Abstract
Mitogen-activated protein (MAP) kinase phosphatases are important negative regulators of the levels and kinetics of MAP kinase activation that modulate cellular responses. The dual-specificity phosphatase MAP KINASE PHOSPHATASE1 (MKP1) was previously shown to regulate MAP KINASE6 (MPK6) activation levels and abiotic stress responses in Arabidopsis thaliana. Here, we report that the mkp1 null mutation in the Columbia (Col) accession results in growth defects and constitutive biotic defense responses, including elevated levels of salicylic acid, camalexin, PR gene expression, and resistance to the bacterial pathogen Pseudomonas syringae. PROTEIN TYROSINE PHOSPHATASE1 (PTP1) also interacts with MPK6, but the ptp1 null mutant shows no aberrant growth phenotype. However, the pronounced constitutive defense response of the mkp1 ptp1 double mutant reveals that MKP1 and PTP1 repress defense responses in a coordinated fashion. Moreover, mutations in MPK3 and MPK6 distinctly suppress mkp1 and mkp1 ptp1 phenotypes, indicating that MKP1 and PTP1 act as repressors of inappropriate MPK3/MPK6-dependent stress signaling. Finally, we provide evidence that the natural modifier of mkp1 in Col is largely the disease resistance gene homolog SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 (SNC1) that is absent in the Wassilewskija accession. Our data thus indicate a major role of MKP1 and PTP1 in repressing salicylic acid biosynthesis in the autoimmune-like response caused by SNC1.
INTRODUCTION
Mitogen-activated protein (MAP) kinase cascades are conserved signal transduction systems in eukaryotic cells that transmit and integrate a multitude of environmental and intracellular signals. These cascades relay signals by sequential phosphorylation and consist of three interlinked kinase components, a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a terminal MAP kinase (MAPK). MAPKs are activated by dual phosphorylation of Thr and Tyr within their T-X-Y consensus sequence by the dual-specificity MAPKKs. The magnitude and duration of MAPK activation determines the outcome of the cellular reaction (Marshall, 1995; Keyse, 2008). Therefore, tight regulation of MAPK activation is a prerequisite for producing specific and adequate physiological responses (McClean et al., 2007). Deregulation of MAPK activity has a broad range of deleterious effects in all eukaryotic organisms, including being a common alteration in human cancers (Wu, 2007).
Regulated dephosphorylation and inactivation of MAP kinases by protein phosphatases directly counterbalances activation through MAPKKs and is a major determinant of the physiological outcome of signaling (e.g., Junttila et al., 2008; Keyse, 2008). Since phosphorylation of both Thr and Tyr residues is required for MAPK activity, dephosphorylation of either amino acid is sufficient for inactivation. This can be achieved by members of the three major classes of protein phosphatases, namely, Tyr-specific phosphatases (PTPs), Ser/Thr-specific phosphatases, or dual-specificity (Thr/Tyr) protein phosphatases (DSPs) (Keyse, 2008). In particular, members of a subgroup of the DSP class are known to be dedicated MAPK phosphatases (MKPs) (Camps et al., 2000). Defects in such dual-specificity MKPs have a broad range of detrimental effects in multicellular eukaryotes (e.g., Martin-Blanco et al., 1998; Ulm et al., 2001; Monroe-Augustus et al., 2003; Naoi and Hashimoto, 2004; Zhang et al., 2004; Christie et al., 2005; Lee and Ellis, 2007).
In Arabidopsis thaliana, members of all three major classes of protein phosphatases were linked to MAPK deactivation, albeit to different degrees. A PP2C-type Ser/Thr phosphatase, ABI1, was found to interact with and inactivate MAP KINASE6 (MPK6), implicating it in abscisic acid–dependent stress signaling (Leung et al., 2006). Another PP2C-type phosphatase, AP2C1, was recently shown to inactivate wound stress–responsive MPK4 and MPK6 in vivo, and its overexpression was found to compromise innate immunity against a necrotrophic fungal pathogen (Schweighofer et al., 2007). Mutant ap2c1 plants, on the other hand, were not significantly affected in their pathogen response; however, they were shown to have an elevated wound response and to be more resistant to herbivorous attack by spider mites (Schweighofer et al., 2007).
PTP1 is the only member of the phosphotyrosine-specific protein phosphatases in Arabidopsis. PTP1 gene expression was found to be altered in response to different environmental stresses (Xu et al., 1998). PTP1 was shown to be able to dephosphorylate and deactivate MPK4 and MPK6 in vitro, and an involvement of PTP1 in oxidative stress signaling was suggested (Huang et al., 2000; Gupta and Luan, 2003), but its role in vivo has not yet been reported.
Five DSP-type phosphatases with described or predicted MKP function are encoded in the genome of Arabidopsis. For one of them, the dual-specificity protein Tyr phosphatase 1 (DsPTP1; Gupta et al., 1998), no in vivo role has been described yet. The other four DSP-type MKPs were genetically linked to abscisic acid/auxin signaling (INDOLE-3-BUTYRIC ACID RESPONSE5; Monroe-Augustus et al., 2003; Lee et al., 2009), microtubule organization (PROPYZAMIDE-HYPERSENSITIVE1; Naoi and Hashimoto, 2004), oxidative stress tolerance (MKP2; Lee and Ellis, 2007), and genotoxic and salt stress tolerance (MKP1; Ulm et al., 2001, 2002).
MKP1 is required for genotoxic stress resistance in Arabidopsis and was found to interact with the stress-activated MPK3, MPK4, and MPK6 in a directed yeast two-hybrid assay (Ulm et al., 2001, 2002). MKP1 was further shown to regulate the genotoxic stress-responsive activity level of MPK6 in vivo (Ulm et al., 2002). The mkp1 mutant, however, does not show general stress sensitivity, as, on the contrary, mkp1 was found to be more tolerant to salt stress (Ulm et al., 2002). Recently, it was described that MKP1 is regulated by calmodulin (CaM) through two CaM binding domains. Binding of CaM increased the phosphatase activity of Arabidopsis MKP1 about twofold, indicating a link between Ca2+ and MKP1-mediated regulation of MAPK signaling (Lee et al., 2008). The tobacco (Nicotiana tabacum) MKP1, however, was not activated by CaM binding but by interaction with its substrate, the SALICYLIC ACID-INDUCED PROTEIN KINASE (SIPK), which is the ortholog of Arabidopsis MPK6 (Yamakawa et al., 2004; Katou et al., 2005). Transient overexpression of N. tabacum MKP1 in tobacco leaves abolished SIPK-mediated cell death (Katou et al., 2005). The rice (Oryza sativa) MKP1 was recently described as a negative regulator of wound responses, and the rice insertional mutant line mkp1 shows a semidwarf phenotype (Katou et al., 2007).
Despite the importance of MAPK pathways in a broad range of stress responses and developmental programs, the contribution of negative regulatory MKPs has been described genetically only in very few cases in multicellular eukaryotes. Similarly, the specific and/or overlapping functions of different classes of MAPK-regulating phosphatases are not well understood. Here, we show that Arabidopsis MKP1 and PTP1 have redundant functions in the suppression of salicylic acid (SA) and camalexin biosynthesis and defense responses. We found that the constitutive defense response in mkp1 and the mkp1 ptp1 double mutant is partially dependent on MPK3 and MPK6 and accumulation of SA. Moreover, the described phenotype of mkp1 and mkp1 ptp1 is only apparent in the Columbia (Col) ecotype, but not in the Wassilewskija (Ws) ecotype. Previous work has identified the Toll Interleukin 1 receptor/nucleotide binding/leucine-rich repeat (TIR-NB-LRR) receptor-like resistance gene homolog SUPPRESSOR OF npr1-1, CONSTITUTIVE1 (SNC1), which induces constitutive defense responses and dwarfed growth when overexpressed or hyperactivated by mutation (Stokes et al., 2002; Zhang et al., 2003; Yang and Hua, 2004; Li et al., 2007). This gene is specific to Col, and there is no closely related ortholog in Ws (Yang and Hua, 2004). Our genetic analysis indeed identified SNC1 as a natural modifier of MKP1 function.
RESULTS
MKP1 and PTP1 Interact with and Dephosphorylate MPK6
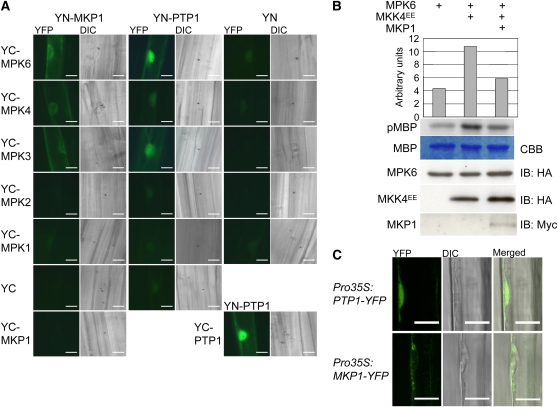
PTP1 was previously shown to dephosphorylate activated MPK6 in vitro (Gupta and Luan, 2003). However, we could not identify direct interaction of PTP1 with MPK6 or other tested MPKs in a directed yeast two-hybrid assay. A possible reason for this might be a rather transient and weak interaction of PTP1 with its substrate. To investigate direct interaction in planta, we used the bimolecular fluorescence complementation (BiFC) assay, which stabilizes a protein–protein interaction after the reconstitution of the C-terminal and N-terminal parts of a split yellow fluorescent protein (YFP) (Kerppola, 2006). Indeed, YN-PTP1 was found to interact with itself (YC-PTP1), YC-MPK6, and to a lesser extent YC-MPK3 in transiently transformed mustard (Sinapis alba) hypocotyl cells (Figure 1A), a well-established system for BiFC assays in planta (e.g., Stolpe et al., 2005; Favory et al., 2009). The YFP fluorescence resulting from cobombardment of YN-PTP1 and YC-MPK4 is very weak; thus, their direct physical interaction remains uncertain (Figure 1A). No sign of interaction could be detected between YN-PTP1 and YC-MPK1, YC-MPK2, or any of the empty vector controls (Figure 1A). Moreover, we could confirm the yeast two-hybrid based interaction of MKP1 with MPKs 3, 4, and 6 (Ulm et al., 2002) in the BiFC assays (Figure 1A). Using a transient protoplast expression system, we could show that MKP1 is able to inactivate coexpressed and activated MPK6 (Figure 1B). However, we could not express PTP1 to detectable levels in the protoplast system to convincingly assay its effect on MPK6 activity.
Figure 1.
MKP1 and PTP1 Interact with and Deactivate MPK6, but Differ in Their Subcellular Localization.
(A) BiFC visualization of interactions in transiently transformed mustard hypocotyl cells. YFP signal indicates reconstitution through direct interaction of the tested protein partners. Combinations with YN and YC designate the empty vector controls; YC-MPK1 and YC-MPK2 are shown as negative controls indicating specificity of the detected interactions. The corresponding differential interference contrast (DIC) image is shown for each fluorescence image. Bars = 20 μm.
(B) Arabidopsis protoplasts were transiently transformed with the indicated combinations of HA-MPK6, HA-MKK4EE (constitutively active form), and myc-MKP1, all expressed under the control of the 35S constitutive promoter. Respective MPK6 activities were determined by phosphorylation of the artificial substrate myelin basic protein (MBP) as shown by autoradiography (pMBP) and the corresponding quantification. The MBP panel shows Coomassie blue (CBB) stained loading. Levels of HA-MPK6, HA-MKK4EE, and myc-MKP1 were determined by immunoblot (IB) analysis. A representative experiment out of three comparable repetitions is shown.
(C) MKP1-YFP localizes mainly to the cytoplasm and PTP1-YFP to both the nucleus and cytoplasm. YFP fluorescence in root cells of Arabidopsis seedlings stably expressing cauliflower mosaic virus 35S promoter-driven MKP1-YFP and PTP-YFP fusion constructs as detected with a LSM 510 confocal laser scanning microscope (Zeiss). Bars = 10 μm.
Interestingly, the interaction of YC-MPK6 with YN-MKP1 was found to be primarily cytoplasmic, whereas the interaction of YC-MPK6 with YN-PTP1 was primarily nuclear (Figure 1A). We thus generated stable transgenic lines expressing MKP1-YFP and PTP1-YFP driven by the cauliflower mosaic virus 35S promoter in mkp1 and ptp1 mutant backgrounds, respectively. In agreement with the BiFC interaction data, confocal microscopy suggests that MKP1-YFP is mainly localized to the cytosol, whereas PTP1-YFP can be detected both in the cytosol and nucleus (Figure 1C).
MKP1 and PTP1 Perform Partially Redundant Functions in Vivo
The overlap of their interaction partners indicated functional redundancy of MKP1 and PTP1. To directly test this possibility, we isolated a T-DNA insertion line disrupting the PTP1 gene (ptp1-1, SALK_118658). In this line, the T-DNA is inserted into exon 4 and results in the absence of wild-type PTP1 mRNA, indicating that it is a functional null mutant (see Supplemental Figure 1 online). The ptp1 mutant, however, did not show any obvious change in its morphology or development under standard growth conditions (Figure 2A; see Supplemental Figure 1 online).
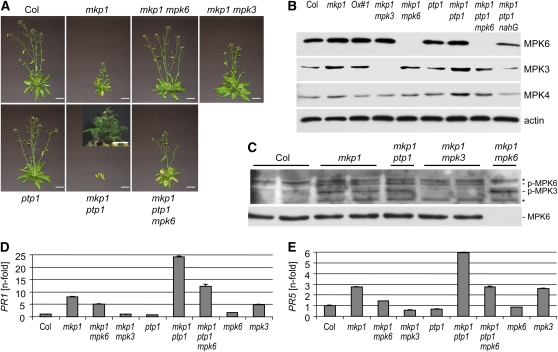
Figure 2.
MKP1 and PTP1 Act Redundantly to Regulate Growth Homeostasis and PR Gene Expression in an MPK3- and MPK6-Dependent Manner.
(A) Photographs of 40-d-old plants grown on soil under standard conditions are shown. Aberrant phenotypes of mkp1(Col) and mkp1 ptp1 are suppressed by mpk3 and mpk6 mutations. Bars = 2 cm. Small inset to mkp1 ptp1 shows a close-up of a different plant with the same genotype (bar = 0.5 cm).
(B) Immunoblot analysis of MPK3, MPK4, MPK6, and actin (loading control) protein levels in 22-d-old soil-grown plants.
(C) MPK3 and MPK6 activation profile in 22-d-old soil grown plants as detected by immunoblotting using anti-phospho-p44/42 MAP kinase antibodies (p-MPK6 and p-MPK3). The immunoblot was reprobed with anti-MPK6 antibody. Two samples from independent biological repetitions were loaded for Col, mkp1, and mkp1 mpk3.
(D) and (E) Quantitative RT-PCR analysis of PR1 (D) and PR5 (E) expression levels in 22-d-old plants compared with wild-type Col. A representative experiment out of three independent biological repetitions is shown. Error bars represent sd of the technical triplicates.
As the only available mkp1 null mutant was in the Ws background (Ulm et al., 2001), we introgressed this allele into the Col accession before crossing it with the ptp1 mutant. Surprisingly, the introgression of mkp1 into Col itself generated plants with altered morphology and development not seen in the mkp1(Ws) mutant. The mkp1(Col) plants showed weak dwarfism, aberrant leaf development, long pedicels, early senescence, stunted and misshaped inflorescences, and low fertility (Figure 2A; see Supplemental Figure 2A online). This mutant phenotype was (1) apparent even after 15 backcrosses to Col, (2) complemented by expression of Polyoma epitope- and YFP-tagged MKP1 (see Supplemental Figures 2B and 2C online; data not shown), and (3) supported by a comparable phenotype of a cosuppression line isolated among ProMKP1:GUS transgenic lines (see Supplemental Figures 2D to 2F online). From now on in the text, we will differentiate between mkp1(Col) and mkp1(Ws) for the single mutants; all double mutants were generated using mkp1(Col).
We then crossed the ptp1 and mkp1(Col) mutants. Strikingly, the mkp1 ptp1 double mutant plants exhibit a severe dwarf phenotype (Figure 2A). These plants show numerous dramatic developmental defects, including early senescence and infertility. The mkp1(Col) and mkp1 ptp1 growth phenotypes become apparent about 2 weeks after germination and are not established at the earlier seedling stage (see Supplemental Figure 3 online). Altogether, we conclude that MKP1 and PTP1 have partially redundant functions under standard growth conditions, with MKP1 having the predominant role.
MPK3 and MPK6 Mediate mkp1 and mkp1 ptp1 Developmental Defects
As both MKP1 and PTP1 regulate MPK6 and likely MPK3, deregulation of MPK6 and/or MPK3 might be the underlying cause for the aberrant phenotype of mkp1(Col) and mkp1 ptp1. Consistently, the double mkp1 mpk6-2 and triple mkp1 ptp1 mpk6-2 knockout mutants show a significantly improved growth phenotype compared with mkp1(Col) and mkp1 ptp1, respectively, although overall plant size was still slightly reduced and early senescence remained (Figure 2A). It should be noted that suppression was very similar with an independent mpk6 allele (mpk6-3; see Supplemental Figure 4 online).
Similarly to mpk6, the mpk3 null mutation partially suppressed the mkp1(Col) phenotype in mkp1 mpk3 double mutants (Figure 2A). Interestingly, however, MPK3 and MPK6 contributed differently to the mkp1(Col) phenotype: (1) mpk3 suppressed the early senescence phenotype of mkp1, whereas mpk6 did not; (2) mpk6 suppressed the inflorescence morphology phenotype of mkp1, whereas mpk3 did not; and (3) mpk6 suppressed the decreased fertility phenotype of mkp1, whereas mpk3 did not. Thus, mutations in mpk3 and mpk6 suppress mkp1 phenotypes, although distinctly and to a different extent.
Protein gel blot analysis using specific antibodies confirmed the absence of MPK3 and MPK6 in the respective lines and indicated that MPK3 and MPK4 protein levels are slightly elevated in mkp1(Col) and mkp1 ptp1 compared with wild-type plants (Figure 2B). More importantly, immunoblot analysis using an antiphospho-p44/42 antibody, which recognizes only the doubly phosphorylated and thus active forms of MPK3 and MPK6, identified elevated levels of the phosphorylated forms of these two kinases in mkp1(Col) and mkp1 ptp1 compared with the wild type under standard growth conditions (Figure 2C). As expected, in mkp1 mpk3 and mkp1 mpk6, the cross-reacting bands of phospho-MPK6 and phospho-MPK3 are absent, respectively (Figure 2C). Thus, the loss-of MKP1 and MKP1/PTP1 is associated with increased levels of active MPK3 and MPK6, and in agreement, the mkp1(Col) and mkp1 ptp1 phenotypes largely depend on the presence of MPK3 and MPK6. We thus conclude that loss of MKP1 and combined loss of MKP1 and PTP1 lead to a multitude of developmental and morphological alterations, several of which could be genetically attributed to MPK3 and/or MPK6 function.
PR Gene Expression Is Elevated in mkp1 and mkp1 ptp1
The phenotypes of mkp1(Col) and mkp1 ptp1 indicate a constitutive stress response, also displayed by mutants with a constitutive pathogen response (e.g., Jirage et al., 2001). This phenotype is often associated with an upregulation of PATHOGENESIS-RELATED (PR) gene expression. Quantitative real-time PCR analysis of PR1 and PR5 transcript abundance in 22-d-old soil-grown plants revealed an up to 25-fold increase of PR1 and 6-fold increase of PR5 transcript levels in mkp1 ptp1 compared with wild-type Col (Figures 2D and 2E). The absence of MPK6 partially suppresses this molecular phenotype of mkp1(Col) and mkp1 ptp1 as well (Figures 2D and 2E). This result is in agreement with partial suppression of growth and developmental aberrations of mkp1 ptp1. Similarly, mpk3 knockout in the mkp1(Col) background reduced PR1 and PR5 transcription back to wild-type levels (Figures 2D and 2E). We conclude that the growth phenotype and PR gene hyperexpression indicate an MPK3/MPK6-dependent, constitutively elevated stress response in mkp1 ptp1 and to a lesser extent in mkp1(Col).
SA Accumulates to High Levels in mkp1 and mkp1 ptp1
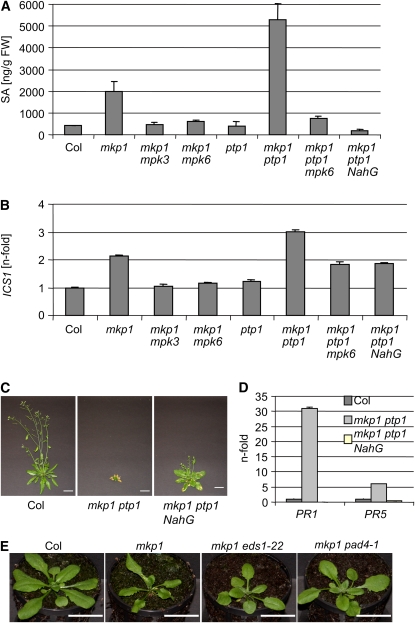
A high level of PR gene expression is often associated with elevated levels of the phytohormone SA. Therefore, we determined the SA levels in rosette leaves of 22-d-old plants grown under standard conditions in soil. In agreement with our phenotypic and molecular analysis, mkp1(Col) shows a 5-fold accumulation of total SA compared with the wild type, and the mkp1 ptp1 double mutant shows a 13-fold increase (Figure 3A). In both cases, the SA hyperaccumulation was dependent on functional mpk6; thus, SA levels were comparable to those in the wild type in mkp1 mpk6 and mkp1 ptp1 mpk6 mutant lines. Similarly, the mkp1 mpk3 double mutant resulted in SA levels comparable to those in the wild type (Figure 3A). It should be noted that separate analysis of free and conjugated SA levels in both cases agreed with the data of total SA (see Supplemental Figure 5 online). To identify a possible molecular cause for the differences in SA levels, we determined the expression levels of the ICS1/SID2 gene, which encodes the isochorismate synthase of the major SA biosynthetic pathway (Wildermuth et al., 2001). In agreement with the phenotypes, PR gene expression as well as SA measurements, we identified a corresponding overexpression of ICS1 in mkp1(Col) and mkp1 ptp1 that was dependent on MPK3 and MPK6 (Figure 3B).
Figure 3.
SA Accumulation Is Largely Responsible for the Aberrant mkp1(Col) and mkp1 ptp1 Phenotypes.
(A) Total SA levels of 22-d-old soil-grown plants, as determined by HPLC. The average of three independent biological samples is shown for each genotype. Error bars represent sd.
(B) Quantitative RT-PCR analysis of ICS1 expression levels. Representative data of three biological replicates are shown. Error bars represent sd of the technical triplicates.
(C) Photographs of 40-d-old soil-grown plants showing that expression of NahG largely suppresses the mkp1 ptp1 growth phenotype (note that the Col and mkp1 ptp1 photographs are identical to those shown in Figure 2A; all photographs were taken in parallel, under identical conditions). Bars = 2 cm.
(D) Quantitative RT-PCR analysis of PR1 and PR5 expression levels in 22-d-old mkp1 ptp1 and mkp1 ptp1 NahG compared with wild-type Col. Representative data of three biological replicates are shown. Error bars represent the sd of the technical triplicates.
(E) Photographs of 22-d-old plants grown on soil under standard conditions showing that eds1 and pad4 mutations suppress the mkp1 growth phenotype. Bars = 2 cm.
[See online article for color version of this figure.]
Elevated SA Levels Are Responsible for the mkp1 and mkp1 ptp1 Phenotypes
To determine the contribution of SA hyperaccumulation to the mkp1(Col) and mkp1 ptp1 phenotypes, we crossed these mutants with a NahG transgenic line expressing a bacterial salicylate hydroxylase that degrades SA in planta (Gaffney et al., 1993). Indeed, expression of NahG dramatically reduced SA levels and largely suppressed the mkp1(Col) and mkp1 ptp1 phenotypes, including PR gene expression (Figures 3A, 3C, and 3D; see Supplemental Figures 5A and 5B online). At later developmental stages, however, a number of phenotypes remained in the NahG suppressor lines, including early leaf senescence, a weak inflorescence phenotype, and infertility. Interestingly, NahG expression also reduced MPK3, MPK4, and MPK6 protein levels, indicating that SA modulates their accumulation (Figure 2B).
The lipase-like proteins ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and PHYTOALEXIN DEFICIENT4 are important regulators of SA synthesis in plant defense signaling (Wiermer et al., 2005). Therefore, we tested if mutations in these two factors are also able to suppress the mkp1(Col) phenotype, which was indeed found to be the case (Figure 3E).
We thus conclude that the phenotypes of mkp1(Col) and mkp1 ptp1 are largely due to increased SA synthesis and accumulation, indicating that a major function of MKP1 and PTP1 is to repress the MAPK pathway, involving MPK3 and MPK6, which leads to SA biosynthesis and expression of PR1 and PR5.
mkp1 Mutants Have Increased Resistance to the Bacterial Pathogen Pseudomonas syringae and Accumulate Higher Levels of the Phytoalexin Camalexin
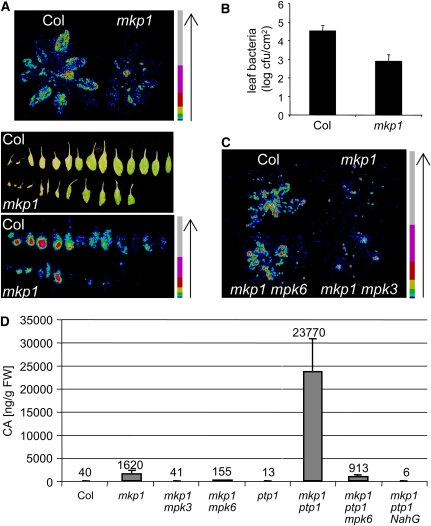
Our data indicate that the mkp1 phenotype is related to constitutive defense responses potentially associated with increased disease resistance. Thus, we assayed disease resistance using the Arabidopsis pathogen P. syringae pv tomato DC3000 strain carrying a chromosomal insertion of the luxCDABE operon from Photorhabdus luminescens, encoding both a luciferase reporter (luxAB) and enzymes for the production of its substrate (luxCDE), under a constitutive promoter to noninvasively determine bacterial growth (Fan et al., 2008). Indeed, in comparison to the wild type, mkp1(Col) allowed much less growth of P. syringae as determined by bioluminescence, which reflects bacterial growth in the infected Arabidopsis leaves (Figure 4A), and leaf bacteria measurement by serial dilution plating (Figure 4B). This enhanced pathogen resistance of mkp1(Col) required MPK6, as mkp1 mpk6 were clearly less resistant than mkp1(Col) and more similar to the wild type (Figure 4C). Surprisingly, however, pathogen resistance of mkp1 mpk3 was similar to that of mkp1 (Figure 4C). Thus, it seems that the elevated pathogen resistance of mkp1 requires MPK6 but not MPK3 and that resistance is not strictly correlated with elevated SA levels and PR gene expression.
Figure 4.
mkp1(Col) Has Increased Resistance to P. syringae Infection and Constitutively Accumulates Elevated Levels of Camalexin.
(A) Bioluminescence representing bacterial growth on rosettes and detached leaves 1 and 4 d, respectively, after spray inoculation with P. syringae pv tomato DC3000 strain carrying the luxCDABE operon. Color scale bars are shown; arrows indicate increasing photon intensity. Cold colors (e.g., blue and green) represent regions of lower photon counts, and warm colors (e.g., yellow and red) represent regions of more intense luminescence.
(B) Leaf bacteria as determined by serial dilution plating 4 days postinoculation. The means ± se (n = 10) are shown. The difference between Col and mkp1(Col) is statistically significant (Student's t test, P < 0.001). Representative data of three independent biological repetitions are shown in (A) and (B).
(C) Bioluminescence representing bacterial growth on plants 3 d after spray inoculation with Pst DC3000 lux. Representative data of two independent repetitions is shown.
(D) Camalexin levels in 22-d-old plants. The average level of three independent samples is shown. Error bars represent sd.
Camalexin is an important phytoalexin in Arabidopsis often associated with pathogen resistance. Moreover, it was previously shown that constitutive activation of MPK3 and MPK6 elicits accumulation of camalexin and thereby strengthens pathogen resistance (Ren et al., 2008). Thus, we determined camalexin levels in mkp1(Col) and mkp1 ptp1 under standard growth conditions. In agreement with our previous results, we found ∼40-fold increases in camalexin levels in mkp1(Col) and 600-fold in mkp1 ptp1 that are dependent on the presence of MPKs 3 and 6 (Figure 4D). Moreover, the absence of camalexin in the mkp1 ptp1 NahG line is consistent with the requirement of SA for its biosynthesis and the reported effect of NahG on camalexin accumulation (Zhao and Last, 1996; Heck et al., 2003) (Figure 4D).
The Col Accession-Specific Receptor-Like R Gene Homolog, SNC1, Is a Natural Modifier of mkp1
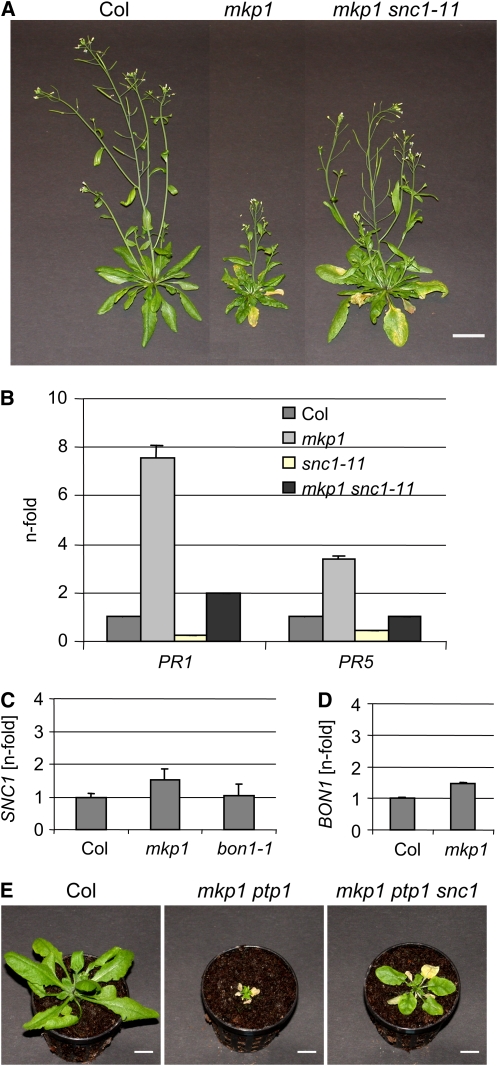
The phenotypes that we describe for mkp1(Col) were reminiscent of the previously described bonsai 1 (bon1) mutation in a copine family member (Hua et al., 2001). In particular, similar to bon1 (Yang and Hua, 2004), the mkp1(Col) growth phenotype is (1) restricted to the Col accession and not apparent in Ws, (2) associated with elevated levels of SA and PR gene expression, (3) suppressed by mutant combination with eds1, pad4, and NahG, and (4) suppressed by elevated temperature (see Supplemental Figure 6 online). Importantly, Yang and Hua (2004) could show that the bon1-1 loss-of-function mutation activates the Col accession-specific TIR-NB-LRR receptor-like SNC1, leading to constitutive defense responses and, consequently, reduced cell growth. Our data indicate a related role of MKP1 in regulating plant growth homeostasis by repressing inappropriate stress signaling. We thus hypothesized that the Col-specific SNC1 resistance gene homolog is responsible for the constitutive defense response apparent in mkp1(Col). To test this, we generated an mkp1 snc1-11 double mutant; indeed, the snc1 mutation could partially suppress the mkp1(Col) phenotype: both growth habit and PR gene expression are more similar to those of the wild type than to those of the single mkp1(Col) mutant (Figures 5A and 5B). It is also of note that the snc1 single mutant exhibited PR1 and PR5 expression levels lower than those in the wild type, indicating a low-level SNC1-mediated defense response in wild-type Col plants (Figure 5B).
Figure 5.
SNC1 Is a Natural Modifier of mkp1.
(A) Photographs of 40-d-old soil-grown plants showing that snc1 largely suppresses the mkp1 growth phenotype (note that the Col and mkp1 photographs are identical to those shown in Figure 2A; all photographs were taken in parallel, under identical conditions). Bars = 2 cm.
(B) to (D) Quantitative RT-PCR analysis of PR1 and PR5 (B), BON1 (C), and SNC1 (D) gene expression. Representative data of three independent experiments are shown. Error bars represent the sd of the technical triplicates.
(E) Photographs of 34-d-old soil-grown plants showing that snc1 partially suppresses the mkp1 ptp1 growth phenotype. Bars = 1 cm.
[See online article for color version of this figure.]
Previously, it was shown that the SNC1-dependent constitutive defense responses and dwarf phenotype are associated with transcriptional upregulation of SNC1 in bon1 mutants as well as in the epigenetic variant bal (Stokes et al., 2002; Yang and Hua, 2004; Li et al., 2007). Similarly, mutational hyperactivation and transgenic overexpression of SNC1 leads to an SA-dependent constitutive defense response and dwarfed growth (Stokes et al., 2002; Zhang et al., 2003; Li et al., 2007). Therefore, we tested SNC1 expression levels in mkp1(Col) but could not identify a statistically significant upregulation (Figure 5C). However, under our growth conditions, we could also not detect a significant increase of SNC1 expression in bon1-1 (Figure 5C), even though it showed a strong growth phenotype. Similarly, BON1 mRNA levels were not strongly affected in mkp1(Col) (Figure 5D).
The overlapping functions of MKP1 and PTP1 described in this work indicated that PTP1 might also be involved in SNC1-mediated responses. To test this, we generated an mkp1 ptp1 snc1-11 triple mutant. As expected, the snc1 mutation could partially suppress the mkp1 ptp1 growth phenotype as well (Figure 5E).
We conclude that SNC1 is a natural modifier of mkp1 and thus plays a major role in its accession-dependent growth phenotype. Altered SNC1 gene expression, however, is unlikely the cause of the detrimental effects detected in mkp1(Col), but rather the SNC1 signaling pathway is likely sensitized in the absence of MKP1.
DISCUSSION
The fitness of all organisms depends largely on their resistance to adverse environmental stress conditions. In particular, sessile plants are inevitably exposed to rapidly changing environmental cues, which have to be recognized and integrated to launch proper responses. Signal transmission and integration often uses MAPK pathways that are regulated by a balance of kinase and phosphatase action. This balance determines the magnitude and duration of MAPK activation and thereby the final biological response. This study demonstrates that (1) MKP1 and PTP1 have redundant functions in regulating plant growth homeostasis by repressing inappropriate stress signaling; (2) this repression is based on ensuring proper MPK3 and MPK6 regulation; (3) SA biosynthesis is regulated by the MKP1-PTP1 regulated MAPK pathway; and (4) the TIR-NB-LRR-type resistance gene homolog SNC1 is a natural modifier of mkp1. Our observations thus place MKP1 and PTP1 as crucial repressors of plant SA-dependent autoimmune-like responses. This conclusion was confirmed by genetic analysis with mutants compromised in SA accumulation and disease resistance.
MKP1 plays an important role in the integration and fine-tuning of plant responses to various environmental challenges (Ulm et al., 2002). Under nonstressed conditions, however, growth of the original mkp1 mutant in the Ws background is indistinguishable from that of the wild type; however, the mkp1 mutant has a slightly reduced fertility (Ulm et al., 2001). By contrast, here, we show that loss of MKP1 in the Col background results in a constitutive stress response phenotype causing aberrant growth and development. It should be noted here that the cause for the semidwarf phenotype described for the rice mkp1 mutant might be related (Katou et al., 2007). The constitutive stress response phenotype of mkp1(Col) is exaggerated in the mkp1 ptp1 double mutant, demonstrating an overlapping role of MKP1 and PTP1 as crucial negative regulators of defense responses. However, the presence of MKP1 in the cytoplasm and of PTP1 in the cytoplasm and nucleus indicate that they act in both distinct and overlapping subcellular localizations. The cytosolic localization might indicate an involvement in regulating basal MPK activity levels and/or in early events directly after MPK activation, before translocation of the activated MPKs to the nucleus. Future experiments in which the mutants are complemented with constructs that add artificial nuclear localization and nuclear export signals should be able to address the importance of MKP1 and PTP1 subcellular localization. Notwithstanding this possibility, the functional redundancy between MKP1 and PTP1 seems rather specific, as double mutants of ptp1 or mkp1 with mutants of the other four DSP-type MKPs did not lead to any aberrant phenotype under standard growth conditions (S. Bartels and R. Ulm, unpublished results). Interestingly, an analysis using the Arabidopsis coexpression tool (Manfield et al., 2006) suggests that PTP1 is also the most closely coexpressed gene of MKP1 across a large microarray dataset, in comparison with the other four DSP-type MKPs (see Supplemental Figure 7 online). However, higher-order combinatorial mutants will be required to fully understand the potential functional overlap and specificities among the MAPK inactivating phosphatases. Due to the common MAPK substrates, it will also be of particular interest to determine the potential redundancy between MKP1 and AP2C1 function.
MKP1 and PTP1 have partially overlapping functions as repressors of SA biosynthesis and related defense responses. Consequently, mkp1(Col) and mkp1 ptp1 mutants show enhanced levels of SA, camalexin, PR gene expression, dwarf growth, and early senescence. Interestingly, these phenotypes are largely suppressed by mpk3 and mpk6 mutations. This indicates that MPK3 and 6 are positive regulators of SA-dependent defense responses under negative regulation by MKP1 and PTP1. In agreement, pathogen-induced synthesis of the phytoalexin camalexin was found to be dependent on the activation of MPK3 and MPK6 (Ren et al., 2008). Moreover, pathogen resistance was found to be compromised in MPK6-silenced plants (Menke et al., 2004). Interestingly, the Arabidopsis pathogen P. syringae counteracts the defense-related MPK3 and MPK6 activation by secreting an effector protein with a unique phosphothreonine lyase activity that removes the phosphate group from phosphothreonine to suppress immunity (Zhang et al., 2007a). Altogether, this indicates that MPK3/MPK6 activation is an important node in SA-mediated defense responses, kept under check by MKP1 and PTP1.
Elevated SA-mediated defense responses are usually associated with elevated resistance to biotrophic pathogens. Indeed, we found that mkp1(Col) is more resistant to the bacterial pathogen P. syringae and that this phenotype is suppressed by mpk6, correlating with SA, camalexin, and PR gene expression levels. Interestingly, however, even though these constitutive resistance markers are similarly suppressed in mkp1 mpk3, this double mutant still showed elevated tolerance to P. syringae. This indicates that the elevated resistance of mkp1 is not strictly correlated with its constitutive stress response phenotype but rather with the presence of MPK6. It is also of note that reciprocal inhibition between MPK6 and MPK3 seems to occur (Wang et al., 2008), which could imply that loss of one enhances the activity of the remaining MAPK. An added complexity is apparent by the fact that MPK3, MPK4, and MPK6 protein levels are modulated by SA levels, as indicated by the reduced levels in NahG plants and elevated levels in mkp1(Col) and mkp1 ptp1 (Figure 2B). This observation is in agreement with a recent report on the effect of a functional SA analog (Beckers et al., 2009). Nevertheless, MPK6 is at least partially responsible for the enhanced resistance of mkp1 and likely of mkp1 mpk3, at least in the latter case independent of the constitutively elevated levels of SA and camalexin. However, the exact role of MPK6 in conferring pathogen resistance remains to be determined.
Arabidopsis MKP1 was recently found to be a Ca2+/CaM binding protein, and its phosphatase activity seems to be regulated by this interaction (Lee et al., 2008). Interestingly, Ca2+ signaling was recently found to regulate SA-mediated immunity through the Arabidopsis CaM binding transcription factor SIGNAL RESPONSIVE1 (SR1) that is a repressor of EDS1 expression (Du et al., 2009). Strikingly, the atsr1 null mutant plants exhibit growth and molecular phenotypes associated with elevated SA levels that are very similar to those of mkp1(Col) described here (Du et al., 2009). Thus, it is intriguing to speculate that there is a connection between Ca2+ signaling and MAPK regulation in defense responses that involves CaM-mediated MKP1 regulation, in parallel with SR1 regulation.
The five DSP-type MKPs are potential negative regulators of MAPK pathways, which consist of the 20 MAPKs, 10 MAPKKs, and >60 MAPKKKs present in the Arabidopsis genome (Ichimura et al., 2002). Six MAPKKs were linked to the activation of MPK3 and MPK6, namely, MKKs 2, 3, 4, 5, 7, and 9 (Asai et al., 2002; Teige et al., 2004; Takahashi et al., 2007; Yoo et al., 2008; Popescu et al., 2009; Zhou et al., 2009). This already complex network was recently extended by an extensive in vitro protein interaction screen to include MKK6 for activation of MPK3 and 6, and MKK1 for MPK6 (Popescu et al., 2009). Consistent with this multitude of activators and pathways, the stress-activated MPKs 3 and 6 were implicated in a broad range of biological responses, including numerous biotic and abiotic stress factors (Colcombet and Hirt, 2008). This suggests that the MPKs have different biological functions depending on the particular activator. In agreement with our data on knockouts of the negative regulatory phosphatases, overexpression of the MPK3 and 6 activators MKK7 and 9 was shown to result in cell death in transiently transformed Nicotiana benthamiana (Popescu et al., 2009). MKK7 overexpression in Arabidopsis results in mild growth alterations in combination with SA accumulation, constitutive PR gene transcription, and elevated pathogen resistance (Zhang et al., 2007b). Similarly, a recent report demonstrated a role of the MKK9-MPK6 cascade in regulation of leaf senescence, with overexpression of a constitutively active MKK9 resulting in dwarfism and early senescence (Zhou et al., 2009). These phenotypes very much resemble the phenotype of mkp1(Col) and mkp1 ptp1 described here, and the MKK9 overexpression phenotypes were also suppressed by the mpk6 mutation (Zhou et al., 2009). Thus, MKP1 and PTP1 likely serve as crucial regulators of the MKK7/MKK9-MPK3/MPK6 signaling pathways that regulate stress responses and senescence.
It is of note that knockouts in MEKK1, MKK1/MKK2, and MPK4 also result in dwarf growth due to a constitutive stress response phenotype and elevated levels of SA (Petersen et al., 2000; Ichimura et al., 2006; Nakagami et al., 2006; Qiu et al., 2008). Similar to mkp1(Col) and mkp1 ptp1, the mpk4 phenotype was partially suppressed by eds1 and pad4 mutations, as well as by the NahG transgene (Brodersen et al., 2006). Thus, MPK4 is a negative regulator of SA-dependent defense responses, whereas MPKs 3 and 6 are positive regulators. This opposing effect of MPK4 to MPK3/MPK6 in the regulation of plant defense responses is reminiscent of the inhibition of the ERK pathway by stress-activated p38/JNK MAP kinase signaling in mammals (Junttila et al., 2008). Interestingly, we found that the mpk6 mutation could not suppress the dwarf phenotype of mpk4 (see Supplemental Figure 8 online), indicating that this pathway is genetically distinct from the MKP1/PTP1-regulated MPK6 pathway. It thus remains to be determined how the apparent crosstalk between the negative regulatory MEKK1-MKK1/MKK2-MPK4 and the positive regulatory MKK9/MKK7-MPK3/MPK6 pathways in defense responses are mechanistically interconnected and what exact role MKP1/PTP1-mediated repression of the latter pathway plays in this.
The presence and absence of a constitutive mkp1 phenotype in the Col and Ws ecotype backgrounds, respectively, is largely due to the Col-specific TIR-NB-LRR receptor-like protein SNC1. Interestingly, the presence of SNC1 in Col seems to increase PR gene expression, as their basal levels are reduced in the snc1 mutant. This indicates that SNC1 constantly triggers defense responses at very low and nondetrimental levels even without pathogen attack. In the absence of MKP1 and PTP1, the response threshold is lowered, resulting in an overreaction of the plant. Similarly, epigenetically caused overexpression of SNC1 (such as in the bal mutant), mutational hyperactivation, or loss of its repressor BON1 is sufficient to trigger constitutive defense responses (Stokes et al., 2002; Zhang et al., 2003; Yang and Hua, 2004). It is of note that the bon1 mutant has a similar phenotype to that of mkp1(Col) and mkp1 ptp1 described here. BON1 and BON1-ASSOCIATED PROTEIN1 negatively regulate SNC1 by a yet unknown mechanism (Yang et al., 2006). It will thus be of interest to further determine the link between BON1 function and MAPK pathways, and MKP1/PTP1 in particular.
In general, resistance genes are known to rapidly duplicate and mutate to create new specific resistance to constantly evolving pathogens (Noel et al., 1999). The advantage of multiple resistance genes is counterbalanced by potentially detrimental misexpression or misactivation that might lead to autoimmune-type responses. Thus, these rapidly evolving clusters are potent activators of plant defense and need to be tightly regulated. Recently, clusters of TIR-NB-LRR disease resistance genes have been shown to elicit hybrid necrosis, a type of deleterious autoimmune response resulting from incompatibility of Arabidopsis ecotypes. It was thus suggested that such a mechanism might create gene flow barriers in evolution (Bomblies et al., 2007; Alcazar et al., 2009). Our data suggest that MKP1/PTP1-regulated MAPK pathways are likely involved in the regulation of such intra- and interspecific compatibilities, which ensures an optimal balance between growth and pathogen resistance by modulating SA biosynthesis.
METHODS
Plant Material and Growth Conditions
bon1-1, eds1-22 (SALK_071051), mpk3-1 (SALK_151594), mpk4 (SALK_056245), mpk6-2 (SALK_073907), mpk6-3 (SALK_ 127507), pad4-1 (N3806), ptp1-1 (SALK_118658), snc1-11, and the NahG transgenic lines are all in the Col ecotype (Gaffney et al., 1993; Zhou et al., 1998; Alonso et al., 2003; Yang and Hua, 2004). The mkp1-1 mutation (Ulm et al., 2001) was introgressed from its Ws background into Col by crossing for at least nine times.
To generate double mutants, single mutants were crossed and the double mutants identified by PCR genotyping in the F2 generation. Plant genomic DNA for PCR analysis was prepared according to Edwards et al. (1991). T-DNA- and gene-specific primers were used as described in Supplemental Table 1 online.
Arabidopsis thaliana seeds were surface-sterilized with sodium hypochlorite and plated on half-strength Murashige and Skoog medium (Duchefa) supplemented with 1% sucrose and 0.5% Phytagel (Sigma-Aldrich). Seeds were stratified for at least 2 d at 4°C and germinated aseptically at 24°C in a standard growth chamber (MLR-350; Sanyo) with a 12-h/12-h light/dark cycle. Standard growth on soil in a phytochamber was under a 16-h/8-h light/dark cycle with 21°C/19°C, if not otherwise indicated.
Generation of Transgenic Arabidopsis Lines
The MKP1 and PTP1 coding regions were cloned by the Gateway BP reaction into pDONR207 using primers attB1-MKP1/attB2-MKP1-STOP and attB1-PTP1/attB2-PTP1-STOP (see Supplemental Table 2 online), respectively, and verified by sequencing. Gateway-based cloning was then used to insert PTP1 into the binary destination vector pB7YWG2 and MKP1 into pH7YWG2 (Karimi et al., 2002). Arabidopsis plants were transformed by Agrobacterium tumefaciens using the floral dip method (Clough and Bent, 1998). The resulting transgenic lines described in this work were genetically determined to have the transgene integrated at a single locus.
BiFC Assay and Epifluorescence Microscopy
Sinapis alba–based BiFC assays were performed as described previously (Stolpe et al., 2005). A Pro35S:CFP control plasmid was always cobombarded to identify transformed cells prior to the analysis of YFP fluorescence. PCR fragments for MKP1, PTP1, and MPK coding sequences (including stop codons) were generated with the primers listed in Supplemental Table 2 online and cloned with the Gateway BP reaction into pDONR207 generating entry clones. Gateway LR reaction was then used to clone MKP1 and PTP1 into the pE-SPYNE-GW destination vector and the MPKs into pE-SPYCE-GW. The empty vectors used as negative controls were generated by recombination with an empty pENTRY3C clone, which was produced by EcoRI digestion and self-ligation to remove the ccdB gene. The Gateway-compatible BiFC binary vectors pE-SPYNE-GW and pE-SPYCE-GW were kindly provided by Caroline Carsjens and Wolfgang Dröge-Laser (University of Göttingen).
Transient Expression in Arabidopsis Protoplasts and Immunocomplex Kinase Assays
The open reading frames of MPK6 and MKP1 were cloned into the plant expression vector pGREEN and constitutively active MKK4EE (with both putative phosphorylation sites changed to Glu residues: T224E and S230E) into pRT100 and fused at their C-terminal end either to a triple HA epitope (MPK6 and MKK4EE) or to a c-myc epitope (MKP1), as described before (Doczi et al., 2007). Arabidopsis protoplast transient expression assays and protein extraction were performed as described (Ouaked et al., 2003). For immunocomplex kinase assays MPK6 was isolated from the total protein extract using an anti-MPK6–specific antibody incubated for 2 h at 4°C together with Sepharose-A beads. Kinase reactions of the immunoprecipitated MPK6 proteins were performed at 30°C for 30 min in 15 μL of kinase buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 5 mM EGTA, and 1 mM DTT) containing 1.5 μg of MBP, 0.1 mM ATP, and 2 μCi of [γ-32P]ATP. The kinase reactions were stopped by adding 4× SDS loading buffer, and proteins were separated by 15% SDS-PAGE. MBP phosphorylation was analyzed using a phosphor imager and quantified using ImageQuant analysis software (GE Healthcare).
Generation of Antibodies, Immunoprecipitation Assays, and Protein Gel Blot Analysis
Rabbit polyclonal antibodies were generated against synthetic peptides derived from the MPK3 protein sequence (MNTGGGQYTDFPAVEC), MPK4 (CFGSSGDQSSSKGVA), and MPK6 (MDGGSGQPAADTEMTC) and were affinity purified against the peptide (Eurogentec).
For protein gel blot analysis, total cellular proteins (10 μg) were separated by electrophoresis in 10% SDS-polyacrylamide gel and electrophoretically transferred to a polyvinylidene fluoride membrane according to the manufacturer's instructions (Bio-Rad). We used polyclonal primary antibodies against MPK3, MPK4, MPK6, actin (Sigma-Aldrich), and phospho-p44/42 MAP kinase (Cell Signaling Technologies) and monoclonal anti-GFP (BAbCO), with horseradish peroxidase–conjugated anti-rabbit, anti-goat, and anti-mouse immunoglobulins (DAKO) as secondary antibodies, as required. Signal detection was performed using the ECL Plus Western Detection Kit (GE Healthcare).
Quantitative Real-Time PCR
Arabidopsis total RNA was treated with DNaseI according to the manufacturer's specifications (Qiagen). Per PCR reaction, cDNA was synthesized from 25 ng of RNA with a 1:1 mixture of random hexamers and oligo(dT) using the TaqMan reverse transcription reagents kit (Applied Biosystems). Quantitative RT-PCR was performed in a 96-well format using a 7300 real-time PCR system (Applied Biosystems). PCR reactions were performed using the ABsolute SYBR Green Rox Mix Kit following the manufacturer's instructions (Thermo Scientific). The gene-specific probes and primers were as follows: SNC1 with SNC1_for (5′-AGGAATTAGATCTTGTTGGATGC-3′) and SNC1-rev (5′-CCCCTCACATTGAGAAAAGC-3′); BON1 with BON1_for (5′-CCTTGCTTCTAAGATTTCAACAG-3′) and BON1_rev (5′-GAATTGGAGTTCCATGCTCTAC-3′); PR1 with PR1_for (5′-AGAGTGTATGAGTCTGCAGTTG-3′) and PR1_rev (5′-CTCTTGTAGGTGCTCTTGTTC-3′); MKP1 with MKP1_for (5′-CCATTTTGTGTCAGATGGACTTG-3′) and MKP1_rev (5′-TGCTAGCAACTCTGTCTGATC-3′); and ICS1 with ICS1_for (5′-CCGTCTCTGAACTCAAATCTC-3′) and ICS1_rev (5′-ATTCTGGGCTTGAAGCCAATC-3′). For PR5, TaqMan quantitative RT-PCR was performed using the gene-specific probe 6-FAM-ACAGACTTCACTCTAAGGAACAATTGCCCT-TAMRA with primers PR5_for (5′-CTCTTCCTCGTGTTCATCAC-3′) and PR5-rev (5′-TCCTTGACCGGCGAGAGT-3′) as described before (Favory et al., 2009). cDNA concentrations were normalized to the 18S rRNA transcript levels as standard using the Eukaryotic 18S rRNA Kit (Applied Biosystems). Average results of three biological replicates, each measured as triplicates, are given.
SA and Camalexin Extraction and Quantification by HPLC
Samples (500 mg) were taken from soil-grown 22-d-old plants, combining rosette from three to four plants per sample. SA and camalexin were extracted and quantified by HPLC as previously described (Garcion et al., 2008). Average results from at least three different samples per genotype are given.
Pseudomonas syringae Growth Assays and Luciferase Imaging
Plants were maintained at 22°C day/20°C night with 9-h daylength. Inoculation of plants with P. syringae pv tomato (Pst) was performed essentially as described (Zipfel et al., 2004). Prior to inoculation, the bioluminescent strain of Pst DC3000 containing the LuxCDABE operon (Fan et al., 2008) was streaked onto a King's B agar containing 50 μg/mL kanamycin and 60 μg/mL rifampicin. After 3 d, bacteria were scraped from the plate, resuspended to a final OD600 = 0.1 in sterile water containing 0.04% Silwet, and sprayed onto the upper surface of leaves. Plants were covered for 24 h postinfection to maintain higher humidity. Luciferase signal from whole rosettes or detached leaves was detected using the Photek HRPCS4 photon detection camera (Photek). Counting of leaf bacteria by serial dilution plating was performed as previously described (Katagiri et al., 2002). Prior to imaging detached leaves or sampling for serial dilution plating, leaf tissue was surface sterilized with 70% ethanol for 15 s and rinsed with water.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers AT1G10210 (MPK1), AT1G59580 (MPK2), AT1G71860 (PTP1), AT1G74710 (ICS1/SID2), AT1G75040 (PR5), AT2G14610 (PR1), AT2G43790 (MPK6), AT3G45640 (MPK3), AT3G55270 (MKP1), AT4G01370 (MPK4), AT4G16890 (SNC1), and AT5G61900 (BON1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the ptp1-1 T-DNA Insertion.
Supplemental Figure 2. Comparison of mkp1(Ws) versus mkp1(Col), Complementation of mkp1(Col) with a Pyo-MKP1 Transgene, and mkp1-Cosuppression (cs) Line.
Supplemental Figure 3. Growth Phenotypes of mkp1(Col) and mkp1 ptp1 Appear Rather Late in Development.
Supplemental Figure 4. Suppression of the mkp1(Col) Growth Phenotype by the mpk6-3 Allele.
Supplemental Figure 5. Levels of Free and Conjugated Salicylic Acid.
Supplemental Figure 6. Suppression of mkp1(Col) and mkp1 ptp1 Phenotypes by Elevated Temperatures (28°C).
Supplemental Figure 7. Cocorrelation Scatterplot of MKP1 and PTP1 Coexpression Compared with IBR5, PHS1, MKP2, and DsPTP1.
Supplemental Figure 8. mpk6 Mutation Does Not Suppress the mpk4 Dwarf Growth Phenotype.
Supplemental Table 1. Primer Sequences Used for Mutant Genotyping.
Supplemental Table 2. Primer Sequences Used for Molecular Cloning.
Supplementary Material
Acknowledgments
We thank Andreas Hiltbrunner (University of Tübingen) for critical reading of the manuscript, Markus Funk (University of Freiburg) for excellent technical assistance, and Tonia Oberlin for contributions to the initial phase of this work. We also thank Jian Hua (Cornell University) for providing bon1-1 seeds, Christiane Nawrath (University of Lausanne) for NahG transgenic seeds, and the Nottingham Arabidopsis Stock Centre for SALK mutant lines. J.-P.M. and A.B. were supported by the Swiss National Science Foundation (Grant 3100A0-104224 to J.-P.M.), J.C.A. by the Human Frontier Science Program Grant RGP22/2006, S.B. by the Graduiertenkolleg GRK1305 “Signal Systems in Plant Model Organisms,” M.A.G.B. by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School), and R.U. by the Emmy Noether Programme of the German Research Foundation (DFG, UL341/1-1).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Roman Ulm (roman.ulm@biologie.uni-freiburg.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Alcazar, R., Garcia, A.V., Parker, J.E., and Reymond, M. (2009). Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. USA 106 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Beckers, G.J., Jaskiewicz, M., Liu, Y., Underwood, W.R., He, S.Y., Zhang, S., and Conrath, U. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J.L., and Weigel, D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Bjorn Nielsen, H., Zhu, S., Newman, M.A., Shokat, K.M., Rietz, S., Parker, J., and Mundy, J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47 532–546. [DOI] [PubMed] [Google Scholar]
- Camps, M., Nichols, A., and Arkinstall, S. (2000). Dual specificity phosphatases: A gene family for control of MAP kinase function. FASEB J. 14 6–16. [PubMed] [Google Scholar]
- Christie, G.R., Williams, D.J., Macisaac, F., Dickinson, R.J., Rosewell, I., and Keyse, S.M. (2005). The dual-specificity protein phosphatase DUSP9/MKP-4 is essential for placental function but is not required for normal embryonic development. Mol. Cell. Biol. 25 8323–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet, J., and Hirt, H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413 217–226. [DOI] [PubMed] [Google Scholar]
- Doczi, R., Brader, G., Pettko-Szandtner, A., Rajh, I., Djamei, A., Pitzschke, A., Teige, M., and Hirt, H. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L., Ali, G.S., Simons, K.A., Hou, J., Yang, T., Reddy, A.S., and Poovaiah, B.W. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457 1154–1158. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J., Crooks, C., and Lamb, C. (2008). High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 53 393–399. [DOI] [PubMed] [Google Scholar]
- Favory, J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754–756. [DOI] [PubMed] [Google Scholar]
- Garcion, C., Lohmann, A., Lamodiere, E., Catinot, J., Buchala, A., Doermann, P., and Metraux, J.P. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., Huang, Y., Kieber, J., and Luan, S. (1998). Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 16 581–589. [DOI] [PubMed] [Google Scholar]
- Gupta, R., and Luan, S. (2003). Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol. 132 1149–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, S., Grau, T., Buchala, A., Metraux, J.P., and Nawrath, C. (2003). Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36 342–352. [DOI] [PubMed] [Google Scholar]
- Hua, J., Grisafi, P., Cheng, S.H., and Fink, G.R. (2001). Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 15 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Li, H., Gupta, R., Morris, P.C., Luan, S., and Kieber, J.J. (2000). ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 122 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, K., Casais, C., Peck, S.C., Shinozaki, K., and Shirasu, K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281 36969–36976. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., et al. (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7 301–308. [DOI] [PubMed] [Google Scholar]
- Jirage, D., Zhou, N., Cooper, B., Clarke, J.D., Dong, X., and Glazebrook, J. (2001). Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 26 395–407. [DOI] [PubMed] [Google Scholar]
- Junttila, M.R., Li, S.P., and Westermarck, J. (2008). Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 22 954–965. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Katagiri, F., Thilmony, R., and He, S.Y. (2002). The Arabidopsis thaliana-Pseudomonas syringae interaction. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Katou, S., Karita, E., Yamakawa, H., Seo, S., Mitsuhara, I., Kuchitsu, K., and Ohashi, Y. (2005). Catalytic activation of the plant MAPK phosphatase NtMKP1 by its physiological substrate salicylic acid-induced protein kinase but not by calmodulins. J. Biol. Chem. 280 39569–39581. [DOI] [PubMed] [Google Scholar]
- Katou, S., Kuroda, K., Seo, S., Yanagawa, Y., Tsuge, T., Yamazaki, M., Miyao, A., Hirochika, H., and Ohashi, Y. (2007). A calmodulin-binding mitogen-activated protein kinase phosphatase is induced by wounding and regulates the activities of stress-related mitogen-activated protein kinases in rice. Plant Cell Physiol. 48 332–344. [DOI] [PubMed] [Google Scholar]
- Kerppola, T.K. (2006). Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse, S.M. (2008). The regulation of stress-activated MAP kinase signalling by protein phosphatases. Topics Curr. Genet. 20 33–49. [Google Scholar]
- Lee, J.S., and Ellis, B.E. (2007). Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 282 25020–25029. [DOI] [PubMed] [Google Scholar]
- Lee, J.S., Wang, S., Sritubtim, S., Chen, J.G., and Ellis, B.E. (2009). Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 57 975–985. [DOI] [PubMed] [Google Scholar]
- Lee, K., Song, E.H., Kim, H.S., Yoo, J.H., Han, H.J., Jung, M.S., Lee, S.M., Kim, K.E., Kim, M.C., Cho, M.J., and Chung, W.S. (2008). Regulation of MAPK phosphatase 1 (AtMKP1) by calmodulin in Arabidopsis. J. Biol. Chem. 283 23581–23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Orfanidi, S., Chefdor, F., Meszaros, T., Bolte, S., Mizoguchi, T., Shinozaki, K., Giraudat, J., and Bögre, L. (2006). Antagonistic interaction between MAP kinase and protein phosphatase 2C in stress recovery. Plant Sci. 171 596–606. [Google Scholar]
- Li, Y., Yang, S., Yang, H., and Hua, J. (2007). The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors. Mol. Plant Microbe Interact. 20 1449–1456. [DOI] [PubMed] [Google Scholar]
- Manfield, I.W., Jen, C.H., Pinney, J.W., Michalopoulos, I., Bradford, J.R., Gilmartin, P.M., and Westhead, D.R. (2006). Arabidopsis Co-expression Tool (ACT): Web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 34 W504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, C.J. (1995). Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 80 179–185. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco, E., Gampel, A., Ring, J., Virdee, K., Kirov, N., Tolkovsky, A.M., and Martinez-Arias, A. (1998). puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean, M.N., Mody, A., Broach, J.R., and Ramanathan, S. (2007). Cross-talk and decision making in MAP kinase pathways. Nat. Genet. 39 409–414. [DOI] [PubMed] [Google Scholar]
- Menke, F.L., van Pelt, J.A., Pieterse, C.M., and Klessig, D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe-Augustus, M., Zolman, B.K., and Bartel, B. (2003). IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami, H., Soukupova, H., Schikora, A., Zarsky, V., and Hirt, H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281 38697–38704. [DOI] [PubMed] [Google Scholar]
- Naoi, K., and Hashimoto, T. (2004). A semidominant mutation in an Arabidopsis mitogen-activated protein kinase phosphatase-like gene compromises cortical microtubule organization. Plant Cell 16 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van Der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Ouaked, F., Rozhon, W., Lecourieux, D., and Hirt, H. (2003). A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22 1282–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Popescu, S.C., Popescu, G.V., Bachan, S., Zhang, Z., Gerstein, M., Snyder, M., and Dinesh-Kumar, S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J.L., Zhou, L., Yun, B.W., Nielsen, H.B., Fiil, B.K., Petersen, K., Mackinlay, J., Loake, G.J., Mundy, J., and Morris, P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D., Liu, Y., Yang, K.Y., Han, L., Mao, G., Glazebrook, J., and Zhang, S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer, A., et al. (2007). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L., Kunkel, B.N., and Richards, E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe, T., Susslin, C., Marrocco, K., Nick, P., Kretsch, T., and Kircher, S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226 137–146. [DOI] [PubMed] [Google Scholar]
- Takahashi, F., Yoshida, R., Ichimura, K., Mizoguchi, T., Seo, S., Yonezawa, M., Maruyama, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige, M., Scheikl, E., Eulgem, T., Doczi, R., Ichimura, K., Shinozaki, K., Dangl, J.L., and Hirt, H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15 141–152. [DOI] [PubMed] [Google Scholar]
- Ulm, R., Ichimura, K., Mizoguchi, T., Peck, S.C., Zhu, T., Wang, X., Shinozaki, K., and Paszkowski, J. (2002). Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., Revenkova, E., di Sansebastiano, G.P., Bechtold, N., and Paszkowski, J. (2001). Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 15 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Liu, Y., Bruffett, K., Lee, J., Hause, G., Walker, J.C., and Zhang, S. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer, M., Feys, B.J., and Parker, J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Wu, G.S. (2007). Role of mitogen-activated protein kinase phosphatases (MKPs) in cancer. Cancer Metastasis Rev. 26 579–585. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Fu, H.H., Gupta, R., and Luan, S. (1998). Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 10 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, H., Katou, S., Seo, S., Mitsuhara, I., Kamada, H., and Ohashi, Y. (2004). Plant MAPK phosphatase interacts with calmodulins. J. Biol. Chem. 279 928–936. [DOI] [PubMed] [Google Scholar]
- Yang, S., and Hua, J. (2004). A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Yang, H., Grisafi, P., Sanchatjate, S., Fink, G.R., Sun, Q., and Hua, J. (2006). The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J. 45 166–179. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D., Cho, Y.H., Tena, G., Xiong, Y., and Sheen, J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., et al. (2007. a). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Dai, Y., Xiong, Y., DeFraia, C., Li, J., Dong, X., and Mou, Z. (2007. b). Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 52 1066–1079. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Blattman, J.N., Kennedy, N.J., Duong, J., Nguyen, T., Wang, Y., Davis, R.J., Greenberg, P.D., Flavell, R.A., and Dong, C. (2004). Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature 430 793–797. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., and Last, R.L. (1996). Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell 8 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Cai, Z., Guo, Y., and Gan, S. (2009). An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.