Abstract
Impeded DNA replication or a deficiency of its control may critically threaten the genetic information of cells, possibly resulting in genome alterations, such as gross chromosomal translocations, microsatellite instabilities, or increased rates of homologous recombination (HR). We examined an Arabidopsis thaliana line derived from a forward genetic screen, which exhibits an elevated frequency of somatic HR. These HR events originate from replication stress in endoreduplicating cells caused by reduced expression of the gene coding for the catalytic subunit of the DNA polymerase δ (POLδ1). The analysis of recombination types induced by diverse alleles of polδ1 and by replication inhibitors allows the conclusion that two not mutually exclusive mechanisms lead to the generation of recombinogenic breaks at replication forks. In plants with weak polδ1 alleles, we observe genome instabilities predominantly at sites with inverted repeats, suggesting the formation and processing of aberrant secondary DNA structures as a result of the accumulation of unreplicated DNA. Stalled and collapsed replication forks account for the more drastic enhancement of HR in plants with strong polδ1 mutant alleles. Our data suggest that efficient progression of DNA replication, foremost on the lagging strand, relies on the physiological level of the polymerase δ complex and that even a minor disturbance of the replication process critically threatens genomic integrity of Arabidopsis cells.
INTRODUCTION
The duplication of the genome is a key step of cell proliferation, intrinsically bearing a high mutagenic risk. Therefore, DNA replication of all organisms is safeguarded by the concerted action of evolutionarily conserved cell cycle regulators, DNA integrity checkpoints, repair mechanisms, and a faithful replication machinery (reviewed in Nyberg et al., 2002; Bartek et al., 2004; Toueille and Hübscher, 2004). At replication forks of eukaryotic organisms, coordinated DNA synthesis of leading and lagging strands is achieved by the three replication DNA polymerase complexes α, δ, and ε (Polα, Polδ, and Polε) (reviewed in Hübscher et al., 2002). Polδ is anticipated to be the main replicative DNA polymerase that upon homodimerization synthesizes both nascent strands in a coordinated fashion (Alberts, 2003). Thereby, the looped structure of single-stranded DNA (ssDNA) on the lagging strand is coated by the replication protein A complex (RPA). Due to the 5′-3′ polarity of DNA synthesis by all polymerases, leading strand synthesis is continuous, whereas discrete DNA stretches (Okazaki fragments) are formed on the lagging strand, each of which requires priming by the Polα/Primase complex. Facilitated by a versatile interaction network of the auxiliary proteins PCNA, RF-C and RPA, the resulting oligonucleotides are elongated by the more processive Polδ holoenzyme that replaces Polα (the so-called “pol switch”) (Waga and Stillman, 1994).
The accuracy of the DNA copying process is guaranteed by the proofreading activity of Polδ and Polε, by the postreplicative mismatch repair, and by mechanisms that ensure the structural maintenance and the progression of replication forks (reviewed in Hoeijmakers, 2001). Physical obstacles, such as DNA lesions, intrinsic interference with DNA synthesis, and collision with RNA polymerases, eventually cause stalling or even collapsing of forks, which can result in DNA loss or gross chromosomal rearrangements. These mutagenic events derive from inaccurate recombinational repair of DNA double-strand breaks (DSBs) at collapsed forks. A fairly well studied and evolutionary conserved example is the stalling of replication forks at defined sites in the rDNA (Urawa et al., 2001; Burkhalter and Sogo, 2004; Calzada et al., 2005). In addition, replication stress triggers a checkpoint response mediated by ATR (Mec1 in yeast), which is believed to prevent forks from collapsing and to facilitate the ultimate resumption of DNA synthesis (reviewed in Nyberg et al., 2002; Branzei and Foiani, 2007).
In view of the considerable evolutionary conservation of eukaryotic DNA replication and repair, comparable mechanisms can be expected in plants (Bray and West, 2005; Shultz et al., 2007). However, only fragmentary information is available. Apart from basic descriptions of rice (Oryza sativa) genes coding for the homologs of the catalytic subunit of Polα, of two subunits of Polδ, and of the replication accessory proteins FEN1 and PCNA (Yokoi et al., 1997; Kimura et al., 2001; Uchiyama et al., 2002), most functional studies of core replication genes, including mutational analysis, were performed in Arabidopsis thaliana. Of the three replicative DNA polymerases, the catalytic subunit of Polα (INCURVATA2) (Barrero et al., 2007) and two of the four subunits of Polε (Jenik et al., 2005; Ronceret et al., 2005; Yin et al., 2009) were characterized. In addition, paralogs of components of the single-strand binding protein RPA complex were identified as modifiers of epigenetic gene regulation (Elmayan et al., 2005; Xia et al., 2006). Furthermore, the Arabidopsis ATR paralogs were shown to fulfill a function in the replication checkpoint (Culligan et al., 2004, 2006; Friesner et al., 2005; Vespa et al., 2005). Nevertheless, mutational analysis of core replication factors requires further investigation.
In this study, we describe Arabidopsis plants mutated in the POLδ1 gene, which codes for the catalytic subunit of the DNA polymerase δ holoenzyme. The reduced expression of POLδ1 resulted in greater genome instability, as measured by the frequency of somatic HR in reliable transgene-based HR substrates lines. These have proven to be a powerful tool to analyze the impact of endogenous and environmental factors on the genome stability in situ. Such substrates consist of two nonfunctional parts with sequence homology of a plant expression unit coding for the firefly luciferase (LUC) or the bacterial β-glucuronidase (GUS) enzymes (Schuermann et al., 2005). Error-free recombination between the homologous sequences can be visualized by an in situ enzymatic assay detecting the restoration of the functional reporter gene in a single cell. In the context of a genetic screen for enhanced frequencies of homologous recombination (HR) using such a substrate line, the crucial role of Arabidopsis Polδ levels in the prevention of replication-dependent genome instabilities became apparent. Genetic analysis of the plants with reduced expression of POLδ1, in combination with chemical inhibition of DNA replication, suggests that at least two mechanisms contribute to the observed genome instability. We propose a model according to which the reduced cellular pool of Polδ in S-phase leads to impeded replication and thereby to an accumulation of ssDNA on the lagging strand. This unreplicated DNA triggers the formation of aberrant secondary DNA structures and fork stalling, which eventually result in chromosomal breaks that are repaired by HR.
RESULTS
Arabidopsis Plants Mutated in the POLδ1 Gene Show Enhanced Frequencies of Somatic Intramolecular HR
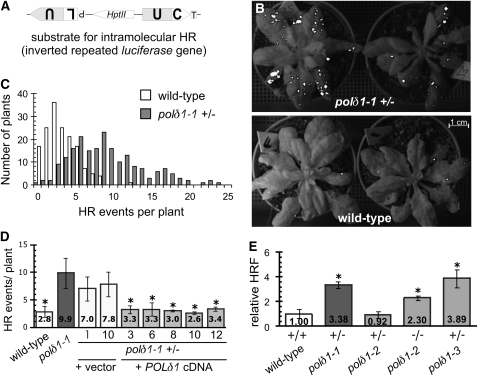
An Arabidopsis HR substrate line, which marks intramolecular recombination events between inverted repeats (IRs) of the luciferase gene (Figure 1A), was mutagenized by random integration of Agrobacterium tumefaciens T-DNA elements (see Supplemental Figure 1 online). The T2 offspring were scored for altered somatic HR frequencies (HRFs). Among others, we identified an insertional mutation in the POLδ1 gene (Arabidopsis Genome Initiative identifier At5g63960) that encodes the sole Arabidopsis homolog of the catalytic subunit of the Polδ complex (see Supplemental Figure 2 online). Hemizygous polδ1-1 mutant plants revealed increased occurrence of HR events in a dominant manner (Figures 1B and 1C). We note that plants homozygous for the mutation could not be obtained, suggesting that POLδ1 is an essential gene. In a population of ∼200 hemizygous polδ1-1 mutant plants in the 8 to 12 leaf developmental stage, the distribution of the number of HR events per plant differed significantly from that of wild-type plants. Whereas a wild-type plant exhibited approximately three HR events on average, a mean frequency of more than nine events were counted in hemizygous polδ1-1 plants (Figure 1C).
Figure 1.
Rosette Stage polδ1 Mutant Plants Exhibit Enhanced Frequencies of Homologous Recombination.
(A) Scheme of the HR substrate locus in the Luciferase-based reporter line 50B that allows the detection of HR repair events within the same DNA molecule (intramolecular HR). The duplicated region of a firefly luciferase expression unit (gray boxes, U) provided as inverted repeats serves as the target for HR. The two nonfunctional luciferase parts are separated by a spacer region encoding the selectable hygromycin resistance marker (HptII). P and T indicate the promoter and the terminator sequences, respectively.
(B) HR events (white dots) of hemizygous polδ1-1 (top panel) and wild-type plants (bottom panel), detected by the restoration of the functional Luciferase gene.
(C) Distribution of plants with a given number of somatic HR events in hemizygous polδ1-1 (+/−) and in wild-type (+/+) populations (n > 200), which were analyzed by the two-tailed Student's t test for unequal variances (P value = 3.16E-101).
(D) polδ1-1 plants were complemented with vector control or with the cDNA of POLδ1 driven by a strong viral promoter. In selected T2 lines, hemizygous polδ1-1 plants ectopically expressing the POLδ1 cDNA were statistically analyzed for the mean number of HR events per plant. Error bars indicate standard deviations of independent experiments (n ≥ 3). Asterisks indicate significantly different numbers of HR compared with hemizygous polδ1-1 plants.
(E) Assessment of the intramolecular HRF of Arabidopsis plants with hemizygous (+/−) or homozygous (−/−) mutant alleles of POLδ1 compared with wild-type controls. Asterisks indicate significant differences of HRFs compared with wild-type plants. Error bars indicate standard errors of independent experiments (n ≥ 3).
We complemented the polδ1-1 mutation by ectopically expressing the POLδ1 cDNA (GenBank accession number DQ160246) driven by a viral promoter (Cestrum yellow leaf curling virus; see Supplemental Methods online) in the hemizygous mutant background. We observed a suppression of the enhanced HRF in several independent lines (Figure 1D), which proves the genetic linkage between phenotype and mutation. The ectopically expressed POLδ1 cDNA also permitted the growth of homozygous polδ1-1 plants that still revealed severe growth defects (see Supplemental Figure 4A online) and high luciferase activity. A direct comparison of the HR frequency between the small homozygous polδ1-1 and the phenotypically normal appearing wild-type and hemizygous mutant plants was impeded; these plants were therefore excluded from the analysis of the HR frequency.
We obtained two additional Arabidopsis lines with T-DNA insertions in the POLδ1 gene from public collections (see Supplemental Figure 2 online) and crossed them into the HR reporter lines. Homozygous polδ1-2 and hemizygous polδ1-3 plants exhibited a 2.3- and 3.9-fold increased HRF in the Luciferase-based IR substrate line, respectively (Figure 1E). To exclude a genetic or epigenetic interaction between the POLδ1 and the HR substrate locus, we crossed the mutant plants with a reporter line carrying the same spatial arrangement of IRs but based on the GUS gene and integrated into an independent genetic locus. GUS reporter lines also allow the visualization of HR events, but in contrast with LUC reporter lines, they permit the analysis of HR events on the level of single cell and thus their precise localization. Comparable with the findings in the LUC-based line, we found a 5.1- and a 2.5-fold enhancement of the HRF for hemizygous polδ1-1 and homozygous polδ1-2 plants, respectively (Table 1). Comparison of complemented and allelic mutant plants confirmed that the phenotype of increased HR reflects the response of plants to mutations in the POLδ1 gene.
Table 1.
The Effect of polδ1 Alleles on Intramolecular HRF in Plants Containing Substrates with Inverted Repeats
| Rel. HR Increase in LUC and GUS Substrates |
||||||
|---|---|---|---|---|---|---|
| polδ1 Allele | Type of Allele | HR Class | LUC | (± SEM) | GUS | (±se) |
| polδ1-1, +/− | Insertional (T-DNA) | WHR | 3.38 | (±0.27)** | 5.1 | (±0.3)** |
| polδ1-2, −/− | Insertional (T-DNA) | WHR | 2.3 | (±0.18)** | 2.5 | (±0.4)* |
| polδ1-3, +/− | Insertional (T-DNA) | WHR | 3.89 | (±0.72)** | na | – |
| RNAi-POLδ1-1 | Epigenetic (Pmas, hairpin RNA) | WHR | na | – | 5.3 | (±1.4)** |
| RNAi-POLδ1-2 | Epigenetic (Palc, hairpin RNA) | SHR | na | – | 9.6 | (±3.2)** |
| RNAi-POLδ1-3 | Epigenetic (Palc, hairpin RNA) | SHR | na | – | 47.4 | (±11.5)** |
| RNAi-POLδ1-4 | Epigenetic (Palc, hairpin RNA) | SHR | na | – | 54.1 | (±7.8)** |
| RNAi-POLδ1-5 |
Epigenetic (PU6, antisense RNA) |
SHR |
na |
– |
99.8 |
(±22.6)** |
Arabidopsis mutant plants with reduced POLδ1 expression were generated by T-DNA insertion mutagenesis or by RNAi-mediated downregulation driven by various promoter sequences (Pmas, mannopine synthase; Palc, recombinant alcohol-inducible promoter; PU6, Arabidopsis U6 snRNP promoter). The HRF of plants was assessed in the same homozygous HR substrate line IR1, and the HR increase relative to wild-type plants was calculated. Mutant alleles were classified according to their HR phenotypes: weak, WHR; strong, SHR. *, **: statistically significance level P ≤ 0.05 or P ≤ 0.01, respectively, analyzed by ANOVA (n ≥ 3 independent experiments).
A Reduction of POLδ1 Expression Enhances the Frequency of HR
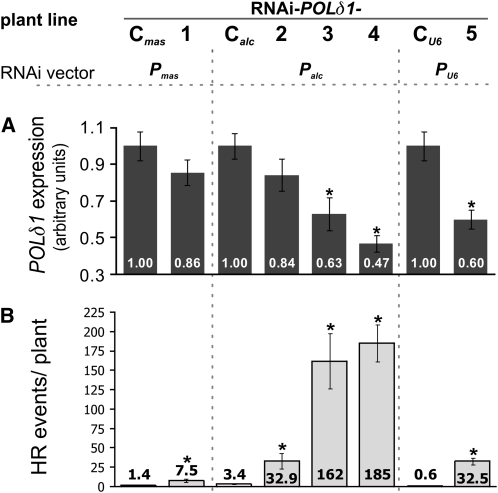
The POLδ1 expression data of the insertional mutant lines did not allow a firm conclusion on the cause of the dominant HR phenotype: a reduced endogenous transcript level encoding the full-length POLδ1 protein was observed in all lines, but aberrant transcripts also were detected (see Supplemental Figure 3 online). The latter could potentially lead to the production of a protein with a dominant-negative influence on the correct function of POLδ1. To distinguish between these two possibilities, we generated RNA interference (RNAi) mutant plants with a moderate level of POLδ1 downregulation, taking into account that POLδ1 is crucial for plant growth. We therefore used different promoters to drive the expression of the RNAi-mediating transcripts in planta. We could generate only a few plants that harbored RNAi-inducing transcripts driven by the ubiquitously expressed agrobacterial mannopine synthase or by the Arabidopsis U6 snRNP promoter (Table 1). They mostly exhibited either severely affected growth or absence of downregulation of POLδ1 expression; this points to a counterselection for plants with strong RNAi alleles for the essential gene. By contrast, phenotypically normal plants were obtained when the RNAi transcript was driven by a synthetic alcohol-inducible promoter preferentially expressing in somatic cells (Figure 2A; see Supplemental Figure 4C online). We note that the downregulation of the POLδ1 transcripts did not require an induction by alcohol and therefore most likely reflects leaky expression of this promoter in the absence of the inducing agent. All experimental data derives from plants without induction.
Figure 2.
Arabidopsis Plants with Reduced POLδ1 Expression Show Increased HRFs.
Arabidopsis lines with epigenetic POLδ1 alleles were generated by the integration of three different RNAi-mediating vector constructs (top panel) into a GUS-based HR reporter line with inverted repeats (line IR1). Pmas, mannopine synthase; Palc, recombinant alcohol-inducible promoter; PU6, Arabidopsis U6 snRNP promoter. Two weeks after germination, the HRF and the POLδ1 expression level were assessed.
(A) The steady state levels of POLδ1 transcripts were analyzed by quantitative RT-PCR. The expression data of mutant lines was plotted relative to their respective vector controls (C lines). Note: induction of the Palc promoter was not required to reduce POLδ1 expression.
(B) Recombination events per plant were assessed in independent plant lines and are shown relative to their respective vector controls (C lines). Error bars indicate standard error of independent experiments (n ≥ 3). Asterisks indicate statistically significant differences compared with controls.
For the analysis of HR levels, we again excluded morphologically abnormal RNAi lines to avoid pleiotropic effects. We measured steady state levels of POLδ1 transcripts in total RNA pools of whole RNAi plants by quantitative RT-PCR and found a reduction, ranging from 10 to 50% compared with wild-type plants (Figure 2A). We observed higher numbers of HR events in these lines; in some plants, up to several hundred GUS-positive cells were counted (Figure 2B). The relative enhancement ranged from ∼5 to 100 times higher HRF than in the control plants (Table 1) and correlates well with the reduction of POLδ1 transcript levels (Figure 2). We propose that the increased HR phenotypes of mutant lines with insertional or epigenetic alleles reflect downregulation of POLδ1 expression, which presumably leads to a lower cellular level of DNA polymerase δ. Thus, the increase in HRF of the insertional polδ1-1 and -3 mutants is likely to be caused by haploinsufficiency rather than by dominant-negative POLδ1 variants. To facilitate the interpretation of subsequent experimental outcomes, the mutant alleles were categorized into two subclasses, representing weak and strong HR phenotypes (WHR/SHR) (Table 1).
The Increased HRF of Mutant Plants Does Not Derive from Failure of DNA Repair
So far, we established the correlation of POLδ1 expression levels and increased HRF, but the underlying mechanism remained ambiguous. Considering the generally high functional conservation of DNA transaction processes in eukaryotes, the malfunctioning of at least two fundamental, DNA polymerase δ-dependent, cellular mechanisms could be responsible for the observed genetic instability: (1) DNA repair and (2) DNA replication (Bray and West, 2005; Garcia-Diaz and Bebenek, 2007). To test these activities, we compared the transcriptome of hemizygous polδ1 plants with that of wild-type plants and found the expression of only three genes to be significantly changed by a factor of >1.5 (Table 2). Originating from the activation tag of the mutagenizing T-DNA, a strong transcriptional upregulation was found for the gene At5g63950 located adjacent to POLδ1. Molecular and genetic analyses showed that this DNA repair gene belonging to the RAD54 family has no effect on HRF (see Supplemental Figure 5 online). The transcriptional analysis, together with the developmentally and morphologically normal appearance (see Supplemental Figure 4 online), suggest that polδ1 mutant plants suffer neither from widespread physiological stress nor from upregulated DNA repair activities (see Supplemental Data Set 1 online), which could have caused an increase in HR events.
Table 2.
Expression Analyses of polδ1 Mutant Plants: Whole-Genome Transcription Analysis of Hemizygous polδ1-1 Plants
| Relative Expression | Gene |
|||
|---|---|---|---|---|
| Name | ID | Probe Set ID | Gene Description | |
| 63.22* | RAD26-like | At5g63950 | 247310_at | Putative SNF2/RAD54 family DNA repair and recombination protein |
| 1.87* | KNAT6 | At1g23380 | 263013_at | Homeobox transcription factor |
| 0.55* |
FTSH9 |
At5g58870 |
247766_at |
ATP-dependent peptidase |
Steady state transcript levels of hemizygous polδ1 mutant and wild-type plants were analyzed by Affymetrix ATH1 full-genome GeneChip, and relative expression was calculated. Asterisks indicate statistically significant expression changes.
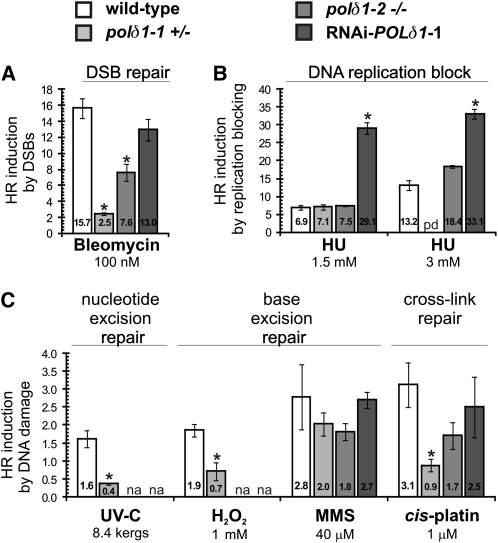
Several reports suggest that DNA damage accumulating in mutant plants deficient in a certain repair step are repaired by alternative pathways. For instance, an increased level of somatic HR events was found in Arabidopsis mutants with impaired nonhomologous end-joining (Gallego et al., 2003), with a reduced efficiency of nucleotide excision repair (Molinier et al., 2004a) or with an inactivated photolyase enzyme (Ries et al., 2000). Since resynthesis of DNA is anticipated to be performed by the DNA polymerase δ in virtually all repair pathways, we hypothesized that reduced levels of POLδ1 might lead to an increased frequency of DNA repair failure and thereby to more DSBs. We first investigated how SHR class mutant plants respond to the induction of DSBs (see Supplemental Figure 6 online) and calculated the ratio of HR events of challenged relative to untreated plants of the same genotype (Figure 3A). The stimulation of the HRF was found to be weaker in polδ1 mutants than in wild-type plants, indicating that the repair of DSBs by HR is impaired by the reduction of POLδ1 level. We also tested the HR response of SHR plants exposed to various physical or chemical treatments that result in the production of DNA lesions that may serve as substrates for nucleotide excision, for base excision, or for DNA cross-link repair but only indirectly as substrates for HR. The exposure of wild-type and most polδ1 mutant plants to these DNA-damaging conditions resulted in increased levels of HR events (see Supplemental Figure 6 online). Comparable to the findings of DSB induction, a reduced stimulation of HR was observed in the three assessed WHR class polδ1 mutants (Figure 3B), excluding a substantial contribution of elevated levels of intermediates of other repair mechanisms to the hyperrecombination phenotype of polδ1 mutant plants. Sustaining this notion, mutant plants were not found to be hypersensitive to a wide variety of tested genotoxic agents, whereas such hypersensitivity is a general feature of plants with defective DNA repair (Fidantsef et al., 2000; Gallego et al., 2000, 2003; Liu et al., 2000; West et al., 2002; Osakabe et al., 2006; Wang and Liu, 2006). Also, the expression of genes, transcriptionally upregulated upon induction of DSBs (Chen et al., 2003; Culligan et al., 2006), was only slightly altered (Table 3). For instance, the expression of RAD51, critical for HR, did not change. On the other hand, about twice as many PARP2 transcripts were detected in plants with epigenetic polδ1 alleles, which might indicate a constitutively increased level of DNA lesions. This is similar to what was described for bru1 mutant plants (Takeda et al., 2004). In addition, using the expression profiling data of plants with the weak polδ1-1 allele, an extensive list of DNA repair and replication-related genes was consulted, and only minor and nonsignificant variations were detected (see Supplemental Data Set 1 online).
Figure 3.
The Interference with DNA Replication but Not the Induction of DNA Damage Stimulates the HRF in polδ1 Mutant Background.
(A) and (B) Wild-type and polδ1 mutant plantlets (in IR1 HR substrate background) were exposed to the DSB-inducing chemical Bleomycin (A) and to various DNA damaging agents (B) that induce first and foremost types of DNA damage that are taken care of by the indicated repair pathways but only accidentally lead to repair or replication intermediates repaired by HR. Photoproducts induced by UV-C are repaired by the nucleotide excision repair pathway. Oxidized and alkylated DNA provoked by H2O2 and by methyl methanesulfonate (MMS), respectively, is mended by the base excision repair pathway. Cis-platin treatment leads to the covalent linkage of both DNA strands, requiring the complex cross-link repair machinery to be restored.
(C) Stalling of replication forks was induced by the drug HU. The induction of intramolecular HR in the different genetic backgrounds was calculated as ratio of HR events of treated and untreated plants. Asterisks indicate statistically differences by analysis of variance (ANOVA). Error bars indicate standard error of independent experiments (n ≥ 4). na, not assessed; pd, plants showed necrosis under these experimental conditions and were excluded from analysis.
Table 3.
Expression Analyses of polδ1 Mutant Plants: Expression of DNA Repair and Checkpoint Factors by Quantitative RT-PCR Analysis
| Relative Expression (±se) in polδ1 Mutant Plants |
||||||
|---|---|---|---|---|---|---|
| Gene |
WHR Class Mutants |
SHR Class Mutants |
||||
| Name | ID | polδ1-1 (+/−) | RNAi-POLδ1-1 | RNAi-POLδ1-3 | RNAi-POLδ1-4 | RNAi-POLδ1-5 |
| RAD51 | At5g20850 | 1.20 (±0.065) | 1.12 (±0.017) | 1.16 (±0.043) | 1.00 (±0.037) | 1.00 (±0.098) |
| BRCA1 | At4g21070 | 0.83 (±0.097) | 1.18 (±0.149) | 1.70 (±0.196) | 1.40 (±0.077)* | 1.17 (±0.169) |
| PARP2 | At4g02390 | 1.18 (±0.273) | 2.00 (±0.411) | 1.74 (±0.208)* | 1.66 (±0.181)* | 1.84 (±0.45) |
| ATR | At5g40820 | 1.05 (±0.439) | 1.34 (±0.512) | 1.84 (±0.117) | 1.88 (±0.157) | 0.73 (±0.095) |
| RAD17 | At5g66130 | 1.24 (±0.045) | 0.83 (±0.076) | 0.83 (±0.013) | 1.36 (±0.119) | 1.14 (±0.128) |
|
RNR2A |
At3g23580 |
2.19 (±0.231) |
1.25 (±0.079) |
1.60 (±0.103)* |
1.25 (±0.388) |
0.65 (±0.061) |
For polδ1 mutant plants, the steady state transcription levels of selected DNA transaction-related genes were analyzed by quantitative RT-PCR. Asterisks indicate statistically significant changes comparing experimental replica (n ≥ 3) of wild-type and mutant plants by the Student's t test.
Most polδ1-Dependent Recombination Events Arise in Differentiated Endoreduplicating Cells
As the previous experiments excluded abortive DNA repair as the major cause of the increased HRF in polδ1 plants, we investigated the impact of disturbed replication on genome stability. Unlike DNA-damaging agents, the DNA replication blocking compound hydroxyurea (HU) stimulated the HRF of the mutant lines more than that of wild-type plants (Figure 3C). For instance, the polδ1-1 and -2 mutant plants exhibited an equal or even more pronounced stimulation by HU, whereas they revealed less stimulation of HR upon exposure to Bleomycin, UV-C, H2O2, and cis-platin pointing to a reduced response to genotoxic stimuli (Figures 3A and 3B). A stronger induction of HR than in the wild type was observed in WHR class RNAi-POLδ1-1 plants. This implies that lowered POLδ1 expression and DNA replication blockage by HU induced molecular events in a cooperative fashion and suggests that hindered DNA replication is the major cause of the increased rates of HR events.
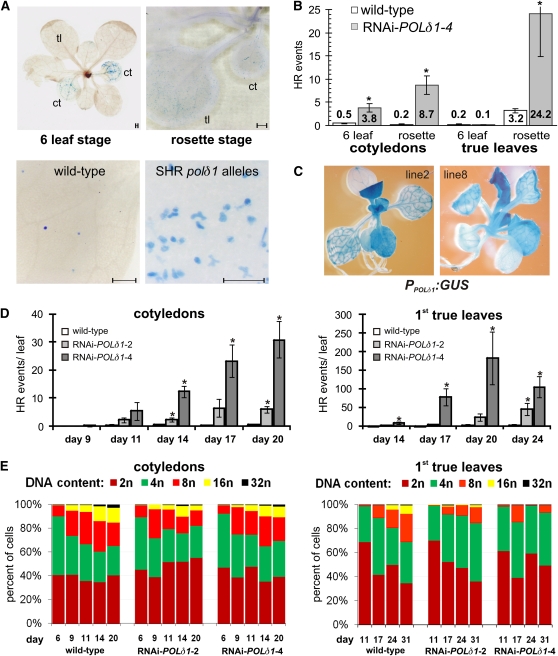
In plants, DNA replication takes place in distinct somatic cell types: in meristematic and differentiated cells. Whereas meristematic DNA replication is followed by cell division that is a prerequisite for plant growth and morphogenesis, some terminally differentiated cells multiply their genome without cytokinesis, a process called endoreduplication (reviewed in Sugimoto-Shirasu and Roberts, 2003). We investigated the contribution of those two cell type–specific kinds of replication to the enhanced HRFs of polδ1 mutants. At the six leaf stage, SHR class plants almost exclusively display HR events in the cotyledons but not in the already formed first true leaves (Figures 4A and 4B). In bolting plants, we detected many HR events in older true leaves and a further increase in the cotyledons. This observation suggests that the polδ1-dependent HR events preferentially take place in differentiated tissue. Indeed, the POLδ1 promoter was found to be more active in young emerging leaves than in mature leaves (Figure 4C), suggesting that the cellular level of Polδ declines with progressing age of the tissue.
Figure 4.
The HR Events of Mutant Plants with Reduced POLδ1 Levels and of Wild-Type Plants Originate in Endoreduplicating Somatic Cells.
The spatial and temporal appearance of HR events was analyzed in representative mutant plants with strong polδ1 alleles (SHR class) in the GUS-based inverted repeat background (line IR1).
(A) Top panel: At early developmental stages, HR events are mostly observed in cotyledons (ct); with progressing age, many of the HR events also occur on true leaves (tl). Bottom panel: Close-up of HR events in true leaves of rosette stage wild-type and SHR class mutant plants, predominantly appearing as staining of single cells. Bars = 1 mm.
(B) Incidences of HR in cotyledons and true leaves were statistically analyzed in line RNAi-POLδ1-4 at an early (six leaf) and late (rosette) developmental stage (n ≥ 6 plants).
(C) Transcriptional activity of POLδ1 promoter fused to GUS reporter gene, monitored in two independent Arabidopsis lines.
(D) The occurrence of HR events in cotyledons and in the first true leaves of wild-type and two RNAi-POLδ1 mutant plant lines was followed over time (days after germination). Error bars indicate standard error of HR events/plant in the assessed plant population (n ≥ 12).
(E) In parallel, nuclear DNA content was measured and the degree of endoreduplication cycles was estimated by the relative distribution of ploidy levels (n ≥ 2). Asterisks indicate significant differences between mutant and wild-type plants.
To correlate the temporal appearance of HR events with endoreduplication, we assessed the enhancement of the HRF and the expansion of nuclear DNA content over time in cotyledons and in the first true leaves. Cotyledons of plants with SHR class polδ1 alleles did not have significantly more HR events than those of wild-type plants until day 9 after germination. Later on, they revealed a gradual enhancement of HRF, which was most pronounced for both tested mutants at day 20 (Figure 4D, left panel). During the same period of time, nuclear DNA content of wild-type cotyledons steadily increased, signifying several rounds of endoreduplication (Figure 4E, left panel). In polδ1 mutant plants, endoreduplication was not impeded but fewer cells with a nuclear DNA content of more than 16n were observed from day 14 on. Similarly, a correlation between endoreduplication cycles and the increasing number of HR events was apparent in true leaves (Figures 4D and 4E, right panels), and nuclei with more than 4n were less frequently counted in mutant plants. In RNAi-POLδ1 plants, cells with 8n to 32n nuclei were more reduced in true leaves than in cotyledons, correlating well with the total number of HR events found in the respective tissues. Moreover, we found that the stronger the HR enhancement of a line was, the earlier it became evident. This implies that the moderate reduction of POLδ1 expression impairs the process of endoreduplication in differentiated somatic cells, which in turn results in more HR events.
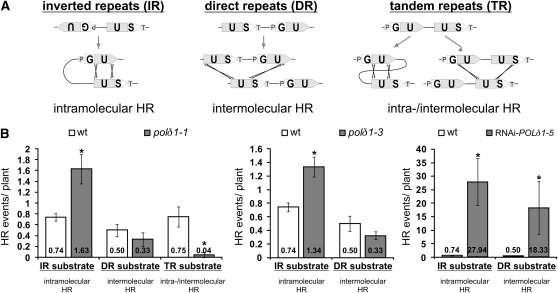
HR Substrates with Inverted Repeats Leads to an Increase in the HR Frequency
To investigate in more detail how HR events emerge, we analyzed WHR and SHR class POLδ1 mutant plants in Arabidopsis substrate lines that allow, depending on the spatial arrangement of the GUS repeats, the discrimination between intra- and intermolecular HR (Figure 5A). Recombination events that only involve DNA sequences within the same molecule result in the functional restoration of the reporter consisting of IRs (Schuermann et al., 2005). Intermolecular HR is the only way to restore a scorable reporter gene between nontandem direct repeats (DRs), whereas DSBs can be repaired by intra- or intermolecular HR when substrates with tandem repeats (TRs) are provided.
Figure 5.
Weak polδ1 Alleles Enhance HRF in a Substrate-Specific Manner.
(A) Scheme of GUS-based HR substrates differing in the arrangement of the repeats with respect to each other. Intra- and intermolecular interactions between IRs and DRs, respectively, give rise to the restoration of a functional marker gene through HR. By contrast, TRs allow repair of breaks by both intra- and intermolecular HR as well as by single-strand annealing.
(B) Plants with either WHR (left and central panels) or SHR (right panel) polδ1 alleles were crossed with substrate lines. HR events were assessed in 2-week-old F1 offspring, which were hemizygous for both the mutation and the substrate locus. Error bars indicate standard error of HR events/plant (n ≥ 6 plants). Asterisks indicate significant differences between mutant and wild-type control plant population by ANOVA.
We crossed WHR plants (Table 1) with the various substrate lines and analyzed the HRF in the offspring hemizygous for both the recombination substrate and the polδ1 allele. These plants revealed a polδ1-dependent increased HRF solely in IR substrate lines but not in DR substrate lines (Figure 5B). In addition, an increase in HRF was not observed when the weak polδ1-1 allele was crossed into a substrate line with TRs. This spatial arrangement also allows the repair of DSBs by the plant's favorite mode of HR, single-strand annealing, employing homologous sequences located on the same but also on another DNA molecule (Puchta, 2005). These data suggest that the HR increase depends on substrates with inverted repeats. By contrast, SHR plants yielded strongly increased HRF in both intra- and intermolecular HR substrate lines (Figure 5B). This implies that those HR events are caused by a mode of action that differs from that of weak alleles. We propose that, in addition to IR-dependent breaks, hampered DNA replication in plants with strong alleles leads to more widespread genome instability.
Stalling of Replication Forks Contributes to the Strong Induction of HRFs
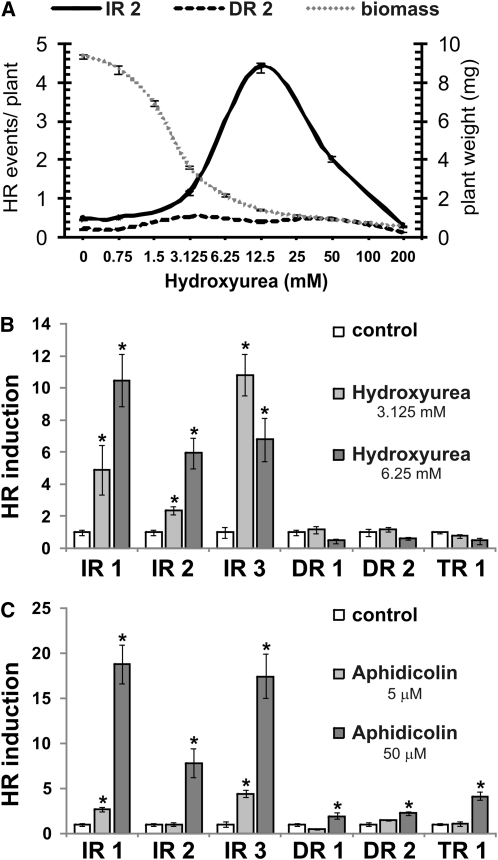
The creation of DSBs and the recruitment of the HR machinery are anticipated to be involved in the cellular response to stalled forks, preventing their detrimental collapse (discussed in Carr, 2002). We therefore tested whether artificially stalled replication forks in wild-type, POLδ1-proficient plants provoke chromosomal breakage, the repair of which may be measured by our HR substrate lines. We assessed the effect of the widely used replication blocker HU on plant growth and on HR induction and observed a dose-dependent enhancement of HRF solely in a substrate line with IR (Figure 6A). The most pronounced induction was found at HU concentrations that severely reduced plant growth.
Figure 6.
Pharmacological Stalling of Replication in Wild-Type Plants Phenocopies the Substrate-Specific Increase of HRF in polδ1 Plants.
(A) The induction of HR and the effect on growth were assessed in 2-week-old wild-type plantlets treated with increasing concentration of hydroxyurea. Intra- and intermolecular recombination events were measured by GUS-based substrates consisting either of IRs or DRs.
(B) and (C) Two-week-old plants of various substrate lines bearing distinct spatial arrangements of the homologous sequences were challenged with the replication blocking agent HU (B) or with Aphidicolin, a specific inhibitor of the replication polymerases Polα, Polδ, and Polε (C). The induction of the HRF relative to untreated plants was calculated. Error bars indicate standard error of independent experiments (n ≥ 2). Asterisks indicate significant differences of the relative HR induction analyzed by ANOVA.
To exclude line-specific differences, we exposed several independent substrate lines, containing different spatial arrangement of the homologous sequences (Figure 5A) to HU. All three reporter lines with IR revealed an up to 10-fold stimulation of the HRF by HU, while no significant changes were found in lines with DR or TR (Figure 6B). HU blocks DNA replication in an indirect manner by depleting the free nucleotide pool, which is thought to provoke or inhibit a variety of cellular processes, such as cell cycle progression and repair synthesis. Therefore, we confirmed the induction of HR by replication block with Aphidicolin, an inhibitor of the replication DNA polymerases α, δ, and ε. At low Aphidicolin concentrations, a slightly increased HRF was observed only in plants with IR substrate (Figure 6C). By contrast, exposure to higher concentrations of Aphidicolin yields more HR events independently of substrate type, resembling HR enhancements of treatments with DSB-inducing chemical compounds (see Supplemental Figure 7 online). Nevertheless, the HR increase was found to be much more pronounced in IR lines. The pharmacological interference with replication results in DSBs, and these breaks occur more often in genomic regions with IR. This dose- and substrate-dependent increase in HR mimics the different HR phenotypes of polδ1 plants, suggesting that stalling of replication forks contributes to HR events in SHR plants.
DISCUSSION
In all organisms, elementary processes, such as growth, cell division, tissue regeneration, or reproduction, require the faithful copying of genetic and also epigenetic information during the S-phase. In view of the size and the complexity of higher eukaryotic genomes, environmental and endogenous disturbance of DNA replication comprise a high intrinsic risk for the genome integrity of the next cell generation. Consequently, the efficiency and accuracy of the DNA synthesis machinery needs to be under firm surveillance by various DNA repair and checkpoint pathways (Nyberg et al., 2002; Bartek et al., 2004; Toueille and Hübscher, 2004). We show that even a slightly reduced expression of the catalytic domain of the replicative DNA polymerase δ results in considerable genome instability, measured as increased frequency of HR in somatic Arabidopsis cells (Table 1, Figures 1 and 2). Chromosomal regions containing IR sequences are particularly affected, leading to a more pronounced enhancement of the HRF (Figure 5). A chromatin structure-independent HR increase was observed when POLδ1 expression was strongly reduced or when stalling of replication forks was induced (Figures 5 and 6).
In line with the essential role of replicative DNA polymerases, two subunits of Polε (POL2a and DPB2) (Jenik et al., 2005; Ronceret et al., 2005) and the catalytic subunit of the Primase/Polα complex (INCURVATA2) (Barrero et al., 2007) turned out to be prerequisites for cell division. Similarly, we found that the deletion of the POLδ1 gene does not allow plant growth (see Supplemental Figures 2 and 4 online). The increased HR phenotype originated from endoreduplicating differentiated cells and was thus unlinked from the essential function in meristematic cells (Figure 4).
Lack of Replicative DNA Polymerases Results in Stalled Replication Forks
In Arabidopsis plants, genome instability can be induced either by pharmacological inhibition or by mutations in the catalytic subunits of Polδ and Polε (POL2a/ABO4) or in the single-strand binding protein RPA (this study; Yin et al., 2009) and therefore seems to be a consequence of interference with DNA replication. Since the replication machinery is also involved in virtually all synthesis-dependent repair pathways, it is often argued that those instabilities originate from unrepaired DNA lesions or repair intermediates that are converted to DSBs when cells enter S-phase. To explain the increased HR phenotype of polδ1 plants, however, we favor a hypothesis according to which the DNA breaks are directly created by uncoordinated and faulty DNA synthesis due to the lack of replication factors. This mode of action was previously suggested to be the cause of gross chromosomal instabilities observed in budding yeast (Saccharomyces cerevisiae) (Chanet and Heude, 2003; Galli et al., 2003) and mouse (Mus musculus) cells (Venkatesan et al., 2007), carrying mutations in the catalytic subunit of Polδ.
In favor of this hypothesis, we observed neither an increased sensitivity to genotoxic agents nor a stimulation of the HRF in polδ1 mutant plants exposed to agents that do not primarily induce DSBs but would lead to an accumulation of unrepaired DNA lesions (Figure 3). On the contrary, Polε (pol2a/abo4) mutant plants were shown to be hypersensitive to methyl methanesulfonate and UV-B, generally indicating a repair deficiency (Yin et al., 2009). Mimicking situations after DSB induction, pol2a/abo4 plants revealed a transcriptional response of repair and checkpoint signaling genes. Plants with polδ1 mutations did not reveal a transcriptional response of these genes or at least did not exhibit a strong response (Table 3; see Supplemental Data Set 1 online). This suggests that the DSBs derived from stalled forks are immediately repaired during S-phase. Checkpoint and HR proteins are abundant during replication; they are closely associated with replisomes and organized in large repair centers to ensure the correction of arising replication errors, as shown in yeast (Lisby et al., 2004). It remains to be further investigated whether the diverging transcriptional responses and sensitivities to genotoxins of polymerase δ and ε mutant plants indicate different mechanisms leading to the increased frequency of HR. In yeast, notably, the POL2a ortholog was shown to play a role in S-phase checkpoint signaling in addition to its catalytic function (Navas et al., 1995; Kesti et al., 1999). The hyperrecombination phenotype of pol2a plants may thus resemble those of polδ1 mutants, and the hypersensitivity could result from the failure of sensing or signaling of replication problems, especially when the plants are exposed to genotoxic agents.
A Tight Control of DNA Replication Ensures the Preservation of Genomes
A central role in dealing with hampered replication is assigned to the S-phase checkpoint kinase ATR and members of the RecQ-like DNA helicase family, correlating with human cancer predisposition syndromes (Hickson, 2003; Bartek et al., 2004). These proteins promote the stabilization of the replisome, prevent the detrimental collapse or processing of the fork, and allow the resumption of DNA synthesis. Activated by ATR, the mammalian RecQ family member Bloom syndrome protein and the structure-specific nuclease Mus81 were found to be compulsory for the active formation of temporal DSBs that might be required for the resumption of replication or the resolution of stalled forks (Shimura et al., 2008).
In Arabidopsis, some of the six RecQ-like helicases were found to modulate genome integrity (Bagherieh-Najjar et al., 2005; Hartung et al., 2007). Plants (Friesner et al., 2005; Vespa et al., 2005) and mammalian cells (Brown and Baltimore, 2000, 2003) lacking ATR were reported to exhibit genome instabilities, checkpoint deficiencies, and futile cell cycle regulation. Moreover, atr Arabidopsis plants exhibited hypersensitivity to replication inhibition by HU and Aphidicolin, emphasizing the role of ATR as S-phase checkpoint kinase (Culligan et al., 2004). Even in wild-type plants, we find that HU and Aphidicolin treatments lead to more HR events (Figure 6), most likely resulting from processed or collapsed replication forks. In addition, some polδ1 mutant lines revealed a more pronounced stimulation of the HRF than wild-type plants when challenged with HU (Figure 3). This implies that inhibited DNA synthesis caused by the reduction of POLδ1 and by HU synergistically leads to more stalled replication forks or DSBs, which may be resolved by HR.
Structural Hindrance of DNA Replication Can Cause Genome Instabilities
Apart from the aforementioned reduction of the essential replication proteins or nucleotides, the smooth operation of DNA synthesis can be hindered when a replication fork encounters the transcription machinery, strongly DNA-bound protein complexes, or secondary DNA structures. Indeed, WHR class plants with weak polδ1 alleles established elevated HRFs only in substrate lines that contain IR, in contrast with strong ones that generated more intra- and intermolecular HR events (Figure 5). This suggests that a minor reduction of the POLδ1 level does not provoke the collapse of replication forks as discussed before but interferes with replication in a DNA structure-specific manner.
We assume that during replication, short stretches of ssDNA on the lagging strand accumulate transiently, base pair with complementary sequences as present in IR substrate lines and form an aberrant hairpin structure. Immediate nucleolytic processing converts them to DSBs that are repaired by HR and thus cause the observed hyperrecombination phenotype. This mechanism was previously proposed to explain the chromosomal instabilities in budding yeast with reduced Polα levels, occasionally preventing the priming of lagging strand DNA synthesis (Lemoine et al., 2005). Alternatively, hairpin structures facilitated by the presence of IR may even be present before replication, presenting a structural obstacle for DNA synthesis and thus may lead to the slowing down of fork progression, eventually to fork stalling and to strand breaks (Nag and Cavallo, 2007; Voineagu et al., 2008). A reduced POLδ1 protein pool might exaggerate the frequency of such events. It is difficult to discriminate between these two explanations for the occurrence of DNA breaks in palindromic sequences or other parts of DNA that can form secondary structures. In any case, the increased potential of IR to threat the genome integrity seems to be a consequence of reduced processivity of DNA replication due to misregulation of internal or external factors.
Speculative Model for the Increased Somatic Homologous Recombination Frequency of Plants with Altered POLδ1 Expression
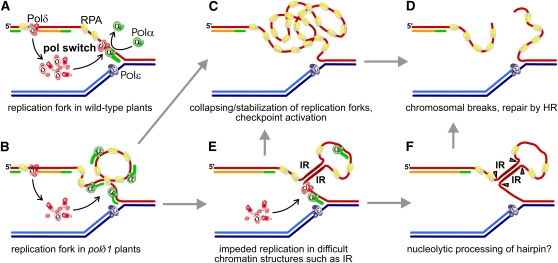
In yeast, although still debated (Pursell et al., 2007), there is evidence that the polymerase δ holoenzyme primarily synthesizes the Okazaki fragments of the lagging strand replacing the less processive and accurate primase/Polα complex (Figure 7A). Retarded switches between the Polα and Polδ may lead to the accumulation of extended stretches of RPA-coated ssDNA as observed microscopically at HU-induced stalled forks in yeast (Lopes et al., 2001; Sogo et al., 2002). These nucleoprotein complexes are considered an initial sensor of unreplicated or damaged DNA, eventually triggering an S-phase checkpoint response and providing a docking platform for downstream checkpoint and repair factors (Zou and Elledge, 2003; Harrison and Haber, 2006).
Figure 7.
Tentative Models for the Molecular Mechanisms Underlying the Genome Instabilities in Arabidopsis Cells with Reduced Levels of DNA Polymerase δ.
(A) Synchronous DNA synthesis on the leading and the lagging strand in wild-type cells requires an efficient exchange (pol switch) of Polα (green) with Polδ (red).
(B) The reduced expression of POLδ1 lowers the availability of Polδ holoenzymes, which delays the pol switch. Correlating with the strength of POLδ1 reduction, replication forks with unreplicated RPA-coated lagging strands accumulate.
(C) Mutant plants carrying strong polδ1 alleles (SHR class) reveal a more pronounced reduction of POLδ1 expression, which may result in high levels of RPA-coated ssDNA, induction of DNA damage signaling cascades, and stalling of replication forks in a sequence-independent manner.
(D) The collapse of the fork or the HR-dependent mechanisms of fork stabilization lead to chromosomal breaks that are substrates for HR.
(E) In WHR class polδ1 plants (weak polδ1 alleles), an increased frequency of HR was only observed in substrate lines with IRs. The moderate reduction of POLδ1 allows secondary structures such as hairpins to be formed, which in turn might boost the stalling of forks (C).
(F) Alternatively, hairpin structures might be subject to nucleolytic processing, ultimately leading to chromosomal breaks (D).
In polδ1 mutant plants, the cellular pool of functional Polδ complexes is likely to be reduced due to transcriptional misregulation, haploinsufficiency, or posttranscriptional gene silencing (Table 1), which may slow down lagging strand DNA synthesis and lead to an accumulation of ssDNA at replication forks (Figure 7B). In Arabidopsis polδ1 mutant plants of the SHR class, HR is implicated either in the stabilization of such POLδ1-depleted replication forks or, if already collapsed, in the repair of breaks or the eventual reassembly of forks and replication restart (Figures 7C and 7D; Branzei and Foiani, 2005). These events are largely independent of the sequence context and located randomly in the genome. This is in line with our data; in plants treated with high doses of the replicative polymerase inhibitor Aphidicolin or with strong epigenetic polδ1 alleles, we observed enhanced frequencies of HR, independent of the spatial arrangement of substrate repeats or genomic locus (Figures 5 and 6).
By contrast, Arabidopsis plants with weak alleles, treated with HU or low concentrations of Aphidicolin, revealed an increased frequency of HR events only when the substrate was provided as IR, emphasizing a sequence-specific induction of breaks. We suggest that these arise from base pairing of the homologous sequences in the unreplicated ssDNA, forming aberrant hairpin structures (Figure 7E). Such secondary structure might impede the smooth progression of DNA replication and trigger eventually the stalling of the fork and chromosomal breakage (Figures 7C and 7D). Alternatively, hairpins could be processed by structure-specific endonucleases (Figure 7F) as previously reported for mammalian cells under replication stress (Shimura et al., 2008).
We conclude that the reduction of core DNA replication factors, such as POLδ1, thus leads to breaks in DNA in somatic Arabidopsis cells. Particular features of chromatin are found to be more prone to break, but stalling of replication forks is likely to be the major reason for the genome instability. The IR-dependent increase of the HR frequencies in the WHR class polδ1 mutant plants could also be explained by a higher sensitivity of the intramolecular HR substrate lines. However, we consider the replication slowing effect of chromatin structure a more likely explanation as it was previously shown that a certain region of the rDNA repeats enhances the HR frequency in Arabidopsis (Urawa et al., 2001).
Mutations in Core Replication Genes Strongly Induce Frequencies of Somatic Homologous Recombination
In recent years, a wide variety of Arabidopsis mutants with altered frequency of HR have been reported to improve the efficiency of gene targeting and thereby to facilitate the genetic manipulation of plants (Britt and May, 2003; Hanin and Paszkowski, 2003; Reiss, 2003). Highlighting the impact on genome stability, the most enhanced HR rates have so far been reported for mutations in Arabidopsis genes tightly interconnected with the process of DNA replication: the catalytic subunit of Polε (POL2a/ABO4), a subunit of RPA (RPA2A/ROR1) (Yin et al., 2009), and two subunits of the chromatin assembly factor 1 (FAS1 and FAS2) (Endo et al., 2006; Kirik et al., 2006). We augmented this list with the catalytic subunit of the DNA polymerase δ, POLδ1, the expression level of which has an immense effect on the HR frequency (Figures 1 and 2). However, it remains to be elucidated whether these replication genes are suitable for the improvement of the gene targeting efficiency since they are mostly essential, their absence provokes uncontrollable genome instability, and their manipulation is often associated with severe morphological alterations. Indicating the broad impact of replication on the physiology of plants, some of these core replication genes were isolated in the course of genetic screenings for phenotypes that are not directly related to DNA replication, such as hormonal signaling and transcriptional gene silencing (Xia et al., 2006; Barrero et al., 2007; Yin et al., 2009).
In conclusion, we suggest that the lowered availability of POLδ1 and most likely other core replication factors leads to DNA breaks that can be detected in situ by systems reporting HR. This highlights the fact that processive and coordinated DNA synthesis on the lagging strand is crucial to prevent severe genome alteration in plants as well as in all other eukaryotic organisms. Studying replication-dependent genome instabilities is challenging due to their tight connection with cell division. Endoreduplication in differentiated plant cells unlinks growth from replication and thus facilitates the functional analysis of the mostly essential replication proteins. We would like to emphasize that the Arabidopsis plants with reduced POLδ1 expression presented here offer an attractive genetic system to investigate mechanisms and proteins that are involved in safeguarding DNA replication. Notably, the depletion of ATR or components of the HR machinery were found to be detrimental in mammals but did not impede plant growth (Brown and Baltimore, 2000; Culligan et al., 2004). Thus, their function and genetic interaction with factors in DNA replication can be easily assessed in the context of a whole multicellular organism without interfering with cell viability.
METHODS
Plant Material
All plant lines used in this study were of the Arabidopsis thaliana ecotype Columbia-0. Mutant plants were obtained from a genetic screen (polδ1-1) or from the public Arabidopsis T-DNA knockout mutant collection at the SALK Institute (polδ1-2, SALK_053085; polδ1-3, SALK_030272) (Alonso et al., 2003). Initially, hemizygous polδ1-1 plants were backcrossed twice with the luciferase-based HR substrate line 50B (see below) or with wild-type plants to clean up the genetic background or to get rid of the HR substrate, respectively. Plants with mutant alleles were identified by PCR analysis (see Supplemental Table 1 online) of small populations or by sulfonamide selection (15 mg/L), when a large number of plants was required. Plants with the polδ1-2 allele were crossed with the respective HR substrate lines. In the F2 and F3 generations, homozygous mutant and out-segregating wild-type plants in a homozygous HR substrate background were identified by PCR and by segregation analysis (polδ1-2, 50 mg/L kanamycin; HR substrate, 10 mg/L hygromycin), respectively. The polδ1-3 allele was introduced to the HR substrate lines using a similar strategy. Hemizygous mutant plants were identified by PCR-based genotyping since the selectable marker gene of the T-DNA conferring resistance to kanamycin was found to be inactive.
Arabidopsis transformation was achieved by the floral dip method as described by Fritsch et al. (2004). We used the Agrobacterium tumefaciens strain C58CIRifR containing the disarmed Ti-plasmid GV3101 and a binary plasmid with the desired construct. For the complementation experiment and the generation of RNAi alleles, binary vectors were constructed with standard molecular methods, as described in more detail in Supplemental Methods online. For the hairpin-spliced RNAi constructs, POLδ1-specific sequences of the 3′ region of the cDNA (gi: DQ160246, position 2550 to 2872) were cloned downstream of the agrobacterial mannopine synthase or the synthetic alcohol-inducible promoter, giving rise to RNAi-POLδ1 lines 1 and 2 to 4, respectively. (Note: all the analyses of RNAi lines 2 to 4 were done in the noninduced state. We assume that the leakiness of the promoter was sufficient to posttranscriptionally downregulate POLδ1 mRNA.) The line RNAi-POLδ1-5 was generated using the Arabidopsis U6 snRNA promoter to drive a 323-nucleotide-long POLδ1 antisense RNA (gi: DQ160246, position 2550 to 2872). To generate Arabidopsis PPOLδ1:GUS fusion lines, the putative promoter region (chr5: 25'598'402- 25'599'599) between the translation initiation codons of the POLδ1 and the neighboring at5g63950 gene was cloned between the bidirectional promoter-less expression units for GUS and GFP in a pCambia2300-derived binary vector. See Supplemental Table 1 online for primer sequences.
Somatic HRF was assessed in plants carrying HR substrates for intra- and/or intermolecular events. A genetic screen for enhanced HRF was conducted in the LUC-based line 50B with IR (Fritsch et al., 2004). Genetic and epigenetic polδ1 mutants as well as pharmacologically treated plants were analyzed in the GUS-based reporter lines 1415, 1445, and 1406 (lines IR1-3) (Gherbi et al., 2001; Fritsch et al., 2004), lines IC-6C and IC-9C (lines DR1,2) (Molinier et al., 2004b), and line 1418 (line TR1) (this study). We note that molecular reassessment of the HR substrate line 1406 used in our lab, originally described as TR line, revealed an IR-type spatial arrangement of the substrate.
Growth Conditions and Pharmacological Treatments
Plants were grown either on standard potting soil or in vitro on Murashige and Skoog (MS) medium (Duchefa) supplemented with 1% sucrose. They were kept in a conditioned growth chamber with 70% relative humidity and 16 h light/21°C and 8 h darkness/16°C cycles.
For pharmacological induction assays, seedlings were grown on solid MS for 8 d and adapted to liquid MS for 1 d. They were incubated in liquid MS supplemented with the indicated concentration of the respective chemical (Sigma-Aldrich) for 3 d and then grown in chemical-free MS/1% sucrose for 4 d and finally assessed for their HRFs.
Assessment of the HR Frequencies
Plants carrying the luciferase substrate were grown on soil and assessed for their HRF at the rosette stage. Fifteen to thirty plants of control and mutant populations were sprayed with 1 mM Luciferin (Biosynth)/ 0.05% Extravon (Novartis). Dark incubation for 30 min ensured the uptake of the Luciferin and reduced background fluorescence. Light emission was recorded twice for a 15-min period by a CCD camera. Using an arithmetic picture merging method, only signals of Luciferase activity present on both pictures were extracted, assigned to individual plants, and counted visually.
Arabidopsis lines carrying a GUS-based HR substrate were grown in vitro to the indicated developmental stage. The restoration of the reporter gene was visualized by histochemical GUS staining according to the standard protocol (Jefferson et al., 1987). HR events of individual plants were assessed visually using a stereomicroscope.
The HRF of the original HR substrate lines and of out-segregating hemizygous polδ1-1 and -3 siblings were found to be similar. In general, HRFs of sulfonamide-selected hemizygous polδ1-1 mutants were compared with HRFs of the original substrate line and the offspring of segregated wild-type plants. Controls for polδ1-3 plants were out-segregating siblings and parental HR substrate lines. Homozygous polδ1-2 mutants were compared with the offspring of out-segregated wild-type population. Plants with epigenetic polδ1 alleles were routinely compared with parental HR substrate lines and a vector-transformed population. The number of HR events per plant in wild-type and mutant populations (30 to 100 plants) was analyzed for distribution and for statistically significant differences. Average HRFs of each population, obtained from independent experiments or from large populations, were calculated and statistically analyzed by the single factor ANOVA function of MS Excel, defining 5% as significance level.
Expression Analyses
Total RNA was isolated from in vitro–grown plants of the indicated age by the Plant RNeasy Miniprep system (Qiagen) according to the manufacturer's instructions, routinely including an on-column DNase treatment.
For genome-wide expression analysis, all lines were profiled in duplicate (grown independently at 2-week intervals). Because the transcriptomes of HR substrate line 50B and wild-type plants were found to be identical, we compared the expression profiles of the polδ1-1 mutant plants with 50B and wild-type plants without distinction. We thus used four replicates of the wild-type condition (twice line 50B, twice wild-type) and two replicates for the mutant allele. Ten micrograms of RNA isolated from 2-week-old in vitro–grown plantlets (20 to 30 plants per line) was reverse transcribed, labeled, and hybridized to Affymetrix ATH1 full genome GeneChip as described by Fritsch et al. (2004). Chip data analysis was performed with the Affymetrix Microarray Suite v5 and GeneSpring 5.0 (Silicon Genetics). First, data with low signal (raw data <50) or unreliable detection statistics were discarded. Changes in gene expression were assessed by looking for concordant changes between replicates using a signed Wilcoxon rank test (as recommended by Affymetrix). The change P value threshold was <0.003 for increase and >0.997 for decrease. The list of significantly changed genes was filtered for a 1.5-fold change cutoff and submitted to the one-way ANOVA test.
Quantitative RT-PCR analysis was performed with the QuantiTect SYBR Green PCR kit (Qiagen) on a Rotor-Gene RG-3000 real-time PCR system (Corbett Research) according the manufacturers' recommendations (for more detailed information, see Supplemental Methods online). For each target gene, quantitative PCR data of at least three replicates were standardized with those of Actin7 (ACT7), preferentially expressed in young and dividing tissues. Expression levels relative to control plants were calculated for each replicate and subjected to two-sided Student's t test assuming unequal variances and a 5% significance level.
Temporal Assessment of HR and Nuclear DNA Content
Plants were germinated in vitro on solid MS. At indicated time points, the cotyledons or the 1st true leaves were dissected; one of them was subjected to the analysis of HR, and the other was used to assess nuclear DNA content by fluorescence-activated cell sorting as described by Galbraith et al. (1983). Leaf material of six independent plants was pooled, submerged with 45 mM MgCl2, 30 mM sodium citrate, 20 mM MOPS, 0.1% (w/v) Triton X-100 supplemented with 50 μg/mL DNase-free RNase (Roche Diagnostics), and 50 μg/mL propidium iodide (Sigma-Aldrich), and chopped into small pieces with a razor blade. The homogenate was filtered by passing through a 50-μm nylon mesh, and nuclear DNA content was assessed with the FACSCalibur flow cytometer (BD Biosciences). Propidium iodide fluorescence was excited with 530 mW at 514 nm and measured in the FL2 channel using a 580/42-nm band-pass filter. In at least two independent experiments, the nuclear DNA content of 104 cells was assessed with the CellQuest software package and its relative distribution was calculated.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: the sequences of the full-length POLδ1 cDNA and the deduced polypeptide can be found under the GenBank accession numbers DQ160246 and ABA41487. The POLδ1 gene corresponds to the Arabidopsis Genome Initiative locus identifier At5g63960.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of the Mutagenizing T-DNA.
Supplemental Figure 2. T-DNA Mutant Alleles of the POLδ1 Gene.
Supplemental Figure 3. POLδ1 Expression Analysis of T-DNA Mutant Plants.
Supplemental Figure 4. Phenotypic Comparison of polδ1 Mutant and Wild-Type Plants.
Supplemental Figure 5. The Rad26-Like Gene At5g63950 Does Not Alter the HR Frequency.
Supplemental Figure 6. Induction of HR in polδ1 Mutant and Wild-Type Plants.
Supplemental Figure 7. The Induction of HR by DNA Damage in Substrate Lines with Different Spatial Arrangement of Repeats.
Supplemental Table 1. List of Oligonucleotides
Supplemental Data Set 1. Whole-Genome Transcription Analysis of polδ1-1 Mutants.
Supplemental Methods. Generation of Complementation and RNAi-Inducing Vectors and Detailed Description of the qRT-PCR Conditions and Oligonucleotides.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Bernd Reiss for kindly providing the activation-tagging vector. We thank Edward Oakeley and Hubertus Kohler for their help with the flow cytometry and the transcriptome analysis, respectively. We thank Ortrun Mittelsten Scheid, Jennifer Cobb, Jean Molinier, Kenji Shimada, Holger Puchta, Primo Schär, and Bettina Freymüller for the critical reading and discussion of this manuscript. This work was financially supported by the Novartis Research Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David Schuermann (david.schuermann@unibas.ch).
Online version contains Web-only data.
References
- Alberts, B. (2003). DNA replication and recombination. Nature 421 431–435. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bagherieh-Najjar, M.B., de Vries, O.M., Hille, J., and Dijkwel, P.P. (2005). Arabidopsis RecQl4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 43 789–798. [DOI] [PubMed] [Google Scholar]
- Barrero, J.M., Gonzalez-Bayon, R., del Pozo, J.C., Ponce, M.R., and Micol, J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA Polymerase α and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19 2822–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek, J., Lukas, C., and Lukas, J. (2004). Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5 792–804. [DOI] [PubMed] [Google Scholar]
- Branzei, D., and Foiani, M. (2005). The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17 568–575. [DOI] [PubMed] [Google Scholar]
- Branzei, D., and Foiani, M. (2007). Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst.) 6 994–1003. [DOI] [PubMed] [Google Scholar]
- Bray, C.M., and West, C.E. (2005). DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 168 511–528. [DOI] [PubMed] [Google Scholar]
- Britt, A.B., and May, G.D. (2003). Re-engineering plant gene targeting. Trends Plant Sci. 8 90–95. [DOI] [PubMed] [Google Scholar]
- Brown, E.J., and Baltimore, D. (2000). ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14 397–402. [PMC free article] [PubMed] [Google Scholar]
- Brown, E.J., and Baltimore, D. (2003). Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhalter, M.D., and Sogo, J.M. (2004). rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol. Cell 15 409–421. [DOI] [PubMed] [Google Scholar]
- Calzada, A., Hodgson, B., Kanemaki, M., Bueno, A., and Labib, K. (2005). Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19 1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, A.M. (2002). Checking that replication breakdown is not terminal. Science 297 557–558. [DOI] [PubMed] [Google Scholar]
- Chanet, R., and Heude, M. (2003). Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae. Curr. Genet. 43 337–350. [DOI] [PubMed] [Google Scholar]
- Chen, I.P., Haehnel, U., Altschmied, L., Schubert, I., and Puchta, H. (2003). The transcriptional response of Arabidopsis to genotoxic stress - A high-density colony array study (HDCA). Plant J. 35 771–786. [DOI] [PubMed] [Google Scholar]
- Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K.M., Robertson, C.E., Foreman, J., Doerner, P., and Britt, A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48 947–961. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., Proux, F., and Vaucheret, H. (2005). Arabidopsis RPA2: A genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 15 1919–1925. [DOI] [PubMed] [Google Scholar]
- Endo, M., Ishikawa, Y., Osakabe, K., Nakayama, S., Kaya, H., Araki, T., Shibahara, K., Abe, K., Ichikawa, H., Valentine, L., Hohn, B., and Toki, S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25 5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidantsef, A.L., Mitchell, D.L., and Britt, A.B. (2000). The Arabidopsis UVH1 gene is a homolog of the yeast repair endonuclease RAD1. Plant Physiol. 124 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner, J.D., Liu, B., Culligan, K., and Britt, A.B. (2005). Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 16 2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch, O., Benvenuto, G., Bowler, C., Molinier, J., and Hohn, B. (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16 479–485. [DOI] [PubMed] [Google Scholar]
- Galbraith, D., Harkins, K., Maddox, J., Ayres, N., Sharma, D., and Firoozabady, E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220 1049–1051. [DOI] [PubMed] [Google Scholar]
- Gallego, F., Fleck, O., Li, A., Wyrzykowska, J., and Tinland, B. (2000). AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J. 21 507–518. [DOI] [PubMed] [Google Scholar]
- Gallego, M.E., Bleuyard, J.Y., Daoudal-Cotterell, S., Jallut, N., and White, C.I. (2003). Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 35 557–565. [DOI] [PubMed] [Google Scholar]
- Galli, A., Cervelli, T., and Schiestl, R.H. (2003). Characterization of the hyperrecombination phenotype of the pol3-t mutation of Saccharomyces cerevisiae. Genetics 164 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz, M., and Bebenek, K. (2007). Multiple functions of DNA polymerases. CRC Crit. Rev. Plant Sci. 26 105–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi, H., Gallego, M.E., Jalut, N., Lucht, J.M., Hohn, B., and White, C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin, M., and Paszkowski, J. (2003). Plant genome modification by homologous recombination. Curr. Opin. Plant Biol. 6 157–162. [DOI] [PubMed] [Google Scholar]
- Harrison, J.C., and Haber, J.E. (2006). Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40 209–235. [DOI] [PubMed] [Google Scholar]
- Hartung, F., Suer, S., and Puchta, H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson, I.D. (2003). RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer 3 169–178. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers, J.H. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411 366–374. [DOI] [PubMed] [Google Scholar]
- Hübscher, U., Maga, G., and Spadari, S. (2002). Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71 133–163. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., Jurkuta, R.E., and Barton, M.K. (2005). Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell 17 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti, T., Flick, K., Keranen, S., Syvaoja, J.E., and Wittenberg, C. (1999). DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3 679–685. [DOI] [PubMed] [Google Scholar]
- Kimura, S., Suzuki, T., Yanagawa, Y., Yamamoto, T., Nakagawa, H., Tanaka, I., Hashimoto, J., and Sakaguchi, K. (2001). Characterization of plant proliferating cell nuclear antigen (PCNA) and flap endonuclease-1 (FEN-1), and their distribution in mitotic and meiotic cell cycles. Plant J. 28 643–653. [DOI] [PubMed] [Google Scholar]
- Kirik, A., Pecinka, A., Wendeler, E., and Reiss, B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18 2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, F.J., Degtyareva, N.P., Lobachev, K., and Petes, T.D. (2005). Chromosomal translocations in yeast induced by low levels of DNA polymerase: A model for chromosome fragile sites. Cell 120 587–598. [DOI] [PubMed] [Google Scholar]
- Lisby, M., Barlow, J.H., Burgess, R.C., and Rothstein, R. (2004). Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118 699–713. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Hossain, G.S., Islas-Osuna, M.A., Mitchell, D.L., and Mount, D.W. (2000). Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J. 21 519–528. [DOI] [PubMed] [Google Scholar]
- Lopes, M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C.S., and Foiani, M. (2001). The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 557–561. [DOI] [PubMed] [Google Scholar]
- Molinier, J., Ramos, C., Fritsch, O., and Hohn, B. (2004. a). CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 16 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier, J., Ries, G., Bonhoeffer, S., and Hohn, B. (2004. b). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D.K., and Cavallo, S.J. (2007). Effects of mutations in SGS1 and in genes functionally related to SGS1 on inverted repeat-stimulated spontaneous unequal sister-chromatid exchange in yeast. BMC Mol. Biol. 8 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas, T.A., Zhou, Z., and Elledge, S.J. (1995). DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80 29–39. [DOI] [PubMed] [Google Scholar]
- Nyberg, K.A., Michelson, R.J., Putnam, C.W., and Weinert, T.A. (2002). Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36 617–656. [DOI] [PubMed] [Google Scholar]
- Osakabe, K., Abe, K., Yoshioka, T., Osakabe, Y., Todoriki, S., Ichikawa, H., Hohn, B., and Toki, S. (2006). Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J. 48 827–842. [DOI] [PubMed] [Google Scholar]
- Puchta, H. (2005). The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 56 1–14. [DOI] [PubMed] [Google Scholar]
- Pursell, Z.F., Isoz, I., Lundstrom, E.B., Johansson, E., and Kunkel, T.A. (2007). Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, B. (2003). Homologous recombination and gene targeting in plant cells. Int. Rev. Cytol. 228 85–139. [DOI] [PubMed] [Google Scholar]
- Ries, G., Buchholz, G., Frohnmeyer, H., and Hohn, B. (2000). UV-damage-mediated induction of homologous recombination in Arabidopsis is dependent on photosynthetically active radiation. Proc. Natl. Acad. Sci. USA 97 13425–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret, A., Guilleminot, J., Lincker, F., Gadea-Vacas, J., Delorme, V., Bechtold, N., Pelletier, G., Delseny, M., Chaboute, M.E., and Devic, M. (2005). Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 44 223–236. [DOI] [PubMed] [Google Scholar]
- Schuermann, D., Molinier, J., Fritsch, O., and Hohn, B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21 172–181. [DOI] [PubMed] [Google Scholar]
- Shimura, T., Torres, M.J., Martin, M.M., Rao, V.A., Pommier, Y., Katsura, M., Miyagawa, K., and Aladjem, M.I. (2008). Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J. Mol. Biol. 375 1152–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz, R.W., Tatineni, V.M., Hanley-Bowdoin, L., and Thompson, W.F. (2007). Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 144 1697–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, J.M., Lopes, M., and Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297 599–602. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6 544–553. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Tadele, Z., Hofmann, I., Probst, A.V., Angelis, K.J., Kaya, H., Araki, T., Mengiste, T., Mittelsten Scheid, O., Shibahara, K., Scheel, D., and Paszkowski, J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toueille, M., and Hübscher, U. (2004). Regulation of the DNA replication fork: A way to fight genomic instability. Chromosoma 113 113–125. [DOI] [PubMed] [Google Scholar]
- Uchiyama, Y., Hatanaka, M., Kimura, S., Ishibashi, T., Ueda, T., Sakakibara, Y., Matsumoto, T., Furukawa, T., Hashimoto, J., and Sakaguchi, K. (2002). Characterization of DNA polymerase delta from a higher plant, rice (Oryza sativa L.). Gene 295 19–26. [DOI] [PubMed] [Google Scholar]
- Urawa, H., Hidaka, M., Ishiguro, S., Okada, K., and Horiuchi, T. (2001). Enhanced homologous recombination caused by the non-transcribed spacer of the rDNA in Arabidopsis. Mol. Genet. Genomics 266 546–555. [DOI] [PubMed] [Google Scholar]
- Venkatesan, R.N., Treuting, P.M., Fuller, E.D., Goldsby, R.E., Norwood, T.H., Gooley, T.A., Ladiges, W.C., Preston, B.D., and Loeb, L.A. (2007). Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol. Cell. Biol. 27 7669–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa, L., Couvillion, M., Spangler, E., and Shippen, D.E. (2005). ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 19 2111–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu, I., Narayanan, V., Lobachev, K.S., and Mirkin, S.M. (2008). Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 105 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga, S., and Stillman, B. (1994). Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369 207–212. [DOI] [PubMed] [Google Scholar]
- Wang, C., and Liu, Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, C.E., Waterworth, W.M., Story, G.W., Sunderland, P.A., Jiang, Q., and Bray, C.M. (2002). Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 31 517–528. [DOI] [PubMed] [Google Scholar]
- Xia, R., Wang, J., Liu, C., Wang, Y., Wang, Y., Zhai, J., Liu, J., Hong, X., Cao, X., Zhu, J.K., and Gong, Z. (2006). ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H., Zhang, X., Liu, J., Wang, Y., He, J., Yang, T., Hong, X., Yang, Q., and Gong, Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase ε mutation in Arabidopsis. Plant Cell 21 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi, M., Ito, M., Izumi, M., Miyazawa, H., Nakai, H., and Hanaoka, F. (1997). Molecular cloning of the cDNA for the catalytic subunit of plant DNA polymerase alpha and its cell-cycle dependent expression. Genes Cells 2 695–709. [DOI] [PubMed] [Google Scholar]
- Zou, L., and Elledge, S.J. (2003). Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300 1542–1548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.