious regulation may be very common and may represent situations where a DNA binding factor is inactive until a cofactor or signal is present or until chromatin remodeling or another event occurs. At other time points or developmental contexts, a different subset of AGL15-bound sites may respond to AGL15 accumulation. Also, microarrays are not sensitive enough to detect some genes that may have low expression in a subset of the cells sampled, as we found for the embryo B3 genes.
Finally, the small percentage of directly bound genes that are responsive are those with a consistent pattern of expression in response to AGL15. Other genes showed perturbations in transcript accumulation, but transcript was increased or decreased in response to both loss and gain of function of AGL15 (and AGL18 in the mutant). One possible explanation for this result is redundancy of gene function and the likely complexity of transcriptional networks. Other genes showed less than a twofold change in response to loss or gain of function of AGL15, but the change was still statistically significant.
AGL15 May Participate in Direct Regulation of the Embryo B3 Domain Encoding Genes
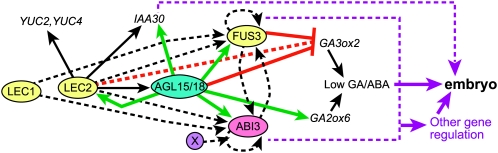
To et al. (2006) performed an elegant series of experiments to investigate redundancy among the LEC/ABI3 transcriptional regulators. Results indicated that FUS3 and ABI3 are regulated by LEC2, but whether this regulation is direct or indirect is not known. Neither FUS3 nor ABI3 was identified as being expressed in response to LEC2, at least within the 4-h timeframe examined postinduction of LEC2, indicating that they may not be direct targets of LEC2 (Braybrook et al., 2006). AGL15 was found to be upregulated by 1 h postinduction of LEC2 and, due to RY motifs (binding site for B3 domain proteins that include LEC2) within the regulatory regions of AGL15, AGL15 may be a direct target of LEC2 (Braybrook et al., 2006). In this study, FUS3 and ABI3 were found to be direct targets of AGL15. AGL15 also appears to regulate LEC2 expression, in what may be a feedback loop, possibly limited by the fact that AGL15 negatively regulates itself (Zhu and Perry, 2005; please note that the apparent expression of AGL15 as listed in Table 1 is due to loss of transcript in the mutant and increased transcript in the 35Spro:AGL15). Thus, AGL15/18 may be part of this intricate network. However, because neither the agl15 nor the agl15 agl18 mutants phenocopy the lec or abi3 mutants, there must be higher levels of redundancy. More than 40 members of the MADS family are expressed in embryo tissue (Lehti-Shiu et al., 2005), so high levels of redundancy are possible.
Despite possible redundancy, changes in transcript accumulation were reproducibly observed in the loss and gain of function of AGL15 relative to the wild type. While it is possible that this simply coincided with early, although not yet visible, somatic embryo development in the SAM SE system, the trend was the same in developing seeds where morphogenesis proceeds at the same rate in all genotypes (Fernandez et al., 2000; Lehti-Shiu et al., 2005). Whether AGL15 was necessary and/or sufficient for changes in transcript accumulation depended on the gene. Figure 7 shows a model modified from To et al. (2006), incorporating AGL15 and some key downstream target genes. Intriguingly, putative orthologs of ABI3 and FUS3 from other species (G. max, Medicago truncatula, Populus trichocarpa, and Oryza sativa) have CArG motifs of similar type and location as found for Arabidopsis in their regulatory regions (see Supplemental Figure 2 online). This conservation may indicate an important role for these cis-elements in control of gene expression.
Figure 7.
Working Model Summarizing the Interaction between the LEC Genes, ABI3, and AGL15/18 and Potentially Other Redundant MADS Factors.
Dotted black lines indicate interactions described by To et al. (2006) that may be direct or indirect. Solid black arrows indicate genes potentially directly regulated by LEC2 (Braybrook et al., 2006; Stone et al., 2008). For direct targets of AGL15, arrows represent induction, and lines with bars represent repression of transcript accumulation.
[See online article for color version of this figure.]
Putative AGL15-Direct Targets and Hormonal Regulation of Embryogenesis
We previously found that a GA catabolic enzyme Arabidopsis GA2ox6 is directly upregulated in response to AGL15 (Wang et al., 2004). A decrease in biologically active GA was correlated with competence for somatic embryo development and decreased seed dormancy (Wang et al., 2004). FUS3 and LEC2 also impact on GA metabolism. FUS3 is thought to directly and LEC2 directly or indirectly repress the GA biosynthetic enzyme GA3ox2, and the GA/ABA ratio determines whether an embryonic or adult leaf develops (Curaba et al., 2004; Gazzarrini et al., 2004; Lumba and McCourt, 2005). We found that AGL15 is also likely to directly repress GA3ox2 expression, and increased transcript was detected in agl15 agl18 compared with in the wild type. Loss of repression of a GA biosynthetic enzyme in response to loss of AGL15/18 or increased expression of a GA catabolic enzyme in response to ectopic AGL15 both impact the accumulation of GA and can thereby affect development in embryo mode.
While GA/ABA may determine whether tissue develops as embryonic or postembryonic tissue, induction of somatic embryogenesis commonly requires treatment of explants with auxin, usually the synthetic auxin 2,4-D. In view of the general requirement for auxin to induce somatic embryogenesis, it is intriguing that posttranslational induction of LEC2 via a glucocorticoid receptor domain induces expression of the auxin biosynthetic genes YUC2 and 4 (Stone et al., 2008). However, YUCCA gene expression alone is not sufficient to promote embryogenesis because overexpression of these genes does not induce SE (Braybrook and Harada, 2008). Therefore, other factors must determine competence to respond to auxin in somatic embryo development.
If and how AGL15 promotes competence for somatic embryogenesis will require further investigation. However, we have shown that ectopic AGL15 accumulation can upregulate IAA30, which has been proposed as conferring competency for somatic embryogenesis (Braybrook et al., 2006). An iaa30 mutant from the SALK confirmed that homozygous knockout lines in Col and 35Spro:AGL15 backgrounds have significantly fewer seedlings showing SAM SE development. IAA30 was also identified as a rapidly responsive and potentially direct (or possibly indirect, through AGL15) target of LEC2 (Braybrook et al., 2006) and is unusual among the IAA proteins in Arabidopsis in that IAA30 lacks domain II, which is involved in binding auxin F-box receptors in response to auxin perception, leading to degradation of the IAA and release of ARFs to mediate the auxin response. IAA20 also lacks domain II and is also a potentially upregulated target of AGL15. Consequently, these IAA proteins have a longer half-life than the canonical IAA proteins (Sato and Yamamoto, 2008). Upregulation of IAA30 would be expected to disrupt auxin responsiveness. Interestingly, several other genes involved in auxin response were identified as being direct targets of AGL15, with significant changes in expression in response to AGL15 (Tables 1 and 2), and some of these are regulated in a manner to limit auxin responses. These include IAA16, which is expressed in response to AGL15, and IBR3, which is involved in oxidation of IBA to IAA (Zolman et al., 2007), and ARF3/ETTIN, both of which are repressed in response to AGL15. Why would AGL15, a promoter of somatic embryogenesis, appear to be involved, at least to some extent, in limiting auxin function? AGL15 is itself upregulated in response to auxin, but this does not appear to be an immediate response, as 1 to 2 d of treatment with 2,4-D or IAA is needed to see a response (Zhu and Perry, 2005). While auxin treatment is important for induction of embryogenesis, cells that are resistant to auxin may be capable of developing as embryos (Emons, 1994). Therefore, it is possible that AGL15 functions to maintain competence for somatic embryo development in a subset of cells by limiting auxin responses. In at least some tissues, auxin signaling leads to GA accumulation by activation of GA biosynthetic enzymes and/or deactivation of GA catabolic enzymes (Weiss and Ori, 2007). Perhaps upregulation of IAAs that are resistant to auxin-induced degradation by proteolysis may lead to decreased GA accumulation, at least in a subset of cells, and thereby promote somatic embryogenesis. However, TAR2 was identified as a potential direct target of AGL15 that is upregulated in response to AGL15 (Table 1), and the TAR2 gene product may have a role in auxin production and ethylene response (Stepanova et al., 2008).
We have only begun to scratch the surface of the crosstalk in regulatory networks involved in embryogenesis, but the picture that is forming is one of complex interaction and coregulation of genes by multiple factors (Figure 7). Further work investigating how select genes and hormones impact on somatic embryogenesis and how higher-order mutants may impact zygotic development should be revealing for deciphering the mechanisms involved in embryogenesis.
METHODS
Plant Material
Arabidopsis thaliana wild-type, insertional loss-of-function alleles (agl15-4, agl18-1, the agl15-4 agl18-1 double mutant, and iaa30) and 35Spro:AGL15 plants (all Col ecotype) were sown on GM (Murashige and Skoog, 1962; supplemented with 10 g L−1 sucrose, 0.5 g L−1 MES, and 7 g L−1 agar, pH 5.6 to 5.7), with 50 μg mL−1 kanamycin for 35Spro:AGL15 seed, chilled for 2 d at 4°C, and transferred to a growth room with a 16-h-light/8-h-dark cycle. At ∼10 d, seedlings were transferred to potting mix (ProMix BX; Premier Brands) and grown in a chamber with a 16-h-light (20°C)/8-h-dark (18°C) cycle. To stage seed, flowers were tagged on the day that they opened and seeds collected and flash frozen in liquid nitrogen for RNA at 7 to 8 and 9 to 10 d after anthesis. Seeds for SAM SE were allowed to develop to dry seed and SAM SE performed as described by Harding et al. (2003). For expression arrays and qRT-PCR, tissue was collected at 10 d after start of culture and flash frozen. To score for embryo production, tissue was examined at 21 d after start of culture.
The embryonic culture tissue (ECT) used as a tissue source for the ChIP experiments has been described previously (Harding et al., 2003). Additionally, ECT was initiated from transgenic Arabidopsis expressing a form of AGL15 that included a C-terminal TAP tag consisting of a calmodulin binding peptide, a TEV protease cleavage site, and two IgG binding domains from Staphylococcus aureus protein A.
ChIP-chip and Data Analysis
ECT (described by Harding et al., 2003) was fixed in MC buffer (10 mM potassium phosphate, pH 7, 50 mM NaCl, and 0.1 M sucrose) with 1% formaldehyde for 1 h on ice under vacuum. The reaction was quenched with cold glycine, added to 0.125 M, and incubation on ice for 10 to 30 min, after which the tissue was washed with cold MC buffer and flash frozen. ChIP was performed as described (Wang et al., 2002) except that the preparation of crude nuclei was done as by Bowler et al. (2004). For experiments using the TAP-tag, after solubilizing the chromatin, an equal volume of IPP150 buffer was added (10 mM Tris, pH 8, 150 mM NaCl, and 0.1% Nonidet P-40; Puig et al., 2001), and an aliquot saved for total input DNA. The solubilized chromatin was mixed gently on a rotating wheel at 4°C for 1 h before pelleting any insoluble material, for 5 min in a microcentrifuge, and moving the supernatant to a new tube. IgG-Sepharose (100 μL of a 50% slurry; Amersham) was added and incubated for 2 to 3 h at 4°C, with gentle mixing. Washing was as described (Wang et al., 2002) but using cold IPP buffer. Elution of the drained beads was performed by incubating in 225 μL of 1% SDS in Tris-EDTA, pH 8, for 15 min at 65 to 70°C. Elution was repeated once, the combined eluants centrifuged for 2 min, and the top 400 μL was used for DNA analysis as described by Wang et al. (2002).
Linear amplification and incorporation of dUTP was performed as detailed in the Affymetrix Chromatin Immunoprecipitation Assay Protocol (http://www.affymetrix.com/products/arrays/specific/arab_tiling.affx), known targets were tested using multiplex enrichment tests to confirm maintenance of enrichment in the immune precipitated sample compared with the controls after amplification, and DNA was sent to the UK Microarray Core Facility for fragmentation, labeling, hybridization of the Affymetrix GeneChip Arabidopsis tiling 1.0R array, and scanning.
Partek GS (Genome Suite; http://www.partek.com/partekgs) software was used to analyze results using the ChIP-on-chip workflow (Downey, 2006). A robust multichip average background correction and quantile normalization without summarization were performed and the data logged (base 2). Perfect match only was used. For each immune precipitation, the cognate preimmune control was subtracted at each probe for each biological replicate to normalize the data to the baseline. Regions with thresholds >0 were detected by statistically examining a sliding window of ∼250 bp (the probe on which the window is centered, and three probes to the left and three to the right) and performing a one-sample t test to determine if neighboring probes have a threshold >0. Regions bound by AGL15 were detected by looking for regions of at least 250 bp where contiguous probes have a P value < 0.01. Overlap in binding sites was detected between biological replicates and reported as overlap on all three replicates of the experiment or on at least two of the three replicates.
CisGenome was also used to analyze the data where test statistics for each probe are computed based on a hierarchical empirical Bayes model and then the test statistics combined in a genomic region using moving average to determine if the region is bound (Ji and Wong, 2005; Ji et al., 2008). Perfect match-mismatch was used to compute probe intensity, and peak detection performed using a moving average cutoff of 2.5 and minimum region of 250 bp. CisGenome identified 3708 peaks (false discovery rate < 0.01).
Enrichment Test and qPCR
For enrichment tests, oligonucleotides for a suspected AGL15 target and for a nonbound control were used in multiplex PCR reactions with independently generated ChIP populations. Products were resolved on 1% agarose gels and imaged using a Gel Doc XR system (Bio-Rad).
For qPCR, 0.5 μL of recovered DNA from ChIP or controls or 1 μL of input DNA diluted 125-fold was added to a reaction consisting of 40,000× diluted SYBR Green I (Invitrogen), 50 mM KCl, 0.2 mM deoxynucleotide triphosphate, 0.5 μM of each oligonucleotide, and 2 to 2.4 units Klentaq in 1× PC2 buffer (Ab Peptides). PCR was performed in an iCycler (Bio-Rad) with an initial 95°C, 2 min denaturation followed by 40 cycles of 95°C, 30 s; 55°C, 30 s; 72°C, 30s, and a final 72°C, 5 min, followed by a melt curve determination.
Quantitation involved normalization of each immune (I) precipitation (or control) sample Ct to the input DNA sample Ct to obtain a ΔCt and then either subtracting ΔCt of the preimmune control precipitation from the ΔCt of the I or subtracting the ΔCt of the nonbound control in the I from the ΔCt of the target in the same I precipitation to obtain ΔΔCt values. 2(−ΔΔCt) for the former gives fold enrichment above background value, whereas for the latter indicates differential site occupancy or binding relative to control fragments (Haring et al., 2007; Mukhopadhyay et al., 2008; SuperArray Bioscience, http://www.superarray.com/manuals/chipqpcrpresentation.pdf). Oligonucleotides used for these experiments are listed in Supplemental Table 3 online.
Expression Arrays and qRT-PCR
TRIZOL Reagent (Invitrogen) was used to isolate total RNA from ∼50 to 100 mg of 10-d-old seedlings grown in liquid media containing 2,4-D (Mordhorst et al., 1998). The RNA was further purified using the RNeasy plant mini kit (Qiagen). Three biological replicates of each genotype (wild type, 35Spro:AGL15, and agl15 agl18) were performed. Preparation of probe for hybridization to Affymetrix ATH1 arrays was as directed by the manufacturer (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Partek GS was used to identify gene expression changes using the gene expression workflow that included a robust multichip average background correction and quantile normalization, after which the log2 of the probe intensity was calculated and one-way analysis of variance performed to generate lists with P < 0.01. These lists were further sorted to identify genes with at least a twofold change in the wild type compared with overexpressor or mutant compared with the wild type with a consistent or no change for the other comparison (e.g., 35Spro:AGL15/Col ≤ 0.5 and agl15 agl18/Col ≥ 1; or 35Spro:AGL15/Col ≤ 1 and agl15 agl18/Col ≥ 2).
For real-time qRT-PCR, 1.0 μg of total RNA was treated with DNase I (Invitrogen) and used for first-strand cDNA synthesis. Reverse transcription was performed using A-MLV reverse transcriptase system (Promega). An aliquot (0.5 μL) of each first-strand cDNA reaction was amplified by specific primer pairs in a reaction containing 1× PCR buffer, 2.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate, 0.5 μM of each oligonucleotide primer, 40,000× diluted SYBR Green I (Invitrogen), and 1.25 units of Platinum Taq (Invitrogen) in a final volume of 20 μL. Amplification was performed in an iCycler (Bio-Rad) as follows: 2 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 55°C, 30 s at 72°C, and 5 min at 72°C. Amplification was followed by a melt curve determination as follows: 180 cycles, each cycle persisting for 10 s and the first cycle at 60°C, with an increase of 0.2°C each cycle after cycle 2. Oligonucleotides are shown in Supplemental Table 3 online. Data analysis was performed using REST software (Pfaffl et al., 2002).
Accession Numbers
AGI loci identifiers are provided in the tables. The data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE17742 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17742).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confirmation of Knockout Alleles of IAA30.
Supplemental Figure 2. Conservation of CArG Motifs within the Regulatory Regions of Genes with Homology to Arabidopsis (At) ABI3 and FUS3.
Supplemental Table 1. Presence of Previously Known AGL15 Targets and Controls as Bound in ChIP-chip.
Supplemental Table 2. DNA Fragments Identified as Being Bound by AGL15 Using ChIP-chip and Verified by Enrichment Tests.
Supplemental Table 3. Oligonucleotides Used to Confirm Binding by AGL15 and Response to AGL15.
Supplemental Data Set 1. All Sites Identified as Present on All Three Replicates of the ChIP-chip Experiment and Annotated to a Nearby Gene by Partek GS.
Supplemental Data Set 2. All Sites Identified as Present on All Three Replicates of the ChIP-chip Experiment and Annotated by Cisgenome.
Supplemental Data Set 3. ATH Expression Results: Genes Responsive at a Cutoff of P < 0.01 and a Twofold Change for One Comparison with Consistent or No Change for the Other Comparison.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (IBN-9984274 and IOS-0922845), the USDA National Research Initiative (2005-35301-15699), and the National Institutes of Health (NIH-P20 RR16481) and by the University of Kentucky. We thank Donna Wall and the UK Microarray Facility for probe generation, array hybridization, and data collection. We also thank Kate Mitchell for assistance maintaining cultures and Randy Dinkins and our anonymous reviewers for valuable comments on the manuscript. We thank Donna Fernandez for providing the single and double mutants for agl15 and agl18 and the ABRC for the iaa30 alleles. We are grateful to Hongkai Ji for assistance with CisGenome. This article (09-06-063) is published with the approval of the Director of the Kentucky Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sharyn E. Perry (sperr2@email.uky.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Adamczyk, B.J., Lehti-Shiu, M.D., and Fernandez, D.E. (2007). The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50 1007–1019. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 653 653–657. [DOI] [PubMed] [Google Scholar]
- Benhamed, M., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56 493–504. [DOI] [PubMed] [Google Scholar]
- Boutilier, K., Offringa, R., Sharma, V.K., Kieft, H., Ouellet, T., Zhang, L., Hattori, J., Liu, C.-M., van Lammeren, A.A.M., Miki, B.L.A., Custers, J.B.M., and van Lookeren Campagne, M.M. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A.V., Tariq, M., and Paszkowski, J. (2004). Chromatin techniques for plant cells. Plant J. 39 776–789. [DOI] [PubMed] [Google Scholar]
- Braybrook, S.A., and Harada, J.J. (2008). LECs go crazy in embryo development. Trends Plant Sci. 13 624–630. [DOI] [PubMed] [Google Scholar]
- Braybrook, S.A., Stone, S.L., Park, S., Bui, A.Q., Le, B.H., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 103 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon, S., Ireland, A., Mungall, C.J., Shu, S., Marshall, B., and Lewis, S. (2009). AmiGO Hub, Web Presence Working Group. AmiGO: Online access to ontology and annotation data. Bioinformatics 25 288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y.-H., Sung, Z.R., and Goodrich, J. (2004). Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development 131 5263–5276. [DOI] [PubMed] [Google Scholar]
- Curaba, J., Moritz, T., Blervaque, R., Parcy, F., Raz, V., Herzog, M., and Vachon, G. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey, T. (2006). Analysis of a multifactor microarray study using Partek genomics solution. Methods Enzymol. 411 256–270. [DOI] [PubMed] [Google Scholar]
- Edgar, R., Domrachev, M., and Lash, A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emons, A.M.C. (1994). Somatic embryogenesis: Cell biological aspects. Acta Bot. Neerl. 43 1–14. [Google Scholar]
- Fernandez, D.E., Heck, G.R., Perry, S.E., Patterson, S.E., Bleecker, A.B., and Fang, S.-C. (2000). The embryo MADS domain factor AGL15 acts postembryonically: Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj, M.D., Zhang, S., Harada, J.J., and Lemaux, P.G. (2005). Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222 977–988. [DOI] [PubMed] [Google Scholar]
- Gallois, J.-L., Nora, F.R., Mizukami, Y., and Sablowski, R. (2004). WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., and McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7 373–385. [DOI] [PubMed] [Google Scholar]
- Harding, E.W., Tang, W., Nichols, K.W., Fernandez, D.E., and Perry, S.E. (2003). Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 133 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring, M., Offermann, S., Danker, T., Horst, I., Peterhansel, C., and Stam, M. (2007). Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, V., Vielle-Calzada, J.P., Hartog, M.V., Schmidt, E.D., Boutilier, K., Grossniklaus, U., and de Vries, S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127 803–816. [PMC free article] [PubMed] [Google Scholar]
- Heck, G.R., Perry, S.E., Nichols, K.W., and Fernandez, D.E. (1995). AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K., Wang, H., and Perry, S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53 172–185. [DOI] [PubMed] [Google Scholar]
- Huang, H., Mizukami, Y., Hu, Y., and Ma, H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene Agamous. Nucleic Acids Res. 21 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H., Jiang, H., Ma, W., Johnson, D.S., Myers, R.M., and Wong, W.H. (2008). An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H., and Wong, W.H. (2005). TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics 21 3629–3636. [DOI] [PubMed] [Google Scholar]
- Kaufmann, K., Muiño, J.M., Jauregui, R., Airoldi, C.A., Smaczniak, C., Krajewski, P., and Angenent, G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7 e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong, R.W., Bui, A.Q., Lee, H., Kwong, L.W., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., and Deng, X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu, M.D., Adamczyk, B.J., and Fernandez, D.E. (2005). Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol. Biol. 58 89–107. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A.L., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lumba, S., and McCourt, P. (2005). Preventing leaf identify theft with hormones. Curr. Opin. Plant Biol. 8 501–505. [DOI] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke, D.W., Meinke, L.K., Showalter, T.C., Schissel, A.M., Mueller, L.A., and Tzafrir, I. (2003). A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol. 131 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordhorst, A.P., Voerman, K.J., Hartog, M.V., Meijer, E.A., van Went, J., Koornneef, M., and de Vries, S.C. (1998). Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, A., Deplancke, B., Walhout, A.J.M., and Tissenbaum, H.A. (2008). Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nakaminami, K., Hill, K., Perry, S.E., Sentoku, N., Long, J.A., and Karlson, D.T. (2009). Arabidopsis cold shock domain proteins: Relationships to floral and silique development. J. Exp. Bot. 60 1047–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Cheng, J.-C., Sung, Z.R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94. [DOI] [PubMed] [Google Scholar]
- Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., and Choi, G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, S.E., Lehti, M.D., and Fernandez, D.E. (1999). The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol. 120 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, S.E., Nichols, K.W., and Fernandez, D.E. (1996). The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell 8 1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W., Horgan, G.W., and Dempfle, L. (2002). Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, F.B., and Meeks-Wagner, D.R. (1995). Seeing double: Appreciating genetic redundancy. Plant Cell 7 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Séraphin, B. (2001). The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24 218–229. [DOI] [PubMed] [Google Scholar]
- Rose, R.J., and Nolan, K.E. (2006). Invited review: Genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula. In Vitro Cell. Dev. Biol. Plant 42 473–481. [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, A., and Yamamoto, K.T. (2008). Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 133 397–405. [DOI] [PubMed] [Google Scholar]
- Stepanova, A.N., Robertson-Hoyt, J., Yun, J., Benavente, L.M., Xie, D.Y., Dolezal, K., Schlereth, A., Jürgens, G., and Alonso, J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Braybrook, S.A., Paula, S.L., Kwong, L.W., Meuser, J., Pelletier, J., Hsieh, T.-F., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 105 3151–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Wang, H.H.Y., and McCarty, D.R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., Kikuchi, A., and Kamada, H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., and Perry, S.E. (2003). Binding site selection for the plant MADS domain protein AGL15: An in vitro and in vivo study. J. Biol. Chem. 278 28154–28159. [DOI] [PubMed] [Google Scholar]
- Thakare, D., Tang, W., Hill, K., and Perry, S.E. (2008). The MADS-domain transcriptional regulator AGAMOUS-Like 15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol. 146 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen, F., Wu, H., Richmond, T., Redman, J.C., Johnson, C., Green, R., Arias, J., and Town, C.D. (2006). Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 47 152–162. [DOI] [PubMed] [Google Scholar]
- To, A., Valon, C., Savino, G., Guilleminot, J., Devic, M., Giraudat, J., and Parcy, F. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M.L., Guillaume, E., Buisine, N., Gagnot, S., Martienssen, R.A., Coupland, G., and Colot, V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir, I., Dickerman, A., Brazhnik, O., Nguyen, Q., McElver, J., Frye, C., Patton, D., and Meinke, D. (2003). The Arabidopsis SeedGenes Project. Nucleic Acids Res. 31 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, G. (2005). What don't we know? How does a single somatic cell become a whole plant. Science 309 86. [DOI] [PubMed] [Google Scholar]
- Wang, H., Caruso, L.V., Downie, A.B., and Perry, S.E. (2004). The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Tang, W., Zhu, C., and Perry, S. (2002). A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS-domain protein that preferentially accumulates in embryos. Plant J. 32 831–843. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Guo, J.H., Lambert, K.N., and Lin, Y. (2007). Developmental control of Arabidopsis seed oil biosynthesis. Planta 226 773–783. [DOI] [PubMed] [Google Scholar]
- Wang, X.C., Niu, Q.W., Teng, C., Li, C., Mu, J.Y., Chua, N.H., and Zuo, J.R. (2009). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19 224–235. [DOI] [PubMed] [Google Scholar]
- Weiss, D., and Ori, N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J.J., and Young, R.A. (2002). Deciphering gene expression regulatory networks. Curr. Opin. Genet. Dev. 12 130–136. [DOI] [PubMed] [Google Scholar]
- Zhu, C., and Perry, S.E. (2005). Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J. 41 583–594. [DOI] [PubMed] [Google Scholar]
- Zolman, B.K., Nyberg, M., and Bartel, B. (2007). IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64 59–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.