Abstract
Sister chromatids are often arranged as incompletely aligned entities in interphase nuclei of Arabidopsis thaliana. The STRUCTURAL MAINTENANCE OF CHROMOSOMES (SMC) 5/6 complex, together with cohesin, is involved in double-strand break (DSB) repair by sister chromatid recombination in yeasts and mammals. Here, we analyzed the function of genes in Arabidopsis. The wild-type allele of SMC5 is essential for seed development. Each of the two SMC6 homologs of Arabidopsis is required for efficient repair of DNA breakage via intermolecular homologous recombination in somatic cells. Alignment of sister chromatids is enhanced transiently after X-irradiation (and mitomycin C treatment) in wild-type nuclei. In the smc5/6 mutants, the x-ray–mediated increase in sister chromatid alignment is much lower and delayed. The reduced S phase–established cohesion caused by a knockout mutation in one of the α-kleisin genes, SYN1, also perturbed enhancement of sister chromatid alignment after irradiation, suggesting that the S phase–established cohesion is a prerequisite for correct DSB-dependent cohesion. The radiation-sensitive51 mutant, deficient in heteroduplex formation during DSB repair, showed wild-type frequencies of sister chromatid alignment after X-irradiation, implying that the irradiation-mediated increase in sister chromatid alignment is a prerequisite for, rather than a consequence of, DNA strand exchange between sister chromatids. Our results suggest that the SMC5/6 complex promotes sister chromatid cohesion after DNA breakage and facilitates homologous recombination between sister chromatids.
INTRODUCTION
Double-strand breaks (DSBs), if not repaired, are lethal, at least for dividing cells, and, if misrepaired, may cause chromosome rearrangements, such as reciprocal translocation, insertions, inversions, duplications, and deletions (for review, see Schubert et al., 2004). DSBs are repaired either by homologous recombination (HR) or by nonhomologous end-joining (NHEJ). The gene products involved in these repair pathways are evolutionarily conserved. While NHEJ simply ligates free ends of double-stranded DNA, HR needs an intact homologous duplex to form a heteroduplex for repairing the damaged site by means of the undamaged homologous sequence (reviewed in Kanaar et al., 1998; Barzel and Kupiec, 2008). Since the physical proximity between the donor and acceptor strands is critical for strand exchange events during HR, closely aligned sister chromatids provide a preferred donor for DNA repair via HR (Kadyk and Hartwell, 1992).
STRUCTURAL MAINTENANCE OF CHROMOSOMES (SMC) complexes have multiple functions in sister chromatid cohesion and condensation and repair of eukaryotic chromosomes and are essential for faithful chromosome segregation (for review, see Lehmann, 2005; Nasmyth and Haering, 2005). Together with non-SMC proteins, including kleisin subunits, SMC proteins form multiprotein complexes, such as the cohesin, the condensin, and the SMC5/6 complex. Two large subunits of cohesin, SMC1 and SMC3, form together with an α-kleisin (SISTER CHROMATID COHESION1 [SCC1]/RADIATION-SENSITIVE21 [RAD21] in somatic cells and RECOMBINATION8 [REC8] in meiotic cells) a tripartite ring that establishes cohesion during DNA replication (S phase cohesion) and holds sister chromatids together. Cohesin, together with the SMC5/6 complex, is involved in DSB repair of G2 cells. Budding yeast mutants of cohesin and SMC5/6 complex components display errors in sister chromatid segregation (Uhlmann and Nasmyth, 1998; Torres-Rosell et al., 2005) and are deficient in DSB repair (Sjögren and Nasmyth, 2001; Ünal et al., 2004; De Piccoli et al., 2006). The Scc2/4 complex is needed to load cohesin and Smc5/6 complexes during the S phase onto chromosomes in budding yeast (Saccharomyces cerevisiae; Ciosk et al., 2000; Lindroos et al., 2006). For cohesion establishment in response to DSB formation, the Scc2/4 complex, the cohesion establishment factor (Ctf7/Eco1), Mre11, which acts as a sensor to DSBs in a complex with Rad50 and Xrs2 (Usui et al., 2001), the checkpoint kinases Mec1, Tel1, and Chk1, and Smc6 (probably in the form of the Smc5/6 complex) are required in yeast (Ström et al., 2007; Ünal et al., 2007, Heidinger-Pauli et al., 2008). In human cultured cells, the SMC5/6 complex is involved in recruitment of cohesin to DSB sites (Potts et al., 2006). Mutations or reduced expression of cohesin or SMC5/6 complex components reduce the frequency of HR repair between sister chromatids (Cortes-Ledesma and Aguilera, 2006; De Piccoli et al., 2006; Potts et al., 2006) but has little impact on the frequency of DSB repair by NHEJ or intrachromatid-recombination in budding yeast (Cortes-Ledesma and Aguilera, 2006; De Piccoli et al., 2006). In human cultured cells, DSB repair by NHEJ was even increased after depletion of cohesion or SMC5/6 complex components (Potts et al., 2006). This suggests that cohesin and SMC5/6 complexes keep sister chromatids aligned and facilitate HR repair.
Arabidopsis thaliana homologs of repair factors involved in HR, NHEJ, and DSB signaling have been identified and characterized (Riha et al., 2002; Friesner and Britt, 2003; reviewed in Schuermann et al., 2005). Knowledge about the role of SMC proteins in DSB repair of plants is still limited. Sister chromatids of Arabidopsis are often aligned in a random manner along chromosome arms in interphase nuclei of meristematic and differentiated cells with a 4C or higher DNA content (Schubert et al., 2006, 2008). Accumulating evidence suggests that sister chromatid alignment in Arabidopsis, as in yeast and mammals, is mediated by cohesins (Cai et al., 2003; Schubert et al., 2009). Mutants of cohesin genes, smc1 (titan8) and smc3 (titan7), show aberrant seed development in Arabidopsis (Liu et al., 2002). One of the four α-kleisin paralogs in Arabidopsis, SYN1/DETERMINATE, INFERTILE1, is necessary for sister chromatid cohesion and correct chromosome segregation during meiosis but is expressed also in meristematic tissues (Peirson et al., 1997; Bai et al., 1999; Bhatt et al., 1999; Cai et al., 2003; da Costa-Nunes et al., 2006). The other three paralogs, SYN2/RAD21.1, SYN3/RAD21.2, and SYN4/RAD21.3, are expressed in all plant tissues and may participate in sister chromatid cohesion. γ-Irradiation induces expression of SYN2/RAD21.1, and the T-DNA insertion line for SYN2/RAD21.1 shows higher sensitivity to ionizing radiation and bleomycin than do wild-type plants (da Costa-Nunes et al., 2006; Kozak et al., 2009). A T-DNA insertion line for one of the two SMC6 genes (the mim [for hypersensitive to MMS, irradiation, MMC] mutant line; Mengiste et al., 1999) showed slow growth in early developmental stages, higher sensitivity to DNA damage than wild-ype plants, and a decreased frequency of HR in somatic cells.
To better understand the impact of DNA damage and of loss-of-function mutations of the SMC5/6 complex on interphase chromosome arrangement in plant cells, we analyzed HR and sister chromatid alignment in somatic cells with and without induced DNA damage in Arabidopsis wild-type, smc5/6, syn1, and rad51 mutant plants.
RESULTS
All Homologs of SMC5 and SMC6 Genes Are Expressed in Arabidopsis
Arabidopsis carries one homolog for yeast SMC5 (AT5G15920) and two homologs for SMC6 (Losada and Hirano, 2005). We call one of the two SMC6 homologs (AT5G07660) SMC6A and the other one SMC6B (AT5G61460, MIM; according to Mengiste et al., 1999). The expression of the three genes was examined in wild-type plants. RNA for RT-PCR was isolated from 2-week-old seedlings as well as from rosette leaves and from immature floral buds. All three transcripts were scarce in leaves but abundant in seedlings and floral buds (Figures 1A and 1B). The transcript level of SMC6B was 21-fold higher in seedlings and 6.4-fold higher in flower buds compared to that of SMC6A. However, the transcript level of SMC6A in floral buds was 6.9-fold higher than that in seedlings, while the increase in transcript level of SMC6B was 2.1-fold higher in flower buds than in seedlings (Figure 1C; see Supplemental Figure 1 online for the location of the PCR primers). Thus, SMC6B seems to be the major SMC6 subunit of the SMC5/6 complex in early developmental stages.
Figure 1.
Structure and Expression of SMC5 and SMC6 Genes.
(A) Scheme of the genes with exons (open boxes) and T-DNA insertions of the corresponding SALK lines. The arrows below the boxes indicate the amplified PCR fragments of (B) and (D) and the direction of transcription. Primer sets used for PCR are shown in Supplemental Table 1 online.
(B) Expression analysis of SMC5/6 genes by RT-PCR using RNA samples from seedlings, rosette leaves, and immature floral buds of wild-type plants. Glyceraldehyde-3-phosphate dehydrogenase C (GAPC) cDNA was amplified as a control.
(C) Differential expression of SMC6 genes. Real-time RT-PCR was used to measure the amount of transcripts in the RNA samples from the indicated plant tissues. Transcript levels of SMC6A and SMC6B are shown in relation to that of ACTIN2 in seedlings and floral buds of wild-type plants. White, SMC6A; gray, SMC6B. Vertical bar and crossed bars represent the sd of two biological replicates. Crossed bars are used to indicate sd not large enough to be depicted in a semilogarithmic graph.
(D) Top panel: DNA fragments amplified from SMC6A cDNA of wild-type and smc6a-1 plants. Bottom panel: DNA fragments amplified from SMC6B cDNA of wild-type, smc6b-1, and, smc6b-2 plants. RT-PCR was performed with RNA samples from rosette leaves. GAPC cDNA was amplified as a control.
To analyze the role of the SMC5/6 complex in HR repair in a plant, we studied five T-insertion lines of Arabidopsis accession Columbia (Col): smc5-1, smc5-2, smc6a-1, smc6b-1, and smc6b-2 (Figure 1A). The T-DNA insertions of smc5-1 and smc5-2 occurred within the second exon. In total, eight independent individuals, hemizygous for T-DNA insertion in the SMC5 gene (SMC5/smc5-1 and SMC5/smc5-2), did not yield homozygous mutants among their progeny. Siliques of SMC5/smc5-1 and SMC5/smc5-2 contained ∼25% shrunken seeds (Table 1), indicating that SMC5 is essential for seed development. In smc6a-1, the T-DNA is inserted in the 11th exon and in smc6b-1 in the 19th intron, while smc6b-2 harbors a T-DNA within the 27th exon. RT-PCR revealed the absence of full-length transcripts in homozygous smc6a-1 and smc6b-2 mutants (Figure 1D). Amplified DNA fragments demonstrated that SMC6A transcripts from the smc6a-1 mutant lack the central region around the T-DNA insertion site. PCR amplified two DNA fragments corresponding to the 3′ region of the SMC6A transcript (F3 fragment; Figures 1A and 1D) in the smc6a mutant as well as in the wild type. The larger fragment was amplified from a cDNA synthesized from a SMC6A transcript variant from which the 21st intron (from the position 5570 to 5666 of the genomic sequence of AT5G07660) was not spliced out (GenBank FJ869873), indicating an alternative splicing within the 3′ region of the SMC6A gene. A termination codon within the additional sequence of the longer transcript variant, having a poly(A) tail, would, if the transcript variant is translated, yield a protein that lacks the C-terminal 182 amino acids, which are required to interact with the δ-kleisin, NON-SMC ELEMENT4 (NSE4), in fission yeast (Schizosaccharomyces pombe) (Figure 2; Palecek et al., 2006). A very faint F2 fragment amplified from the smc6a-1 mutant migrates faster than the F2 fragment from the wild-type plant, suggesting irregular splicing caused by the T-DNA insertion. The 3′ region of the SMC6B transcript downstream of the T-DNA insertion is absent in the smc6b-2 mutant. The very faint F3 fragment obtained from the smc6b-1 mutant is apparently due to rare splicing out of the intron that harbors the T-DNA insertion. These results indicate that smc6a-1 and smc6b-2 mutants do not express wild-type SMC6 transcripts, while smc6b-1 plants can generate wild-type transcripts via splicing out of the T-DNA insertion, albeit at a much lower level than wild-type plants.
Table 1.
Shrunken Seeds in smc5 Mutant Plants
| Line | Normal Seeds (%) | Shrunken Seeds (%) | n |
|---|---|---|---|
| Wild type | 96.7 | 3.3 | 514 |
| SMC5/smc5-1 | 73.2 | 26.8** | 935 |
|
SMC5/smc5-2 |
78.0 |
22.0** |
943 |
n, total number of seeds examined. **, The observed segregation ratio is tested by the χ2 test to determine whether it fits a ratio of 3:1 at the probability of >0.05.
Figure 2.
Architecture of SMC5 and SMC6 and Their Interaction with the δ-Kleisin NSE4.
(A) SMC5, SMC6A, and SMC6B have globular domains at both termini, each of which is connected to a hinge domain via a coiled-coil region. Each SMC protein is backfolded, creating a head domain composed of two globular domains at one end and a hinge domain at the other. NTP binding motifs (Walker A and B motifs) are identified in the N-terminal and the C-terminal globular domains of SMC5 and SMC6.
(B) SMC5 and SMC6 form heterodimers via interaction of their hinge domains, while the head domains of SMC5-SMC6 heterodimers associate with NSE4, according to Palecek et al. (2006).
[See online article for color version of this figure.]
Mutations on SMC6A and 6B Genes Tend to Increase Sensitivity to X-Irradiation
Homozygous mutant plants harboring the mim allele of SMC6B are more sensitive to UV-C, x-rays, methyl methanesulfonate, and mitomycin C (MMC) than are wild-type plants (Mengiste et al., 1999). To find possible functional differences between SMC6A and SMC6B, the effect of X-irradiation (100 Gy) of seeds on root growth was analyzed in smc6 mutant plants at various time points after irradiation (Figure 3). Compared with wild-type seedlings, smc6a-1 and smc6b-1 mutants revealed delayed root growth even in nonirradiated samples. All lines tested showed retarded root growth after irradiation. The difference between wild-type and mutant plants became apparent 10 d after irradiation. The smc6a-1 and the smc6b-1 mutant plants were equally sensitive to x-rays, suggesting that both SMC6 genes are involved in DNA repair. MMC treatment results in retarded growth of the smc6b-1 mutants in comparison to wild-type and smc6a-1 mutant plants (see Supplemental Figure 2 online).
Figure 3.
Root Extension in Control and Irradiated Plants.
Seeds on Murashige and Skoog-agar plates were exposed to 100 Gy of x-rays, and root length was measured at the indicated time. The mean values of root length are plotted. Error bars represent the sd for 20 plants per line. Open square, wild type; open diamond, smc6a-1; open circle, smc6b-1. Solid lines, mock-treated plants; dotted lines, irradiated plants.
X-Rays Enhance Sister Chromatid Alignment in Wild-Type Plants
RAD51 homologs are involved in the repair of damaged DNA via HR (reviewed in Kanaar et al., 1998; Shibata et al., 2001). Arabidopsis rad51 mutants show increased sensitivity to the DNA cross-linking agent MMC but not to DSB inducers, such as γ-rays or bleomycin (Bleuyard et al., 2005; Markmann-Mulisch et al., 2007), while mutants defective in NHEJ are hypersensitive to γ-irradiation (Friesner and Britt, 2003). Because MMC apparently induces DSBs only when cross-link repair interferes with DNA replication, HR might be involved mainly in postreplication repair and NHEJ in DSB repair during other phases of cell cycle. Also, smc6 mutants displayed a slightly increased sensitivity to x-rays compared to the wild type. To study interphase chromosome arrangement in response to X-irradiation in Arabidopsis, we applied fluorescent in situ hybridization (FISH) to nuclei of 4C DNA content (4C nuclei) prepared from the irradiated plants. Irradiation with 20 Gy of x-rays was previously found to induce chromosome bridges in 28% of anaphase nuclei in somatic pistil cells of DNA ligase IV–deficient (lig4) plants (a 25-fold increase compared to the 1.1% of anaphase nuclei of the nonirradiated lig4 plants). The number of DSBs induced by 1 to 50 Gy follows a linear dose relationship, and 20 Gy should yield 9 to 26 DSBs in the Arabidopsis genome according a formula by Erixon and Cedervall (1995). In budding yeast, one DSB is sufficient to trigger a genome-wide cohesion establishment (Ström et al., 2007; Heidinger-Pauli et al., 2008). Therefore, we applied 20 Gy of x-rays to wild-type plants as well as to smc5/6, syn1, and rad51 mutants. Because comet assay studies after bleomycin treatment had previously shown that nearly all single- and double-strand breaks are repaired within 60 min after treatment (Menke et al., 2001), we isolated nuclei 10 and 60 min after irradiation (mai).
Pecinka et al. (2004) have shown that somatic pairing of homologs in unchallenged 2C nuclei occurs mainly at random (in 0.8 to 13%, on average 4.9%, of nuclei) along chromosome arms. However, Abdel Halim et al. (2004) described, within human G1 cells, X-irradiated with 4 Gy, an increased homologous pairing frequency of heterochromatic loci, which are preferentially involved in chromosome rearrangements, but not of euchromatic loci. Therefore, we tested first whether X-irradiation enhances allelic pairing of homologs in 2C nuclei of the wild type. To analyze the frequency of positional pairing, we counted the number of hybridization signals after FISH with a single BAC or a BAC pair carrying inserts of adjacent genomic sequences from mid arm positions of chromosomes 1 and 3 on flow-sorted 2C nuclei of nonirradiated and irradiated 2-week-old wild-type seedlings. One FISH signal per BAC was regarded as allelic pairing, two signals were considered as separation of homologs at the corresponding locus (Figure 4A). Nonirradiated 2C nuclei showed 10.7 and 9.94% of positional pairing at the loci represented by BAC clones T7N9/T2P11 and F18C1 on chromosomes 1 and 3, respectively. The pairing frequency at these positions 10 and 60 mai did not differ significantly from that of nonirradiated samples (Table 2; P > 0.05, χ2 test). This indicates that irradiation with 20 Gy of x-rays does not enforce allelic pairing between homologs and that donor sequences for HR repair are not regularly provided by allelic loci in 2C nuclei.
Figure 4.
Positional Pairing and Sister Chromatid Alignment after FISH with Flow-Sorted Arabidopsis Nuclei.
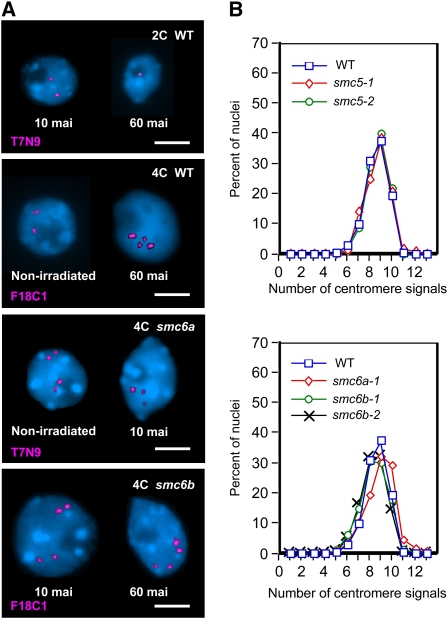
(A) Top: FISH on wild-type 2C nuclei. Left: Separated loci in a nucleus 10 mai. Right: Paired loci in a nucleus, 60 mai. Second row: FISH on wild-type 4C nuclei. Left: Sister chromatid alignment at both homologs in a nonirradiated nucleus. Right: Positional sister chromatid separation at both homologs, 60 mai. Third row: FISH on 4C smc6a-1 nuclei. Left: Positional sister chromatid separation at both homologs in a nonirradiated nucleus. Right: Sister chromatid alignment at one of the homologs, 10 mai. Bottom row: FISH on 4C smc6b-2 nuclei. Left: Positional sister chromatid separation at both homologs, 10 mai. Right: 60 mai. Nuclei are counterstained with 4',6-diamidino-2-phenylindole (blue). Bars = 5 μm.
(B) Centromeric sister chromatid alignment in wild-type and in smc5/6 mutants. Proportion of 4C nuclei with different numbers of FISH signals for centromeric repeats is demonstrated for wild-type plants and for smc5/6 mutants. Top: Wild type versus heterozygotes for the SMC5 gene. Bottom: Wild type versus homozygotes for smc6 genes. The number of 4C nuclei analyzed in each line is as follows: wild type, 310; smc5-1, 209; smc5-2, 237; smc6a-1, 225; smc6b-1, 386; and smc6b-2, 386.
Table 2.
Frequencies of Positional Homologous Pairing in Irradiated and Nonirradiated Arabidopsis 2C Wild-Type Nuclei
| Chromosome 1 |
Chromosome 3 |
|||
|---|---|---|---|---|
| T7N9/T2P11 |
F18C1 |
|||
| mai | Frequency ± sd (%)a | n | Frequency ± sd (%)a | n |
| Nonirradiated | 10.7 | 338 | 9.94 | 352 |
| 10 | 9.45 | 328 | 9.55 | 335 |
| 60 | 11.0 | 328 | 8.71 | 264 |
n, number of nuclei analyzed.
Differences between nonirradiated and irradiated samples are not significant according to the χ2 test (P < 0.05).
Next, we tested completeness of sister chromatid alignment for entire chromosome arms. With a FISH probe covering the top arm of chromosome 1, no completely separated sister chromatid arm territories were found in 264 4C nuclei of the irradiated 2-week-old wild-type seedlings at 10 mai. At 60 mai, 2.3% of 264 nuclei showed completely separated sister chromatid territories for at least one homolog of this chromosome arm. Nonirradiated nuclei display separated sister arm territories in 6.1% of rosette leaf nuclei (Schubert et al., 2006). The frequency of 6.1% is significantly higher than the frequency at 10 mai (P < 0.001, χ2 test) and at 60 mai (P < 0.05, χ2 test), indicating that X-irradiation promotes alignment rather than separation of sister arm territories.
Then we analyzed sister chromatid alignment at homologous loci in 4C nuclei prepared from nonirradiated and irradiated wild-type seedlings by FISH with the same BACs used to examine allelic pairing in 2C nuclei. In nuclei with one or two FISH signals per BAC, sister chromatids are aligned at the corresponding loci, while three or four signals indicate sister chromatid separation in one or both homologs. Ten minutes after irradiation, 4C nuclei showed between 17 and 21% higher frequencies of positional sister chromatid alignment at the two loci (T7N9/T2P11; F18/C1) than nonirradiated nuclei. These differences were highly significant according to the χ2 test (P < 0.001). Sixty minutes after irradiation, the values decreased by 7 to 11% compared to those obtained at 10 mai (Table 3). These results imply that X-irradiation transiently increases cohesion between sister chromatids. Apparently, in Arabidopsis, the breakage-induced enforcement of sister chromatid cohesion relaxes again after completion of DNA repair.
Table 3.
Frequencies of Positional Sister Chromatid Alignment in 4C Arabidopsis Nuclei of Nonirradiated and Irradiated (20 Gy) Wild-Type and Mutant Plants
| Chromosome 1 |
Chromosome 3 |
|||||
|---|---|---|---|---|---|---|
| T7N9/T2P11 |
F18C1 |
|||||
| Plant Material | mai | Frequency (%) | n | Frequency (%) | n | |
| Wild type | Seedlings | Nonirradiated | 54.8 | 704 | 51.4 | 704 |
| 10 | 71.6*** | 880 | 72.7*** | 880 | ||
| 60 | 61.1* | 880 | 65.7*** | 880 | ||
| Wild typea | Seedlings | Nonirradiated | 67.2 | 640 | 65.5 | 640 |
| SMC5/smc5-1a (SALK_107583) | Seedlings | Nonirradiated | 59.7 | 352 | 63.9 | 352 |
| 10 | 63.9 | 352 | 67.6 | 326 | ||
| 60 | 60.8 | 352 | 64.2 | 324 | ||
| SMC5/smc5-2a (SALK_092081) | Seedlings | Nonirradiated | 63.4 | 352 | 67.3 | 352 |
| 10 | 64.2 | 352 | 68.8 | 352 | ||
| 60 | 60.5 | 352 | 64.8 | 352 | ||
| smc6a-1/smc6a-1 (SALK_009818) | Seedlings | Nonirradiated | 48.2 | 704 | 50.7 | 704 |
| 10 | 52.0 | 704 | 50.4 | 702 | ||
| 60 | 50.9 | 704 | 54.5 | 701 | ||
| smc6b-1/ smc6b-1 (SALK_101968) | Seedlings | Nonirradiated | 49.4 | 336 | 54.0 | 302 |
| 10 | 54.3 | 352 | 53.5 | 318 | ||
| 60 | 58.0* | 350 | 60.1 | 336 | ||
| smc6b-2/ smc6b-2 (SALK_135638) | Seedlings | Nonirradiated | 49.7 | 348 | 55.7 | 352 |
| 10 | 55.7 | 352 | 56.0 | 352 | ||
| 60 | 57.7* | 352 | 59.1 | 352 | ||
| Wild type | Rosette leaves | Nonirradiated | 60.7 | 704 | 62.1 | 704 |
| syn1/syn1 (SALK_006687) | Rosette leaves | Nonirradiated | 44.9 | 352 | 52.9 | 346 |
| 10 | 52.3* | 352 | 53.7 | 324 | ||
| 60 | 46.6 | 352 | 44.9 | 316 | ||
| rad51-1/rad51-1 (GABI_134A01)b | Rosette leaves | Nonirradiated | 56.0 | 352 | 65.3 | 352 |
| 10 | 66.2* | 352 | 71.3 | 352 | ||
| 60 |
59.7 |
352 |
67.9 |
352 |
||
n, number of homologous loci analyzed. *, Significance compared with the value for nonirradiated by the χ2 test (P < 0.05). ***, Significance compared with the value for nonirradiated by the χ2 test (P < 0.001).
Cotyledons were clipped off 2 d before preparation of sorted nuclei.
The line GABI_134A01 carries two T-DNA insertions and one of them was segregated out in rad51-1 (Li et al., 2004).
Increased Sister Chromatid Alignment Needs the SMC5/6 Complex
To test the potential role of the Arabidopsis SMC5/6 complex in DSB repair, sister chromatid alignment was analyzed in nonirradiated and irradiated nuclei of heterozygous smc5 and homozygous smc6 mutant lines. First, we compared centromere cohesion in nonirradiated mutant and wild-type seedlings by FISH using the 178-bp centromere-specific probe. Full sister centromere cohesion in 4C nuclei should yield up to 10 FISH signals. More than 80% of nuclei of the wild type as well as of all mutant plants revealed 8 to 11 signals (Figure 4B), indicating no differences in sister centromere cohesion between wild-type and mutant plants and excluding the possibility that reduced sister chromatid alignment after X-irradiation of the mutants is due to precocious sister chromatid separation (before X-irradiation) in interphase nuclei with smc5/6 mutant backgrounds.
To analyze sister chromatid alignment in nuclei from 2-week-old seedlings heterozygous for SMC5, we had to identify heterozygous plants. For genotyping, cotyledons were clipped from seedlings 2 d before preparation of sorted nuclei. Nonirradiated nuclei from the smc5 mutants showed sister chromatid alignment at the tested positions in 60 to 67% of loci. Irradiation did not induce a significant increase in sister chromatid alignment in the smc5 mutant nuclei at 10 mai (0.8 to 4.2% higher in irradiated nuclei than in nonirradiated nuclei), and at 60 mai the alignment frequency returned to the level observed in nonirradiated nuclei (Table 3). The higher alignment frequency in nonirradiated smc5 mutants is apparently caused by cotyledon clipping, since wild-type seedlings showed a similar strong and significant increase of alignment (P < 0.001, χ2 test) when exposed to cotyledon clipping (Table 3). Because of the high basic level of alignment caused by cotyledon clipping, it is difficult to decide whether enhancement of sister chromatid alignment is induced after DSB formation in heterozygous smc5 nuclei or not.
The positional sister chromatid alignment in nonirradiated nuclei of smc6a-1 and both smc6b mutant seedlings was as frequent as that of the wild type at both tested loci. After irradiation, the increase in sister chromatid alignment was delayed and clearly less pronounced in the nuclei of smc6b mutants than in wild-type nuclei. The 4C nuclei of the irradiated smc6 mutants showed at 60 mai 3.4 to 8.7% higher frequencies of sister chromatid alignment than nonirradiated nuclei, and the increase at 60 mai is statistically significant for position T7N9/T2P11 but not for position F18C1 in the smc6b mutants (P < 0.05, χ2 test; Table 3). Although the relative transcript level of SMC6A is ∼20 times less than that of SMC6B in wild-type seedlings, SMC6A seems to be necessary to enhance sister chromatid alignment after X-irradiation. These results suggest that both SMC6A and SMC6B are, likely in a complex together with SMC5, required for establishment of DSB-induced cohesion between sister chromatids.
Disturbed S Phase Cohesion in syn1 Mutants Impairs Establishment of DSB-Mediated Cohesion
SYN1, a REC8/SCC1 homolog, is required in Arabidopsis for sister chromatid cohesion in meiotic as well as in somatic cells (Schubert et al., 2009). Because SYN1 does not contain a Ser residue that becomes phosphorylated in response to DSB formation and is conserved among RAD21/SCC1 homologs (Heidinger-Pauli et al., 2008), SYN1 is likely to be involved in the establishment of S phase cohesion but not of DSB-induced cohesion. To test whether a reduced level of S phase cohesion impairs DNA breakage-mediated increase of sister chromatid alignment, we examined sister chromatid alignment in irradiated 4C nuclei from rosette leaves of a homozygous syn1 mutant (Table 3). Unchallenged syn1 nuclei showed significantly less (P < 0.005, χ2 test) sister chromatid alignment than wild-type rosette leaf nuclei (60.7 and 62.1% in the wild type to 44.9 and 52.9% in syn1; Table 3), indicating that the absence of SYN1 impairs S phase–dependent cohesion. The generally higher frequency of sister chromatid alignment in rosette leaves than in seedlings might be due to changes in nuclear architecture along with development (Mathieu et al., 2003). Sister chromatid alignment was enhanced soon after irradiation in the syn1 nuclei as well as in the wild-type nuclei and the rad51 nuclei (Table 3), but the increase at 10 mai was small and less significant in the syn1 mutant (P < 0.05 at position T7N9/T2P11 and P > 0.05 at F18C1; χ2 test), suggesting that disturbed S phase cohesion impairs establishment of breakage-mediated cohesion in syn1 mutants.
rad51 Mutants Show an Irradiation-Mediated Increase in Sister Chromatid Alignment
Heteroduplex formation between sister chromatids and subsequent DNA synthesis during HR require aligned sister chromatids. Rad51 and its homologs are involved in heteroduplex formation (Kanaar et al., 1998). To see whether mutants of repair components that act downstream of the SMC5/6 complex may influence chromatin dynamics after DNA damage, sister chromatid alignment was analyzed in 4C nuclei prepared from rosette leaves of rad51-1 mutant plants (Table 3). Nonirradiated rad51-1 nuclei showed sister chromatid alignment at a similar frequency as the nonirradiated wild-type nuclei from rosette leaves (P > 0.1, χ2 test). In contrast with the results for smc5/6 mutants, 20 Gy X-rays increased at 10 mai the sister chromatid alignment in rad51-1 nuclei to values similar to those observed in nuclei from wild-type seedlings (71.6 and 72.7% in the wild type compared to 66.2 and 71.3% in rad51; P > 0.05, χ2 test). The increase in sister chromatid alignment in irradiated rad51-1 nuclei is reasonable because cohesin and SMC5/6 complexes are present in the rad51 mutant. The increase indicates that sister chromatid alignment is a prerequisite for, rather than a consequence of, DNA strand exchange between sister chromatids.
Mutations in SMC6A and 6B Genes Cause Reduced HR Frequencies in Somatic Cells
The efficiency of DSB repair via HR was analyzed with the recombination substrate, pDGU.US (Orel et al., 2003). This substrate contains, in direct orientation, two halves of a β-glucuronidase (GUS) gene with a 557-bp overlap separated by an unrelated sequence of 38 nucleotides. After treatment with bleomycin (causes DSBs directly) or with MMC (causes DSBs indirectly during repair of cross-links), a functional GUS gene can be restored by HR. Three different mechanisms can be envisaged (Figure 5): (1) intramolecular single-strand annealing after 5′-break end resection, (2) intermolecular synthesis-dependent strand-annealing, or (3) break-induced replication (BIR) with the sister chromatid or the homologous chromosome as a donor (Malkova et al., 1996; Puchta, 2005). In the case of synthesis-dependent strand-annealing, restoration of the GUS gene can be achieved by two intermolecular template switches: the first within the 557-bp overlap of the GUS sequences and the second behind this region, as both sister chromatids or homologs should be completely homologous distal to the overlap. Alternatively to the second switch, the chromatid might also be copied in toto, resulting in a BIR event (Figure 5). However, BIR would require rereplication licensing for all origins up to the chromosome arm end, which is unlikely to occur in large genomes. In two independent experimental series, the recombination events were monitored by counting GUS-stained blue sectors in seedlings of wild-type and smc6a-1 or wild-type and smc6b-1 mutant plants, both carrying pDGU.US in a homozygous state (Figure 6; see Supplemental Table 2 online). In comparison to the wild-type control, the number of recombination events is reduced to a third in the smc6a-1 and to half in the smc6b-1 mutant. After treatment with bleomycin and mitomycin, the smc6a-1 mutant shows only a third of the recombination events of the wild type, and in the case of smc6b-1, the recombination events are reduced to a forth. Thus, up to three-quarters of the recombination events detected with our assay are due to intermolecular interactions. This might even be an underestimate, as both smc6 mutants, compared to the wild type, still revealed some basic frequency of sister chromatid alignment and a small increase of sister chromatid alignment after irradiation, indicating that the two SCM6 homologs might be able to partially complement each other in HR.
Figure 5.
HR within the DGU.US Reporter Transgene.
Different mechanisms of DSB repair can be envisaged that result in the reconstitution of a functional GUS reporter gene.
(A) Single-strand annealing.
(B) Synthesis-dependent strand annealing via an intermolecular out-of-frame conversion.
(C) Break-induced replication.
Figure 6.
HR in smc6a-1 and smc6b-1 Mutants Compared to the Respective Wild Type.
(A) Analysis of HR events in the smc6a-1 mutant compared to the corresponding wild type.
(B) Analysis of HR events in the smc6b-1 mutant compared to the corresponding wild type.
The wild-type plants were obtained from a segregating population of a cross between a line carrying pDUGU.US and corresponding smc6 mutant plants. Both mutants show a lower rate of HR compared to the wild type without and with DSB induction by application of bleomycin or MMC. HR events depicted for all conditions are mean values of three independent experiments. Vertical bars represent the sd. The data set and additional calculations are summarized in Supplemental Table 2 online.
DISCUSSION
Components of the SMC5/6 Complex Studied So Far Are Essential in Arabidopsis
The SMC5/6 complex is involved in DNA repair in yeasts and human. In yeasts, the SMC5/6 complex comprises two SMC subunits, SMC5 and SMC6, and six non-SMC elements, NSE1 to 6 (Losada and Hirano, 2005; Zhao and Blobel, 2005; Pebernard et al., 2006). Genes for SMC5, SMC6, and NSE1-4 are well conserved in budding and fission yeasts, Drosophila melanogaster, Xenopus laevis, and human. SMC5, 6, and NSE4, a δ-kleisin subunit, form a major tripartite complex interacting with the other NSE proteins (Losada and Hirano, 2005; Palecek et al., 2006). The Arabidopsis genome harbors homologs for these genes, and some of them are duplicated: two homologs for SMC6 and two homologs for NSE4 (NSE4A [AT1G51130] and NSE4B [AT3G20760]; Losada and Hirano, 2005). With RT-PCR, we detected transcripts of the NSE4A but not of the NSE4B gene in seedlings, rosette leaves, and immature floral buds (see Supplemental Figure 3 online), suggesting that NSE4A is a functional gene in somatic cells of Arabidopsis.
SMC5 and SMC6 are each composed of a globular head domain at both termini and a coiled-coil domain interrupted by a hinge domain (Figure 2A). In fission yeast, SMC5 and SMC6 interact with each other through their hinge domains and with NSE4 through their globular head domains (Figure 2B; Sergeant et al., 2005; Palecek et al., 2006). In Arabidopsis, the smc6a-1 mutant expresses SMC6A transcripts lacking its central region around the T-DNA insertion site, which includes the sequence encoding a hinge domain. Therefore, SMC6A of the smc6a-1 mutant might not be able to interact with SMC5. The SMC6B transcripts of smc6b-1 and -2 mutants lack the C-terminal head domain; therefore, the interaction of the mutated SMC6B with NSE4 might be disturbed in both mutants.
The Arabidopsis homolog for NSE1 (AT5G21140; Losada and Hirano, 2005), which encodes a protein with a RING-like motif (Pebernard et al., 2008), is also essential for seed development and the terminal phenotype of the nse1 line emerges at the preglobular embryonic stage (Tzafrir et al., 2004; SeedGenes Project at http://www.seedgenes.org/index.html). The Arabidopsis homologs for NSE2 (AT3G15150), a SUMO ligase-encoding gene, and for NSE3 (AT1G34770), a MAGE (melanoma antigen-encoding) gene, remain to be characterized. The primary structures of NSE5 and NSE6, identified in coprecipitates with the known SMC5/6 components, are quite different even between budding and fission yeasts (Zhao and Blobel, 2005; Pebernard et al., 2006). Functional homologs for these proteins are not yet identified in Arabidopsis.
The Arabidopsis SMC5/6 Complex Is Involved in DSB Repair
Homologous sequences in allelic or ectopic positions are potential donor sequences for DSB repair by HR. In human G1 cells, X-irradiation enforced homologous pairing of pericentromeric heterochromatic loci, which are preferentially involved in chromosome rearrangements but not of euchromatic loci (Abdel Halim et al., 2004). Similarly, in Arabidopsis 2C wild-type nuclei, pairing of homologs at euchromatic regions was not enhanced by irradiation. The constrained movement of DSB ends in mammalian interphase nuclei indicates that search of broken chromosome ends for homology does not extend far beyond the chromosome territory (Soutoglou et al., 2007). Thus, HR in G1 seems to be restricted to homologous sequences, which are by chance in the spatial vicinity of a DSB. Since in Arabidopsis positional pairing between allelic loci occurs at random (i.e., in 0.84 to 13% of unchallenged 2C nuclei; Pecinka et al., 2004) and is not increased after irradiation, most DSBs seem to be repaired by NHEJ rather than by HR in 2C nuclei. Indeed, our previous work indicates that DSB repair using allelic or ectopic homology occurs very rarely, at least in tobacco (Nicotiana tabacum; Puchta, 1999; Gisler et al., 2002).
In budding yeast, the presence of sister chromatids immediately after DNA replication increases cell survival after DSB induction (Kadyk and Hartwell, 1992), and homologous alignment of sister chromatids by cohesin is important for DSB repair in S/G2 phase (Sjögren and Nasmyth, 2001; Ström et al., 2004). Cohesin loaded onto chromatin during S phase accumulates around centromeres (Megee et al., 1999; Bernard et al., 2001) and at defined loci every 5 to 10 kb along chromosome arms (Tanaka et al., 1999; Glynn et al., 2004; Lengronne et al., 2004). Thus, in yeast, sister chromatids are closely aligned during G2 phase. Additionally, cohesin is loaded onto sister chromatids in response to DSB formation and is maintained until the next M phase (Ström et al., 2004, 2007; Ünal et al., 2004, 2007). In contrast with the consistent sister chromatid cohesion along the chromosome arms in yeast, sister chromatid arms are incompletely aligned in Arabidopsis and other plant species (Schubert et al., 2006, 2007, 2008) and probably also in mammals (Volpi et al., 2001; Watrin and Peters, 2006). X-irradiation (and MMC treatment; see Supplemental Table 3 online) significantly increases sister chromatid alignment in 4C wild-type nuclei of Arabidopsis when functional SMC6 genes are present. This suggests that in Arabidopsis as in yeast a tightened sister chromatid cohesion promotes (correct) DSB repair in 4C nuclei. The genotoxin-induced recombination frequency in somatic cells confirmed that the SMC6 genes are required for nearly three-quarters of HR events in Arabidopsis. Thus, we conclude that the SMC5/6 complex enhances sister chromatid alignment after DNA damage and thereby facilitates correct DSB repair via HR between sister chromatids.
Rec8 artificially expressed in mitotic yeast cells binds to chromosome arms and contributes to establishment of S phase cohesion, but it is not deposited to DSB sites (Heidinger-Pauli et al., 2008). Because the Arabidopsis homolog SYN1 is transcribed in somatic cells, and unchallenged syn1 mutant nuclei show reduced sister chromatid alignment and occasional separation of sister centromeres (4.5 to 9.2% of 4C nuclei from different syn1 mutants showed up to 18 centromere-specific FISH signals, while only 1.5% of wild-type 4C nuclei showed up to 12 centromere-specific signals.), SYN1 is thought to be involved in S phase cohesion in somatic cells (Schubert et al., 2009). The syn2/rad21.1 mutant is deficient in DSB repair (da Costa-Nunes et al., 2006; Kozak et al., 2009), though it aligns sister chromatids in unchallenged conditions as frequently as the wild type does (Schubert et al., 2009), suggesting a role of SYN2 in DSB-responsive cohesion. We assume that the S phase cohesion is prerequisite to irradiation-enhanced cohesion establishment by SYN2-containing cohesins. The most prominent feature of multicellular organisms is cell differentiation. Most of the plant cells are differentiated and no longer proliferate. Some differentiated cells undergo endopolyploidization cycles. Expression of the SCC1/RAD21/REC8 homolog SYN1 and of SMC5/6 is lower in mature leaves than in dividing tissues of Arabidopsis (Bhatt et al., 1999; Figure 1B). Similarly, the level of RAD51 expression decreases in developing plants in correlation with a reduction of DSB repair by HR and with a complementary increase in expression of KU70, a component of NHEJ (Boyko et al., 2006). These data suggest that DSBs are preferentially repaired by NHEJ in differentiated cells and by HR between sister chromatids in meristematic cells. The predominance of the more accurate HR mechanism in dividing cells stabilizes the genome in meristematic tissues and ensures the correct transmission of genetic information to daughter cells and to subsequent generations.
METHODS
Plant Materials
The smc5, smc6a, smc6b, and syn1 T-DNA insertion lines (in Col) (Figure 1A) were obtained from the SALK collection (Alonso et al., 2003). The rad51-1 mutant (in Col) was described by Li et al. (2004) and provided by Bernd Reiss. The lig4 mutant in accession Wassilewskija (Friesner and Britt, 2003) was provided by Ann Britt. The seedlings were cultured on germination media (GM)-agar (4.9 g/L Murashige and Skoog micro- and macro-elements, including vitamins [Duchefa], 10 g/L sucrose, pH 5.7, and 8 g/L micro-agar) under long-day conditions (16 h light/8 h dark ) at 22°C for 2 weeks, followed by further cultivation in soil under short-day conditions (8 h light/16 h dark) at 21°C.
PCR-based genotype markers were used to identify the T-DNA insertion mutants. The PCR primers used for genotyping and the left border insertion junctions of these T-DNA insertion lines are listed in Supplemental Tables 4 and 5 online, respectively. PCR using the gene-specific primer sets yielded DNA fragments of ∼1 kb representing the wild-type alleles of SMC5, SMC6A, and SMC6B. The PCR fragment specific for the smc5-1, smc5-2, smc6a-1, smc6b-1, smc6b-2, or syn1 allele was amplified with the primer sets (107583RP, SALK_LB), (092081RP, SALK_LB), (009818RP, SALK_LB), (101968RP, SALK_LB), (135638RP, SALK_LB), or (006687RP, SALK_LB), and the amplification yielded products of ∼0.5 kb. The allele of rad51-1 was identified as described (Li et al., 2004).
X-Irradiation
Plant material was irradiated using the x-ray system YXLON MGC41 (YXLON International) at the Federal Research Center for Cultivated Plants (Quedlinburg, Germany). For analysis of sensitivity to ionizing radiation, seeds were sterilized, plated on Murashige and Skoog-agar media (4.4 g/L Murashige and Skoog micro- and macro-elements, including vitamins [Duchefa], pH 5.7, and 8 g/L micro-agar), and germinated at 4°C. Approximately 48 h later, plants were either mock-irradiated or exposed to 100 Gy (3 Gy/min) and cultured under long-day conditions. For analysis of sister chromatid alignment, plants were irradiated with 20 Gy (0.9 Gy/min) and fixed after the recovery time indicated in Tables 2 and 3. Nuclei were isolated as described below.
MMC Treatment
Seeds were sterilized, plated on GM-agar, and germinated under long-day conditions after 2 d of cold treatment at 4°C. For analysis of sensitivity to MMC, seedlings were transferred into 24-multiwell plastic plates (Falcon) 4 d after germination. Each well contained one seedling in 0.5 mL of liquid GM supplemented with MMC (Sigma-Aldrich) at concentrations of 2.5, 5, 10, 15, or 20 μg/mL. The plates were incubated for another 2 weeks under long-day conditions. For analysis of sister chromatid alignment, 10-d-old seedlings were moistened with water overnight, and the seedlings were mock-treated or exposed to 5 μg/mL of MMC in liquid GM for 30 min, followed by preparation of sorted nuclei.
Preparation of Nuclei, Probe Labeling, and FISH
Nuclei were isolated and flow-sorted according to their ploidy level from 2-week-old seedlings of wild-type, smc5, and smc6 mutants or from leaves of wild-type, syn1, and rad51 mutants as described (Jasencakova et al., 2003).
Isolation of BAC DNA, labeling by nick translation, and FISH were performed according to Jovtchev et al. (2008). BAC clones T2P11 (GeneBank accession number AC005508), T7N9 (AC000348), and F18C1 (AC011620) were labeled with biotin-dUTP, digoxigenin-dUTP, Alexa Fluor 488-5-dUTP, Cy3-dUTP, or Texas Red-12-dUTP. The 178-bp centromeric repeat probe was generated by PCR with specific primers from genomic DNA (Kawabe and Nasuda, 2005) and subsequently labeled with biotin-dUTP. For painting of the chromosome 1 top arm, 15 pools of in total 76 BACs were labeled with biotin-dUTP as described (Pecinka et al., 2004).
Microscopy Evaluation and Image Processing
Fluorescence signals in flow-sorted nuclei were analyzed using an Axioplan 2 (Zeiss) epifluorescence microscope with a ×100/1.4 Zeiss plan apochromat objective. In 4C nuclei, split FISH signals were considered to represent sister chromatid separation when their distance was larger than the signal diameter. Images were acquired separately for each fluorochrome using MetaVue (Molecular Devices) software, a cooled CCD camera (Spot 2e; Diagnostic Instruments), and appropriate excitation and emission filters. Monochromatic images were pseudocolored and merged using Adobe Photoshop 6.0 (Adobe Systems) software.
RNA Analysis
Total RNA was isolated from seedlings, rosette leaves, and floral buds using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Reverse transcription was performed using a first-strand cDNA synthesis kit (Fermentas), oligo(dT)18 primer (Fermentas), and 2 μg of total RNA as starting material. Primers used to amplify cDNA are listed in Supplemental Table 1 online. For conventional RT-PCR, PCR fragments were amplified using iCycler (Bio-Rad) and GoTaq Hot Start polymerase (Promega). All analyses were performed using three independent biological replicates. For quantitative RT-PCR, real-time RT-PCR was run using iCycler iQ (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). Each transcript was quantified twice using two independent biological replicates. As a control, a fragment of ACTIN2 cDNA was amplified for data normalization. The cDNA equivalent to 10 or 100 ng of total RNA was used in a 15-μL PCR reaction to amplify ACTIN2 cDNA or SMC6A and SMC6B cDNAs, respectively. The positions of the primers used to amplify SMC6A and SMC6B cDNAs are depicted in Supplemental Figure 1 online. PCR amplification/cycle graphs are shown to indicate that amplification was in logarithmic phase for each DNA molecule being analyzed (see Supplemental Figure 4 online).
HR Assay
Arabidopsis thaliana seeds were sterilized in 6% sodium hypochlorite solution with a small amount of Tween 20 for 7 min and rinsed five times with sterile water before being stored in sterile agarose solution (1%, w/v) for 1 d at 4°C for stratification. Subsequently, seeds were sown with a pipette on 90-mm Petri dishes (40 seeds per plate) containing GM-agar and were grown in a growth chamber (Percival CU-36L; CLF Laborgeräte) under tightly controlled conditions (16 h light, 24°C/8 h dark, 20°C/100 μmol/m2/s photosynthetic active radiation) for 7 d. Forty seedlings were then transferred with microceps into 90-mm Petri dishes filled with 18 mL of liquid GM and incubated for 24 h in the Percival growth chamber. Subsequently, 2 mL pure MMC (Duchefa) or Bleomycin (Duchefa) containing liquid GM was added to these plantlets, resulting in a final volume of 20 mL and a MMC or Bleomycin concentration of 5 μg/mL, respectively. The plants were then grown for another 5 d until histochemical staining.
Histochemical staining was performed as described (Schmidt-Puchta et al., 2004). Blue sectors were counted using a binocular microscope after the plants had been decolorized with 70% ethanol.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SMC5 (AT5G15920) mRNA, NM_121597; SMC6A (AT5G07660) mRNA, NM_125539; SMC6A mRNA (a transcript variant), FJ869873; SMC6B (AT5G61460) mRNA, NM_120848; NSE4A (AT1G51130) mRNA, NM_103992; and NSE4B (AT3G20760) mRNA, NM_112967.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Position of the Primers Used in Quantitative RT-PCR.
Supplemental Figure 2. Sensitivity of Arabidopsis Lines to Mitomycin C Treatment.
Supplemental Figure 3. Structure and Expression of δ-Kleisin Genes, NSE4A and NSE4B.
Supplemental Figure 4. PCR Amplification/Cycle Graphs in Quantitative Real-Time PCR.
Supplemental Table 1. Oligonucleotide Primers Used for RNA Analysis.
Supplemental Table 2. Homologous Recombination Events in Wild-Type and smc6 Mutants.
Supplemental Table 3. Frequencies of Positional Sister Chromatid Alignment in Mitomycin C–Treated 4C Nuclei.
Supplemental Table 4. Oligonucleotide Primers Used for Genotyping.
Supplemental Table 5. The Sequences of the Left Border Junctions of the T-DNA Insertion Lines.
Supplementary Material
Acknowledgments
We thank Bernd Reiss (Max-Planck-Institut für Züchtungsforschung Köln) for providing the Arabidopsis rad51-1 line, Anne Britt (University of California, Davis) for the lig4 line, Jörg Fuchs (Leibniz-Institut für Pflanzengenetic und Kulturpflanzenforschung Gatersleben) for help with flow-sorting, Ulrich Ryschka and Evelyn Klocke (Julius Kühne Institut, Quedlinburg) for X-irradiation, and Achim Bruder, Martina Kühne, Maren Nitze, and Rita Schubert for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ingo Schubert (schubert@ipk-gatersleben.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abdel-Halim, H.I., Imam, S.A., Badr, F.M., Natarajan, A.T., Mullenders, L.H., and Boei, J.J. (2004). Ionizing radiation-induced instant pairing of heterochromatin of homologous chromosomes in human cells. Cytogenet. Genome Res. 104 193–199. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bai, X., Peirson, B.N., Dong, F., Xue, C., and Makaroff, C.A. (1999). Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzel, A., and Kupiec, M. (2008). Finding a match: How do homologous sequences get together for recombination? Nat. Rev. Genet. 9 27–37. [DOI] [PubMed] [Google Scholar]
- Bernard, P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. (2001). Requirement of heterochromatin for cohesion at centromeres. Science 294 2539–2542. [DOI] [PubMed] [Google Scholar]
- Bhatt, A.M., Lister, C., Page, T., Fransz, P., Findlay, K., Jones, G.H., Dickinson, H.G., and Dean, C. (1999). The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19 463–472. [DOI] [PubMed] [Google Scholar]
- Bleuyard, J.Y., Gallego, M.E., Savigny, F., and White, C.I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41 533–545. [DOI] [PubMed] [Google Scholar]
- Boyko, A., Zemp, F., Filkowski, J., and Kovalchuk, I. (2006). Double-strand break repair in plants is developmentally regulated. Plant Physiol. 141 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, X., Dong, F., Edelmann, R.E., and Makaroff, C.A. (2003). The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116 2999–3007. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Shirayama, M., Shevchenko, A., Tanaka, T., Toth, A., and Nasmyth, K. (2000). Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5 243–254. [DOI] [PubMed] [Google Scholar]
- Cortes-Ledesma, F., and Aguilera, A. (2006). Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 7 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa-Nunes, J.A., Bhatt, A.M., O'Shea, S., West, C.E., Bray, C.M., Grossniklaus, U., and Dickinson, H.G. (2006). Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation. J. Exp. Bot. 57 971–983. [DOI] [PubMed] [Google Scholar]
- De Piccoli, G., et al. (2006). Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 8 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon, K., and Cedervall, B. (1995). Linear induction of DNA double-strand breakage with X-ray dose, as determined from DNA fragment size distribution. Radiat. Res. 142 153–162. [PubMed] [Google Scholar]
- Friesner, J., and Britt, A.B. (2003). Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 34 427–440. [DOI] [PubMed] [Google Scholar]
- Gisler, B., Salomon, S., and Puchta, H. (2002). The role of double-strand break-induced allelic homologous recombination in somatic plant cells. Plant J. 32 277–284. [DOI] [PubMed] [Google Scholar]
- Glynn, E.F., Megee, P.C., Yu, H.G., Mistrot, C., Unal, E., Koshland, D.E., DeRisi, J.L., and Gerton, J.L. (2004). Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2 E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli, J.M., Ünal, E., Guacci, V., and Koshland, D. (2008). The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell 31 47–56. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Soppe, W.J., Meister, A., Gernand, D., Turner, B.M., and Schubert, I. (2003). Histone modifications in Arabidopsis: High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 33 471–480. [DOI] [PubMed] [Google Scholar]
- Jovtchev, G., Watanabe, K., Pecinka, A., Rosin, F.M., Mette, M.F., Lam, E., and Schubert, I. (2008). Size and number of tandem repeat arrays can determine somatic homologous pairing of transgene loci mediated by epigenetic modifications in Arabidopsis thaliana nuclei. Chromosoma 117 267–276. [DOI] [PubMed] [Google Scholar]
- Kadyk, L.C., and Hartwell, L.H. (1992). Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar, R., Hoeijmakers, J.H., and van Gent, D.C. (1998). Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 8 483–489. [DOI] [PubMed] [Google Scholar]
- Kawabe, A., and Nasuda, S. (2005). Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol. Genet. Genomics 272 593–602. [DOI] [PubMed] [Google Scholar]
- Kozak, J., West, C.E., White, C., da Costa-Nunes, J.A., and Angelis, K.J. (2009). Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst.) 8 413–419. [DOI] [PubMed] [Google Scholar]
- Lehmann, A.R. (2005). The role of SMC proteins in the responses to DNA damage. DNA Repair (Amst.) 4 309–314. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., Katou, Y., Mori, S., Yokobayashi, S., Kelly, G.P., Itoh, T., Watanabe, Y., Shirahige, K., and Uhlmann, F. (2004). Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Chen, C., Markmann-Mulisch, U., Timofejeva, L., Schmelzer, E., Ma, H., and Reiss, B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos, H.B., Ström, L., Itoh, T., Katou, Y., Shirahige, K., and Sjögren, C. (2006). Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22 755–767. [DOI] [PubMed] [Google Scholar]
- Liu, C.M., McElver, J., Tzafrir, I., Joosen, R., Wittich, P., Patton, D., Van Lammeren, A.A., and Meinke, D. (2002). Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29 405–415. [DOI] [PubMed] [Google Scholar]
- Losada, A., and Hirano, T. (2005). Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19 1269–1287. [DOI] [PubMed] [Google Scholar]
- Malkova, A., Ivanov, E.L., and Haber, J.E. (1996). Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann-Mulisch, U., Wendeler, E., Zobell, O., Schween, G., Steinbiss, H.H., and Reiss, B. (2007). Differential requirements for RAD51 in Physcomitrella patens and Arabidopsis thaliana development and DNA damage repair. Plant Cell 19 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, O., Jasencakova, Z., Vaillant, I., Gendrel, A.V., Colot, V., Schubert, I., and Tourmente, S. (2003). Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P.C., Mistrot, C., Guacci, V., and Koshland, D. (1999). The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4 445–450. [DOI] [PubMed] [Google Scholar]
- Mengiste, T., Revenkova, E., Bechtold, N., and Paszkowski, J. (1999). An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 18 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, M., Chen, I., Angelis, K.J., and Schubert, I. (2001). DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat. Res. 493 87–93. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and Haering, C.H. (2005). The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74 595–648. [DOI] [PubMed] [Google Scholar]
- Orel, N., Kyryk, A., and Puchta, H. (2003). Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J. 35 604–612. [DOI] [PubMed] [Google Scholar]
- Palecek, J., Vidot, S., Feng, M., Doherty, A.J., and Lehmann, A.R. (2006). The Smc5-Smc6 DNA repair complex: Bridging of the Smc5-Smc6 heads by the kleisin, Nse4, and non-kleisin subunits. J. Biol. Chem. 281 36952–36959. [DOI] [PubMed] [Google Scholar]
- Pebernard, S., Perry, J.J., Tainer, J.A., and Boddy, M.N. (2008). Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol. Biol. Cell 19 4099–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard, S., Wohlschlegel, J., Yates III, J.R., and Boddy, M.N. (2006). The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 26 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka, A., Schubert, V., Meister, A., Kreth, G., Klatte, M., Lysak, M.A., Fuchs, J., and Schubert, I. (2004). Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113 258–269. [DOI] [PubMed] [Google Scholar]
- Peirson, B.N., Bowling, S.E., and Makaroff, C.A. (1997). A defect in synapsis causes male sterility in a T-DNA-tagged Arabidopsis thaliana mutant. Plant J. 11 659–669. [DOI] [PubMed] [Google Scholar]
- Potts, P.R., Porteus, M.H., and Yu, H. (2006). Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25 3377–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H. (1999). Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H. (2005). The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56 1–14. [DOI] [PubMed] [Google Scholar]
- Riha, K., Watson, J.M., Parkey, J., and Shippen, D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Puchta, W., Orel, N., Kirik, A., and Puchta, H. (2004). Intrachromosomal homologous recombination in Arabidopsis thaliana. Methods Mol. Biol. 262 25–35. [DOI] [PubMed] [Google Scholar]
- Schubert, I., Pecinka, A., Meister, A., Schubert, V., Klatte, M., and Jovtchev, G. (2004). DNA damage processing and aberration formation in plants. Cytogenet. Genome Res. 104 104–108. [DOI] [PubMed] [Google Scholar]
- Schubert, V., Kim, Y.M., Berr, A., Fuchs, J., Meister, A., Marschner, S., and Schubert, I. (2007). Random homologous pairing and incomplete sister chromatid alignment are common in angiosperm interphase nuclei. Mol. Genet. Genomics 278 167–176. [DOI] [PubMed] [Google Scholar]
- Schubert, V., Kim, Y.M., and Schubert, I. (2008). Arabidopsis sister chromatids often show complete alignment or separation along a 1.2-Mb euchromatic region but no cohesion “hot spots”. Chromosoma 117 261–266. [DOI] [PubMed] [Google Scholar]
- Schubert, V., Klatte, M., Pecinka, A., Meister, A., Jasencakova, Z., and Schubert, I. (2006). Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, V., Weissleder, A., Ali, H., Fuchs, J., Meister, A., Lermontova, I., and Schubert, I. (2009). Cehesin defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana. Chromosoma 118 591–605. [DOI] [PubMed] [Google Scholar]
- Schuermann, D., Molinier, J., Fritsch, O., and Hohn, B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21 172–181. [DOI] [PubMed] [Google Scholar]
- Sergeant, J., Taylor, E., Palecek, J., Fousteri, M., Andrews, E.A., Sweeney, S., Shinagawa, H., Watts, F.Z., and Lehmann, A.R. (2005). Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol. Cell. Biol. 25 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, T., Nishinaka, T., Mikawa, T., Aihara, H., Kurumizaka, H., Yokoyama, S., and Ito, Y. (2001). Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material. Proc. Natl. Acad. Sci. USA 98 8425–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren, C., and Nasmyth, K. (2001). Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11 991–995. [DOI] [PubMed] [Google Scholar]
- Soutoglou, E., Dorn, J.F., Sengupta, K., Jasin, M., Nussenzweig, A., Ried, T., Danuser, G., and Misteli, T. (2007). Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström, L., Karlsson, C., Lindroos, H.B., Wedahl, S., Katou, Y., Shirahige, K., and Sjögren, C. (2007). Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317 242–245. [DOI] [PubMed] [Google Scholar]
- Ström, L., Lindroos, H.B., Shirahige, K., and Sjögren, C. (2004). Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16 1003–1015. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Cosma, M.P., Wirth, K., and Nasmyth, K. (1999). Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98 847–858. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell, J., Machin, F., Farmer, S., Jarmuz, A., Eydmann, T., Dalgaard, J.Z., and Aragón, L. (2005). SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7 412–419. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., Sweeney, T.C., McElver, J., Aux, G., Patton, D., and Meinke, D. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135 1206–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., and Nasmyth, K. (1998). Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8 1095–1101. [DOI] [PubMed] [Google Scholar]
- Ünal, E., Arbel-Eden, A., Sattler, U., Shroff, R., Lichten, M., Haber, J.E., and Koshland, D. (2004). DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16 991–1002. [DOI] [PubMed] [Google Scholar]
- Ünal, E., Heidinger-Pauli, J.M., and Koshland, D. (2007). DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317 245–248. [DOI] [PubMed] [Google Scholar]
- Usui, T., Ogawa, H., and Petrini, J.H. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7 1255–1266. [DOI] [PubMed] [Google Scholar]
- Volpi, E.V., Sheer, D., and Uhlmann, F. (2001). Cohesion, but not too close. Curr. Biol. 11 R378. [DOI] [PubMed] [Google Scholar]
- Watrin, E., and Peters, J.M. (2006). Cohesin and DNA damage repair. Exp. Cell Res. 312 2687–2693. [DOI] [PubMed] [Google Scholar]
- Zhao, X., and Blobel, G. (2005). A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.