Abstract
A few membrane vesicle trafficking (SNARE) proteins in plants are associated with signaling and transmembrane ion transport, including control of plasma membrane ion channels. Vesicle traffic contributes to the population of ion channels at the plasma membrane. Nonetheless, it is unclear whether these SNAREs also interact directly to affect channel gating and, if so, what functional impact this might have on the plant. Here, we report that the Arabidopsis thaliana SNARE SYP121 binds to KC1, a regulatory K+ channel subunit that assembles with different inward-rectifying K+ channels to affect their activities. We demonstrate that SYP121 interacts preferentially with KC1 over other Kv-like K+ channel subunits and that KC1 interacts specifically with SYP121 but not with its closest structural and functional homolog SYP122 nor with another related SNARE SYP111. SYP121 promoted gating of the inward-rectifying K+ channel AKT1 but only when heterologously coexpressed with KC1. Mutation in any one of the three genes, SYP121, KC1, and AKT1, selectively suppressed the inward-rectifying K+ current in Arabidopsis root epidermal protoplasts as well as K+ acquisition and growth in seedlings when channel-mediated K+ uptake was limiting. That SYP121 should be important for gating of a K+ channel and its role in inorganic mineral nutrition demonstrates an unexpected role for SNARE–ion channel interactions, apparently divorced from signaling and vesicle traffic. Instead, it suggests a role in regulating K+ uptake coordinately with membrane expansion for cell growth.
INTRODUCTION
Vesicle traffic in all eukaryotic cells serves to shuttle membrane material, proteins, and soluble cargo between endomembrane compartments, the plasma membrane, and the extracellular space. Vesicles form by budding, and their delivery at the target membrane is achieved by fusion and intercalation of the lipid bilayers (Brunger, 2005; Sutter et al., 2006a; Lipka et al., 2007). These processes sustain cellular homeostasis and growth in yeast (Ungar and Hughson, 2003), they contribute to neurotransmitter release and nervous signal transmission across the synaptic junctions of nerves (Jahn et al., 2003), and they underpin cell polarity, growth, and development in plants (Campanoni and Blatt, 2007; Grefen and Blatt, 2008). SNARE (soluble N-ethylmaleimide–sensitive factor protein attachment protein receptor) proteins comprise a superfamily, conserved across all eukaryotes, and play important roles in the later stages of vesicle targeting and fusion. SNARE proteins overcome the dehydration forces associated with lipid bilayer fusion in an aqueous environment, and they match vesicles with their destinations for targeting and delivery of specific membrane proteins and cargo. Complementary SNAREs, identified by their core residues (either Arg or Gln) localize to different membranes and interact to form a tetrameric bundle of coiled helices that draws the membrane surfaces together for fusion. In reconstituted membrane preparations, this complex forms a minimal set of proteins required for fusion (Weber et al., 1998; Parlati et al., 1999; Hu et al., 2003). Other components, including the N-ethylmaleimide–sensitive factor, Sec1 and its homologs, affect SNARE conformations and their interactions in vivo (Brunger, 2005; Lipka et al., 2007; Bassham and Blatt, 2008). Nonetheless, the combinatorial specificity of SNARE interactions is thought to contribute significantly to membrane recognition and vesicle targeting (Paumet et al., 2004; Varlamov et al., 2004).
Of the core SNAREs in plants, the syntaxins (members of the Q-SNARE subclass; Sutter et al., 2006a) are the best characterized to date and, functionally, most intriguing. The large number of syntaxin-like SNAREs far outnumbers the identifiable membrane compartments, implying an overlap in trafficking functions and developmental and physiological specialization (Bassham and Blatt, 2008). Consistent with this idea, Arabidopsis thaliana harbors a subclade of Q-SNAREs that show no obvious homologies to any grouping among yeast and mammalian SNAREs but include at least one member that is found at and is functional in traffic to the plasma membrane (Alexandersson et al., 2004; Marmagne et al., 2004; Tyrrell et al., 2007). In many instances, too, the effects of vesicle traffic extend beyond the canonical roles in membrane targeting and vesicle fusion (Grefen and Blatt, 2008). SNARE-related vesicle traffic has been implicated, for example, in the spatial distribution of the auxin efflux carrier PIN1 (Steinmann et al., 1999), with consequences for auxin signaling and development (Dhonukshe et al., 2008), and the vacuolar SNAREs SYP22 and VTI11 are known to play important roles in gravitopism (Kato et al., 2002; Yano et al., 2003). In the latter case, the syp22 and vti11 mutations are associated with an abnormal vacuolar organization, raising the possibility of an indirect effect on the vacuolar membrane structure or composition and, consequently, on gravisensing (Saito et al., 2005), but little is known of the molecular basis for these observations.
SNAREs do have significant impacts on solute transport and its regulation across cellular membranes. Vesicle traffic is known to affect the population of receptors and membrane transport proteins at the plasma membrane and, thus, can be expected to modulate their activities over timescales of minutes to hours. SNARE-mediated trafficking of the mammalian glucose transporter, GLUT4, is one of the best-characterized examples for which delivery and subsequent membrane recycling is critical for insulin-dependent changes in glucose uptake (Bryant et al., 2002). Recent studies have uncovered several instances in which vesicle traffic plays important roles in ion transport, signaling, and response in plants too, notably in basal defense responses to fungal pathogens (Collins et al., 2003) and the bacterial flagellin elicitor flg22 (Robatzek et al., 2006), in constitutive turnover of the BOR1 boron transporter (Takano et al., 2005), and in the delivery, endocytosis, and recycling of the KAT1 K+ channel triggered by abscisic acid (Sutter et al., 2006b, 2007). The latter studies, especially, underscore a remarkable plasticity to posttranslational regulation of ion transport at the plasma membrane.
It is plausible, too, that some plant SNAREs influence membrane ion transport independent of any functions in vesicle traffic. Indeed, a few SNARE proteins in animals are known to interact directly with K+ and Ca2+ channels, notably in neuromuscular and neuroendocrine tissues, to facilitate neurotransmitter release and nervous signal transmission as well as hormone secretion (Jahn et al., 2003; Leung et al., 2007). Previously, we identified the SNAREs SYP121 (=SYR1) of tobacco (Nicotiana tabacum) and its Arabidopsis homolog SYP121 (=SYR1/PEN1) in a screen for signaling elements associated with abscisic acid and drought (Leyman et al., 1999).We found that disrupting SNARE function, by introducing a dominant-negative, cytosolic (so-called) Sp2 fragment in vivo, blocked both K+ and Cl− channel responses to abscisic acid in guard cells (Leyman et al., 1999) and had severe effects on growth, tissue development, and on traffic to the plasma membrane (Geelen et al., 2002; Sutter et al., 2006b). Both tobacco and Arabidopsis SYP121 drive vesicle fusion late in the secretory pathway to the plasma membrane (Geelen et al., 2002), the latter in a canonical SNARE complex (Kwon et al., 2008), and both SNAREs are important for KAT1 K+ channel traffic, distribution, and anchoring within the plasma membrane (Sutter et al., 2006b). However, subsequent work ruled out membrane traffic per se as a mechanism for K+ channel regulation, at least in the short term (Sutter et al., 2007), leaving open a question whether these SNAREs might affect the activities of one or more plant ion channels through direct interaction at the plasma membrane. In exploring alternatives for SNARE-dependent control of plant K+ channels, we screened for protein partners that interact directly with SYP121. Here, we report that SYP121 binds the regulatory (silent) K+ channel subunit KC1, which assembles with different K+ channels to affect their activities. We have focused on its association with the AKT1 K+ channel and show that SYP121 is needed to promote channel gating, AKT1-associated K+ current, K+ uptake at the root epidermis, and for growth when channel-mediated K+ uptake is limiting. These results demonstrate an unexpected role for the SNARE as part of a tripartite protein complex facilitating plant nutrient acquisition, analogous to SNARE-ion channel complexes of mammals but seemingly unrelated to signaling and its coupling to vesicle traffic.
RESULTS
SYP121 Interacts Selectively with KC1
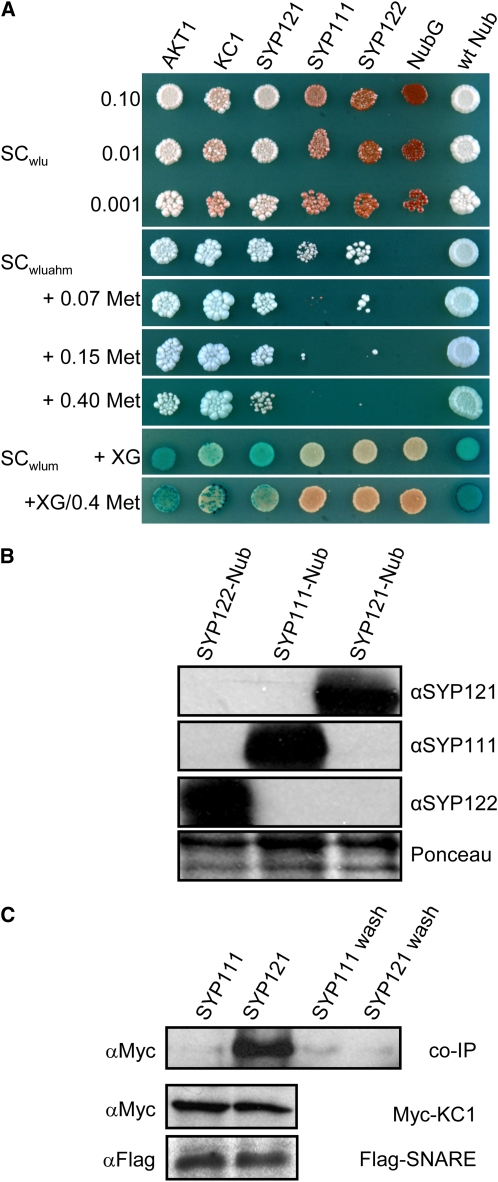
In a search for protein partners of SYP121, we screened a library of 500 integral membrane proteins from Arabidopsis, including all nine Kv-like K+ channels, using a mating-based split-ubiquitin assay for interacting proteins (Obrdlik et al., 2004; Grefen et al., 2009) that gives Ade- and His-independent, Met-sensitive rescue of yeast growth on minimal medium and activation of β-galactosidase activity. Screens were performed in parallel with other Arabidopsis SNAREs as controls for the specificity of interaction. In addition to SYP121, we made use of fusions incorporating the cytokinesis-associated SYP111 (=Knolle) (Jurgens, 2005) and one other plasma membrane SNARE SYP122, the closest homolog to SYP121 with 64% amino acid sequence identity and with which it shares partial functional redundancy in vivo (Assaad et al., 2004; Zhang et al., 2007; Bassham and Blatt, 2008). Of five putative targets identified, the K+ channel subunit KC1 was found to interact strongly and selectively with SYP121, by contrast with SYP111 or SYP122 when expressed at similar levels, and the interaction was evident even when plated on Met concentrations as high as 0.4 mM to suppress expression of the bait construct (Figures 1A and 1B). Conversely, SYP121 interacted preferentially with KC1, by contrast with the Kv-like K+ channel AKT1 (see Supplemental Figure 1 online).
Figure 1.
The Silent K+ Channel KC1 Interacts with the SNARE SYP121 in Arabidopsis.
(A) Yeast mating-based split-ubiquitin assay for interaction with KC1-Cub. Yeast diploids created with Nub-X constructs of AKT1, KC1, SYP121, SYP111, SYP122, and controls (negative, NubG; positive, wild-type Nub) spotted (left to right) on synthetic complete (SC) medium without Trp, Leu, and Ura (SCwlu) to verify crossing and test for adenine synthesis (white colonies, top panel). SC without Trp, Leu, Ura, Ade, His, and Met (SCwluahm) used to verify Ade- and His-independent growth (second panel), and the addition of 0.07, 0.15, and 0.40 mM Met (next three panels) used to verify interaction at lower KC1-Cub expression levels. SC medium without Trp, Leu, Ura, and Met (SCwlum; last two panels) alone and with addition of 0.4 mM Met used with an overlay of X-Gal–containing agarose to assay β-galactosidase activity (Obrdlik et al., 2004). Serial dilutions 0.1, 0.01, and 0.001 of diploid cultures as indicated for spots on SCwlu. Otherwise, only 0.1 dilutions are shown. Note KC1 interaction with AKT1, itself, and SYP121 and the absence of specific interaction with SYP111 and SYP122.
(B) Verification of prey protein expression in diploid yeasts carrying KC1-Cub and Nub-SNAREs. Protein gel blot analysis of total protein extracted from yeast diploids expressing KC1-Cub with Nub-SYP122, Nub-SYP111, and Nub-SYP121 using antibodies specific for SYP122, SYP111, or SYP121 (top panels). Ponceau S stain was used as loading control (bottom panel).
(C) Coimmunoprecipitation of Myc-tagged KC1 by retention with Flag-tagged SYP121 on anti-Flag-coupled Sepharose. Protein gel blot analysis (top, left to right) of eluates coimmunoprecipitating with Flag-tagged SYP111, with SYP121, and the precipitated terminal washes prior to elution from each, respectively, probed with anti-Myc antibody. Control protein gel blots (bottom) show equivalent levels of expression for KC1 and for the SNAREs in each solubilized fraction after coexpression in Sf9 insect cells.
To test the interaction in vitro, we expressed the SNAREs and KC1 in Sf9 insect cells for solubilization and coimmunoprecipitation. Open reading frames were amplified to include N-terminal Flag-tags for the SNAREs and a C-terminal Myc-tag for KC1 before cloning in pVL1393 and incorporation in baculovirus. Baculovirus titers were adjusted to give coexpression with similar levels of the SNAREs and SNARE:KC1 ratios. We harvested and incubated solubilized membrane fractions with antibody-coupled Sepharose. The Sepharose was washed, and bound proteins were eluted. We also collected the washes for precipitation and resolubilization of any released proteins before protein gel blot analysis. Figure 1C illustrates one of five experiments, each of which yielded a similar outcome, in this case showing a selective coimmunoprecipitation with SYP121, but not with SYP111. Immunoprecipitation of SYP121 with Flag-coupled Sepharose yielded a coimmunoprecipitating band of ∼78 kD corresponding to the tagged KC1, which labeled with the anti-Myc antibody. By contrast, immunoprecipitation with the Flag-tagged SYP111 did not yield a band detected by the anti-Myc antibody, although both extracts included similar levels of the epitope-tagged SNARE and KC1 proteins. Included in the protein gel blot analysis shown, the right-most lanes are of total protein precipitated from the final washes prior to elution, confirming the absence of nonspecific retention of the epitopes on the resin (Fletcher et al., 2003).
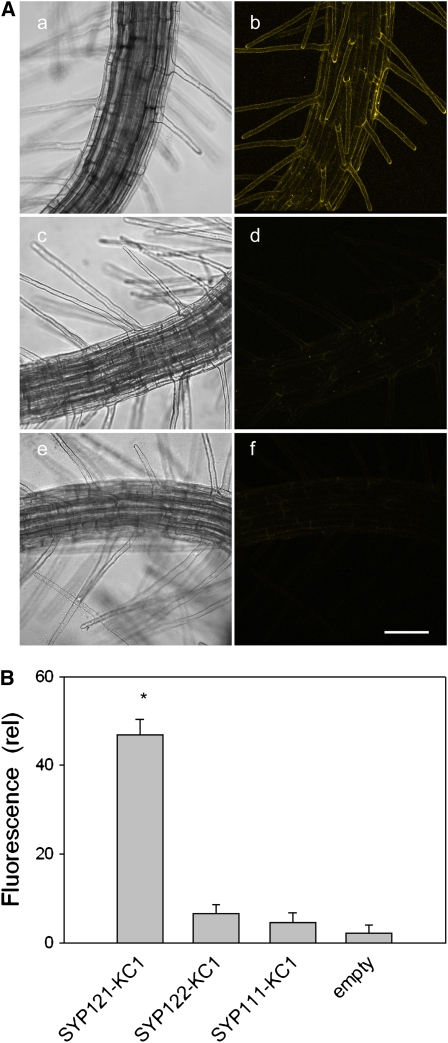
Interactions on heterologous expression in yeast and in vitro following expression in Sf9 insect cells support the idea of an association specific to SYP121 and KC1 but do not rule out the possibility that additional components unique to the plant might be important and support a broader range of SNARE interactions. We therefore made use of a bimolecular fluorescence complementation (BiFC) assay (Walter et al., 2004) to test these interactions in vivo. Constructs incorporating the open reading frames for KC1, SYP121, SYP122, and SYP111 fused to the N- and C-terminal halves of yellow fluorescent protein (YFP) were used to transform Agrobacterium rhizogenes and were transiently expressed in Arabidopsis root epidermis by cocultivation with the Agrobacterium carrying different combinations of the corresponding fusion constructs (Campanoni et al., 2007). Because plant ion channels generally express at levels too low for detection by fluorescence microscopy, expression was driven by the 35S promoter. There is a potential for mistargeting when a protein is overexpressed. Nonetheless, ion channel distributions, and that of most SNAREs, generally align with the native protein in vivo when driven by the constitutive promoter (Uemura et al., 2004; Sutter et al., 2006b, 2007).
We used confocal laser scanning microscopy to quantify and compare fluorescence signals and their distributions obtained on expressing KC1 fused with both the N- and C-terminal halves of YFP with different combinations of SNARE fusions. Confocal stacks were used to derive three-dimensional image reconstructions, and these images were then analyzed for YFP fluorescence intensity after background subtraction. We found pronounced YFP fluorescence in epidermal cells and root hairs when seedlings were transfected with complementary BiFC constructs fused to SYP121 and KC1. By contrast, cocultivation of BiFC fusions with KC1 paired with SYP122 and SYP111, both of which failed to rescue yeast growth in split-ubiquitin assays, yielded fluorescence images comparable to background images obtained from seedlings cocultivated with untransformed Agrobacterium and with mismatched BiFC fusions as controls (Figure 2A). Similar results were obtained in six or more independent experiments for each set of combinations (see Supplemental Figure 2 online). Quantification of YFP fluorescence (Figure 2B) confirmed the absence of significant differences above background when KC1 fusion constructs were expressed with complementary fusions of SYP111 and SYP122 and showed an approximately fivefold enhancement of the fluorescence signal in plants expressing the complementary KC1 and SYP121 fusion constructs. In principle, these results could yield false-positive interactions as a consequence of overexpression. However, the absence of an interaction with close homologs of SYP121 militates against this idea. We found the YFP fluorescence was visibly distinct from the cytoplasm and tonoplast (data not shown; see Figure 7), and it failed to recover after local photobleaching (see Supplemental Figure 2 and Supplemental Movie 1 online), indicating that the complex was not mobile within the cytosol or within a circulating endomembrane compartment. Together with the split-ubiquitin and coimmunoprecipitation assays, these findings led us to conclude that the interaction between SYP121 and KC1 is specific and to discount nonspecific effects associated with enhanced or heterologous expression. We return to these points below.
Figure 2.
Selective BiFC of KC1 with SYP121 Expressed in Arabidopsis Root Epidermis as Fusion Constructs with the N- and C-Terminal Halves of YFP (nYFP and cYFP), Respectively.
(A) Three-dimensional reconstructions from confocal fluorescence image stacks of seedlings expressing (bright-field, left; fluorescence, right) SYP121-nYFP with KC1-cYFP ([a] and [b]), SYP122-nYFP with KC1-cYFP ([c] and [d]), and with the empty (untransformed) Agrobacterium ([e] and [f]). Bar = 50 μm. Images with SYP111-nYFP paired with KC1-cYFP were similar to those of (c) and (d).
(B) Mean fluorescence intensities (arbitrary units) ± se after correction for background of (nontransfected) control measurements (n > 6 independent experiments in each case; * indicates significant difference from the empty control at P < 0.01). See also Supplemental Figure 2 and Supplemental Movie 1 online.
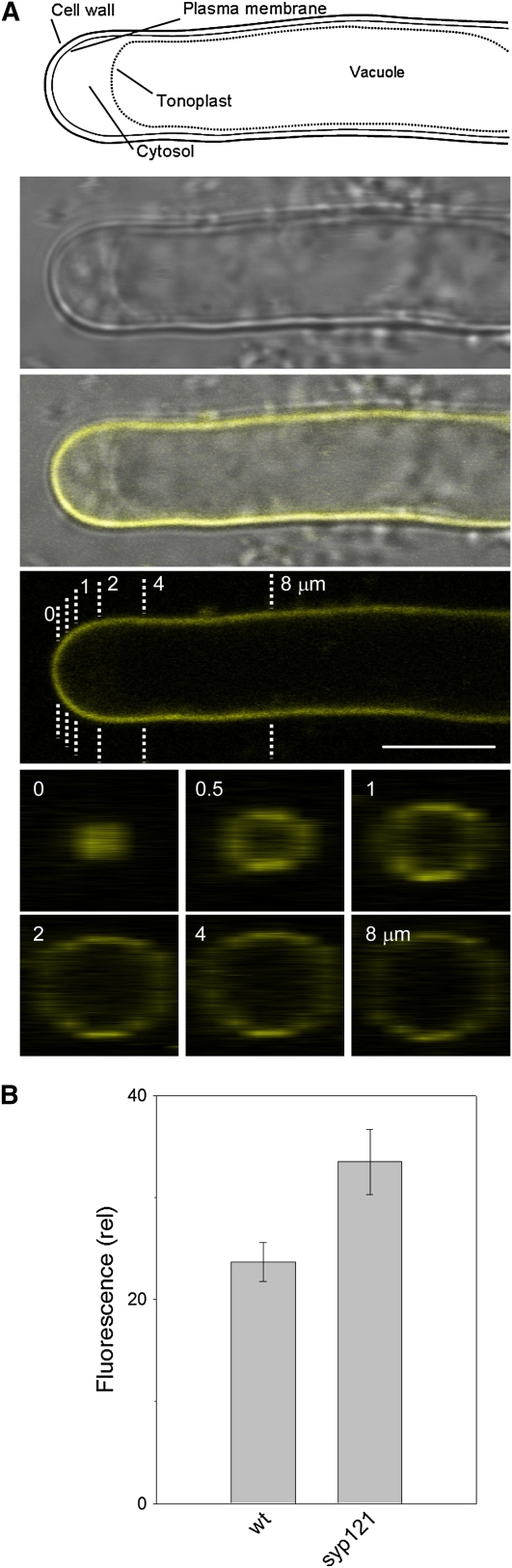
Figure 7.
The syp121-1 Mutation Does Not Affect K+ Channel Expression at the Root Epidermal Plasma Membrane.
(A) Longitudinal optical section (top three panels: bright-field, composite, YFP fluorescence) and three-dimensional transect analysis (bottom six panels) of KC1-YFP distribution in an Arabidopsis syp121-1 mutant root hair. Transects taken at positions (in μm) from the apex as indicated by the dashed lines above. Note the peripheral distribution of the fluorescence and its virtual absence from the dense cytosol and tonoplast boundary behind the tip (top, labeled schematic illustration drawn to scale for reference). Similar results were obtained in each of nine separate experiments with the syp121 mutant and in five experiments with expression in wild-type seedlings (data not shown). Bar = 5 μm.
(B) Mean fluorescence intensities ± se (arbitrary units) of root hairs expressing AKT1-GFP after correction for background of control measurements transfected with nontransformed Agrobacterium (n > 12 independent experiments in each case; means differ significantly at P < 0.05).
SYP121–KC1 Interaction Promotes K+ Channel Gating
To explore the functional impact of SYP121–KC1 interaction, we performed electrophysiological analyses after coexpression with the inward-rectifying K+ channel subunit AKT1. KC1 is a so-called silent K+ channel subunit; expressed on its own, it does not yield measurable K+ currents, but it interacts with different inward-rectifying K+ channel subunits, including AKT1 (Obrdlik et al., 2004; Duby et al., 2008) and affects the voltage dependence of channel gating (Dreyer et al., 1997). AKT1 and KC1 assemble preferentially to form functional channels as heteromers (Duby et al., 2008), and genetic elimination of either subunit suppresses the K+ current in the root epidermis (Hirsch et al., 1998; Reintanz et al., 2002), indicating that in vivo the inward-rectifying K+ channels in Arabidopsis roots are dominated by the heteromeric AKT1-KC1 assembly. When expressed heterologously on its own, AKT1 does yield an inward-rectifying K+ current (Gaymard et al., 1996; Duby et al., 2008). However, a review of the literature (Gaymard et al., 1996; Reintanz et al., 2002; Xu et al., 2006; Duby et al., 2008) shows that its gating, and that of AKT1 expressed together with KC1, differs fundamentally from K+ currents in vivo (see Figure 4 and below), suggesting a missing component needed for gating in the plant.
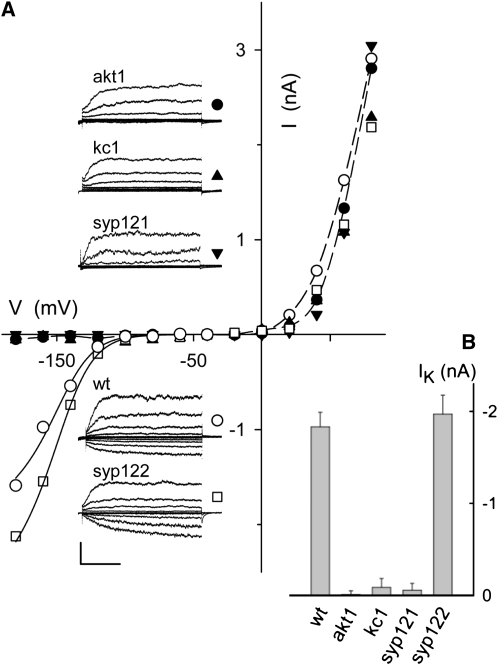
Figure 4.
syp121 and kc1 Mutations Phenocopy the akt1 Mutant in Suppressing Inward-Rectifying K+ Current.
(A) Whole-cell currents (insets) and steady state current-voltage curves from representative Arabidopsis root epidermal protoplasts in 100 mM K+ under voltage clamp, cross-referenced by symbol. Scale bar for current traces (below): 2 nA and 1 s. Clamp cycles: holding voltage, −50 mV; voltage steps, +80 to −180 mV, tailing voltage, −50 mV. Dashed curves in the current voltage plots are empirical polynomial fittings for outward-rectifying K+ currents from wild-type and akt1 mutant protoplasts and are included for visual guidance only. Solid curves are results of joint fittings of inward-rectifying K+ currents to a Boltzmann function (see Figure 3 legend) with V1/2 and δ held in common between wild-type and syp122 mutant protoplasts. The parameter values, δ, −1.97 ± 0.13 and V1/2, −142 ± 5 mV, were statistically equivalent to similar fittings for K+ currents on heterologous expression of AKT1 with KC1 and SYP121 (Figure 3; see also Supplemental Figure 3 online). Note that protoplasts from the akt1, kc1, and syp121 mutants, but not from wild-type and syp122 mutant plants, all showed a loss in K+ current at voltages negative of −100 mV. Records from all protoplasts retain the outward-rectifying K+ current that is nominally dependent on the GORK K+ channel.
(B) Summary of inward-rectifying K+ current amplitudes recorded from wild-type and mutant Arabidopsis root epidermal protoplasts. Data are mean steady state amplitudes ± se obtained at −160 mV from five to seven independent experiments in each case. Means for akt1, kc1, and syp121 mutants are significantly different from the wild type at P < 0.01.
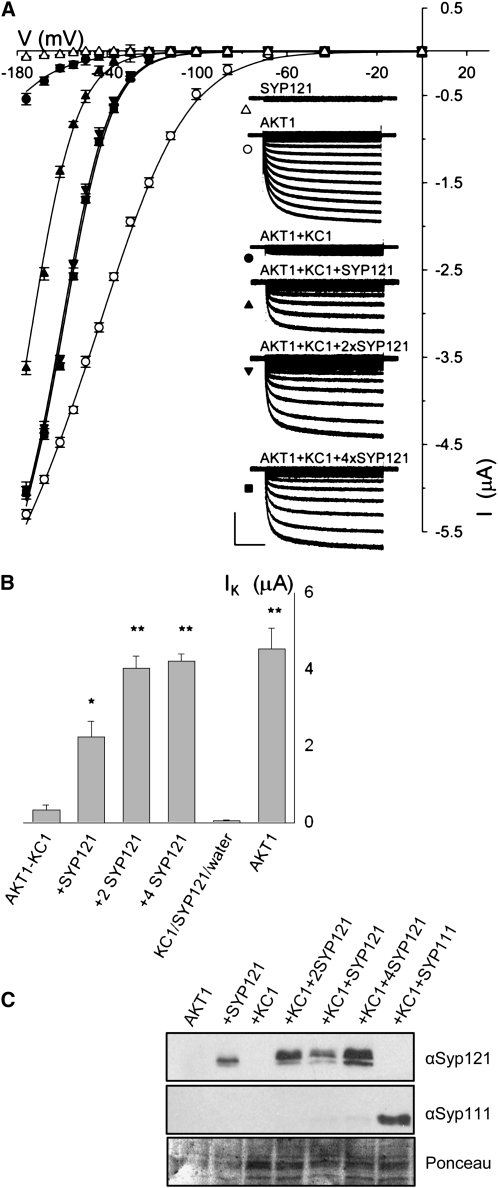
Figure 3 and Table 1 summarize measurements on heterologous expression in Xenopus laevis oocytes and underscores the functional importance of the SYP121–KC1 interaction for the K+ current. Similar results were obtained from baculovirus-transfected Sf9 insect cells (see Supplemental Figure 3 online) using the same constructs employed for the coimmunoprecipitation studies (above). To ensure activation of AKT1 in oocytes, all combinations of channels and SNAREs were coexpressed with the protein kinase CIPK23 and calcineurin-B-like activator CBL1 (Li and Luan, 2006; Xu et al., 2006). However, expression of the K+ current in Sf9 insect cells did not require cotransfection and expression with the CBL and CIPK proteins (Gaymard et al., 1996), presumably because the insect cells include an endogenous protein kinase that activates the K+ channel. In each case, we found that expressing AKT1 together with KC1 gave a K+ current measurable only at voltages negative of −140 mV (Duby et al., 2008), while expression of AKT1 and KC1 with SYP121 yielded a K+ current at voltages near and negative of −100 mV. The current and gating characteristics obtained with the three genes expressed together were statistically equivalent to those observed in the plant. In general, voltage-dependent gating of a channel current in whole-cell recordings can be defined by its characteristic midpoint for channel activation, V1/2, and the gating charge or steepness of its response to a change in voltage, δ (Dreyer and Blatt, 2009). Fitting currents with SYP121 to a Boltzmann function gave a gating charge of 2.18 ± 0.12 (n = 8) in oocytes and 2.16 ± 0.07 in Sf9 insect cells, which compared favorably with a value of 1.97 ± 0.13 (n = 8) obtained in the plant (mean ± se, significant at P < 0.05; cf. Figures 3 and 4, Table 1). Analysis of these K+ currents indicated voltages yielding half-maximal activation (V1/2) near −155 mV, close to the value of −152 ± 5 mV obtained in the plant and comparable to similar measurements from protoplasts of wheat (Triticum aestivum; Gassmann and Schroeder, 1994; Buschmann et al., 2000) and rye (Secale cereale) roots (White and Lemtirichlieh, 1995). We also noted that increasing SYP121:KC1 expression ratios progressively displaced the current characteristic to more positive voltages (Figure 3, Table 1), consistent with a well-defined and saturable stoichiometry of KC1 and SYP121 protein function in modifying channel gating.
Figure 3.
Coexpression with SYP121 Selectively Rescues AKT1-KC1 K+ Current in Xenopus Oocytes.
(A) Current traces and steady state current voltage curves recorded under voltage clamp in 96 mM K+ from oocytes expressing SYP121 alone (triangles), AKT1 alone (open circles), AKT1 with KC1 (closed circles∂, molar ratio 1:1), and AKT1 with KC1 and SYP121 (SYP121:KC1 cRNA molar ratios: 1:1, upright triangle; 2:1, downward triangle; 4:1, square). Clamp cycles: holding voltage, −50 mV; voltage steps, 0 to −180 mV. Insets: Corresponding whole-cell currents cross-referenced by symbol. Scale: 2 μA and 1 s. Currents from oocytes injected with water and with KC1 cRNA only gave results similar to those for SYP121, and currents from oocytes injected with cRNAs for AKT1 together with KC1 and SYP111 or SYP122 were indistinguishable from those injected with cRNAs for AKT1 plus KC1 alone. Currents from oocytes injected with cRNAs for AKT1 and SYP121 showed gating characteristics similar to those for AKT1 alone and expressing KC1 alone or with SYP121 failed to yield a measureable current (data not shown). All measurements performed as coexpressions with CBL1 and CIPK23 essential for AKT1 function in oocytes in 1:1:1 molar ratios with AKT1 (Xu et al., 2006), and no current was observed in the absence of CBL1/CIPK23 expression (data not shown). Final cRNA volume for each oocyte was equal. Similar results were obtained with the same construct combinations when expressed in Sf9 insect cells (see Supplemental Figure 3 online) without the additional expression load of CBL1/CIPK23. Solid curves are the results of joint, nonlinear least squares fitting of the K+ currents (IK) to a Boltzmann function IK = gmax(V − EK)/(1 + eδ(V−V1/2)/RT), where gmax is the conductance maximum, and V, EK, R, and T have their usual meanings. The characteristic voltage dependence (V1/2) indicates the midpoint of the voltage range for gating, and the apparent gating charge (δ) is an unique property of the gate, its sensitivity to voltage changes and the associated conformations. Best fittings were obtained with gmax held in common and with separate, joint values for δ with and without SYP121 expression. Similar results were obtained in each of eight separate analyses. (see Table 1).
(B) Summary of K+ current amplitudes recorded from oocytes expressing AKT with KC1 in combinations with SYP121 (cRNA injection ratios indicated) and expressing AKT1 alone. Data are means ±se obtained at -160 mV from >9 independent measurements in each case (significant difference from AKT1+KC1 ** at P < 0.01 and * at P < 0.05).
(C) Verification of SNARE protein expression in oocytes. Oocytes were injected with AKT1, CIPK23, CBL1 cRNA (AKT1, molar ratios 1:1:1), and with SYP121 (+SYP121, molar ratios 1:1:1:1), KC1 (+KC1, molar ratios 1:1:1:1), KC1 and Syp121 (+KC1+SYP121, molar ratios 1:1:1:1:1; +KC1+2SYP121, molar ratios 1:1:1:1:2; +KC1+4SYP121, molar ratios 1:1:1:1:4), and KC1 and SYP111 (+KC1+SYP111, molar ratios 1:1:1:1:1). Protein gel blot analysis of total membrane protein extracted from oocytes collected after electrical recordings detected with antibodies specific to SYP121 and SYP111. Ponceau S stain was used to normalize SYP121 expression levels for lanes with KC1 and yielded ratios of 1: 2.04:3.4.
Table 1.
Coexpression with SYP121 Recovers Native AKT1-KC1 K+ Channel Gating Characteristics in Xenopus Oocytes
| Expressed Proteins | V1/2 (mV) | δ |
|---|---|---|
| AKT1 | −128 ±5 | −1.21 ±0.03 |
| AKT1+KC1 | −208 ± 7 | |
| AKT1+KC1 +SYP121 (1:1) | −174 ± 5* | −2.18 ±0.12** |
| +KC1 +SYP121 (1:2) | −160 ± 2** | |
| +KC1 +SYP121 (1:4) |
−155 ± 2** |
Results of joint, nonlinear, least squares fitting of K+ currents as described in Figure 3. For purposes of joint fittings, best results were obtained with the gating charge, δ, held in common for currents from oocytes expressing AKT1 and AKT1+KC1 and separately for currents from oocytes expressing combinations of AKT1+KC1 with SYP121. Values for the voltage giving half-maximal conductance, V1/2, were allowed free between data sets. Data are from eight separate analysis sets. Both analyses and visual inspections showed roughly a twofold increase in δ and a saturable shift in V1/2, with SYP121 inclusions as summarized below (means ± se, significant difference from AKT1+KC1: *, P < 0.05; **, P < 0.01), consistent with inward-rectifying K+ channel characteristics in the plant (see Figure 4).
By contrast, expressing AKT1 alone also yielded a current, but showed anomalous characteristics unlike those in the plant. Equivalent results were obtained on expressing AKT1 with SYP121 but without KC1 (data not shown). In each case, recordings showed an appreciable current at voltages positive of −100 mV and fittings yielded values for gating charge and V1/2 of 1.21 ± 0.03 and −128 ± 5 mV (n = 12), respectively (Figure 3, Table 1). Roughly the same relationships were evident on analyzing currents from Sf9 insect cells: expressing AKT1 alone (see Supplemental Figure 3 online) yielded mean V1/2 and gating charge values of −115 ± 3 mV and 1.12 ± 0.06 (n = 6), respectively. Currents obtained with AKT1 and KC1 were well-fitted jointly with the same anomalous gating charge (Figure 3; Supplemental Figure 3 online). Fittings of the AKT1-KC1 current assumed a common conductance maximum and V1/2 beyond the range of measurable currents and must therefore be viewed with caution but are consistent with previous analyses and observations that KC1 coexpression does not affect significantly the gating charge of the K+ channels (Duby et al., 2008). This proviso aside, the findings indicate profound changes to channel gating with SYP121. Given the similarities of the K+ currents in the plant with those of AKT1 expressed with KC1 and SYP121, they also implicate the SNARE as a missing component of the AKT1-KC1 K+ channel complex.
SYP121 and KC1 Activate K+ Channel Current and K+ Uptake
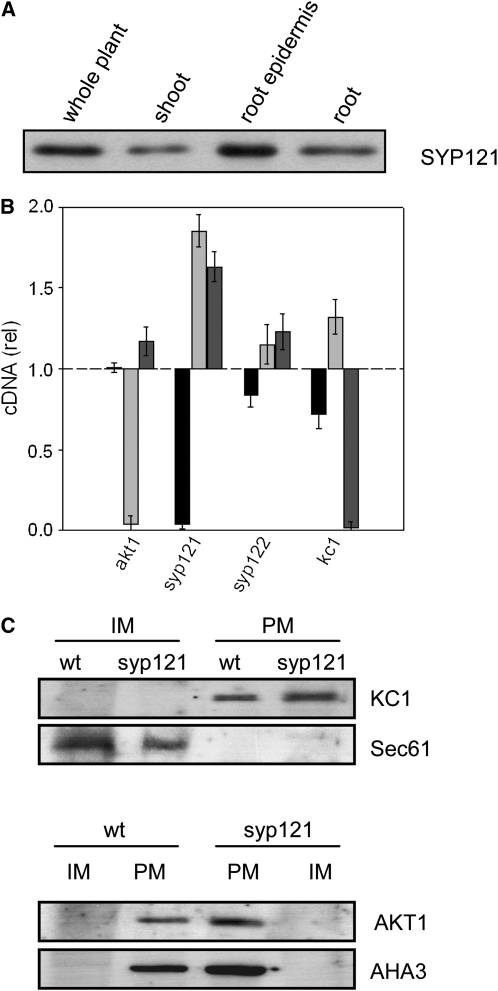
To examine the impact of SYP121 on K+ currents and channel gating in the plant, we made use of patch clamp recordings with protoplasts from seedlings of wild-type and mutant Arabidopsis lines carrying lesions in various SNAREs and the K+ channels. Additionally, we examined the effects of these mutants on seedling growth and K+ nutrition. In Arabidopsis, AKT1 is expressed principally in the root, notably in the epidermis where, together with KC1, it contributes to K+ uptake and nutrition (Lagarde et al., 1996; Reintanz et al., 2002; Xu et al., 2006). SYP121 is found throughout the plant (Leyman et al., 1999; Collins et al., 2003) and is strongly expressed in the root, including the root epidermis (Figure 6A). We therefore performed whole-cell patch clamp measurements with protoplasts derived from the root epidermis, comparing wild-type protoplasts with those from syp121-1 (Collins et al., 2003), kc1-2 (H. Sentenac, personal communication), akt1-1 (Hirsch et al., 1998) mutants and, as a control, syp122-1 (Zhang et al., 2007) mutant plants. For verification, additional measurements made use of the independent alleles kc1-f (Reintanz et al., 2002), syp121-2, syp121-4 (Zhang et al., 2007), and SYP121 and SYP122 complementations of syp121 (Pajonk et al., 2008). The syp121-1, syp121-2, and syp121-4 mutants arise from single point mutations that result in translational terminations. The remaining mutations arise from single-gene, T-DNA (kc1-2, akt1-1, and syp122-1) or transposon (kc1-f) insertions that give null mutations in the corresponding genes. Measurements were performed on protoplasts from both Columbia-0 (Col-0) and Wassilewskija (Ws) wild-type plants, as the SNARE mutants are in the Col-0 background, but the akt1 and kc1-2 mutants are in the Ws background. No substantive differences in K+ current were found between the Col-0 and Ws ecotypes (data not shown) (Gierth et al., 2005), and we pooled these results.
Figure 6.
SNARE and K+ Channel Transcription and Expression in Arabidopsis.
(A) The SNARE SYP121 is expressed strongly in the Arabidopsis root and root epidermis. Protein gel blot analysis of total proteins (10 μg/lane) extracted from whole Arabidopsis, shoot, root, and root epidermis and probed with anti-SYP121 primary antibody. All four lanes yielded a single band close to 37 kD, corresponding to SYP121 (Tyrrell et al., 2007) and consistent with the expression patterns for AKT1 and KC1 (Birnbaum et al., 2003).
(B) Quantification of SYP121 and K+ channel transcripts in the wild type and mutant Arabidopsis. Real-time PCR of SYP121 (black bars), AKT1 (light-gray bars), and KC1 (dark-gray bars) transcript levels in each of the mutant lines syp121-1, syp122-1, akt1-1, and kc1-1 after standardization on ACT2 transcript levels. Data normalized to the corresponding amplification yields in the wild type and are means ± se from three independent experiments. No appreciable decrease was evident for the K+ channel subunits in either of the SNARE mutant or the complementary K+ channel mutant lines. A significant increase (P < 0.05) in relative transcript level was evident in the kc1 and akt1 mutants for AKT1 and KC1 genes, respectively.
(C) KC1 K+ channel localization to the plasma membrane is unaffected in syp121-1 mutant Arabidopsis. Protein gel blot analysis of plasma membrane (PM) and inner membrane (IM) fractions separated by two-phase partitioning of microsomal membranes isolated from roots of wild-type and syp121-1 mutant plants. Parallel SDS-PAGE was run of all fractions (1.3 μg protein/lane), and PVDF membranes were probed with polyclonal antibodies to KC1 and AKT1 (see Supplemental Figure 6 online) before stripping and reprobing with antibodies to the endoplasmic reticulum Sec61 (Yuasa et al., 2005) as a marker for inner membranes and with polyclonal antibody to the H+-ATPase AHA3 (Pardo and Serrano, 1989) as a marker for the plasma membrane. Protein gel blots were visualized by 125I radiotracer phosphor imaging.
Patch clamp records from protoplasts of the wild-type plants uniformly showed both inward- and outward-rectifying currents characteristic of the K+ channels found in these tissues and associated with the AKT1, KC1, and GORK K+ channel genes (Lagarde et al., 1996; Hirsch et al., 1998; Ivashikina et al., 2001; Reintanz et al., 2002). As expected for the K+ channels, these currents were subject to extracellular K+ concentration and were blocked by the K+ channel blockers Cs+ and tetraethylammonium chloride (Very and Sentenac, 2003; Dreyer and Blatt, 2009). Figure 4 shows representative measurements from protoplasts of the wild type and each of several mutant Arabidopsis lines bathed in 100 mM K+. In protoplasts from the wild-type plants, the outward-rectifying K+ channels were characterized by a rising current, evident at clamp voltages positive of 0 mV, that relaxed to steady state with halftimes (t1/2) of 200 to 300 ms. Similar characteristics were observed for the outward-rectifying currents from each of the mutant lines. The wild-type plants also yielded protoplasts with an inward-rectifying K+ current that relaxed with halftimes of 300 to 400 ms and was evident principally at voltages near and negative of −120 mV. Analysis of the gating characteristics in eight independent experiments from wild-type protoplasts yielded a gating charge of 1.97 ± 0.13 and V1/2 of −152 ± 5 mV. A similar inward-rectifying K+ current was found in protoplasts from the syp122-1 mutant. However, the inward-rectifying current was absent in protoplasts from the akt1 mutant, as Hirsch et al. (1998) reported previously. It was also virtually absent in protoplasts from the syp121 and the kc1 mutant plants. Finally, we found that the inward-rectifying K+ current was restored in syp121-1 mutant Arabidopsis, yielding currents equivalent to the wild type (Figure 4, ○) when complemented with fluorescently tagged constructs of SYP121 driven by the SYP121 and 35S promoters. These observations underscore the importance of all three genes, SYP121, KC1, and AKT1, for expression of the K+ current in the plant. The fact that none of the mutants showed an obvious effect on the outward-rectifying K+ current associated with the GORK K+ channel in the same cells indicated that SYP121, KC1, and AKT1 were essential only for the inward-rectifying K+ current. (At first glance, the loss of inward current in the kc1 mutants appeared at odds with one previous study [Reintanz et al., 2002]. However, a detailed examination indicates these differences to be superficial. A brief analysis is provided with the Supplemental Note online)
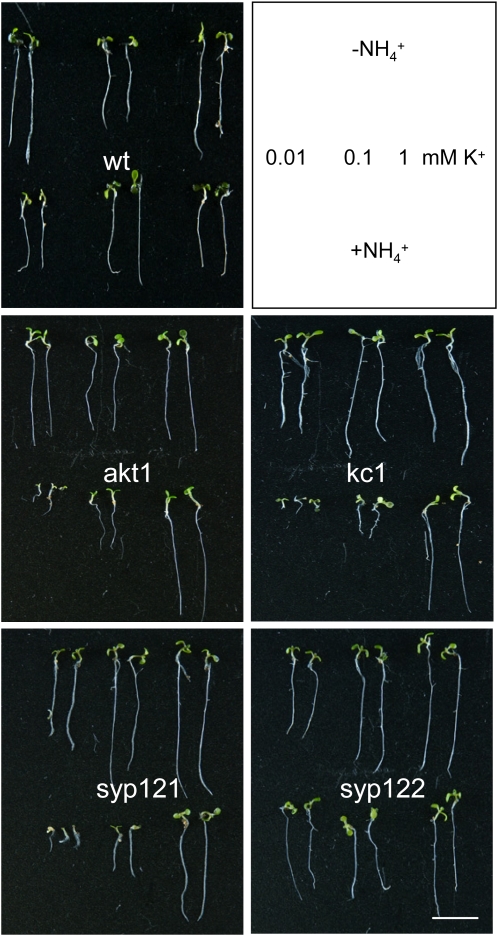
Along with the AKT1-associated K+ channel, Arabidopsis elaborates a number of high-affinity K+ transporters in root tissues (Ashley et al., 2006). Thus, although the K+ ion is a major inorganic osmotic solute essential for plant cell growth and development, eliminating the inward-rectifying K+ channels in the akt1 mutant normally has little or no effect on growth even at submillimolar K+ concentrations. Including NH4+ in the medium exposes a dependence on the AKT1-associated current for K+ uptake and growth by inhibiting K+ flux through these other transporters in Arabidopsis (Hirsch et al., 1998; Spalding et al., 1999). We reasoned that if a complex of SYP121 and KC1 was required for the AKT1-associated K+ channel and K+ uptake, then mutations in these genes should also suppress seedling growth in low K+ concentrations in the presence of NH4+. To examine K+ uptake and plant growth, we germinated seedlings of the wild type and each of the Arabidopsis mutant lines on 10 μM, 100 μM, and 1 mM K+, with and without 2 mM NH4+ for 7 to 10 d before measuring root length and K+ contents. Figure 5 shows the results from one of five replicate experiments. In each case, wild-type plants grown under these conditions showed only marginal reductions in root length and K+ content (see also Supplemental Figures 4A and 4B online), irrespective of the presence or absence of NH4+. As expected, the akt1 mutant showed a similar pattern of growth in the absence of NH4+, but in its presence, root growth was strongly reduced in parallel with the available K+ concentration below 1 mM and K+ content was similarly suppressed. We found that, like the akt1 mutant, root length and K+ content were also strongly reduced in both the syp121-1 and kc1-2 mutants but only when germinated and grown in the presence of NH4+. By contrast, the syp122-1 mutant showed roughly a 20% reduction in root length compared with wild-type plants, but this effect was not associated with NH4+ and was evident at all K+ concentrations. Complementary results were obtained with the kc1-f, syp121-2, and syp121-4 mutants (see Supplemental Figure 4C online), indicating that the NH4+-sensitivity in K+-dependent growth was not allele specific. Finally, we found that NH4+ independence was restored in syp121-1 mutant Arabidopsis complemented with a fluorescently tagged construct of SYP121 but not SYP122 when driven by the SYP121 or 35S promoters (see Supplemental Figure 4C online; Pajonk et al., 2008). These observations, and the electrophysiological analysis summarized above, demonstrate a close mimicry between the functional and phenotypic characteristics of mutations in genes encoding AKT1, SYP121, and KC1 in the plant.
Figure 5.
syp121 and kc1 Mutations Phenocopy the akt1 Mutant in Suppressing NH4+-Sensitive Growth at Submillimolar [K+].
Wild-type and mutant Arabidopsis seedlings germinated and grown in modified Murashige and Skoog (MS) with 0.01, 0.1, and 1.0 mM K+ with and without 2 mM NH4+ for 10 d. Reference to growth conditions indicated in frame (top right). Bar = 1 cm. Statistical analysis of root length and K+ content are summarized in Supplemental Figure 4 online.
SYP121 Forms a Tripartite Complex with AKT1 and KC1, and Its Mutation Does Not Suppress K+ Channel Expression or Membrane Traffic
A plausible explanation for the syp121 and kc1 phenotypes is that these gene products are essential components of a complex with AKT1 and are important for function of the K+ channels. In addition to explaining the similarity of the phenotypes with that of the akt1 mutant, this interpretation also accords with evidence for their interaction, the preferential assembly of KC1 and AKT1, and reconstitution of the native channel gating characteristics when all three components are expressed heterologously. Nonetheless, we examined two alternative explanations for the in vivo data. The first explanation, of a reduction in AKT1 expression, might account for the kc1 and syp121 phenotypes (although without reference to the channel gating characteristics on heterologous expression). The second, of a requirement for SYP121 to deliver channels to the plasma membrane, might also account for a loss of the K+ current. Indeed, membrane traffic itself offers a potential mechanism for posttranslational regulation of transport capacity both in animal (Bryant et al., 2002) and plant cells (Sutter et al., 2007) and conceivably might be tightly coupled to SYP121-mediated vesicle fusion.
To examine the effects of the syp121 and kc1 mutations on AKT1 transcription, we performed quantitative PCR analysis on mRNA-derived cDNA from the wild type and selected Arabidopsis mutant lines using gene-specific primers for AKT1, KC1, and SYP121, including SYP122 for comparison, and primers for the ACT2 actin gene as a control for normalizations. The results (Figure 6B) showed no reduction in AKT1 and KC1 transcript levels in either the syp121-1 or syp122-1 mutant. In fact, transcript levels for the two K+ channel genes actually increased 50 to 80% in the syp121-1 mutant. Furthermore, marginal increases were also evident in AKT1 and KC1 transcript levels in the kc1-2 and akt1 mutants, respectively. Thus, in neither case could an effect of these mutations be explained by suppressed transcription of the K+ channel genes. We also tested for possible effects of the syp121 mutation on channel protein expression and traffic to the plasma membrane. Total microsomal membranes were isolated from roots of hydroponically grown Arabidopsis, and plasma and inner membrane fractions from the wild type and the syp121-1 mutant were separated by two-phase partitioning prior to analysis by protein gel blots (Santoni, 2006). We found expression of the AKT1 and KC1 proteins in microsomal fractions to be similar in the syp121-1 mutant compared with wild-type Arabidopsis (see Supplemental Figure 6 online). Most importantly, the distributions of KC1 and AKT1 exhibited no difference between the wild type and syp121 mutant when analyzed by two-phase separation, and both proteins separated with the plasma membrane fraction (Figure 6C). Partitioning was verified using the AHA3 H+-ATPase epitope (Pardo and Serrano, 1989) as a marker for the plasma membrane and the Sec61 epitope as a marker for the inner membrane (endoplasmic reticulum; Yuasa et al., 2005) fractions.
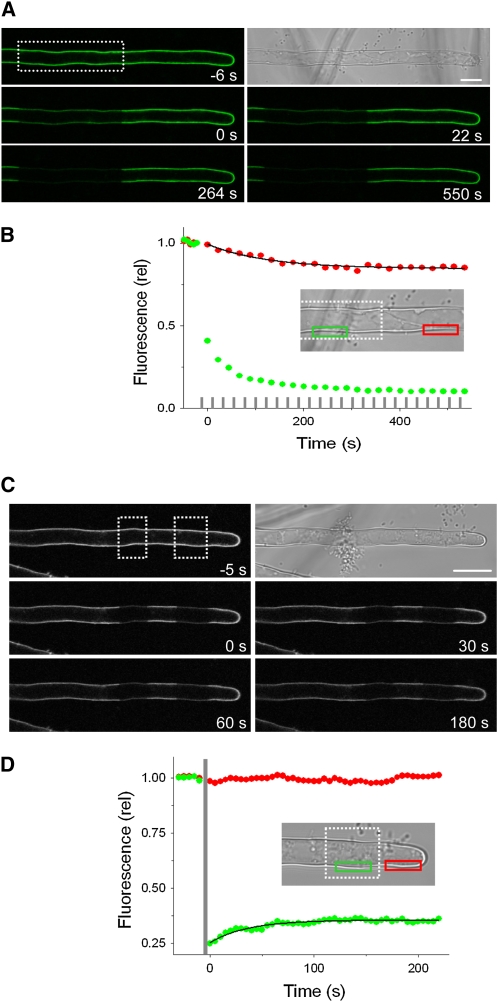
In separate experiments, we transiently expressed fluorescently tagged AKT1 and KC1 in root epidermis of Arabidopsis wild-type and syp121-1 mutant lines (Campanoni et al., 2007) and assessed their distributions by confocal imaging. Figure 7A shows the results from one of nine independent experiments, each of which yielded similar results for KC1 when expressed in the syp121-1 mutant. Equivalent results were obtained for AKT1 when expressed in the mutant SNARE background (see Supplemental Figure 5 online) and for both constructs when expressed in wild-type Arabidopsis (data not shown). In every case, KC1 was peripherally localized, distinct from the cytoplasm and tonoplast. Channel expression was largely unaffected in the syp121-1 mutant, although AKT1 expression was marginally higher in the syp121-1 mutant compared with wild-type Arabidopsis (Figure 7B). We used fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) analysis to examine channel mobility at the cell periphery. Figure 8 (see Supplemental Movies 2 and 3 online) showed a similar predominance of the AKT1-green fluorescent protein (GFP) fluorescence in the immobile fraction, indicating that the syp121-1 mutation did not lead to an appreciable retention of the channel protein within the secretory pathway or mobile in the cytosol. Again, equivalent results were obtained in seedlings expressing KC1-YFP (data not shown). It is of interest that KC1 localization at the plasma membrane appeared independent of AKT1 (see Figures 2 and 7). This latter observation contrasts with a recent report (Duby et al., 2008) suggesting that the protein is retained within the secretory pathway in tobacco unless coexpressed with AKT1. One simple explanation in this case is that the Arabidopsis KC1 protein normally associates with additional factors needed for its traffic that are absent in tobacco. Nonetheless these data, and the results of the two-phase partitioning experiments, show that the loss of K+ current in the syp121-1 mutant cannot be explained by suppressed delivery of the K+ channels to the membrane surface.
Figure 8.
The syp121 Mutation Does Not Affect K+ Channel Mobility at the Cell Periphery Assessed by FRAP and FLIP in Arabidopsis syp121 Mutant Root Hairs.
(A) and (B) AKT1-GFP FLIP in an Arabidopsis syp121-1 mutant root hair.
(A) Bright-field and fluorescence image taken before (top two frames) and at times during repetitive photobleaching (start of photobleach cycles, relative t = 0) at the distal end of the root hair (photobleach area indicated in the first fluorescence image). See Supplemental Movie 3 online for full image sequence.
(B) FLIP analysis of the fluorescence signals taken from the regions indicated (inset, green and red boxes) and corrected for background fluorescence decay shows a limited loss of signal outside the area of photobleaching (photobleach periods indicated by gray bars). The solid curve is the result of nonlinear least squares fitting of the postbleach fluorescence signal to a single exponential function yielding an immobile fraction of 0.84. Similar results were obtained in each of four separate experiments with the syp121 mutant and in four experiments with wild-type seedlings and KC1-YFP in place of AKT1-GFP. Bar = 10 μm.
(C) and (D) AKT-GFP FRAP in an Arabidopsis syp121-1 mutant root hair.
(C) Bright-field and fluorescence image taken before (top two frames) and after photobleaching (end of photobleach, relative t = 0) the distal end of the root hair (photobleach areas indicated in the first fluorescence image). See Supplemental Movie 2 online for full image sequence.
(D) FRAP analysis of the fluorescence signals taken from the regions indicated (inset, green and red boxes) and corrected for background fluorescence decay shows the loss of signal after photobleaching and its limited recovery. Photobleach time indicated by gray bar. The solid curve is the result of nonlinear least squares fitting of the postbleach fluorescence signal to a single exponential function. Similar results were obtained in each of eight separate experiments with the syp121 mutant and in seven experiments with wild-type seedlings, yielding immobile fractions of 0.86 ± 0.04 and 0.84 ± 0.03, respectively. Bar = 10 μm.
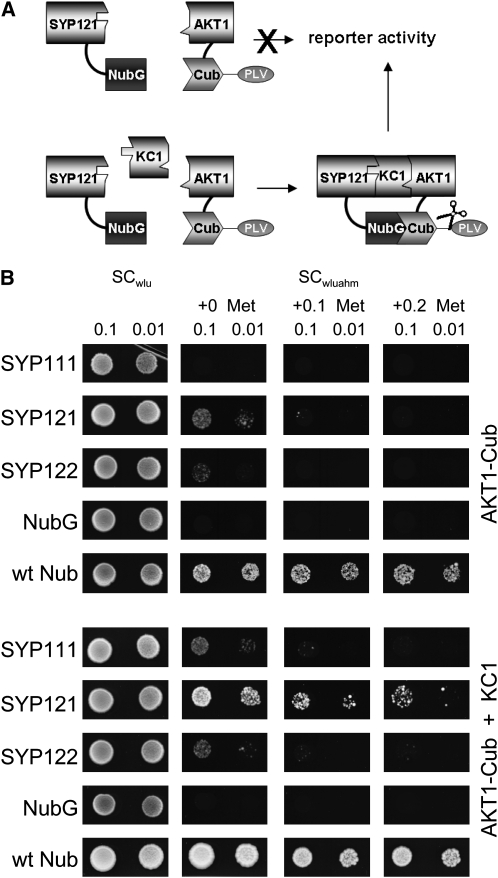
Finally, we used a variation on the split-ubiquitin assay (Grefen et al., 2009), a so-called split-ubiquitin bridge (SUB) assay, to test for formation of a tripartite complex of SYP121 with KC1 and AKT1 in yeast. In this case, we made use of a split-ubiquitin fusion construct of AKT1 with complementary constructs of the SNAREs SYP121, SYP122, and SYP111. Yeast were transformed with the AKT1 construct paired with each of the SNARE constructs either with or without the inclusion of KC1 in a yeast expression vector before dropping on plates to monitor growth. Because KC1 interacts selectively with SYP121 and SYP121 does not interact appreciably with AKT1 (see Supplemental Figure 1 online), we reasoned that yeast growth should be rescued through an AKT1–SYP121 interaction only if the KC1 subunit serves to bridge these two proteins. In fact, yeast growth was evident, even when plated on 0.2 mM Met to suppress expression of the AKT1 bait construct, but only when coexpressed with KC1 and SYP121 (Figure 9). These results confirm the ability of SYP121 to form a tripartite complex with KC1 and AKT1, consistent with requirement for all three gene products to support channel-mediated K+ uptake (Figure 10).
Figure 9.
Tripartite Assembly of SYP121 with AKT1 Requires the KC1 Subunit as a Bridge.
(A) Concept of the yeast SUB assay for formation of a tripartite protein assembly. By analogy with the mating-based split-ubiquitin approach, the SUB assay depends on protein interaction to bring together the two halves of the ubiquitin protein, which leads to cleavage of the VP16 transactivator (PLV) and activation of the reporter gene. In this case, however, assembly of the proteins carrying the split-ubiquitin moieties depends on inclusion of the third protein (KC1) component.
(B) Yeast split-ubiquitin assay for interaction after transformation with AKT1-Cub, the Nub-X fusion constructs of SYP121, SYP122, and SYP111, with (bottom) and without (top) coexpression of KC1. Controls (negative, NubG; positive, wtNub) are included and transformed yeast are spotted (top to bottom) on SC medium without Trp, Leu, and Ura (SCwlu) to verify crossing. Yeast growth on SC without Trp, Leu, Ura, Ade, His, and Met (SCwluahm) was used to verify Ade- and His-independent growth (second panel), and the addition of 0.1 and 0.2 mM Met was used to verify interaction at lower AKT1-Cub expression levels. Serial dilutions (0.1 and 0.01) as indicated for each frame. Note yeast growth on Met only with KC1 coexpression. Protein expression was verified by protein gel blot analysis in each case (data not shown).
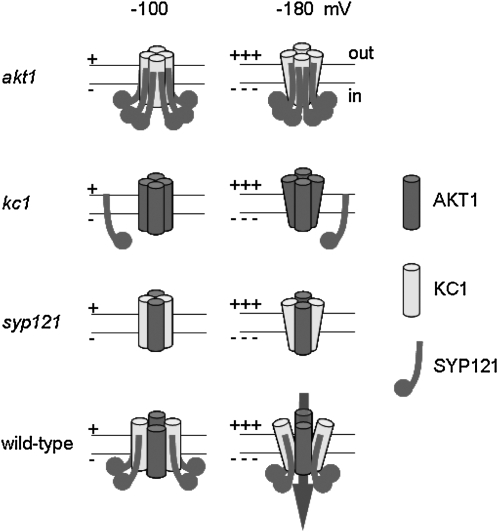
Figure 10.
Tripartite Assembly with SYP121 Determines the Gating of Heteromeric AKT1-KC1 K+ Channels.
Model of channel subunit assembly and gating in the plant based on channel gating characteristics on heterologous expression and in the plant (Figures 3 and 4; see Supplemental Figure 3 online). Eliminating any one of the three proteins AKT1 (akt1), KC1 (kc1) or SYP121 (syp121) prohibits normal gating and K+ flux, either by preventing the assembly of the necessary heteromeric core of AKT1 and KC1 subunits (akt1 and kc1) or by preventing association of SYP121 with the channel core through its binding with KC1 (kc1 and syp121). We assume that AKT1 and KC1 form the core of the channel and its pore, consistent with their structural homologies to other Kv-like K+ channel subunits and the observation that heterologous expression of AKT1 alone and with KC1 yields a current on heterologous expression. The approximate twofold change in the apparent gating charge on coexpression with SYP121 (Figure 3; see also Supplemental Figure 3 online) points to profound changes in the conformation of the channel. Thus, assembly of the heteromeric AKT1-KC1 channel core and binding of at least two SYP121 proteins to each KC1 subunit (wild type) gives a channel gate with native characteristics, underpinning its physiological voltage dependence and K+ flux (arrow).
DISCUSSION
Membrane vesicle traffic and the SNARE proteins that drive it are increasingly recognized as important players in plant cell development and growth, as well as signaling and defense. Vesicle traffic impacts on the steady state complement of membrane proteins and their tissue distribution during development, and at the cellular level it effects the turnover of ion channels and transporters as well as its modulation by hormones and environmental factors (Grefen and Blatt, 2008). These processes clearly play a part in coordinating the ensemble of transport activities at the plasma membrane, although many details are only now beginning to come to light. Additionally, there have been hints of other roles for vesicle trafficking proteins, notably for SNAREs in modulating the gating of K+, Cl−, and Ca2+ channels in plants (Leyman et al., 1999; Sokolovski et al., 2008). However, substantial evidence that could point to such interactions, let alone their consequences in plants, has been lacking. Our results now address this important gap in knowledge, demonstrating a direct and selective interaction of the Arabidopsis SNARE SYP121 with KC1, a regulatory K+ channel subunit, as part of a tripartite SNARE-K+ channel protein complex. We report that KC1 associates selectively with SYP121 over its closest homolog, SYP122, and another related SNARE SYP111. Additionally, we found that SYP121 interacts preferentially with KC1 over other Arabidopsis Kv-like channels and that SYP121 interacts strongly with the AKT1 K+ channel subunit, but only in the presence of KC1. Genetic analysis showed that both the SNARE and KC1 are essential in vivo for K+ current and uptake at the root epidermis associated with the inward-rectifying K+ channel AKT1 and for growth when channel-mediated K+ uptake is limiting. Finally, we uncovered fundamental changes to K+ channel gating that promoted the K+ current and recapitulated the native characteristics in the plant when SYP121 was heterologously coexpressed with the AKT1 and KC1 proteins. These results demonstrate a wholly unexpected role for a SNARE as part of a protein complex facilitating plant mineral nutrition, analogous to SNARE-ion channel complexes of mammals but apparently unrelated to signaling and its coupling to membrane vesicle traffic. Additionally, because KC1 assembles in heteromeric complexes with different channel subunits, they implicate SNARE binding in controlling other K+ channels.
A Functional Requirement for SYP121 and KC1 in K+ Transport and Nutrition
Key to understanding the functional relationship of SYP121 to KC1 was our finding that the several kc1 and syp121 mutants independently suppressed the inward-rectifying K+ currents in root epidermal protoplasts, phenocopying the akt1 mutant characteristics in K+ current as well as K+-sensitive growth and K+ uptake in the presence of NH4+ (Hirsch et al., 1998). Not only do these data rule out allele-specific effects of the mutations, but they indicate that all three proteins, SYP121, KC1, and AKT1, are needed for activity in K+ nutrition and thus provide a functional link to the physiology of the plant. Several observations support a unique association of these gene products in this context. We note (Figures 4 and 5; see also Supplemental Figure 4 online) that none of the mutants showed growth or developmental phenotypes in the absence of NH4+ nor alterations in outward-rectifier K+ current under voltage clamp, so discounting a more general effect of the genetic lesions on transport and K+ channel activities; complementing the syp121-1 mutant with SYP121, but not with SYP122, rescued the mutant phenotype; finally, the null mutant syp122-1, which eliminated the closest homolog to SYP121 (Lipka et al., 2007; Bassham and Blatt, 2008), showed no appreciable effect on the inward-rectifying K+ current nor on the NH4+-sensitive K+ dependence for growth and K+ accumulation. The latter observations, especially, underscore the functional specificity for SYP121 and lend confidence to its physiological interpretation. Additionally, we were able to rule out an indirect effect of SYP121 on the K+ channels at the plasma membrane. Neither kc1 nor syp121 suppressed AKT1 transcription, and syp121-1 had no appreciable effect on channel expression or delivery to the plasma membrane. These findings accord with separate evidence for SYP121-KC1 binding that we revisit below and, hence, with the concept of a functional assembly of the three gene products in vivo (Figures 9 and 10).
Much the same conclusion can be drawn from electrophysiological analyses of the heterologously expressed proteins, and these results merit further scrutiny. We found that coexpressing AKT1 and KC1 gave a K+ current with gating characteristics that differed markedly with SYP121 coexpression. Notably, joint fittings indicated a mean gating charge near 2 with SYP121 coexpression but values near unity in its absence. SYP121 coexpression also displaced the voltage yielding half-maximal conductance, V1/2, to more positive voltages. Because both gating parameters are associated directly with biophysical properties of the K+ channel voltage sensor (Dreyer and Blatt, 2009; Hille, 2001), their sensitivity to SYP121 coexpression is difficult to reconcile with an effect on membrane vesicle traffic or a simple change in the population of K+ channels at the membrane. By the same token, the effects are inconsistent with binding and titration of KC1 by the SNARE that might favor channel assembly solely of the AKT1 subunit and thereby unmask its current characteristics. Simply altering the number of channels at the membrane could be expected to affect current amplitude, but not its intrinsic voltage sensitivity, and titrating KC1 subunits should have yielded gating characteristics equivalent to those recorded with AKT1 alone. The data are, however, readily understood as consequences of changes in the conformations assumed by the channels during gating when associated with the SNARE. Significant, too, is the fact that the gating characteristics recorded from the root epidermal protoplasts (Figure 4) matched closely those recovered on coexpression with all three proteins, AKT1, KC1, and SYP121, but differed markedly from those obtained on expressing AKT1 alone or with KC1 (Figure 3; see also Supplemental Figure 3 online; Duby et al., 2008). This comparison, again, supports the idea that SYP121 is a critical component of the channel protein complex previously missing in heterologous expression studies, and it concurs with a recent study (Duby et al., 2008) suggesting that AKT1 and KC1 preferentially form heterotetramers in vivo. It also implies that heterologous expression of AKT1 on its own yields channels with anomalous characteristics atypical of the K+ channels in the plant.
SYP121–KC1 Interaction Contributes to a Tripartite K+ Channel Complex
Many details of the molecular interaction between SYP121 and KC1 remain to be explored. Nonetheless, the association of these two proteins is clearly unusual. KC1 showed a consistent selectivity for the SYP121 partner over its closest structural and functional homolog SYP122, as well as another related SNARE, SYP111. This selectivity was evident in split-ubiquitin assays with yeast, in coimmunoprecipitation experiments after heterologous expression, and on expression as BiFC fusion constructs in Arabidopsis (Figures 1 and 2). Considered individually, each of these methods raises certain caveats with its application, whether in relation to non-native (heterologous) expression or to constitutive overexpression in vivo (Fletcher et al., 2003; Walter et al., 2004; Grefen et al., 2009). However, their common consensus in this singular result, and the lack of similar interactions among the closely related proteins tested in parallel, finds explanation only in the unique association between SYP121 and KC1. Because KC1 preferentially assembles with other K+ channel subunits, including AKT1 (Dreyer et al., 1997; Obrdlik et al., 2004; Duby et al., 2008), the SYP121-KC1 partnership points to its function in different tripartite K+ channel complexes. Thus, it suggests a paradigm of SNARE action in binding with a single protein that, in turn, assembles with different K+ channels to modulate their activities. For the AKT1 K+ channel, this concept is born out in the unique impact of SYP121 and KC1 expression on the inward-rectifying K+ current when heterologously expressed and in the requirement for all three gene products to recover the inward-rectifying K+ current in vivo.
In fact KC1, like AKT1, is a member of the Kv-like K+ channel family (Dreyer and Blatt, 2009) and comprises six membrane-spanning helices, a K+ channel pore loop, and GYGD K+ channel signature sequence. These channels also include a Kv-like voltage sensor in the fourth transmembrane helix that, with the first three transmembrane helices, is thought to form a lever situated at the periphery of the channel when the subunits assemble as a functional tetramer. Together, AKT1 and KC1 yield a current when expressed in tobacco (Duby et al., 2008), so these subunits appear to assemble a functional channel pore without SYP121 per se. It is easy to envisage, then, that SYP121 interacts with elements of the voltage sensor levers at the periphery of the channel assembly, preferentially forming a ring of four SNAREs adjacent the four voltage sensors of the K+ channel. Such an arrangement is consistent with the 2:1 stoichiometry of SYP121:KC1 (Figure 3) in 1:1 assembly of KC1 and AKT1. The near doubling of the gating charge and changes in voltage dependence on coexpression with SYP121 point to an altered conformation of the K+ channel and to profound changes in the protein dynamics coupling voltage and the channel gate, either through conformations linking the voltage sensor and gate or by affecting the electric field around the voltage sensor itself (Hille, 2001; Dreyer and Blatt, 2009). So it will be of interest now to identify the amino acid residues contributing to SYP121 interaction with KC1. We note that KC1 lacks sequences homologous to the N-terminal cytosolic domain of the mammalian Kv1.1 K+ channel that binds Syntaxin1A, and it also lacks sequences homologous to the SNARE binding domain at the C terminus of the Kv2.1 K+ channel (Tsuk et al., 2005; Leung et al., 2007). Thus, SYP121 may bind to another domain of KC1 altogether to enable the gate.
A Governor Model for SNARE Action on K+ Channel Gating and Channel-Dependent K+ Uptake
A few mammalian SNAREs affect subtly the gating of K+ and Ca2+ channels. These interactions arise principally from associations with mammalian Syntaxin1A in neuromuscular and neuroendocrine tissues and are thought to aid in signal transmission (Leung et al., 2007). The fact that SYP121 should be important for the gating of a K+ channel and inorganic mineral nutrition demonstrates a role for a SNARE apparently divorced from any role in signaling, and it highlights a functional divergence for SNARE–ion channel interactions in plants. So how might we understand the role of SYP121 in a context of K+ acquisition? In plants, potassium is an essential macronutrient and the predominant inorganic, osmotic solute for cell turgor, cell expansion, and development as well as physiological responses to environmental stress. Plants elaborate a number of membrane transporters in the root for K+ uptake from the soil, in Arabidopsis including low- and high-affinity H+-coupled transporters and K+ channels (Ashley et al., 2006; Amtmann and Blatt, 2007). The activity of these transporters is regulated, and their coordination enables plants to survive even when the available K+ outside varies over three to four orders of magnitude in concentration. However, with the exception of the HAK5 transporter, none of the genes encoding K+ transporters in Arabidopsis show changes in transcript levels in response to K+ availability (Maathuis et al., 2003; Gierth et al., 2005), implying translational and posttranslational regulation. The remarkably broad range of available K+ concentrations seen by plant roots also necessitates controls on the gating of K+ channels, on the one hand to avoid futile cycling of K+ across the plasma membrane and, on the other, to coordinate K+ uptake with cell expansion and growth.
One means of compensation lies in the capacity of these channels to assemble in heterotetramers with the different subunits imparting a range of gating properties to the functional channel units (Dreyer and Blatt, 2009). Duby et al. (2008) have suggested that incorporating the KC1 subunit offers a mechanism to compensate AKT1 gating under limiting K+ concentrations. This idea is an attractive one, but it leaves significant experimental and conceptual gaps. Because K+ channels are generally thought to assemble prior to export from the endoplasmic reticulum (Ma et al., 2002; Yuan et al., 2003; Heusser and Schwappach, 2005), it implies substantial translational control in KC1 and AKT1 synthesis or in the availability of these proteins for assembly in the endoplasmic reticulum. It would require a high constitutive or enhanced turnover rate in the population of K+ channels at the plasma membrane in parallel with the available K+ outside. Data that might speak to each of these points are simply not at hand or find little support in the literature (Amtmann and Blatt, 2009). Even if it were so, however, the idea does not address the need to adjust K+ uptake through the channels with the osmotic requirements for cell expansion. Finally, because the apparent gating charge of the K+ channels is largely unaffected by assembly with KC1—an analysis of conductance shows gating charges of 1.2 or less in every case (see Figure 2 of Duby et al., 2008)—it does not address the discrepancy in gating characteristics with the K+ channels recorded in the plant.
We now speculate that the tripartite complex of SYP121 and the K+ channels coordinates K+ uptake with the rate of vesicle fusion and expansion of the plasma membrane. Thus, the SNARE–K+ channel interaction may serve as a molecular governor, analogous to the mechanical invention of James Watt, to keep channel-mediated uptake of the osmotically active K+ ion in check and coordinate its transport with changes in cell volume (Grefen and Blatt, 2008): we suggest that changes in SNARE conformation to the open state required for vesicle fusion (Brunger, 2005) may promote K+ channel activity, thus facilitating K+ uptake in parallel with the delivery of new membrane material. In its favor, this idea affords a plausible mechanism for gating control that acts on the existing population of channels at the membrane, without the need for K+ channel synthesis, assembly, and trafficking per se. It is also consistent with past observations that expressing a competitive (dominant-negative) fragment of the tobacco SYP121 uncouples osmotic solute accumulation from cell growth (Geelen et al., 2002; Sokolovski et al., 2008). Certainly, our findings introduce an entirely new dimension to K+ nutrition in the plant, pointing to a complex of interacting proteins more extensive than previously recognized (Li and Luan, 2006; Xu et al., 2006). Because AKT1 expression occurs primarily in the root epidermis, its overriding impact is on K+ accumulation and growth. Nonetheless, assemblies with other K+ channels may be important in plant pathogen resistance (Blatt et al., 1999; Collins et al., 2003; Kwon et al., 2008) and in the control of guard cell K+ channels that affect leaf water loss and vegetative CO2 exchange with the atmosphere (Hetherington and Woodward, 2003). Thus, we anticipate the interaction of SYP121 with KC1 will prove an important starting point for exploring the molecular mechanics integrating membrane traffic with K+ transport and cellular volume control in a wide range of plant tissues.
METHODS
Molecular Biology
For yeast mating-based split-ubiquitin assays, open reading frames for AKT1 (At2g26650), KC1 (At4g32650), SYP121 (At3g11820), SYP111 (At1g08560), and SYP122 (At3g52400) were amplified with gene-specific primers, including overhangs necessary for recombinatorial in vivo cloning (Obrdlik et al., 2004). For Nub-X fusions in vector pNX-gate32-3HA, a stop codon was included to omit the C-terminal HA-tag. Plasmid DNA for each construct was extracted from single colonies counterselected with G418, retransformed into Escherichia coli Xl1blue (Promega), and sequenced before assays. Glycerol stocks of positive colonies were used to generate diploids. Diploids were grown from saturated cultures in liquid SC medium lacking Trp, Leu, and Ura after dilution to 0.1 OD600 and regrowth to 0.8 OD600. Serial dilutions of 0.1, 0.01, and 0.001 OD600 in water were dropped at 10 μL per spot onto plates. Yeast total protein of dipolids expressing KC1-Cub and Nub-SNAREs were extracted as described previously (Obrdlik et al., 2004). Polyclonal antibodies against SYP122, SYP121, and SYP111 were used at a dilution of 1:40,000.
A variation on the split-ubiquitin assay was used to verify tripartite interactions. For this purpose, KC1 including a stop codon was cloned in the yeast expression vector pVTU-Dest (Grefen et al., 2009) and transformed into the yeast strain THY.AP4 (Obrdlik et al., 2004) together with AKT1-Cub and a corresponding Nub-SNARE fusion construct (SYP111, SYP121, and SYP122) or the negative and positive controls NubG and NubWt. Transformed yeast was selected on plates lacking Trp, Leu, and Ura. After 3 d, pools of five colonies were transferred to liquid media, grown overnight, and then used for a dilution series of OD600 0.1 and 0.01 dropped on SC plates lacking Trp, Leu, Ura, Ade, and His with increasing Met concentrations. Growth was monitored on days 2, 3, and 5. Equal amounts of yeast used for the dilution series were harvested, proteins extracted, and, using polyclonal antibodies, expression verified by protein gel blots (Grefen et al., 2009).
For BiFC, the KC1 open reading frame was amplified with primers containing an XbaI site as overhang and fused as a C-terminal fusion to cYFP in pSPYCE-35S (Walter et al., 2004). N-terminal fusions with nYFP were constructed after modification of pSPYNE-35S to generate pN-SPYNE-35S by SmaI and Ecl136II digestion and subsequent ligation to remove the fluorophore. nYFP was amplifed using pSPYNE-35S as a template with overhangs in the reverse primer that included a Myc-tag and linker (forward 5′–3′ including an XbaI site, GACTAGYTATGGTGAGCAAGGGCGAGGA; reverse 5′–3′ including a SpeI site, GCTCTAGAAAGATCCTCCTAGAAATCAACTTTTGCTCGGCGGTGATATAGACGTTGTGG). The PCR product was double-digested with XbaI and SpeI and cloned into pSPYNE-35S lacking the fluorophore using the XbaI site, thereby reconstituting this site for insertion of the different SNARE coding sequences. To generate a 35S-driven AKT1-GFP construct, the AKT1 open reading frame was amplified by PCR and cloned into the pENTR/D-TOPO(r) Gateway entry vector (Invitrogen) before recombination into the pGWB5 binary vector to recover a C-terminal GFP fusion. KC1-YFP was amplified by PCR to include XbaI and XhoI overhangs and was cloned into pPTbar, kindly provided by K. Schumacher (Heidelberg, Germany), after SalI and XbaI digestion. Plasmid DNA for all constructs was verified by sequencing, subsequently transformed by electroporation into Agrobacterium rhizogenes MSU440, and single colonies stored as glycerol stocks after selective growth and verification of plasmid content.
For electrophysiology and coimmunoprecipitation after expression in SF9 insect cells, KC1, SYP121, SYP122, and SYP111 were amplified with gene-specific primers that included N-terminal Flag- and His-tags for the SNAREs and C-terminal Myc- and His-tags for KC1. AKT1 was amplified with gene-specific primers that included a C-terminal VSVG tag. The primers included EcoRI and NotI restriction sites that were used to clone the PCR products into pVL1393 (BD Biosciences). Baculovirus carrying all constructs were generated and titered using the BaculoGold transfection kit (BD Biosciences) according to manufacturer's instructions. Virus from second amplifications were used to infect Sf9 cells (Invitrogen) grown at cell densities of 107 cells/ mL in Sf9 culture medium (Invitrogen).
For electrical recordings using Xenopus laevis oocytes, AKT1, CIPK23, and CBL1 constructs in pGEMHE were obtained from Xu et al. (2006), and KC1, SYP122, and SYP111 were cloned using the BamHI restriction site into the same vector. SYP121 was cloned via MscI and NcoI into the pGT vector (Johansson et al., 2006). Plasmids were linearized and capped cRNA synthesized in vitro using T7 mMessage mMachine (Ambion). cRNA quality as a single band was confirmed by denaturing gel electrophoresis. cRNA was mixed to ensure equimolar ratios unless otherwise noted. To ensure uniform injections of AKT1 transcript, mixtures were made up to a standard volume as necessary with RNase-free water.
Quantitative PCR
Total RNA was extracted from 12-d-old Arabidopsis thaliana seedlings using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Genomic contaminations were eliminated via RQ1 DNase treatment (Promega), and the total RNA was subsequently purified and concentrated using RNeasy plant mini kit columns (Qiagen). First-strand cDNA was prepared using SuperScript II reverse transcriptase enzyme (Invitrogen) according to the manufacturer's protocol. PCR was performed in an Mx4000 Multiplex Quantitative PCR system machine (Stratagene) with the Brilliant SYBR Green QPCR Master Mix (Stratagene). Gene-specific primers used were as follows: for AKT1, 5′-TCTTTCAGGATTCGAGAAAC-3′ (forward) and 5′-TCTAGCAACTCCTTGAAACTC-3′ (reverse); for KC1, 5′-ATATTGCGATACACAAG-3′ (forward) and 5′-GACCTAACTTCGCTAAT-3′ (reverse) (Szyroki et al., 2001); for SYP121, 5′-CACTGGAGGAAGTCAACC-3′ (forward) and 5′-TCACAAAAACCAAATAAACG-3′ (reverse). Specificity was tested by sequencing the purified PCR products, and contaminating genomic DNA was tested using SYP121 intron-specific primers 5′-AATTGGTTTCCGAACACTG-3′ (forward) and 5′-TGTCGCACCACCACTATG-3′ (reverse). All quantifications were normalized to ACT2 actin cDNA fragments amplified by with gene-specific primers (forward 5′–3′, CTAAGCTCTCAAGATCAAAGGCTTA; reverse 5′–3′, ACTAAAACGCAAAACGAAAGCGGTT).
Coimmunoprecipitation
For coimmunoprecipitation, Sepharose G was equilibrated with Buffer A (130 mM NaCl, 10 mM HEPES, pH 6, 1 mM PMSF, and Complete EDTA-free proteinase inhibitor cocktail; Roche), incubated at 4°C with anti-Flag antibody (Sigma-Aldrich) in Buffer A overnight, and washed of unbound antibody. The antibody-coupled Sepharose was blocked with 1% BSA for 1 h at 20°C, and a 50% suspension was prepared in fresh Buffer A plus 1% BSA and stored with 0.02% NaN3. KC1 and SNARE proteins were obtained from Sf9 cells harvested 42 h after transfection, washed with Buffer A, and collected by centrifugation. The cell pellet was suspended in Buffer A with 1% Nonidet P-40 (Sigma-Aldrich) by pipetting and incubated with gentle agitation overnight at 4°C. After 30 min centrifugation at 16,000g, the supernatant was diluted with a 10-fold excess of Buffer A and mixed with Flag-coupled Sepharose suspension. After 1 h gentle agitation at 20°C, the suspension was washed with 6 volumes of Buffer A, and the last wash fraction collected and incubated with 10 μL Strataclean Sepharose (Stratagene) to precipitate all remaining protein. Proteins bound with the Flag-coupled Sepharose were released by incubation with SDS loading buffer at room temperature for 1 h. Eluted fractions and proteins collected from the final wash were analyzed by protein gel blots using monoclonal anti-Myc antibody (Sigma-Aldrich) and the ECL Advance detection kit (GE Healthcare).
Fractionation and Immunodetection
Total protein was extracted by grinding leaf tissue frozen in liquid N2 and resuspending 1:1 (w/v) in extraction buffer containing 100 mM Tris-HCl, pH 8.0, 1% SDS, 1% sodium deoxycholate, 20 mM EDTA, 1 mM DTT, and 0.2 mM PMSF. For protein gel blot analysis of root tissues, the epidermal layer was stripped and isolated by passage in liquid N2 before grinding. Samples were centrifuged at 10,000g for 10 min at 4°C to remove debris, and sodium dodecylsulfate was removed as necessary by centrifugation at 16,000g for 3 min after mixing with 20 volumes of 100 mM KH2PO4. Total membrane proteins were extracted by resuspending ground tissue in extraction buffer containing 100 mM Tris-HCl, pH 7.5, 300 mM sucrose, 1 mM EDTA, 2.5 mM DTT, and 0.1 mM PMSF. Samples were homogenized, centrifuged at 10,000g at 4°C to remove debris, and microsomes in the supernatant pelleted by centrifugation at 100,000g for 35 min at 4°C. Protein was quantified by Bradford assay (Bio-Rad) and calibrated against BSA. Proteins were analyzed by protein gel blot using commercial monoclonal Myc and Flag antibodies (Sigma-Aldrich) and polyclonal rabbit antibodies at a dilution of 1:4000 for the SNAREs (Tyrrell et al., 2007). Blots were bound with secondary antibody using commercial horseradish peroxidase–coupled goat anti-rabbit antibody (Sigma-Aldrich) and analyzed with the ECL Advance detection kit (GE Healthcare).
Plasma membrane and internal membrane fractions were separated by two-phase partitioning (Santoni, 2006) using root tissue of 7-week-old plants grown hydroponically under a 8:16 (light:dark) short day at 22°C. Membrane proteins from two-phase fractions were enriched using Urea-NaOH (Santoni, 2006). Final pellets were resuspended in 500 μL of solubilization buffer containing 2% (w/v) lysophosphatydyl choline, 7 M urea, 2 M thiourea, 0.5% Triton X-100, 20 mM DTT, and Complete EDTA-free proteinase inhibitor cocktail (Roche). Resuspended pellets were incubated with gentle head-over-end rotation at 4°C for 12 h before centrifugation and precipitation with trichloroacetic acid. This pellet was resuspended in 5% sodium dodecylsulfate, and protein concentrations were determined using the Micro-BCA assay (Thermo Scientific) according to the manufacturer's instructions. Samples were mixed with equal volumes of 2× SDS loading buffer (0.125 mM Trizma base, 20% [w/v] glycerol, 0.2 M DTT, 4% (w/v) sodium dodecylsulfate, and 1% Bromophenol Blue) and incubated at room temperature for 12 h. Aliquots (1.3 μg protein) were separated by SDS-PAGE and transferred to PVDF membrane (Immobilon-P; Millipore) electrophoretically for protein gel blot analysis. Antibodies against AHA3 (Pardo and Serrano, 1989) and Sec61 (Yuasa et al., 2005) were used at 1:100 dilutions. Polyclonal antibodies for AKT1 and KC1 were generated commercially (Agrisera) in rabbits against synthetic peptides corresponding to unique amino acid sequences H594-G608 and C776-V789, respectively, coupled to keyhole limpit hemocyanin. Immune sera from final bleeds were checked by ELISA (Agrisera) and verified for the absence of cross-reactivity in the preimmune sera. Reacting bands of ∼75 kD (KC1) and 80 kD (AKT1) were detected by protein gel blot analysis with solubilized membranes from wild-type Arabidopsis and for the absence of the corresponding band in membranes from kc1-2 and akt1-1 mutant plants (see Supplemental Figure 6 online). Purified antibodies to the channels were used at 1:50 dilutions. For protein gel blot analysis, secondary goat-anti-rabbit whole IgG fraction (Sigma-Aldrich) was radiolabeled with 125I using Pierce precoated iodination tubes (Thermo Scientific) following the manufacturer's instructions. The secondary antibody was used at 1:4000 dilution and detected using a Fujifilm FLA5000 phosphor imager system.
Plant Growth and K+ Accumulation
Sterile wild-type and mutant Arabidopsis were germinated and grown in liquid culture and on 1% agarose plates with a dilute, modified MS medium containing 2 mM NaNO3, 0.2 mM NaH2PO4, 0.1 mM CaCl2, and 1 mM MgSO4, supplemented with MS micronutrients and 0.3% sucrose, and with or without addition of 2 mM NH4Cl in a modification of previously published work (Hirsch et al., 1998). KCl was added to give final concentrations of 0.01, 0.1, and 1.0 mM, and all solutions were filter-sterilized. Plants were harvested after 7 or 10 d, measured, weighed, and dried before extraction in 1 M HNO3 and dilution for K+ content analysis using a Perkin-Elmer ICP-OES 2100DV system. Standards were prepared in parallel using the same acid solution and measurements were performed in triplicate.
Electrophysiology
Xenopus oocytes and stage V and VI oocytes were isolated from mature Xenopus, and the follicular cell layer was digested with 2 mg/mL collagenase (type 1A; Sigma-Aldrich) for 1 h. Injected oocytes were incubated in ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 10 mM HEPES-NaOH, pH 7.4) supplemented with gentamycin (5 μg/mL) at 18°C for 3 d before electrophysiological recordings. Whole-cell currents were recorded under voltage clamp using an Axoclamp 2B (Axon Instruments) two-electrode clamp circuit as described previously (Leyman et al., 1999; Sutter et al., 2006b). Measurements were performed under continuous perfusion with 96 mM KCl, 1.8 mM MgCl2, 1.8 mM CaCl2, and 10 mM HEPES-NaOH, pH 7.2. Oocytes yielding currents were collected and total membrane protein isolated according to Sottocornola et al. (2006) using 20 μL of extraction buffer per oocyte. Polyclonal antibodies against SYP121 and SYP111 were used at dilutions of 1:5000 in combination with the ECL Advance detection kit (GE Healthcare). Protein gel blots were quantified and normalized to the Ponceau S stain using ImageJ (http://rsbweb.nih.gov/ij/).
Sf9 cells were harvested after 2 d incubation posttransfection and, prior to electrophysiological recordings, were transferred to a bath solution containing 100 mM KCl, 5 mM MgCl2, 4 mM CaCl2, 5 mM glucose, and d-sobitol to give an osmolality of 290 mOsm kg−1. Patch electrodes were two-step pulled from Kimax-51 glass capillaries (Kimble Products) and displayed resistances of 7 to 10 MΩ when filled with 150 mM K-D-gluconate, 1 mM EGTA, 2 mM MgCl2, 10 mM HEPES-NaOH, pH 7.4, and 1 mM Mg-ATP (osmolality, 300 mOsm kg−1). Measurements were performed under continuous perfusion and expression verified by protein gel blot analysis.
Arabidopsis root epidermal protoplasts were obtained by enzymatic digestion in 100 mM glycine, 2% Cellulase Onozuka RS (Yakult Honsan), 1.5% Cellulysin (Calbiochem), 0.4% Pectolyase Y-23 (Kikkoman), 10 mM CaCl2, 1 mM MgCl2, and 5 mM MES-NaOH, pH 5.7, adjusted to 300 mOsm with d-sorbitol. Protoplasts were collected after 30 min incubation at 28°C on rotary shaker (10 rpm), washed twice with the same solution minus digestive enzymes, and kept on ice until use. Recordings were performed in bath solutions of 10, 30, and 100 mM KCl, 2 mM MgCl2, and 10 mm MES-NaOH, pH 5.5, adjusted with d-sorbitol to 300 mOsm kg−1. Patch electrodes were filled with 150 mM K-D-gluconate, 1 mM EGTA, 0.5 mM CaCl2 (free [Ca2+], 50 nM), 1 mM Mg-ATP, 1 mM MgCl2, and 10 mM HEPES-NaOH, pH 7.6 (final osmolarity, 350 mOsm kg−1) and showed resistances of 20 to 30 in bath solution. All recordings were analyzed and leak currents subtracted using standard methods (Leyman et al., 1999; Sutter et al., 2006b) with Henry III software (Y-Science; University of Glasgow).
Confocal Microscopy
Arabidopsis seedlings were grown in 0.5× MS, pH 7.2, for 3 to 5 d and transfected by cocultivation with A. rhizogenes MSU440 at 0.5 OD600 for both coexpressed partners as described (Campanoni et al., 2007). Confocal images were obtained as before (Sutter et al., 2006b) on a Zeiss LSM510-UV microscope. GFP fluorescence was excited with the 458- or 488-nm argon laser lines; YFP fluorescence was excited with the 514-nm laser line. Emitted light was collected through a NFT515 dichroic and 505- to 530-nm (GFP) and 535- to 590-nm (YFP) band-pass filters. Pinholes were set to 1 airy unit. Bright-field images were collected with a transmitted light detector. Laser intensity, photomultiplier gain, and offset were standardized.
Statistics
Statistical analysis of independent experiments is reported as means ± se as appropriate with significance determined by Student's t test or analysis of variance.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number At3g18780 (ACT2).
Author Contributions
R.P. performed initial split-ubiquitin screens and, along with M.P., generated constructs and SNARE antibodies. I.J., S.S., and A.H. generated constructs for oocyte and baculovirus expression. S.S. performed Sf9 and Arabidopsis electrophysiology. Z.C. performed electrical recordings from SYP121 rescue lines. P.C. analyzed gene transcription. A.H. performed split-ubiquitin and coimmunoprecipitation assays, designed and performed BiFC studies, plant growth, two-phase experiments, and oocyte electrophysiology, and designed and generated the KC1 and AKT1 antibodies. C.G. developed and performed bridge assays. M.R.B. designed studies, performed plant growth and K+ content analysis, performed some electrophysiology and imaging studies and their analysis, and wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. AKT1 Interacts with Itself and the KC1 K+ Channel Subunit but Not with the SNAREs SYP121 and SYP111 of Arabidopsis.
Supplemental Figure 2. FRAP Analysis in a Root Hair Expressing BiFC Fusion Constructs of SYP121 and KC1 with the N- and C-Terminal Halves of YFP (nYFP and cYFP), Respectively.
Supplemental Figure 3. SYP121 Expression Alters Channel Gating of the AKT1-KC1 K+ Current in Sf9 Insect Cells.
Supplemental Figure 4. syp121 and kc1 Mutants of Arabidopsis Show Reduced Growth and K+ Accumulation at Low External [K+] in the Presence of NH4+.
Supplemental Figure 5. The syp121 Mutation Does Not Affect AKT1 K+ Channel Expression at the Root Epidermal Plasma Membrane.
Supplemental Figure 6. Polyclonal Antibodies Generated against Synthetic Peptide Antigens Corresponding to Unique Amino Acid Sequences H594-G608 of KC1 and C776-V789 of AKT1 Recognize the Corresponding K+ Channel Proteins.
Supplemental Movie 1. SYP121 and KC1 Interact in a Nonmobile Fraction at the Cell Periphery as Indicated by the Lack of FRAP of the BiFC Product Formed of the SYP121-nYFP and KC1-cYFP Fusion Proteins.
Supplemental Movie 2. The syp121 Mutation Does Not Affect AKT1 K+ Channel Expression at the Root Epidermal Plasma Membrane as Assessed by AKT1-GFP FRAP in an Arabidopsis syp121 Mutant Root Hair.
Supplemental Movie 3. The syp121 Mutant Does Not Affect AKT1 K+ Channel Expression at the Root Epidermal Plasma Membrane as Assessed by AKT1-GFP FLIP in an Arabidopsis syp121 Mutant Root Hair.
Supplemental Note.
Supplemental References.
Supplementary Material
Acknowledgments
We are grateful to Wolf Frommer (Stanford, USA), Petr Obrdlik (Iongate Biosciences, Germany), J.L. Revuelta and C. Vilarino (Salamanca, Spain) for support with split-ubiquitin assays, J. Kudla (Tübingen), H. Sentenac (Montpellier), W.-H. Wu (Beijing), Hans Thordahl (Copenhagen) and Andreas Nebenführ (Tennessee) for constructs and Arabidopsis lines. Andrew Love and Patrick Armengaud (Glasgow) advised on RT-PCR analysis. J. Christie, P. Jones and G. Sibbet (Glasgow) helped establish the Sf9 insect cell cultures. This work was supported by grants from the Biotechnology and Biological Sciences Research Council, the Leverhulme Trust, and by a John Simon Guggenheim Memorial Fellowship to MRB.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael R. Blatt (m.blatt@bio.gla.ac.uk).
Online version contains Web-only data.
References
- Alexandersson, E., Saalbach, G., Larsson, C., and Kjellbom, P. (2004). Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45 1543–1556. [DOI] [PubMed] [Google Scholar]
- Amtmann, A., and Blatt, M.R. (2007). Regulation of ion transporters. In Plant Solute Transport, A.R. Yeo and T. Flowers, eds (Oxford, UK: Blackwell), pp. 99–132.
- Amtmann, A., and Blatt, M.R. (2009). Regulation of macronutrient transport. New Phytol. 181 35–52. [DOI] [PubMed] [Google Scholar]
- Ashley, M.K., Grant, M., and Grabov, A. (2006). Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 57 425–436. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Qiu, J.L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., Wanner, G., Peck, S.C., Edwards, H., Ramonell, K., Somerville, C.R., and Thordal-Christensen, H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham, D.C., and Blatt, M.R. (2008). SNAREs - Cogs or coordinators in signalling and development? Plant Physiol. 147 1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blatt, M.R., Grabov, A., Brearley, J., Hammond-Kosack, K., and Jones, J.D.G. (1999). K+ channels of Cf-9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor-dependent signal transduction. Plant J. 19 453–462. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T. (2005). Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys. 38 1–47. [DOI] [PubMed] [Google Scholar]
- Bryant, N.J., Govers, R., and James, D.E. (2002). Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 3 267–277. [DOI] [PubMed] [Google Scholar]
- Buschmann, P.H., Vaidyanathan, R., Gassmann, W., and Schroeder, J.I. (2000). Enhancement of Na+ uptake currents, time-dependent inward- rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 122 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanoni, P., and Blatt, M.R. (2007). Membrane trafficking and polar growth in root hairs and pollen tubes. J. Exp. Bot. 58 65–74. [DOI] [PubMed] [Google Scholar]
- Campanoni, P., Sutter, J.-U., Craig, S., Littlejohn, G., and Blatt, M.R. (2007). A generalized method for transfecting root epidermis uncovers endosomal dynamics in Arabidopsis root hairs. Plant J. 51 322–330. [DOI] [PubMed] [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J.L., Huckelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dreyer, I., Antunes, S., Hoshi, T., Müller-Röber, B., Palme, K., Pongs, O., Reintanz, B., and Hedrich, R. (1997). Plant K+ channel alpha-subunits assemble indiscriminately. Biophys. J. 72 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer, I., and Blatt, M.R. (2009). What makes a gate? The ins and outs of Kv-like K+ channels in plants. Trends Plant Sci. 14 383–390. [DOI] [PubMed] [Google Scholar]
- Duby, G., Hosy, E., Fizames, C., Alcon, C., Costa, A., Sentenac, H., and Thibaud, J.B. (2008). AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J. 53 115–123. [DOI] [PubMed] [Google Scholar]
- Fletcher, S., Bowden, S.E.H., and Marrion, N.V. (2003). False interaction of syntaxin 1A with a Ca2+-activated K+ channel revealed by co-immunoprecipitation and pull-down assays: Implications for identification of protein-protein interactions. Neuropharmacology 44 817–827. [DOI] [PubMed] [Google Scholar]
- Gassmann, W., and Schroeder, J.I. (1994). Inward-rectifying K+ channels in root hairs of wheat - A mechanism for aluminum-sensitive low-affinity K+ uptake and membrane potential control. Plant Physiol. 105 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard, F., Cerutti, M., Horeau, C., Lemaillet, G., Urbach, S., Ravallec, M., Devauchelle, G., Sentenac, H., and Thibaud, J.B. (1996). The baculovirus/insect cell system as an alternative to Xenopus oocytes - First characterization of the AKT1 K+ channel from Arabidopsis thaliana. J. Biol. Chem. 271 22863–22870. [DOI] [PubMed] [Google Scholar]
- Geelen, D., Leyman, B., Batoko, H., Di Sansabastiano, G.P., Moore, I., and Blatt, M.R. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: Evidence from competition with its cytosolic domain. Plant Cell 14 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth, M., Maser, P., and Schroeder, J.I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation- induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen, C., and Blatt, M.R. (2008). SNAREs - Molecular governors in signalling and development. Curr. Opin. Plant Biol. 11 600–609. [DOI] [PubMed] [Google Scholar]
- Grefen, C., Obrdlik, P., and Harter, K. (2009). The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 479 1–17. [DOI] [PubMed] [Google Scholar]
- Hetherington, A.M., and Woodward, F.I. (2003). The role of stomata in sensing and driving environmental change. Nature 424 901–908. [DOI] [PubMed] [Google Scholar]
- Heusser, K., and Schwappach, B. (2005). Trafficking of potassium channels. Curr. Opin. Neurobiol. 15 364–369. [DOI] [PubMed] [Google Scholar]
- Hille, B. (2001). Ionic Channels of Excitable Membranes. (Sunderland, MA: Sinauer Press).
- Hirsch, R.E., Lewis, B.D., Spalding, E.P., and Sussman, M.R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280 918–921. [DOI] [PubMed] [Google Scholar]
- Hu, C., Ahmed, M., Melia, T.J., Sollner, T.H., Mayer, T., and Rothman, J.E. (2003). Fusion of cells by flipped SNAREs. Science 300 1745–1749. [DOI] [PubMed] [Google Scholar]
- Ivashikina, N., Becker, D., Ache, P., Meyerhoff, O., Felle, H.H., and Hedrich, R. (2001). K+ channel profile and electrical properties of Arabidopsis root hairs. FEBS Lett. 508 463–469. [DOI] [PubMed] [Google Scholar]
- Jahn, R., Lang, T., and Sudhof, T.C. (2003). Membrane fusion. Cell 112 519–533. [DOI] [PubMed] [Google Scholar]
- Johansson, I., Wulfetange, K., Poree, F., Michard, E., Gajdanowicz, P., Lacombe, B., Sentenac, H., Thibaud, J.B., Mueller-Roeber, B., Blatt, M.R., and Dreyer, I. (2006). External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J. 46 269–281. [DOI] [PubMed] [Google Scholar]
- Jurgens, G. (2005). Cytokinesis in higher plants. Annu. Rev. Plant Biol. 56 281–299. [DOI] [PubMed] [Google Scholar]
- Kato, T., Morita, M.T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M., and Tasaka, M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451 835–840. [DOI] [PubMed] [Google Scholar]
- Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., and Grignon, C. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9 195–203. [DOI] [PubMed] [Google Scholar]
- Leung, Y.M., Kwan, E.P., Ng, B., Kang, Y., and Gaisano, H.Y. (2007). SNAREing voltage-gated K+ and ATP-sensitive K+ channels: Tuning beta-cell excitability with syntaxin-1A and other exocytotic proteins. Endocr. Rev. 28 653–663. [DOI] [PubMed] [Google Scholar]
- Leyman, B., Geelen, D., Quintero, F.J., and Blatt, M.R. (1999). A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283 537–540. [DOI] [PubMed] [Google Scholar]
- Li, L.G., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 12625–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V., Kwon, C., and Panstruga, R. (2007). SNARE-Ware: The role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23 147–174. [DOI] [PubMed] [Google Scholar]
- Ma, D., Zerangue, N., Raab-Graham, K., Fried, S.R., Jan, Y.N., and Jan, L.Y. (2002). Diverse trafficking patterns due to multiple traffic motifs in G protein-activated inwardly rectifying potassium channels from brain and heart. Neuron 33 715–729. [DOI] [PubMed] [Google Scholar]
- Maathuis, F.J.M., et al. (2003). Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J. 35 675–692. [DOI] [PubMed] [Google Scholar]
- Marmagne, A., Rouet, M.A., Ferro, M., Rolland, N., Alcon, C., Joyard, J., Garin, J., Barbier-Brygoo, H., and Ephritikhine, G. (2004). Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3 675–691. [DOI] [PubMed] [Google Scholar]
- Obrdlik, P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajonk, S., Kwon, C., Clemens, N., Panstruga, R., and Schulze-Lefert, P. (2008). Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J. Biol. Chem. 283 26974–26984. [DOI] [PubMed] [Google Scholar]
- Pardo, J.M., and Serrano, R. (1989). Structure of a plasma membrane H+-ATPase gene from the plant Arabidopsis thaliana. J. Biol. Chem. 264 8557–8562. [PubMed] [Google Scholar]
- Parlati, F., Weber, T., Mcnew, J.A., Westermann, B., Sollner, T.H., and Rothman, J.E. (1999). Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc. Natl. Acad. Sci. USA 96 12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumet, F., Rahimian, V., and Rothman, J.E. (2004). The specificity of SNARE-dependent fusion is encoded in the SNARE motif. Proc. Natl. Acad. Sci. USA 101 3376–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz, B., Szyroki, A., Ivashikina, N., Ache, P., Godde, M., Becker, D., Palme, K., and Hedrich, R. (2002). AtKC1, a silent Arabidopsis potassium channel alpha-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. USA 99 4079–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, C., Morita, M.T., Kato, T., and Tasaka, M. (2005). Amyloplasts and vacuolar membrane dynamics in the living graviperceptive cell of the Arabidopsis inflorescence stem. Plant Cell 17 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni, V. (2006). Plasma membrane protein extraction and solubilization for proteomic studies. In Plant Proteomics: Methods and Protocols, H. Thiellement, M. Zivy, C. Damerval, and V. Mechin, eds (New York: Academic Press), pp. 93–109.
- Sokolovski, S., Hills, A., Gay, R., and Blatt, M.R. (2008). Functional interaction of the SNARE protein NtSyp121 in Ca2+ channel gating, Ca2+ transients and ABA signalling of stomatal guard cells. Mol. Plant 1 347–358. [DOI] [PubMed] [Google Scholar]
- Sottocornola, B., Visconti, S., Orsi, S., Gazzarrini, S., Giacometti, S., Olivari, C., Camoni, L., Aducci, P., Marra, M., Abenavoli, A., Thiel, G., and Moroni, A. (2006). The potassium channel KAT1 is activated by plant and animal 14-3-3 proteins. J. Biol. Chem. 281 35735–35741. [DOI] [PubMed] [Google Scholar]
- Spalding, E.P., Hirsch, R.E., Lewis, D.R., Qi, Z., Sussman, M.R., and Lewis, B.D. (1999). Potassium uptake supporting plant growth in the absence of AKT1 channel activity - Inhibition by ammonium and stimulation by sodium. J. Gen. Physiol. 113 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Sutter, J.U., Campanoni, P., Blatt, M.R., and Paneque, M. (2006. a). Setting SNAREs in a different wood. Traffic 7 627–638. [DOI] [PubMed] [Google Scholar]
- Sutter, J.U., Campanoni, P., Tyrrell, M., and Blatt, M.R. (2006. b). Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18 935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter, J.U., Sieben, C., Hartel, A., Eisenach, C., Thiel, G., and Blatt, M.R. (2007). Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17 1396–1402. [DOI] [PubMed] [Google Scholar]
- Szyroki, A., Ivashikina, N., Dietrich, P., Roelfsema, M.R.G., Ache, P., Reintanz, B., Deeken, R., Godde, M., Felle, H., Steinmeyer, R., Palme, K., and Hedrich, R. (2001). KAT1 is not essential for stomatal opening. Proc. Natl. Acad. Sci. USA 98 2917–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, J., Miwa, K., Yuan, L.X., von Wiren, N., and Fujiwara, T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuk, S., Michaelevski, I., Bentley, G.N., Joho, R.H., Chikvashvili, D., and Lotan, I. (2005). Kv2.1 channel activation and inactivation is influenced by physical interactions of both syntaxin 1A and the syntaxin 1A/soluble N-ethylmaleimide-sensitive factor-25 (t-SNARE) complex with the C terminus of the channel. Mol. Pharmacol. 67 480–488. [DOI] [PubMed] [Google Scholar]
- Tyrrell, M., Campanoni, P., Sutter, J.-U., Pratelli, R., Paneque-Corralles, M., and Blatt, M.R. (2007). Selective targeting of plasma membrane and tonoplast traffic by inhibitory (dominant-negative) SNARE fragments. Plant J. 51 1099–1115. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Ueda, T., Ohniwa, R.L., Nakano, A., Takeyasu, K., and Sato, M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29 49–65. [DOI] [PubMed] [Google Scholar]
- Ungar, D., and Hughson, F.M. (2003). SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19 493–517. [DOI] [PubMed] [Google Scholar]
- Varlamov, O., Volchuk, A., Rahimian, V., Doege, C.A., Paumet, F., Eng, W.S., Arango, N., Parlati, F., Ravazzola, P., Orci, L., Sollner, T.H., and Rothman, J.E. (2004). i-SNAREs: Inhibitory SNAREs that fine-tune the specificity of membrane fusion. J. Cell Biol. 164 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Very, A.A., and Sentenac, H. (2003). Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 54 575–603. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Weber, T., Zemelman, B.V., Mcnew, J.A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T.H., and Rothman, J.E. (1998). SNAREpins: Minimal machinery for membrane fusion. Cell 92 759–772. [DOI] [PubMed] [Google Scholar]
- White, P.J., and Lemtirichlieh, F. (1995). Potassium currents across the plasma membrane of protoplasts derived from rye roots - A patch clamp study. J. Exp. Bot. 46 497–511. [Google Scholar]
- Xu, J., Li, H.D., Chen, L.Q., Wang, Y., Liu, L.L., He, L., and Wu, W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360. [DOI] [PubMed] [Google Scholar]
- Yano, D., Sato, M., Saito, C., Sato, M.H., Morita, M.T., and Tasaka, M. (2003). A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. USA 100 8589–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H.B., Michelsen, K., and Schwappach, B. (2003). 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr. Biol. 13 638–646. [DOI] [PubMed] [Google Scholar]
- Yuasa, K., Toyooka, K., Fukuda, H., and Matsuoka, K. (2005). Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41 81–94. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.G., Feechan, A., Pedersen, C., Newman, M.A., Qiu, J.L., Olesen, K.L., and Thordal-Christensen, H. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49 302–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.