Abstract
Agrobacterium tumefaciens causes crown gall disease by transferring and integrating bacterial DNA (T-DNA) into the plant genome. To examine the physiological changes and adaptations during Agrobacterium-induced tumor development, we compared the profiles of salicylic acid (SA), ethylene (ET), jasmonic acid (JA), and auxin (indole-3-acetic acid [IAA]) with changes in the Arabidopsis thaliana transcriptome. Our data indicate that host responses were much stronger toward the oncogenic strain C58 than to the disarmed strain GV3101 and that auxin acts as a key modulator of the Arabidopsis–Agrobacterium interaction. At initiation of infection, elevated levels of IAA and ET were associated with the induction of host genes involved in IAA, but not ET signaling. After T-DNA integration, SA as well as IAA and ET accumulated, but JA did not. This did not correlate with SA-controlled pathogenesis-related gene expression in the host, although high SA levels in mutant plants prevented tumor development, while low levels promoted it. Our data are consistent with a scenario in which ET and later on SA control virulence of agrobacteria, whereas ET and auxin stimulate neovascularization during tumor formation. We suggest that crosstalk among IAA, ET, and SA balances pathogen defense launched by the host and tumor growth initiated by agrobacteria.
INTRODUCTION
Agrobacterium tumefaciens is a pathogenic bacterium that causes crown gall disease, a plant tumor affecting a wide range of plant species. Crown galls develop upon transfer of a portion of the tumor-inducing (Ti) plasmid, the transfer-DNA (T-DNA), into the genome of the bacterium's plant hosts (Chilton et al., 1980). T-DNA transfer is initiated when Agrobacterium detects phenolic molecules released from actively growing cells in a plant wound. These phenolics induce expression of multiple virulence (vir) genes, encoding products responsible for processing and transferring the single-stranded T-DNA across the bacterial membrane system into the plant cell, where it becomes integrated into the genome at an essentially random location (McCullen and Binns, 2006). Genes encoded by the T-DNA are expressed and subsequently alter plant hormone levels, leading to uncontrolled cell division and tumor formation. Although the elucidation of plant factors supporting the transformation process has been crucial to our understanding of this interaction (Gelvin, 2003; Citovsky et al., 2007) little is known about the timing and type of responses that plants mount against Agrobacterium and how those compare with responses elicited by other pathogens and symbionts.
Plants have evolved efficient mechanisms to respond to microorganisms that infect their hosts (Hammond-Kosack and Jones, 1996; Nimchuk et al., 2003). The perception of pathogen-associated molecular patterns (PAMPs) leads to a rapid activation of defense mechanisms, such as a localized burst of reactive oxygen species and programmed plant cell death (the hypersensitive response) at infection sites. It also causes stimulation of basal defenses that are regulated by a network of interconnecting signal transduction pathways, in which salicylic acid (SA) and jasmonic acid (JA) together with ethylene (ET) function as key signaling molecules (Glazebrook, 2001; Thomma et al., 2001; Pieterse et al., 2009). JA and ET accumulate in response to pathogen infection or herbivore damage, resulting in the activation of distinct sets of pathogenesis-related genes (PR). It has been reported that along with auxin and cytokinin (Weiler and Schroeder, 1987; Zambryski et al., 1989; Malsy et al., 1992), the phytohormone ET is a limiting factor of crown gall morphogenesis because ET deficiency or insensitivity leads to inhibition of tumor growth (Aloni et al., 1998; Wachter et al., 2003). SA-mediated defense responses provide protection from biotrophic fungi, oomycetes, and bacteria, including Erysiphe orontii, Peronospora parasitica, and Pseudomonas syringae. Mutant plants, such as sid2 (SA induction-deficient) and eds5 (enhanced disease susceptibility), that are deficient in SA accumulation upon pathogen challenge are more susceptible to pathogen infection than wild-type plants (Nawrath and Metraux, 1999; Wildermuth et al., 2001). The SID2 gene encodes a putative chloroplast-localized isochorismate synthase, and mutant plants are therefore defective in SA synthesis and systemic acquired resistance (SAR) activation and exhibit enhanced susceptibility to pathogens. SA depletion in transgenic plants expressing the bacterial nahG gene, a salicylate hydroxylase, also impairs induction of basal defenses, although nahG expression has pleiotropic effects due to catechol accumulation (Heck et al., 2003; van Wees and Glazebrook, 2003). The ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) protein and its interacting partner, PHYTOALEXIN DEFICIENT4 (PAD4) are also required for accumulation of the plant defense-potentiating molecule SA (Feys et al., 2001). Recent results point to a fundamental role of EDS1 and PAD4 in transducing redox signals in response to certain biotic and abiotic stresses. These intracellular proteins are essential regulators of basal resistance to invasive obligate biotrophic and certain hemibiotrophic pathogens. These proteins are important activators of SA signaling and also mediate antagonism between the JA and ET defense response pathways (Wiermer et al., 2005). SA-induced PR gene expression and SAR occur primarily through a signaling pathway involving the transcriptional activator NONEXPRESSER OF PR1 (NPR1). Mutant npr1-1 plants are defective in this signaling and exhibit decreased PR gene expression (Cao et al., 1997). Levels of SA, however, accumulate to those seen in wild-type plants in response to infection by various pathogens. Previous studies have uncovered a role for SA on Agrobacterium by inhibiting vir gene induction (Yuan et al., 2007, Anand et al., 2008) and thereby affecting agrobacterial virulence. By contrast, defense reactions against many necrotrophic fungi do not involve SA, but rely on ET and JA accumulation and signaling.
Plant defenses could be launched at any step of Agrobacterium-mediated tumorigenesis, starting with (1) the attachment of agrobacteria to the plant cell, followed by (2) the stable integration of the T-DNA into the plant genome and (3) ending with tumor growth. In this study, genome-wide gene expression analysis and measurements of stress signaling molecules were integrated to present a comprehensive overview of the defense signaling pathways throughout the different stages of crown gall development.
RESULTS
Once Agrobacterium has invaded the plant, it sustains a long-term association with the plant cell. We set out to explore whether the plant activates defense reactions against this virulent pathogen at any stage of tumor development. These stages can be roughly defined as time points when (1) agrobacteria come into close contact with the plant cell at initiation of infection, (2) the T-DNA is transferred into the plant cell and the encoded oncogenes are expressed, and (3) morphological changes indicate the development of a tumor. For these studies, the bases of main inflorescence stalks, still attached to intact Arabidopsis thaliana plants, were infected right above the rosette leaves in order to maintain conditions close to nature. The advantage of this experimental system is that the host plant response can be analyzed without phytohormone pretreatment. The development of Agrobacterium–induced plant tumors primarily depends on excessive production of auxin and cytokinin by the T-DNA–encoded oncogenic enzymes. By contrast, calli or suspension cell cultures are cultivated in the presence of exogenous supplied auxins and cytokinins, which makes it difficult to analyze the crosstalk between hormones derived from T-DNA–encoded gene products and those produced by the host during the course of crown gall development. Furthermore, translocation of nutrients and signaling molecules from the host into the tumor still can take place and influences the physiological state of tumors.
Rationale for Choosing Time Points for Analysis
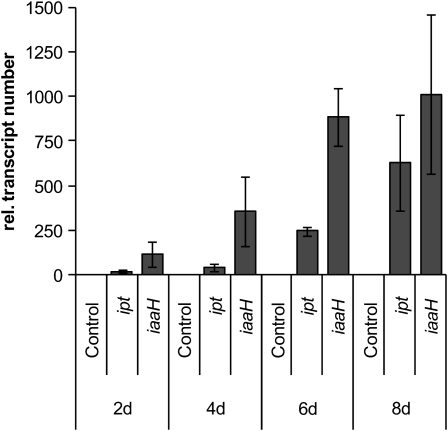
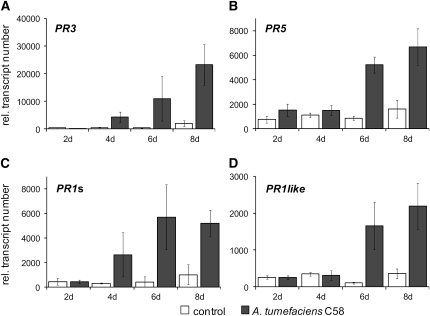
In order to elucidate general responses of Arabidopsis to agrobacteria, we set out to analyze the three stages described above that mark characteristic steps in the course of crown gall development. We chose to define the two early steps in Arabidopsis–Agrobacterium interaction by the appearance of transcripts of the T-DNA–encoded genes ipt (for isopentenyl-transferase) and iaaH (for indoleacetamide hydrolase) in Arabidopsis inflorescence stalk tissue. Transcripts were assayed by quantitative real-time PCR (qRT-PCR). Young inflorescence stalks of Arabidopsis (ecotype Wassilewskija [Ws-2]) were wounded and inoculated with Agrobacterium (strain C58) at the base just above the rosette without induction of virulence beforehand (Figure 1A). Since it is not known when after infection the T-DNA is present in the host cells of intact plants, we collected inflorescence stalk segments at 1, 3, 6, 12, and 24 h and 2, 4, 6, and 8 d postinoculation. Wounded but uninfected stalks served as a control. Transcripts of the ipt and iaaH genes could not be detected within the first 24 h postinoculation (data not shown) but were observed after 2 d of infection, albeit in very low numbers. Both ipt and iaaH transcripts accumulated significantly after 6 d of inoculation (Figure 2). Transcripts of Arabidopsis PR genes (PR3, PR5, PR1s, and PR1-like), which are strongly elevated in 35-d-old Arabidopsis tumors (Deeken et al., 2006), were not increased within the first 24 h of infection with the virulent Agrobacterium strain C58 (data not shown). Later on, transcripts of the PR genes accumulated in a time frame similar to that for the T-DNA–encoded ipt gene. The chitinase PR3 (Figure 3A; 10-fold) and PR1s (Figure 3C; 13-fold) appeared 4 d postinoculation. Transcripts of PR5, encoding an antimicrobial thaumatin-like protein (Figure 3B; 3.5-fold), and PR1-like (Figure 3D; sevenfold) increased only at 6 d postinoculation. Thus, by 6 d postinoculation, transcripts of oncogenes and PR genes were easily detectable, but stalk morphology was not yet affected; for this reason, we chose 6 d postinoculation as our middle time point for transcriptome analysis and determination of signaling molecules. The earliest time point studied was 3 h postinoculation, since we reasoned that the host needs some time to respond to the just invading pathogen. Furthermore, transcripts of the ipt or iaaH gene or other T-DNA–encoded genes have not been detected before 6 h postinoculation with agrobacteria (Veena et al., 2003), an observation we confirmed by qRT-PCR. The final step of a successful plant cell transformation is the development of a tumor (Figure 1B). For this stage, we analyzed 35-d-old tumors, as we had in our previous microarrays studies (Deeken et al., 2006).
Figure 1.
Tumor Induction and Visualization of H2O2 Production on Arabidopsis Inflorescence Stalks (Ecotype Ws-2) upon Infection with the Agrobacterium Strains C58 (nocc, No. 284, Max Planck Institute for Plant Breeding, Cologne, Germany) and GV3101 (pMP90, Koncz and Schell, 1986).
(A) The black frame indicates the area of wounding and/or infection at the base of an inflorescence stalk, just above the rosette.
(B) A representative tumor, 35 d postinoculation (dpi) with strain C58.
(C) and (E) A brownish color emerges after treatment with DAB, indicating H2O2 production at 3 h (C) or 6 d (E) after wounding without inoculation of agrobacteria (control).
(D), (F), and (G) No H2O2 production was visible when wounded inflorescence stalks were inoculated with the strains C58 3 hpi (D) and 6 dpi (F) or GV3101 6 dpi (G).
(H) The fully developed tumor, but not the tumor-free area of the inflorescence stalk, stained with DAB exhibited H2O2 production (brownish color) 35 dpi with the strain C58.
Figure 2.
Increase in Transcription of the T-DNA–Encoded Oncogenes ipt and iaaH.
Arabidopsis (ecotype Ws-2) inflorescence stalks were wounded and inoculated at the base just above the rosette with Agrobacterium strain C58 for the indicated time points. Wounded, but not inoculated, stalks served as control. The number of transcripts was calculated from qRT-PCR data and normalized relative to 10,000 molecules of ACTIN2/8. Results shown represent mean values ± se from at least three independent experiments.
Figure 3.
Transcriptional Activation of PR Genes.
Arabidopsis inflorescence stalks (ecotype Ws-2) were wounded and inoculated with Agrobacterium strain C58 at the base just above the rosette. The number of transcripts of (A) PR3 (At3g12500), (B) PR5 (At1g75040), (C) PR1s (At2g19970), and (D) PR1like (At2g19990) was determined by qRT-PCR and normalized relative to 10,000 molecules of ACTIN2/8 at the indicated time points. Wounded, but not inoculated, stalks served as control. Results shown represent mean values ± se from at least three independent experiments.
The Oncogenic Strain C58 Affected Four Times as Many Arabidopsis Genes as the Disarmed Strain GV3101
Gene expression changes at 3 h postinoculation, 6 d postinoculation, and 35-d-old tumors were studied with 32 ATH1 genome chips of Arabidopsis (Affymetrix; Table 1) and statistically analyzed as described by Deeken et al. (2006) with a few adaptations (see Methods section). To determine whether Arabidopsis genes respond to the T-DNA–encoded oncogenes or to bacterial effector proteins codelivered by agrobacteria into the plant cell, two different Agrobacterium strains were used for inoculation: (1) the oncogenic strain C58 and (2) a T-DNA–deficient derivate of C58, GV3101, which only lacks the T-DNA, but not the proteinaceous virulence factors, such as VirD2, VirE2, VirE3, and VirF (Vergunst et al., 2000, 2003), or any other effector proteins. The fold changes of differentially expressed genes were calculated from agrobacteria-treated samples versus wounded, but noninfected inflorescence stalk tissue (control). Only fold changes of genes ≥2-fold or ≤0.5, which met the significance criteria of P value ≤ 0.01 are presented here (see Supplemental Data Set 1, data sheet 1, online). Genes with signal intensities close to background levels (<200) in one of the two treatments (infected or wounded) were excluded. Randomly selected genes were also analyzed by qRT-PCR to assess the validity of the microarray data. The Pearson's correlation coefficients calculated from the comparison of microarray and qRT-PCR data were 0.9923 for C58 (+T-DNA) and 0.9932 for GV3101 (−T-DNA). Thus, the fold changes detected by both methods correlated well for the randomly selected genes (see Supplemental Figure 1 online).
Table 1.
Time Points of Treatment of Arabidopsis Inflorescence Stalks (Ecotype Ws-2) with the Two Agrobacterium Strains, C58 and GV3101, and Number of Microarrays Analyzed per Treatment
| Treatment | Label | Agrobacterium Strain | Number of Microarrays |
|---|---|---|---|
| 3 h postinfection | 3 h postinoculation | C58 | 6 |
| 3 h postinfection | 3 h postinoculation | GV3101 | 3 |
| 3 h postwounding | Control | – | 6 |
| 6 d postinfection | 6 d postinoculation | C58 | 3 |
| 6 d postinfection | 6 d postinoculation | GV3101 | 3 |
| 6 d postwounding | Control | – | 3 |
| 35 d postinfectiona | Tumor | C58 | 4 |
| 35 d postwoundinga |
Control |
– |
4 |
Microarray data previously published by Deeken et al. (2006).
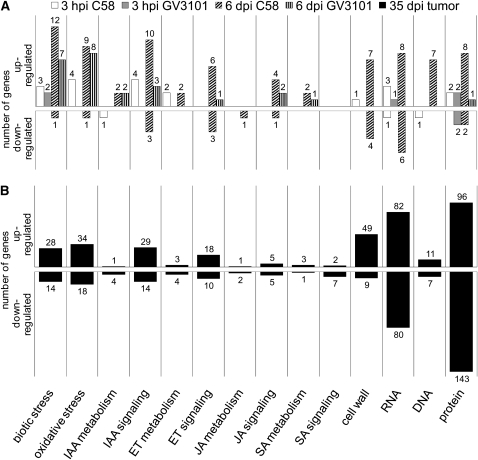
Upon inoculation with the oncogenic strain C58, 35 genes were transcriptionally changed at 3 h postinoculation. By contrast, only eight genes were affected by strain GV3101 lacking a T-DNA (see Supplemental Figure 2A online). The transcription of five genes was influenced by both strains (see Supplemental Figure 2B online). After T-DNA integration at 6 d postinoculation, 196 genes responded to strain C58 and 48 genes to a treatment with strain GV3101 (see Supplemental Figure 2A online). The majority of the 48 genes influenced by strain GV3101 also responded to strain C58 (36 genes; see Supplemental Figure 2C online). In 35-d-old tumors, the transcription of 2076 genes was changed (see Supplemental Figure 2A online). Taken together, strain C58, harboring a T-DNA, affected four times as many genes as strain GV301 during the early stages of Arabidopsis-Agrobacterium infection.
At Early Stages of Infection, Only a T-DNA–Bearing Strain Triggers Transcription of Genes Involved in Changes of Host Morphology
For functional characterization of the differentially expressed genes, the pathway analysis program MapMan (http://gabi.rzpd.de/projects/MapMan, Version 2.2.0, July 2008) was used. This program refers to the database TAIR for annotation of the genes (ftp://ftp.Arabidopsis.org/home/tair/Microarrays/Affymetrix:%20affy_ATH1_array_elements-2008-5-29.txt). According to this program, the functional category “stress” was the largest class of genes affected by both strains at both time points (see Supplemental Figure 3 online). The majority of genes in this category were involved in pathogen defense, encoding, for example, disease resistance (R) proteins, PR proteins, defensins, chitinases, and proteinase inhibitors. The second largest category was “hormone,” containing genes involved in hormone metabolism and signaling. Genes of this category did not respond nearly as well to strain GV3101 (see Supplemental Figures 3B and 3D online) as to C58 (see Supplemental Figures 3A and 3C online). Genes of the functional category “RNA,” with mainly transcription factors, and the category “cell wall,” comprising expansins and xyloglucosyl transferases, in addition to genes involved in modification of DNA, proteins, and lipid metabolism also responded only to strain C58 at initiation of infection (3 h postinoculation) and at the time of T-DNA transfer (6 d postinoculation). Thus, only strain C58 triggers transcription of genes needed for changes in morphology and initiation of tumor development. The difference profiles between strains C58 and GV3101 show that the number of functional categories and the number of genes therein was clearly higher at both early time points when the oncogenic strain was applied (see Supplemental Figure 4 online). Genes of 12 categories at 3 h postinoculation (see Supplemental Figures 4A and 4B online) and 18 categories at 6 d postinoculation responded only to the oncogenic strain C58 (see Supplemental Figures 4C and 4D online). By contrast, only two categories at 3 h postinoculation and three at 6 d postinoculation were specific for strain GV3101.
Pathogen Defense Genes Responded Predominantly to the Oncogenic Strain C58
In response to pathogen attack, early signaling results in increased expression of genes encoding antimicrobial PR proteins (van Loon et al., 2006a; Sels et al., 2008). Since this study aims to dissect pathogen defense responses of Arabidopsis to agrobacteria, we analyzed the category “biotic stress,” a subcategory of the MapMan category “stress” in more detail. As early as 3 h postinoculation, two genes encoding chitinases (At2g43620 and At4g01700), a proteinase inhibitor (At1g17860), and MLO12 (At2g39200) were induced either by the tumorigenic strain C58 or T-DNA–depleted strain GV3101 (see Supplemental Data Set 2, data sheet 1, online). At 6 d postinoculation, the number of genes involved in pathogen defense that were upregulated multiplied in response to both strains: 12 genes by strain C58 and seven by GV3101 (Figure 4A; biotic stress category). Among them were PR5, PR1S, and PR1-like, three genes for which elevated transcription had already been detected by qRT-PCR in C58-infected tissues at 6 d postinoculation (Figure 3). In addition, two chitinases (At2g43590 and PR3) and two defensins, PDF1.2 (At5g44420) and PDF1.2b (At2g26020), also responded specifically to strain C58. PR4 (At3g04720), PYK10 (At3g09260), encoding a β-glycosidase putatively involved in defense responses, and three chitinases (At2g43610, At2g43620, and At2g43570) exhibited increased transcription in response to both agrobacteria strains (see Supplemental Data Set 2, data sheet 1, online). The number of differentially expressed genes related to pathogen defense increased further during tumor development (Figure 4B, biotic stress category). In fully developed tumors (35 d postinoculation), 28 genes were upregulated, and the transcription of 14 genes was downregulated. Among the latter were proteinase inhibitors and several of the coiled coil or toll/interleukin1 receptor nucleotide binding site leucine-rich repeat class of disease resistance (R) genes. R genes are known to monitor the action of isolate-specific pathogen effectors and can trigger hypersensitive response (Robatzek and Saijo, 2008). In summary, genes involved in pathogen defense signaling were expressed at all time points analyzed but responded predominantly to the oncogenic strain C58. Moreover, the genes affected by both strains are candidates for genes that may respond to agrobacterial effectors, rather than to the T-DNA–encoded oncogenes.
Figure 4.
Number of Arabidopsis Genes Either Up- or Downregulated within the Indicated Functional Categories According to MapMan (http://gabi.rzpd.de/projects/MapMan, Version 2.2.0, July, 2008).
The following experiments are presented: (A) Differentially expressed genes of inflorescence stalks treated with Agrobacterium strain C58 (3 h postinoculation [hpi] C58) or GV3101 (3 hpi GV3101) for 3 h or 6 d (6 d postinoculation [dpi] C58 or 6 dpi GV3101), as well as of (B) mature tumors induced by strain C58 (35 dpi tumor). The category “biotic stress” comprises pathogen defense genes, that of “oxidative stress” genes involved in redox regulation, those of phytohormone metabolism categories genes involved in phytohormone biosynthesis and degradation, and phytohormone signaling included all genes involved in perception or in signaling or are activated by the respective phytohormone. The category “cell wall” encompasses genes involved in cell wall synthesis, degradation, and modification and the “RNA” category mainly transcription factors as well as genes involved in RNA transcription, processing, and binding. The genes of the categories “DNA” and “protein” are involved in DNA and protein modifications. Annotated genes are listed in Supplemental Data Set 2 online.
To identify the Arabidopsis genes that have been shown to respond to the agrobacterial effector elf26, we compared our 3 h postinoculation microarrays with those treated with an elf26 peptide (Zipfel et al., 2006). This peptide derivative represents a fragment of the elongation factor EF-Tu, a highly conserved motif of one of the most abundant proteins in microbes, including agrobacteria. Elf26 peptides induce PAMP-triggered innate immunity responses, associated with disease resistance in Arabidopsis (Schwessinger and Zipfel, 2008). The comparison of differentially expressed genes revealed that 28 out of the 35 Arabidopsis genes affected by strain C58 responded in a similar manner to elf26 (see Supplemental Data Set 1, data sheet 6, online). The eight genes influenced by strain GV3101 in Arabidopsis at initiation of infection responded to this PAMP, too. The elf26 peptide induces 948 Arabidopsis genes (Zipfel et al., 2006), while the virulent Agrobacterium strain C58 induces expression of just 35 genes. These data suggest that agrobacteria, like other microbes, seem to be able to dampen host responses.
H2O2 Accumulation Is Prevented at the Beginning of the Infection and Transformation Process
Reactive oxygen species such as hydrogen peroxide (H2O2) act as messengers in signaling cascades activated by diverse external stimuli, such as wounding or pathogen attack. Inflorescence stalks of Arabidopsis synthesize H2O2 3 h and 6 d after wounding as indicated by diaminobenzidine (DAB) staining (Figures 1C and 1E). A reddish-brown precipitate caused by H2O2 accumulation was generated in wounded areas. In wounded inflorescence stalks with agrobacteria, however, no H2O2 was detected at 3 h postinoculation (Figure 1D) and 6 d postinoculation (Figures 1F and 1G). Strong DAB staining was observed in tumors (Figure 1H), indicating that agrobacteria were able to suppress H2O2 accumulation early, but not late, in the infection process.
Several genes encoding enzymes that function in the cellular protection against oxidative stress and toxic compounds were upregulated at the three time points analyzed (Figures 4A and 4B, oxidative stress). While the oncogenic strain C58 activates transcription of the glutathionine S-transferase gene, GSTU24 (At1g17170), and two peroxidases (At4g08770 and At5g64120) as early as 3 h postinoculation, strain GV3101, lacking a T-DNA, did not (see Supplemental Data Set 2, data sheet 2, online). Finally, in tumors, some genes involved in oxidative stress were transcriptionally activated, but several were also downregulated (Figure 4B, oxidative stress category).
The Phytohormones Auxin, ET, and SA, Rather Than JA Regulate Tumor Development
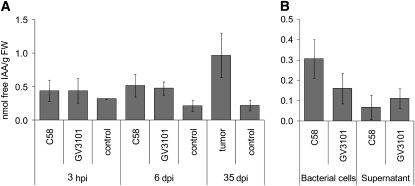
The phytohormones indole-3-acetic acid (IAA), JA, ET, and SA play a role in plant–pathogen interactions and trigger the expression of defense genes. We thus monitored the phytohormone levels throughout infection. We found that free IAA was not significantly higher in infected plants than in mock-treated plants at 3 h postinoculation but was elevated more than twofold at 6 d postinoculation in response to strain C58 or GV3101 (Figure 5A). Mature tumors accumulated 4.4 times more auxin compared with control tissue. The virulent strain C58 was previously reported to elicit production of twice as much auxin as a strain without T-DNA–encoded oncogenes (Kutacek and Rovenska, 1991). We confirmed this finding under our experimental settings (C58, 0.31 nmol/g fresh weight versus GV3101 and 0.16 nmol/g fresh weight). In addition, both strains secreted auxin into the culture medium (Figure 5B).
Figure 5.
Content of Free IAA.
(A) Arabidopsis inflorescence stalks (ecotype Ws-2) harvested 3 h (3 h postinoculation [hpi]), 6 d (6 d postinoculation [dpi]), or 35 dpi with either Agrobacterium strain C58 or GV3101 were compared with wounded but not inoculated stalks (control). Results are given in nmol per g fresh weight (FW).
(B) Pellet and supernatant of strain C58 and GV3101 grown overnight in rich medium (YEB). Bars represent mean values (±sd) of three independent experiments.
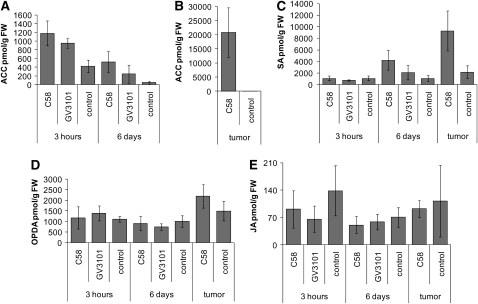
The precursor of ET, 1-amino-cyclopropane-1-carboxylate (ACC), was elevated 2.8-fold and 2.2-fold upon infection with Agrobacterium strain C58 and GV3101, respectively, already at 3 h postinoculation. At 6 d postinoculation, the levels of ACC were significantly higher than in wounded plants when strain C58, but not when strain GV3101 was inoculated (Figure 6A), whereas in tumors, ACC accumulated to exceptionally high levels (Figure 6B). SA levels increased fourfold at 6 d upon infection with the oncogenic strain C58 and 4.3-fold in fully developed tumors (Figure 6C). By contrast, neither the levels of JA nor of its precursor 12-oxo-phytodienoic acid (OPDA) were significantly different at any stage of the Arabidopsis–Agrobacterium interaction (Figures 6D and 6E). This indicates that the signaling molecule ET plays a role before and after T-DNA integration and SA only after T-DNA integration. JA, by contrast, does not seem to act as signaling molecule during the time course of tumor development in the Arabidopsis–Agrobacterium interaction.
Figure 6.
Content of Signaling Molecules Involved in Pathogen Defense after Inoculation of Agrobacterium.
Arabidopsis inflorescence stalks were inoculated with either strain C58 or GV3101 for 3 h, 6 d, and 35 d (tumor).
(A) and (B) Levels of ACC, a precursor for ET biosynthesis. Note the different scale of the ordinate in graph (B).
(C) to (E) Levels of SA (C), OPDA (D), a precursor of JA biosynthesis, and JA (E). Results are given in pmol per g fresh weight (FW). Bars represent mean values (±sd) of five independent experiments.
To extend this analysis of signaling molecule accumulation, we also examined the transcription of genes implicated in the synthesis, modification, and/or perception of these signals. At initiation of infection, transcription of host genes involved in IAA, JA, and SA metabolism were not elevated (Figure 4A). Two genes, encoding enzymes for auxin/camalexin biosynthesis (CYP71A13, At2g30770; CYP17B2, At4g39950) were found to be induced only at 6 d postinoculation by both Agrobacterium strains and in mature tumors. This correlated with an increase in auxin levels (Figure 5A) and also with higher T-DNA–encoded oncogene transcript levels of iaaH (Figure 2). Thus, auxin derived from the T-DNA–encoded oncogenes augments the endogenous host auxin levels. Consistent with our measurements of elevated ACC levels, transcripts of genes involved in ACC (ASC6, At4g11280; ASC8, At4g37770) or ET biosynthesis (ACO1, At2g19590) were elevated at all time points only in response to the oncogenic strain C58. The elevated SA levels at 6 d postinoculation and in tumors were accompanied by the induction of two genes coding for Adenosyl-l-methionine:salicylic acid carboxyl methyltransferases (At5g38020 and At1g66690), engaged in SA methylation (Ross et al., 1999). Genes involved in SA biosynthesis, such as phenylalanine ammonia-lyases or isochorismate synthases (ICS), remained unchanged. This may point to regulation of SA biosynthesis on the posttranscriptional level. EDS5-mRNA (At4g39030) coding for a multidrug and toxin extrusion transporter that is known to be involved in pathogen-dependent accumulation of SA (Nawrath et al., 2002) was found to be elevated in tumors (see Supplemental Data Set 2, data sheet 10, online).
Our genome-wide expression studies of Arabidopsis genes involved in phytohormone signal transduction revealed that four auxin-inducible genes were already induced at 3 h postinoculation; these included two of the early auxin-responsive GH3 family (GH3.3, At2g23170; GH3.5/WES1, At4g27260) involved in auxin inactivation by conjugation, the auxin-responsive transcriptional regulator IAA5 (At1g15580), and the auxin-inducible ACC synthase 8 (ACS8, At4g37770; see Supplemental Data Set 2, data sheet 4, online). After T-DNA integration, at 6 d postinoculation, 10 genes involved in auxin signaling were transcriptionally activated by strain C58 and only three by strain GV3101 (Figure 4A). In 35-d-old tumors, 29 IAA signaling-related genes responded to the presence of strain C58 (Figure 4B). PR genes and distinct genes of the phytohormone pathways are known as markers of the classical stress response in host plants, such as VSP2 (At5g24770) for JA, PDF1.2 (At5g44420) and PR3 for JA/ET, PR4 for ET, and PR1 (At2g14610) and PR2 (At3g57260) for SA. Among them only genes of the ET signaling pathway, PDF1.2, PR3 (confirmed by qRT-PCR; Figure 3A), and PR4, were induced at 6 d postinoculation (but not at 3 h postinoculation) by the oncogenic strain C58. PR4 was the only gene that responded to both strains at 6 d postinoculation. In addition to the classical PR genes, the transcription of genes encoding the ET receptor ETR2 (At3g23150) and an ET response factor (At5g25190) responded only to strain C58 after T-DNA integration (see Supplemental Data Set 2, data sheet 6, online). In the tumor, several genes involved in ET perception and signaling were activated. By contrast, genes involved in JA signaling, such as JAZ family members, COI1 (At2g39940; Katsir et al., 2008; Staswick, 2008), or MYC2 (At1g32640; Lorenzo et al., 2004) as well as the two classical marker genes of the SA-induced SAR response pathway, PR1 (At2g14610; Laird et al., 2004) and NPR1 (At1g64280), were never found to be activated. This observation suggests that the auxin and ET signaling pathways, rather than SA-induced SAR, seems to be induced in the host during Arabidopsis–Agrobacterium interaction.
Mutant Plants with High SA Levels Are Resistant to Agrobacterium, while Those with Low Levels Promote Tumor Growth
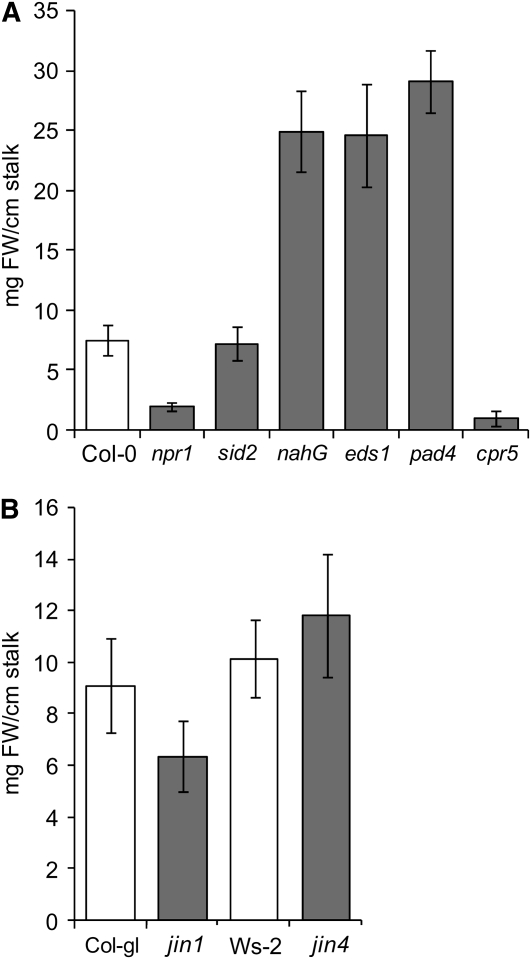
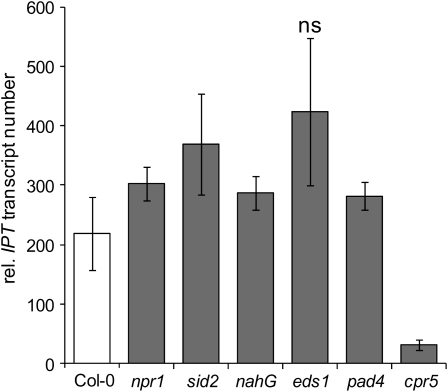
Since SA accumulated at 6 d postinoculation and in tumors, mutants and transgenic plants impaired in SA-biosynthesis, accumulation, and signaling were analyzed for tumor formation ability. Inflorescence stalks were inoculated with the tumor-inducing strain C58. Tumor growth on SA-deficient nahG plants (van Wees and Glazebrook, 2003) was increased by ∼3.4-fold compared with the wild type Columbia-0 (Col-0) (Figure 7A). Similarly, eds1 and pad4 mutant plants, with defects in SA accumulation upon pathogen attack, were also more susceptible to tumor growth (3.8- and 3.2-fold, respectively). Tumor development on the SA biosynthesis mutant sid2 (Nawrath and Metraux, 1999), which lacks a functional ICS1 and is unable to accumulate SA after infection with pathogens, was indistinguishable from that of the wild type Col-0. The SA signaling mutant, npr1, which accumulates higher levels of SA as the wild type upon infection with avirulent bacteria (Shah et al., 1997), developed much smaller tumors. Furthermore, on cpr5 plants, a mutant with high levels of SA (Bowling et al., 1997) and constitutive SA- and ET/JA-induced PR gene expression (Clarke et al., 2000) tumor growth was strongly impaired. Only four out of 22 cpr5 plants analyzed developed very small tumors compared with the parental line Col-0 (Figure 7A). We determined the transcript numbers of the T-DNA–encoded ipt gene upon inoculation of npr1, sid2, nahG, eds1, pad4, and cpr5 plants with the oncogenic strain C58 6 d postinoculation. Transcription of the ipt gene was strongly repressed in cpr5 plants only (Figure 8). npr1, sid2, nahG, eds1, or pad4 plants expressed similar numbers of ipt transcripts as the wild type Col-0. Thus, in plants with constitutively high levels of SA, tumor growth is impaired, particularly if pathogen defense signaling is also activated as in the cpr5 mutant.
Figure 7.
Tumor Development on Inflorescence Stalks of Arabidopsis Mutant Plants Impaired in SA, ET, and JA Signaling or Biosynthesis Pathways.
Tumor development was induced on (A) plants with altered levels of SA (sid2, nahG, eds1, and pad4) and/or SA-mediated signaling (npr1 and cpr5) as well as on (B) JA/ET-signaling mutants (jin1 and jin4). Tumor development was induced on wounded inflorescence stalks upon inoculation with Agrobacterium strain C58. Tumors were removed from the inflorescence stalks after 35 d and weighed separately (mg fresh weight [FW] per cm stalk). Values represent means of n = 33 npr1, n = 14 nahG, n = 19 eds1, n = 19 pad4, n = 12 sid2, n = 22 cpr5, n = 19 jin1, n = 10 jin4, n = 15 Ws-2, n = 21 Col-gl, and n = 22 Col-0 plants (±sd).
Figure 8.
Relative Expression Levels of the Agrobacterial Oncogene ipt in Arabidopsis Wild-Type and Mutant Plants.
Inflorescence stalks of mutant or transgenic Arabidopsis plants with altered levels of SA (sid2, nahG, eds1, and pad4) and/or SA-mediated signaling (npr1 and cpr5) were wounded and inoculated with Agrobacterium strain C58 for 6 d. Transcript numbers were quantified with real-time PCR and normalized relative to 10,000 molecules of ACTIN2/8. Numbers represent means of four independent experiments ±sd. ns, not significant (P > 0.05) according to one-way analysis of variance with Bonferroni post-hoc test.
To elucidate the impact of JA on tumor development, we analyzed tumor growth on jin1 (JASMONATE INSENSITIVE1, MYC2) and jin4 (jar1-1) mutants. The mutant jin1 lacks the basic helix-loop-helix leucine-zipper transcription factor MYC2 (Lorenzo et al., 2004), whereas jin4 is impaired in conjugation of JA to an amino acid (Staswick and Tiryaki, 2004). However, none of the JA-insensitive mutants had a significant effect on tumor growth (Figure 7B).
DISCUSSION
Agrobacterium-mediated transformation of the plant cell has been extensively studied (Gelvin, 2003; Citovsky et al., 2007), whereas a comprehensive knowledge about plant host defense responses to agrobacteria is still limited. In recent years compelling evidence has demonstrated that reactive oxygen species like hydrogen peroxide (H2O2) and hormones such as SA, JA, and ET are the primary signals inducing defense responses through recognized defense signaling pathways (Lopez et al., 2008). When plants encounter an invading pathogen, gene expression and responses signaled through defense hormones are activated to restrict pathogen invasion. In addition, pathogens also trigger the modulation of pathways involving hormones that control developmental processes, such as auxin and cytokinin.
We have focused our studies of Agrobacterium-induced defense responses in Arabidopsis on three time points with quite a time interval that revealed distinct differences in signaling molecule levels (Figures 5 and 6) and gene expression profiles (see Supplemental Figures 4A and 4B online). These differences revealed that the host responded to both agrobacterial strains as early as 3 h postinoculation and that the oncogenic strain C58 incites a much stronger pathogen defense response than strain GV3101 at both early time points. Furthermore, our studies documented that the development of Agrobacterium-induced tumors on Arabidopsis was accompanied by elevated production of abscisic acid (ABA) (Efetova et al., 2007), IAA, SA, and ET, but not JA. This was in agreement with previous findings that the development of Agrobacterium-induced tumors on Ricinus communis involves the production of the phytohormones JA, auxin, cytokinin, ET, and ABA (Veselov et al., 2003). These phytohormones are successively required for vascularization and successful tumor development. Our hormone and transcriptome profiles revealed that the IAA and ET phytohormone pathways are important at initiation of the Arabidopsis–Agrobacterium interaction. After T-DNA integration, the host accumulated SA, which controls tumor formation. The observation that the oncogenic strain C58 incites a much stronger pathogen defense response than strain GV3101 without T-DNA (see Supplemental Figure 4 online) indicates that the T-DNA–encoded gene products are involved in the induction of host defenses against agrobacteria.
Auxin and ET Are Involved in the Initiation of Infection with Agrobacteria
Three hours postinoculation with Agrobacterium strain C58 or GV3101, very few genes involved in pathogen defense were induced and may reflect the activation of innate immunity responses in host plants to PAMPs (see Supplemental Data Set 1, data sheet 6, online). The PAMP elf26, however, induces a much stronger response in Arabidopsis when applied as pure peptide; 948 differentially expressed genes according to Zipfel et al. (2006) versus 35 and eight genes were transcriptionally affected by strain C58 and GV3101, respectively. Thus, agrobacteria seems to be able to dampen host responses elicited by the general PAMP. Furthermore, the H2O2 signal appeared to be suppressed. At initiation of infection, the agrobacterial catalase (Xu and Pan, 2000) seems to degrade H2O2 produced by the host plant. Later on, at 6 d postinoculation, transcription of several peroxidases and to GSTs might prevent H2O2 accumulation (see Supplemental Figures 3C and 3D online). PAMP-triggered immunity, activated by P. syringae in Arabidopsis, involves the mitogen-activated protein kinase pathway, which includes MPK3 and MPK6 (Bethke et al., 2009; Boller and He, 2009). These genes were not affected by the virulent Agrobacterium strain C58.
An independent investigation demonstrated that defense response genes were activated in tobacco (Nicotiana tabacum) BY2 cell suspensions upon infection with different Agrobacterium strains within 3 to 6 h (Veena et al., 2003). With the onset of T-DNA transfer between 1 and 2 d after infection, the transcription of these defense genes decreased in BY2 cells. At the time point of T-DNA integration (6 d postinoculation) in Arabidopsis plants, however, the number of defense genes affected increased fourfold, with a fold change in transcription comparable to that at 3 h postinoculation. In a microarray-based study with suspension-cultured cells of Arabidopsis infected with the wild-type Agrobacterium strain A348, transcript levels did not change within 4 to 24 h (Ditt et al., 2006). Forty-eight hours after infection, genes involved in phytohormone signaling biosynthesis were either downregulated or less than twofold induced (auxin, ET, and ABA). The discrepancy between the two previous studies and ours may originate from the different plant systems used. Veena et al. (2003) and Ditt et al. (2006) worked with suspension cultured cells with high transformation efficiency, while we inoculated intact Arabidopsis plants, which do not require phytohormone pretreatment and allow monitoring of induced hormone profiles.
The levels of the ET precursor, ACC, were already elevated at 3 h postinoculation (Figure 6A), very likely representing a wounding response (Boller and Kende, 1980). Higher levels of ET in inoculated tissues compared with the control at 3 h postinoculation, however, seem to originate from the agrobacterial infection. The overproduction of ACC in infected Arabidopsis tissues correlated with the induction of genes involved in ACC biosynthesis, like ACS6 and ACS8, by the T-DNA–harboring strain C58 exclusively. This strain contained twice as much auxin compared with strain GV3101 (Figure 5B). Since auxin is known to induce the transcription of ACS8 and the conversion of Met to ET (Yu and Yang, 1979), Arabidopsis-derived auxin together with the auxin released by agrobacteria may stimulate ACC biosynthesis. Surprisingly, genes encoding ET receptors, ET-responsive factors, or PR proteins, which are marker genes for ACC treatment (van Loon et al., 2006b), were not activated at initiation of infection. This may indicate that ET is not sensed or signaled by the host at this time point of agrobacterial infection (3 h postinoculation). Instead, ET may control agrobacterial virulence. Recently it was reported that ET production in plants suppresses vir gene expression in Agrobacterium during the course of transformation (Nonaka et al., 2008). These and our data indicate that the host plant is capable of controlling vir gene expression already at the start of infection.
In addition to ET, auxin also affects pathogen virulence (Spoel and Dong, 2008). Several bacteria and fungi produce auxin to modulate the hormone balance of their host. Auxin promotes susceptibility to the bacterial disease (Robert-Seilaniantz et al., 2007; Lopez et al., 2008). Exogenous application of synthetic auxin to plants enhances susceptibility to P. syringae, whereas mutant plants impaired in auxin signaling exhibit enhanced resistance (Navarro et al., 2006; Chen et al., 2007; Wang et al., 2007). At initiation of infection, the amount of auxin produced by oncogenic agrobacteria may perturb the balance of this phytohormone in infected Arabidopsis tissues. Conjugation of free auxin has been proposed to function in permanent inactivation and temporary storage of auxin as well as in detoxifying excess IAA and protecting the free acid against peroxidative degradation, whereas none of the genes involved in hydrolysis of auxin conjugates generates bioactive free IAA (LeClere et al., 2002). Two genes of the early auxin-responsive GH3 family (GH3.3 and GH3.5/WES1), which are involved in conjugating free auxin, were found to be induced, but none were involved in IAA hydrolysis (see Supplemental Data Set 2, data sheet 4, online). Preliminary studies revealed that the vast majority of auxin measured at the three time points in the course of Arabidopsis tumor development was conjugated and thus inactive (J. Ludwig-Müller, unpublished results). Furthermore, free IAA has been proposed to stimulate the frequency of tumor induction (Morris, 1986). In our plant samples treated with agrobacteria for 3 h, the levels of free IAA were slightly, although not significantly, increased (Figure 5A). Therefore, it seems likely that the additional auxin released by agrobacteria at the beginning of infection may be counteracted by the host expression of GH3 genes in order to dampen tumor induction. However, further studies with Arabidopsis auxin biosynthesis and signaling mutants are required to confirm this hypothesis.
After T-DNA Integration and in the Tumors Auxin, SA, ET, and H2O2 Control Tumor Development
At 6 d postinoculation, when the T-DNA–encoded transcripts ipt and iaaH markedly increased in Arabidopsis inflorescence stalk tissue, the levels of IAA (Figure 5A), ET, and SA, but not JA (Figure 6), increased. The transcription of several genes involved in pathogen defense, IAA and ET, but not SA, signaling was also higher. More genes responded to strain C58 compared with the T-DNA–deficient strain GV3101 (Figure 4A). This suggested that the host strengthens pathogen defense signaling more strongly in response to T-DNA transformation and expression of oncogenes.
The lack of upregulation of genes encoding JA signaling components, such as COI1 and JAZ proteins in response to strain C58, correspond to the unchanged JA levels. Usually an elevated JA level is reflected in upregulation of these genes by a positive feedback loop in JA biosynthesis and signaling (Wasternack, 2007).
SA plays a central role in plant disease resistance. Exogenous application of SA induces a set of PR genes that leads to SAR (Uknes et al., 1992). However, in our system, the elevated levels of SA and S-adenosyl-l-methionine:carboxyl methyltransferase transcripts at 6 d postinoculation in C58-infected tissues and in tumors did not cause activation of the PR genes known to be markers for SAR signaling. Treatment of plants with synthetic auxin was recently shown to repress defense genes induced by SA (Wang et al., 2007). This might explain the lack of induction of any gene involved in SA-dependent signaling in Agrobacterium-infected Arabidopsis tissues, despite the accumulation of SA (Figure 6C). The elevated auxin levels at 6 d postinoculation and in tumors may promote agrobacteria invasion by suppression of SA-mediated host defenses. Recently it has been shown that SA has an inhibitory effect on virulence of agrobacteria (Yuan et al., 2007; Anand et al., 2008). SA directly inhibits the expression of the vir regulon of the Ti plasmid that is essential for the transfer and integration of the T-DNA into the host genome. In our studies, SA accumulated only after T-DNA integration (6 d postinoculation) when the virulent strain C58 was inoculated (Figure 6C). It seems likely that the elevated SA levels in plants infected with agrobacteria do not induce pathogen defense signals of the host. Instead, SA may exert a direct effect on the Agrobacterium's virulence machinery. This was also supported by our findings and those of others (Yuan et al., 2007) that mutant plants with low SA levels promote, whereas those with high SA levels inhibit, tumor growth (Figure 7A). Furthermore, constitutively high levels of SA and PR gene expression interfered with the expression of oncogenes, since the cpr5 mutant expressed only very low numbers of ipt transcripts at 6 d postinoculation compared with wild-type plants (Figure 8). Taken together, these data indicate that plants challenged with agrobacteria accumulate SA after T-DNA integration, which acts directly on oncogenic agrobacteria leading to reduced virulence.
For successful tumor development, morphological adaptations are essential: these include neovascularization to supply the growing tumor with nutrients (Ullrich and Aloni, 2000) and suberization of the outer cell layers to protect the disrupted tumor surface against drought stress (Efetova et al., 2007). Hydrogen peroxide production in fully developed tumors (Figure 1H) and the expression of 34 genes involved in oxidative stress (Figure 4B) are at least partly associated with suberization of outer tumor cell layers. The polymerization of suberin monomers involves peroxidases for which H2O2 is the electron donor. Suberization together with ET-triggered ABA production (Veselov et al., 2003) induces drought protective mechanisms in tumors (Efetova et al., 2007).
It has been demonstrated that auxin and ET control vascular development in plants as well as in tumors (Aloni et al., 1998, 2003), and it is well known that high auxin levels induce ET emission. Besides their action in pathogen defense during initiation of infection with agrobacteria, this may imply an additional role for these phytohormones in regulating vascular development at later stages of tumor development. This hypothesis is substantiated by the expression of several transcription factors involved in vascular development (e.g., MONOPTERUS/IAA24, At1g19850; DOF2.5, At2g46590; REVOLUTA, At4g32880) only in tumors (see Supplemental Data Set 1, data sheet 1, online).
CONCLUSIONS
The comparison of gene expression patterns between an oncogenic and a nononcogenic strain revealed a highly specific response for the oncogenic strain, whereas most of the genes regulated in the interaction with the nononcogenic strain GV3101 could be attributed to PAMP signaling. Furthermore, our findings suggest that auxin modulates ET- and SA-dependent responses during Arabidopsis–Agrobacterium interaction. Genes involved in auxin biosynthesis and signaling responded only to the oncogenic strain C58. This Agrobacterium strain produces twice as much auxin and induces auxin production in the host after oncogene expression. At initiation of infection (3 h postinoculation), auxin may stimulate pathogen defense by promoting ET production, which did not induce ET-dependent signaling in the host. Instead, ET may reduce virulence of agrobacteria as was recently suggested by Nonaka et al. (2008). At this stage of Arabidopsis–Agrobacterium interaction, morphological adaptations are not yet initiated. In this model, at later stages of T-DNA-transfer and integration (6 d postinoculation) when morphological adaptations begin, auxin again stimulates ET production which is then, together with auxin, required to induce vascular differentiation. At the same time, the levels of SA increase, but SA-dependent signaling in the host is not activated. It is possible that SA signaling is repressed by auxin. SA and ET seem to control agrobacterial virulence and thereby T-DNA transfer and integration. The sequential and concerted action of IAA, ET, and SA may stabilize a balance between pathogen defense launched by the host and tumor growth initiated by agrobacteria throughout the course of tumor development. This balance allows a long-term existence of agrobacteria in the host and prevents an uncontrolled growth of crown galls on the host plant.
METHODS
Arabidopsis thaliana Ecotypes, Agrobacteria Strains, and Inoculation Procedure
Plant cultivation and tumor induction were performed as described by Deeken et al. (2003). Wild-type Arabidopsis (cv Ws-2; Col-0; Col-gl1) and mutant plants (jin1, jin4, pad4, eds1, nahG, sid2, npr1, and cpr5) were inoculated at the base of inflorescence stalks. Plants were infected with either the nopaline-using Agrobacterium tumefaciens strain C58 nocc (nopaline catabolism construction, number 584; Max Planck Institute for Plant Breeding, Cologne, Germany) or with the nontumorigenic Agrobacterium strain GV3101 (pMP90) lacking the T-DNA but not vir genes (Holsters et al., 1980; Koncz and Schell, 1986). Agrobacteria were cultivated on YEB-agar plates (0.5% [w/v] yeast extract, 0.5% [w/v] tryptone, 0.5% [w/v] sucrose, 50 mM MgSO4, and 1.5% agar, pH 7.0) overnight. Bacteria were scratched from the surface of the plate and directly transferred onto wounded inflorescence stalk areas, thereby preventing contamination with YEB medium. Acetosyringone was not added because agrobacterial virulence should only be induced by host factors at the time point of inoculation. Inoculated and wounded, but noninfected, inflorescence stalk areas were collected at 1, 3, 6, 12, and 24 h postinoculation and 2, 4, 6, and 8 d postinoculation, frozen in liquid nitrogen, and stored at −80°C.
Characterization of H2O2 Formation
DAB staining for H2O2 detection was performed as described (Thordal-Christensen et al., 1997).
RNA Isolation, Reverse Transcription, and qRT-PCR
Total RNA was isolated with the RNeasy plant mini kit (Qiagen) and treated with DNase I (3 units/μL; AppliChem) according to the manufacturer's protocols to remove any DNA contamination. Single-stranded cDNA was synthesized with Superscript RT (Gibco BRL) from 2.5 μg of total RNA. For quantification, 2 μL of a 1:20 dilution of the single-stranded cDNA reaction mix in water was used. PCRs were performed by applying a LightCycler carousel-based system and the LightCycler-FastStart DNA Master SYBR Green I kit (Roche Molecular Biochemicals). With this system, PCR products were detected fluorescently, using the intercalating fluorescent dye SYBR Green I. The increase in fluorescent signal, measured at each amplification cycle, correlates with the amount of PCR product formed. Relative concentrations of cDNA present during the exponential phase of the reaction were determined by plotting fluorescence against cycle number on a logarithmic scale. The initial template concentration of each sample was calculated by LightCycler software (Roche Molecular Biochemicals). Amounts of the cDNA of interest were determined by comparing the results to a standard curve produced by real-time PCR of serial dilutions from (20, 2, 0.2, and 0.02 fg/μL) of a known amount of that cDNA (10 ng/μL) as external standard. To normalize for possible variation in the amount and quality of cDNA between different samples, the Arabidopsis housekeeping genes ACTIN2 and ACTIN8 served as internal standard. Their cDNA concentrations were determined in parallel in each sample. All transcript numbers were finally normalized to 10,000 molecules of ACTIN2/8. All relative transcript numbers represent the mean calculated from three to five independent experiments. For each experiment, tumor material from a minimum of five plants was used. Error bars represent mean values ±se. Primers used for qRT-PCR are listed in Supplemental Table 1 online.
Microarray Analysis
In general, microarrays (ATH1; Affymetrix) were analyzed as described by Deeken et al. (2006), with the statistical software R (R Development Core Team, 2009; http://www.R-project.org) and add-on packages for microarray analysis from Bioconductor (Gentleman et al., 2004). For normalization, the variance stabilization algorithm by Huber et al. (2002) was applied. The normalization procedure was slightly adapted compared with the one described by Deeken et al. (2006) because the microarrays were not all hybridized at the same time. The first set of microarrays hybridized consisted of three chips for each of the two experiments: (1) 3 hpi C58, (2) 3-h control, and (3) all but one control of the 6-d time point. The second set again consisted of three microarrays for each of the three experiments: (1) 3 hpi C58, (2) 3 hpi GV3101, (3) 3-h controls, and one control array of 6 d. For each of the two experimental time points (3 h postinoculation and 6 d postinoculation), the first set of microarrays was normalized, and their model parameters were used to normalize the second data set of that time point to reduce any influence that could arise from the two time points of hybridization as described in the following package documentation: http://bioconductor.org/packages/2.4/bioc/vignettes/vsn/inst/doc/likelihoodcomputations.pdf. Thus, the first data set served as reference data set to normalize the one from the second time point. To summarize individual probes of an Affymetrix probe set, the median polish algorithm from the RMA normalization (Robust Multichip Average) was used (Irizarry et al., 2003). Differentially expressed genes were assessed with a linear model approach implemented in the R package LIMMA (Linear Models for Microarray Analysis; Smyth, 2004). Two separate models for the two time points, 3 h postinoculation and 6 d postinoculation, were fit with the coefficients C58, GV3101, and control for each probe set. In contrast with the procedure described by Deeken et al. (2006), microarray weights according to Ritchie et al. (2006) were applied for analysis of differential gene expression, giving higher weights to microarrays that better fit the linear models for individual probe sets. To estimate microarray weights, the function arrayWeights from the LIMMA package fits a heteroscedastic model for the expression values of each probe set. In the heteroscedastic model, variance depends on a probe set and the microarray. The inverse of the microarray variance factor is then used as microarray quality weight. All P values of the differential gene expression analysis were corrected for multiple testing with the false discovery rate from Benjamini and Hochberg (2000).
Extraction and Quantitative Analysis of JA and OPDA
Plant material pooled from at least 20 plants was taken in triplicates, frozen in liquid nitrogen, and stored until use. For extraction, 0.5 g fresh weight of each tissue was homogenized in a mortar under liquid nitrogen and extracted with 10 mL methanol and appropriate nanograms of (2H6)JA or (2H5)OPDA as internal standards. The homogenate was filtered, and the elute was evaporated and acetylated with 200 μL pyridine and 100 μL acetic acid anhydride at 20°C overnight. The extract was dried, dissolved in 2 mL ethyl acetate, passed through a chromabond-SiOH column, 500 mg (Macherey-Nagel), and the column was washed with further 3 mL ethyl acetate. Combined liquids were evaporated and dissolved with 10 mL of methanol and placed on a column filled with 3 mL DEAE-Sephadex A25 (Amersham Pharmacia Biotech) (Ac—-form, methanol); the column was washed with 3 mL methanol. After washing with 3 mL 0.1 M acetic acid in methanol, eluents with 3 mL of 1 M acetic acid in methanol and 3 mL of 1.5 M acetic acid in methanol were collected, evaporated, and separated on preparative HPLC using a Eurospher 100-C18 column, (5 μm, 250 × 4 mm), solvent A: methanol, solvent B: 0.2% acetic acid in water and a gradient of 40% solvent A and 60% B to 100% solvent A in 25 min. Fractions at retention times of 13 to 14.5 min (JA) and 21.75 to 22.5 min (OPDA) were combined and evaporated. Evaporated samples from the HPLC were dissolved in 200 μL CHCl3/N,N-diisopropylethylamine (1:1,v/v) and derivatized with 10 μL pentafluorobenzylbromide at 20°C overnight. After evaporation, samples were dissolved in 5 mL n-hexane and passed through a Chromabond-SiOH column, 500 mg (Machery-Nagel). The pentafluorobenzyl esters were eluted with 7 mL n-hexane/diethylether (1:1, v/v). Eluates were evaporated, dissolved in 100 μL CH3CN, and analyzed by gas chromatography–mass spectrometry (GC-MS).
Extraction, Purification, and Quantification of SA
Fresh plant material (10 to 500 mg) was homogenized with 10 mL methanol and appropriate amounts of (2H6)SA (500 ng per 500 mg plant material) as internal standard (Campro Scientific). The homogenate was centrifuged and the pH was adjusted to alkaline conditions by addition of NH4OH and subsequently evaporated. Samples were dissolved in 500 μL methanol and 5 mL water, and the solution was passed through a LiChrolut C18 cartridge (500 mg; Merck). The pH of the eluent was adjusted to alkaline conditions with NH4OH and the sample evaporated and separated by preparative HPLC.
HPLC (Eurospher 100-C18; 5 μm, 250 × 4 mm; Knauer) was performed with solvent A (methanol) and B (0.2% [v/v] acetic acid in water) and a gradient of 40% A and 60% B to 100% A in 25 min. Fractions from 8.00 to 10.00 min were collected, and the pH was adjusted to alkaline conditions by addition of ∼50 μL N,N-diisopropylethylamine, evaporated, and derivatized to pentafluorobenzyl esters (PFB-esters).
For derivatization, evaporated samples were dissolved in 200 μL chloroform/N,N-diisopropylethylamine (1:1; v/v) and derivatized with 10 μL pentafluorobenzylbromide at 50°C for 1 h. The evaporated samples were dissolved in 5 mL n-hexane and passed through a Chromabond-SiOH column, 500 mg (Machery-Nagel). The pentafluorobenzyl esters were eluted with 7 mL n-hexane/diethylether (1:1) for SA-PFB esters. Elutes were evaporated up to the last traces of solvent, dissolved in 100 μL acetonitrile, and analyzed by GC-MS.
For GC-MS (Polaris Q; Thermo-Finnigan), the following conditions were used: 100 eV, negative chemical ionization, ionization gas NH3, ion source temperature 200°C, column Rtx-5MS (Restek), 15 m × 0.25 mm, 0.25 μm film thickness, cross-bond 5% diphenyl to 95% dimethyl polysiloxane, injection temperature 220°C, interface temperature 250°C; helium 1 mL min−1; splitless injection; the column temperature program was: 1 min 60°C, 25°C min−1 to 180°C, 5°C min−1 to 270°C, 10°C min−1 to 300°C, 10 min 300°C; the retention times of PFB-esters were: SA 6.99 min, (2H4)SA 6.96 min; MS fragments at m/z 137 (native SA) and at m/z 141 (standard) were used for quantification
Extraction, Purification, and Quantification of ACC
Plant material (10 to 500 mg) was homogenized with 10 mL methanol and appropriate amounts (1 ng standard per 5 mg plant material) of (2H4)ACC (CDN Isotopes) as internal standard. The homogenate was filtered and placed on a column filled with 3 mL DEAE-Sephadex A25 (Amersham Pharmacia Biotech). The column was washed with 3 mL methanol and 3mL of 0.1 n acetic acid in methanol. Combined elutes were evaporated and resuspended in 5 mL water using short ultrasonification and were subsequently passed through a 500 mg LiChrolut RP-18 cartridge (Merck). The elute was evaporated and derivatized into PFB-amid-PFB-esters.
For derivatization, evaporated samples were dissolved in 200 μL chloroform3/N,N-diisopropylethylamine (1:1, v/v) and derivatized with 10 μL pentafluorobenzylbromide at 50°C for 1 h. The evaporated samples were dissolved in 5 mL n-hexane and passed through a Chromabond-SiOH column, 500 mg (Machery-Nagel). The pentafluorobenzyl esters (PFB-amide-PFB-ester) were eluted with 7 mL n-hexane/diethylether (2: 1, v/v). The eluates were evaporated, dissolved in 100 μL MeCN, and analyzed by GC-MS.
For GC-MS, the following conditions were used with a Polaris Q instrument (Thermo-Finnigan): 100 eV, negative chemical ionization, ionization gas NH3, ion source temperature 200°C, the column Rtx-5MS (Restek), 15 m × 0.25 mm, 0.25-μm film thickness, cross-bond 5% diphenyl to 95% dimethyl polysiloxane, injection temperature 220°C, interface temperature 250°C; helium 1 mL min−1; splitless injection; the column temperature program was 1 min 60°C, 25°C min−1 to 180°C, 5°C min−1 to 270°C, 10°C min−1 to 300°C, 10 min 300°C; the retention times of PFB-amide-PFB-esters were: 8.71 min for (2H4)ACC and 8.74 min for ACC; MS fragments at m/z 284 (standard ACC) and at m/z 280 (native ACC) were used for quantification.
Extraction, Purification, and Quantification of IAA
Free IAA was extracted from ∼100 mg fresh weight of plant tissue using 2-propanol/glacial acetic acid (95:5, v/v) with a mortar and pestle. To each extract 100 ng (13C6)-IAA (Cambridge Isotope Laboratories) was added. For each sample, three independent extractions were performed. The samples were incubated under continuous shaking (500 rpm) for 2 h at 4°C and then centrifuged for 10 min at 10,000g. The supernatant was removed, and 200 μL water were added. The organic phase was evaporated under a stream of N2, the aqueous phase was adjusted to 2.5, and the free IAA was extracted twice with equal volumes of ethyl acetate. The organic phases were combined, evaporated under N2, and the residue dissolved in 100 μL ethyl acetate. Methylation of all samples was performed with diazomethane (Cohen, 1984). The methylated samples were dissolved in 30 μL ethyl acetate for GC-MS analysis of which 2.5 μL were injected.
For GC-MS, the following conditions were used applying a Varian Saturn 2100 ion-trap (Varian): 70 eV, electron impact ionization, column Phenomenex ZB-5 column (Phenomenex), 30 m × 0.25 mm, 0.25-μm film thickness, injection temperature 250°C, trap temperature 200°C (Campanella et al., 2003); helium 1 mL min−1; splitless injection; for higher sensitivity, the μSIS mode was used (Wells and Huston, 1995); the column temperature program was: 1 min 70°C, 20°C min−1 to 280°C, 5 min 280°C, the retention time of methyl ester of IAA was10.81 min. MS fragments at m/z 136 (standard IAA) and at m/z 130 (native IAA) were used for quantification.
Accession Numbers
Accession numbers for sequences used in this article are presented in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version for this article.
Supplemental Figure 1. Verification of Differentially Expressed Genes Based on Microarray Analysis by Quantitative Real-Time PCR.
Supplemental Figure 2. Number of All Genes Differentially Transcribed in Arabidopsis Inflorescence Stalks upon Inoculation with Agrobacterium.
Supplemental Figure 3. Functional Categorization of All Differentially Expressed Genes.
Supplemental Figure 4. Difference Profiles of Arabidopsis Gene Expression between the Oncogenic Strain C58 or the T-DNA–Depleted Strain GV3101.
Supplemental Table 1. Primers Used for qRT-PCR.
Supplemental Data Set 1. List of Differentially Expressed Arabidopsis (Ws-2) Genes shown in Supplemental Figure 2 and Supplemental Figure 4 as well as Comparison of Agrobacteria and elf26 Peptide-Responsive Genes.
Supplemental Data Set 2. Differentially Expressed Arabidopsis (Ws-2) Genes of the Functional Categories Presented in Figure 4.
Supplementary Material
Acknowledgments
We thank O. Miersch (Leibniz Institute of Plant Biochemistry, Halle, Germany) for initial help in determination of JA and OPDA, S. Heinze (Institute of Botany, Technische Universität, Dresden) for helping with the extraction of IAA samples, and J. Schwarz and T. Latz (Julius-von-Sachs-Institute, University of Wuerzburg, Germany) for excellent technical assistance. Many thanks to S. Berger (Julius-von-Sachs-Institute) for supplying us with Arabidopsis mutant seeds and B. Usadel (Max-Planck-Institute for Molecular Plant Physiology, Golm, Germany) for the MapMan applications. Finally, we are very grateful to L. Banta (Williams College, Williamstown, MA) for intense discussions and critically reading the manuscript. For generous financial support, we thank the Deutsche Forschungsgemeinschaft (SFB567, project B5; GRK1342, project A7).C.-W.L. was supported by the DAAD STIBET Fellowship program of the University of Wuerzburg Graduate Schools.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the instructions for Authors (www.plantcell.org) is: Rosalia Deeken (deeken@botanik.uni-wuerzburg.de).
Online version contains Web-only data.
References
- Aloni, R., Schwalm, K., Langhans, M., and Ullrich, C.I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853. [DOI] [PubMed] [Google Scholar]
- Aloni, R., Wolf, A., Feigenbaum, P., Avni, A., and Klee, H.J. (1998). The never ripe mutant provides evidence that tumor-induced ethylene controls the morphogenesis of Agrobacterium tumefaciens-induced crown galls on tomato stems. Plant Physiol. 117 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, A., Uppalapati, S.R., Ryu, C.M., Allen, S.N., Kang, L., Tang, Y., and Mysore, K.S. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (2000). On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25 60–83. [Google Scholar]
- Bethke, G., Unthan, T., Uhrig, J.F., Poschl, Y., Gust, A.A., Scheel, D., and Lee, J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T., and He, S.Y. (2009). Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T., and Kende, H. (1980). Regulation of wound ethylene synthesis in plants. Nature 286 259–260. [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y.D., Klessig, D.F., and Dong, X.N. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella, J.J., Ludwig-Mueller, J., Bakllamaja, V., Sharma, V., and Cartier, A. (2003). ILR1 and sILR1 IAA amidohydrolase homologs differ in expression pattern and substrate specificity. Plant Growth Regul. 41 215–223. [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Agnew, J.L., Cohen, J.D., He, P., Shan, L., Sheen, J., and Kunkel, B.N. (2007). Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 104 20131–20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton, M.D., Saiki, R.K., Yadav, N., Gordon, M.P., and Quetier, F. (1980). T-DNA from Agrobacterium Ti plasmid is in the nuclear DNA fraction of crown gall tumor cells. Proc. Natl. Acad. Sci. USA 77 4060–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V., Kozlovsky, S.V., Lacroix, B., Zaltsman, A., Dafny-Yelin, M., Vyas, S., Tovkach, A., and Tzfira, T. (2007). Biological systems of the host cell involved in Agrobacterium infection. Cell. Microbiol. 9 9–20. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.D. (1984). Convenient apparatus for the generation of small amounts of diazomethane. J. Chromatogr. A 303 193–196. [Google Scholar]
- Deeken, R., Engelmann, J.C., Efetova, M., Czirjak, T., Muller, T., Kaiser, W.M., Tietz, O., Krischke, M., Mueller, M.J., Palme, K., Dandekar, T., and Hedrich, R. (2006). An integrated view of gene expression and solute profiles of Arabidopsis tumors: A genome-wide approach. Plant Cell 18 3617–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditt, R.F., Kerr, K.F., de Figueiredo, P., Delrow, J., Comai, L., and Nester, E.W. (2006). The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 19 665–681. [DOI] [PubMed] [Google Scholar]
- Efetova, M., Zeier, J., Riederer, M., Lee, C.W., Stingl, N., Mueller, M., Hartung, W., Hedrich, R., and Deeken, R. (2007). A central role of abscisic acid in drought stress protection of Agrobacterium-induced tumors on Arabidopsis. Plant Physiol. 145 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin, S.B. (2003). Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis–2001 status. Curr. Opin. Plant Biol. 4 301–308. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, S., Grau, T., Buchala, A., Metraux, J.P., and Nawrath, C. (2003). Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36 342–352. [DOI] [PubMed] [Google Scholar]
- Holsters, M., Silva, B., Van Vliet, F., Genetello, C., De Block, M., Dhaese, P., Depicker, A., Inze, D., Engler, G., Villarroel, R., van Montagou, M., and Schell, J. (1980). The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3 212–230. [DOI] [PubMed] [Google Scholar]
- Huber, W., Von Heydebreck, A., Sültmann, H., Poustka, A., and Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 (suppl. 1): S96–S104. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Katsir, L., Schilmiller, A.L., Staswick, P.E., He, S.Y., and Howe, G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kutacek, M., and Rovenska, J. (1991). Auxin synthesis in Agrobacterium tumefaciens and A. tumefaciens-transformed plant tissue. Plant Growth Regul. 10 313–327. [Google Scholar]
- Laird, J., Armengaud, P., Giuntini, P., Laval, V., and Milner, J.J. (2004). Inappropriate annotation of a key defence marker in Arabidopsis: Will the real PR-1 please stand up? Planta 219 1089–1092. [DOI] [PubMed] [Google Scholar]
- LeClere, S., Tellez, R., Rampey, R.A., Matsuda, S.P.T., and Bartel, B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277 20446–20452. [DOI] [PubMed] [Google Scholar]
- Lopez, M.A., Bannenberg, G., and Castresana, C. (2008). Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr. Opin. Plant Biol. 11 420–427. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsy, S., Vanbel, A.J.E., Kluge, M., Hartung, W., and Ullrich, C.I. (1992). Induction of crown galls by Agrobacterium tumefaciens (strain C58) reverses assimilate translocation and accumulation in Kalanchoe daigremontiana. Plant Cell Environ. 15 519–529. [Google Scholar]
- McCullen, C.A., and Binns, A.N. (2006). Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22 101–127. [DOI] [PubMed] [Google Scholar]
- Morris, R.O. (1986). Genes specifying auxin and cytokinin biosynthesis in phytopathogens. Annu. Rev. Plant Physiol. 37 509–538. [Google Scholar]
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., and Jones, J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439. [DOI] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Metraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt III, B.F., and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Nonaka, S., Yuhashi, K., Takada, K., Sugaware, M., Minamisawa, K., and Ezura, H. (2008). Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol. 178 647–656. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M., Leon-Reyes, A., Van der Ent, S., and Van Wees, S.C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316. [DOI] [PubMed] [Google Scholar]
- Ritchie, M.E., Diyagama, D., Neilson, J., van Laar, R., Dobrovic, A., Holloway, A., and Smyth, G.K. (2006). Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics 7 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Saijo, Y. (2008). Plant immunity from A to Z. Genome Biol. 9 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz, A., Navarro, L., Bari, R., and Jones, J.D. (2007). Pathological hormone imbalances. Curr. Opin. Plant Biol. 10 372–379. [DOI] [PubMed] [Google Scholar]
- Ross, J.R., Nam, K.H., D'Auria, J.C., and Pichersky, E. (1999). S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367 9–16. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B., and Zipfel, C. (2008). News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 11 389–395. [DOI] [PubMed] [Google Scholar]
- Sels, J., Mathys, J., De Coninck, B.M., Cammue, B.P., and De Bolle, M.F. (2008). Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 46 941–950. [DOI] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10 69–78. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed]
- Spoel, S.H., and Dong, X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3 348–351. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13 66–71. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Penninckx, I.A., Broekaert, W.F., and Cammue, B.P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13 63–68. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localisation of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Uknes, S., Mauchmani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired-resistance in Arabidopsis. Plant Cell 4 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, C.I., and Aloni, R. (2000). Vascularization is a general requirement for growth of plant and animal tumours. J. Exp. Bot. 51 1951–1960. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C., Geraats, B.P., and Linthorst, H.J. (2006. b). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11 184–191. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C., Rep, M., and Pieterse, C.M. (2006. a). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33 733–742. [DOI] [PubMed] [Google Scholar]
- Veena, Jiang, H., Doerge, R.W., and Gelvin, S.B. (2003). Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 35 219–236. [DOI] [PubMed] [Google Scholar]
- Vergunst, A.C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C.M., Regensburg-Tuink, T.J., and Hooykaas, P.J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290 979–982. [DOI] [PubMed] [Google Scholar]
- Vergunst, A.C., van Lier, M.C., den Dulk-Ras, A., and Hooykaas, P.J. (2003). Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 133 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselov, D., Langhans, M., Hartung, W., Aloni, R., Feussner, I., Gotz, C., Veselova, S., Schlomski, S., Dickler, C., Bachmann, K., and Ullrich, C.I. (2003). Development of Agrobacterium tumefaciens C58-induced plant tumors and impact on host shoots are controlled by a cascade of jasmonic acid, auxin, cytokinin, ethylene and abscisic acid. Planta 216 512–522. [DOI] [PubMed] [Google Scholar]
- Wachter, R., et al. (2003). Vascularization, high-volume solution flow, and localized roles for enzymes of sucrose metabolism during tumorigenesis by Agrobacterium tumefaciens. Plant Physiol. 133 1024–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Pajerowska-Mukhtar, K., Culler, A.H., and Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wasternack, C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, E.W., and Schroeder, J. (1987). Hormone genes and crown gall disease. Trends Biochem. Sci. 12 271–275. [Google Scholar]
- Wells, G., and Huston, C. (1995). High-resolution selected ion monitoring in a quadrupole ion trap mass spectrometer. Anal. Chem. 67 3650–3655. [Google Scholar]
- Wiermer, M., Feys, B.J., and Parker, J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Xu, X.Q., and Pan, S.Q. (2000). An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35 407–414. [DOI] [PubMed] [Google Scholar]
- Yu, Y.B., and Yang, S.F. (1979). Auxin-induced ethylene production and its inhibition by aminoethyoxyvinylglycine and cobalt ion. Plant Physiol. 64 1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z.C., Edlind, M.P., Liu, P., Saenkham, P., Banta, L.M., Wise, A.A., Ronzone, E., Binns, A.N., Kerr, K., and Nester, E.W. (2007). The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. USA 104 11790–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski, P., Tempe, J., and Schell, J. (1989). Transfer and function of T-DNA genes from agrobacterium Ti and Ri plasmids in plants. Cell 56 193–201. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J.D., Boller, T., and Felix, G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.