Abstract
Transcription of mitochondrial genes in animals, fungi, and plants relies on the activity of T3/T7 phage-type RNA polymerases. Two such enzymes, RPOTm and RPOTmp, are present in the mitochondria of eudicotyledonous plants; RPOTmp is additionally found in plastids. We have characterized the transcriptional role of the dual-targeted RNA polymerase in mitochondria of Arabidopsis thaliana. Examination of mitochondrial transcripts in rpoTmp mutants revealed major differences in transcript abundances between wild-type and rpoTmp plants. Decreased levels of specific transcripts were correlated with reduced abundances of the respiratory chain complexes I and IV. Altered transcript levels in rpoTmp were found to result from gene-specific transcriptional changes, establishing that RPOTmp functions in distinct transcriptional processes within mitochondria. Decreased transcription of specific genes in rpoTmp was not associated with changes in promoter utilization; therefore, RPOTmp function is not promoter specific but gene specific. This implies that additional gene-specific elements direct the transcription of a subset of mitochondrial genes by RPOTmp.
INTRODUCTION
The evolution of mitochondria from the bacterial endosymbiont was accompanied by a loss of the bacterial-type RNA polymerase and replacement by a T3/T7 phage-type RNA polymerase in a common ancestor of almost all extant eukaryotes (reviewed in Tracy and Stern, 1995; Gray and Lang, 1998; Hess and Börner, 1999). In Saccharomyces cerevisiae, the nuclear RPO41 gene encodes a phage-type RNA polymerase operating as the catalytic subunit of the mitochondrial transcription machinery (Greenleaf et al., 1986; Masters et al., 1987). A similar enzyme functions as the core RNA polymerase in human mitochondria (Tiranti et al., 1997; Falkenberg et al., 2002). Genes encoding T3/T7 phage-like RNA polymerases, which are commonly designated RPOT genes, have also been identified in the nuclear genomes of various photosynthetic organisms. According to in vitro and in vivo import studies, a small family of three RPOT genes in Arabidopsis thaliana encodes a mitochondrial (RPOTm) and a plastidial (RPOTp) RNA polymerase and an enzyme imported into both mitochondria and plastids (RPOTmp) (Hedtke et al., 1997, 1999b, 2000). Subcellular targeting of RPOT gene products to mitochondria, plastids, and both organelles is likewise seen in other eudicotyledonous plants, such as Nicotiana species (Kobayashi et al., 2001; Hedtke et al., 2002). By contrast, cereals harbor only two RPOT genes that code for a mitochondrial and a plastidial RNA polymerase, respectively (Chang et al., 1999; Ikeda and Gray, 1999; Emanuel et al., 2004; Kusumi et al., 2004).
Distinct functions of RPOTm and RPOTmp in mitochondria are yet to be assigned. The RPOTm and RPOTmp genes in Arabidopsis have been reported to display overlapping expression patterns in different tissues and at different developmental stages (Emanuel et al., 2006); RPOTm and RPOTmp were therefore proposed to recognize different types of mitochondrial promoters. An alternative picture of RPOTm and RPOTmp functions has been suggested by a study of an Arabidopsis line lacking RPOTmp (Baba et al., 2004). Based on a developmental analysis of RPOT transcript levels and on the observation that in the mutant, the induction of several plastid genes in dark-grown seedlings upon illumination was delayed, Baba et al. (2004) proposed RPOTmp to be the key RNA polymerase transcribing organellar genes during early seedling development and favored a role of both RPOTm and RPOTp at a later developmental stage. However, plants lacking RPOTmp displayed no changes in the accumulation of several mitochondrial transcripts tested (Baba et al., 2004). RPOTm, but not RPOTmp, has been shown to specifically recognize mitochondrial promoters in vitro (Kühn et al., 2007). The transcriptional role of RPOTmp in mitochondria is thus unclear from the current literature. RPOTmp and RPOTp were suggested to have partially redundant roles in plastids as plants lacking either RPOTmp or RPOTp showed similar morphological and developmental alterations when compared with the wild type; a double mutant disrupted in both of these enzymes was seedling lethal (Hricova et al., 2006).
Recent studies of transgenic lines lacking RPOTmp or RPOTp provided evidence for two distinct plastidial promoters being used by the dual-targeted enzyme (Courtois et al., 2007; Swiatecka-Hagenbruch et al., 2008). Of these promoters, the rrn16 Pc promoter is active only during germination and early seedling development (Courtois et al., 2007). Its decreasing importance at subsequent stages has been reported to coincide with RPOTmp becoming attached to thylakoids via an intrinsic thylakoid membrane protein (Azevedo et al., 2008). However, the RPOTmp-specific plastidial promoter PclpP-58 (Swiatecka-Hagenbruch et al., 2008) is active throughout plant development (Zoschke et al., 2007).
Plant mitochondria transcribe their genomes from numerous promoters (Tracy and Stern, 1995). Frequently, multiple promoters are active in the upstream region of one gene (Lupold et al., 1999; Kühn et al., 2005). Using transcriptionally competent mitochondrial extracts, promoter sequences of up to 25 nucleotides comprising the transcription start site have been defined to be required for correct and efficient initiation of transcription in vitro (Rapp and Stern, 1992; Rapp et al., 1993; Caoile and Stern, 1997; Dombrowski et al., 1999). Plant mitochondrial promoter sequences are highly variable, and in contrast with the abundant information on plastid promoters (Hajdukiewicz et al., 1997; Liere et al., 2004; Courtois et al., 2007; Swiatecka-Hagenbruch et al., 2008), there is very little data on the recognition of these promoters by different RNA polymerases. Comparisons of plastidial and mitochondrial promoters in Arabidopsis have identified no common motifs that might direct transcription initiation by RPOTmp in both organelles (Kühn et al., 2005; Swiatecka-Hagenbruch et al., 2007).
To define distinct RPOTmp- and RPOTm-dependent transcriptional mechanisms in mitochondria, we studied Arabidopsis lines with disrupted rpoTmp or rpoTm alleles. Disruption of RPOTm was found to be lethal. In plants lacking RPOTmp, the transcription of a subset of mitochondrial genes was decreased, leading to greatly reduced levels of respiratory chain complexes I and IV.
RESULTS
Disruption of RPOTm Is Lethal
We were unable to isolate plants homozygous for a T-DNA insertion in RPOTm from two independent T-DNA lines, SALK_005875 (allele rpoTm-1) and GABI_350F01 (rpoTm-2). Genotyping the offspring of self-pollinated plants heterozygous for an rpoTm-1 or rpoTm-2 allele identified fewer heterozygous individuals than expected for an embryo-lethal knockout. In reciprocal backcrosses, we found transmission of rpoTm alleles by both male and female gametes to be reduced by two-thirds, which could be due to decreased rpoTm gamete fitness, such as partial gamete lethality, or decreased fitness of heterozygous embryos (see Supplemental Figure 1 online).
Mitochondrial Transcript Abundances Are Altered in rpoTmp Mutants
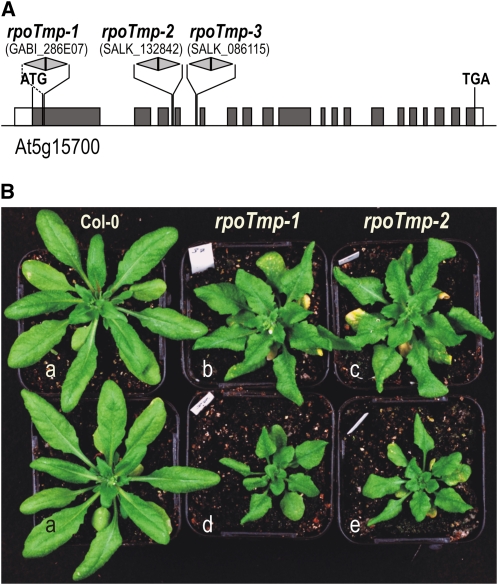
Three independent rpoTmp lines lacking a functional RPOTmp gene were isolated from T-DNA insertion lines GABI_286E07 (rpoTmp-1), SALK_132842 (rpoTmp-2), and SALK_086115 (rpoTmp-3) (Figure 1A). These T-DNA insertion lines were used previously to study Arabidopsis plants lacking RPOTmp (Baba et al., 2004; Courtois et al., 2007), but no molecular defects were detected in mitochondria. We found all three rpoTmp mutant lines to be delayed in their development from germination onwards (Figure 1B), as reported in the earlier studies. The mutants showed wrinkly rosette leaves, consistent with observations by Baba et al. (2004).
Figure 1.
Isolation of Arabidopsis rpoTmp Mutants.
(A) RPOTmp gene organization and positions of T-DNA insertions in the three rpoTmp mutant alleles rpoTmp-1, rpoTmp-2, and rpoTmp-3. RPOTmp gene exons are represented as boxes (dark gray, coding sequences; white, untranslated regions). Gray arrowheads indicate T-DNA insertions and point toward the T-DNA left border. Note that double arrowheads represent two T-DNA copies inserted as inverted repeats, with left borders facing outward. Through PCR with primers annealing to the T-DNA left border and the flanking RPOTmp sequence, all insertions were found to be present as inverted repeats of the T-DNA. Insertion sites were verified by sequencing of PCR products.
(B) Phenotypes of wild-type plants (Col-0, plants labeled a) and rpoTmp-1 (b and d) and rpoTmp-2 mutant plants (c and e) grown in a 16-h photoperiod. A comparison of 4.5-week-old wild-type and mutant plants (a, d, and e) illustrates the developmental delay of rpoTmp mutants. Mutants grown for 6 weeks (b and c) are approximately at the same developmental stage and of a similar size as 4.5-week-old wild-type plants.
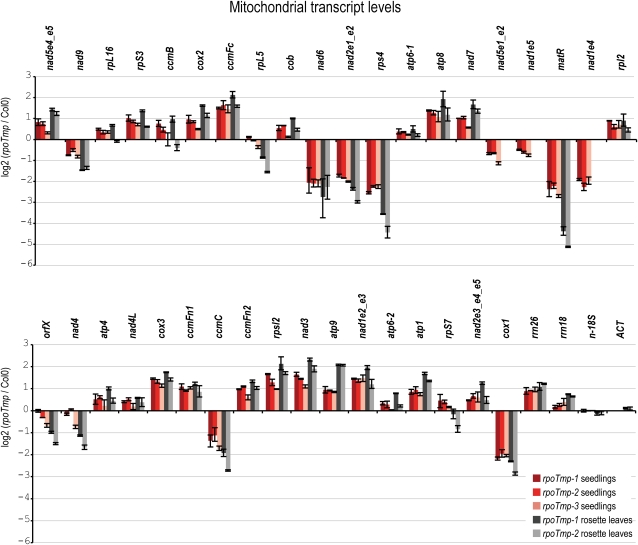
The rosette leaf morphology displayed by rpoTmp plants is reminiscent of the leaf phenotype described for mutants impaired in mitochondrial function (Falcon de Longevialle et al., 2007; Reichheld et al., 2007; Van Aken et al., 2007). We therefore investigated the accumulation of mitochondrial transcripts in rpoTmp seedlings and rosette leaves using a quantitative RT-PCR (qRT-PCR) screen. This survey, which covered all annotated protein-encoding and rRNA genes of the Arabidopsis mitochondrial genome, showed changes in the abundances of numerous mitochondrial transcripts in mutant lines compared with the wild type (Figure 2). To account for the independent transcription of exons of the trans-spliced nad1, nad2, and nad5 mRNAs, transcripts nad1e2_e3 (the transcript comprising exons 2 and 3 of nad1; other nad transcripts are named accordingly), nad1e4, nad1e5, nad2e1_e2, nad2e3_e4_e5, nad5e1_e2, and nad5e4_e5 were examined separately in the qRT-PCR screen. In 7-d-old seedlings, a fourfold reduction in transcript levels was measured for matR, nad1e4, nad2e1_e2, nad6, cox1, and rps4 in all three mutant lines. Less marked reductions in transcript levels were seen for nad9 and ccmC. Numerous other transcripts were found to be more abundant in mutant lines, with the most elevated (ccmFc) being increased threefold. The measured transcript levels were consistent between rpoTmp-1, rpoTmp-2, and rpoTmp-3. This strongly indicates that the altered mitochondrial transcript abundances in mutant plants were due to the absence of a functional RPOTmp gene.
Figure 2.
Mitochondrial Steady State Transcript Levels in rpoTmp Mutants.
Transcript levels are depicted as the log2 ratio of transcript levels in mutants compared with levels in wild-type (Columbia-0 [Col-0]) plants. Transcript abundances of all mitochondrial protein-encoding and rRNA genes were determined by qRT-PCR in both mutant and wild-type plants at the seedling and at the rosette stage. Different transcripts were targeted for the trans-spliced nad1, nad2, and nad5 mRNAs; data points are labeled according to the exons amplified by qRT-PCR (e.g., nad1e1_e2 for a cis-spliced transcript comprising exons 1 and 2 of the nad2 gene). In the assays done on nad1, nad2, and nad5, the mature mRNA plus the pre-mRNAs that are not yet trans-spliced are measured. Because of the low trans-splicing efficiencies, the measured transcripts are partly or even mostly pre-mRNAs and reflect the levels of the independently transcribed mRNA parts. Three technical replicates were averaged per genotype; standard errors are indicated. The nuclear 18S rRNA gene (seedlings) or the nuclear 18S rRNA and ACT genes (rosette leaves) were used for data normalization.
Similar differences between RNA levels in mutant and wild-type plants were seen when transcripts extracted from rosette leaves of older plants were analyzed (Figure 2). For selected mRNAs, such as matR, the differences were even more pronounced at the later developmental stage. The observed differences in transcript abundances between mutant and wild-type plants indicate a developmentally persistent molecular defect in plants lacking a functional RPOTmp gene.
No differences were seen when comparing plastidial transcript levels between rpoTmp and wild-type seedlings under our growth conditions and at the developmental stages that we studied (see Supplemental Figure 2 online).
Transcript Levels but Not Sizes Differ between rpoTmp Mutants and Wild-Type Plants
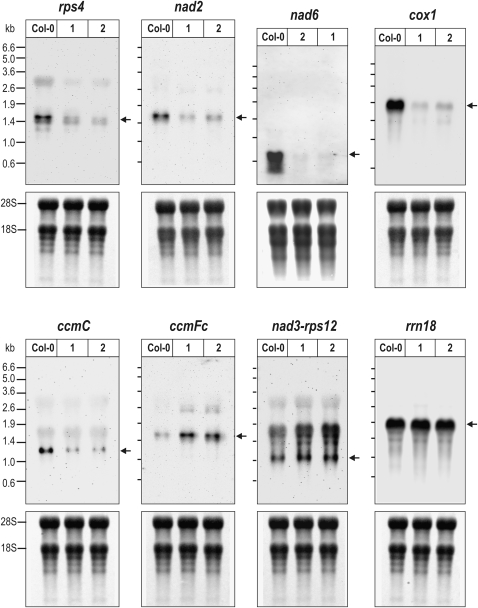
As a previous study of rpoTmp mutants reported no changes in mitochondrial transcript abundances (Baba et al., 2004), we examined the accumulation of selected mitochondrial transcripts through RNA gel blots to validate the qRT-PCR survey. Using probes complementary to transcripts rps4, nad2e1_e2, nad6, cox1, ccmC, ccmFc, and the nad3-rps12 cotranscript, we confirmed reduced rps4, nad2, nad6, cox1, and ccmC mRNA abundances as well as the enhanced accumulation of the ccmFc and nad3-rps12 mRNAs in rpoTmp mutants (Figure 3). In agreement with qRT-PCR results, the 18S rRNA accumulates to similar levels in mutant and wild-type mitochondria (Figure 3). Additionally, transcripts in mutant and wild-type mitochondria were examined using probes specific to nad1e4, nad1e5, and nad5e2 (see Supplemental Figure 3 online). The RNA gel blot showed the nad6 mRNA in rpoTmp to be even more diminished than determined by qRT-PCR. This may be related to qRT-PCR not being strand specific, as opposed to the strand-specific hybridization technique. The synthesis of antisense transcripts has been documented to occur in Arabidopsis mitochondria (Holec et al., 2006). nad6 transcripts consistently appeared more diminished when examined using strand-specific techniques (RNA gel blot and RNase protection assay) than when non-strand-specific methods were applied (qRT-PCR, run-on transcription assays using double-stranded DNA probes; cf. Figures 2 and 4 with 3 and 7). While transcript levels differed between rpoTmp and the wild type for those genes examined by RNA gel blots, no differences were seen in transcript patterns (i.e., no transcripts of unusual size were seen to accumulate in mutants).
Figure 3.
RNA Gel Blot Hybridizations Confirming Mitochondrial Transcript Changes in rpoTmp.
Probes specific to the coding sequence of rps4, nad2 (exon 1), nad6, cox1, ccmC, ccmFc (exon1), nad3, and to the mitochondrial 18S rRNA were hybridized to 15 μg (nad6) or 10 μg (others) of filter-immobilized total RNA isolated from rpoTmp-1 (lanes 1), rpoTmp-2 (2), and wild-type (Col-0) seedlings (top panels). RNA size markers were run alongside samples; marker sizes are indicated on the left of each blot. Signals corresponding in size to expected mature transcripts are indicated by arrows to the right of each panel; sizes have been inferred from transcript 5′ and 3′ ends defined by Forner et al. (2007). No monocistronic nad3 or rps12 mRNAs are detected in Arabidopsis mitochondria (Forner et al., 2007). A higher molecular weight RNA visible in the rps4 blot corresponds to an rps4-nad2e1_e2 cotranscript and was also seen in the nad2 blot when overexposed. The larger band seen in the mutants for ccmFc is the unspliced transcript. The same filters were stained with methylene blue and are shown as a loading control in the bottom panels.
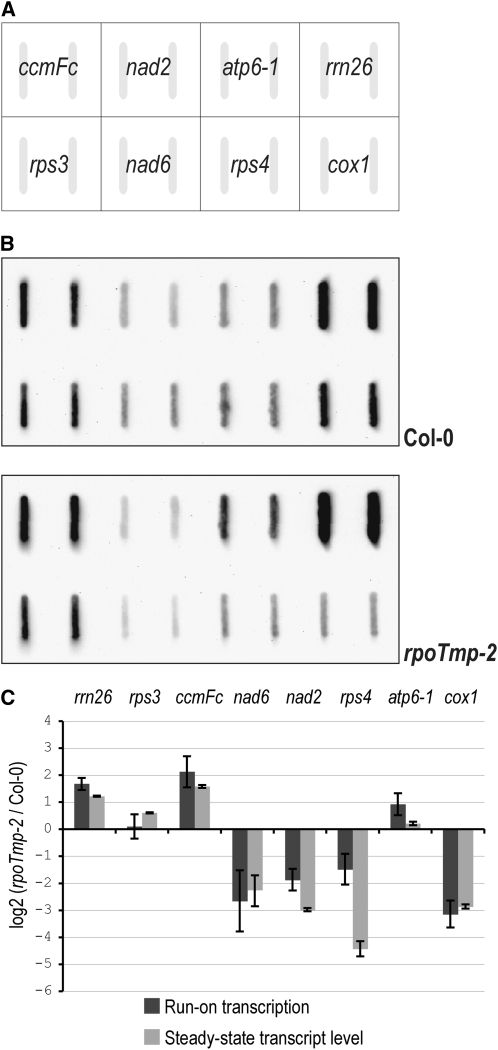
Figure 4.
Run-On Transcription Assay of Mitochondrial Transcription in rpoTmp.
(A) Layout of mitochondrial gene slot blots probed with run-on transcripts; probes were applied to membranes in duplicates.
(B) Filters probed with run-on transcripts derived from mitochondria isolated from wild-type (Col-0) and rpoTmp-2 rosettes.
(C) Mitochondrial run-on transcription rates determined for rpoTmp-2 (dark gray) and steady state transcript levels measured for rpoTmp-2 rosettes (light gray; taken from Figure 2). Transcription rates are depicted as the log2 ratio of a labeled transcript synthesized by mutant mitochondria compared with labeled transcript synthesized by wild-type mitochondria over the assay period of 10 min. Transcription rates as shown in the diagram were determined as means from three (ccmFc, nad6, and rps4) or four (all other genes) independent run-on assays; error bars represent standard deviations.
Figure 7.
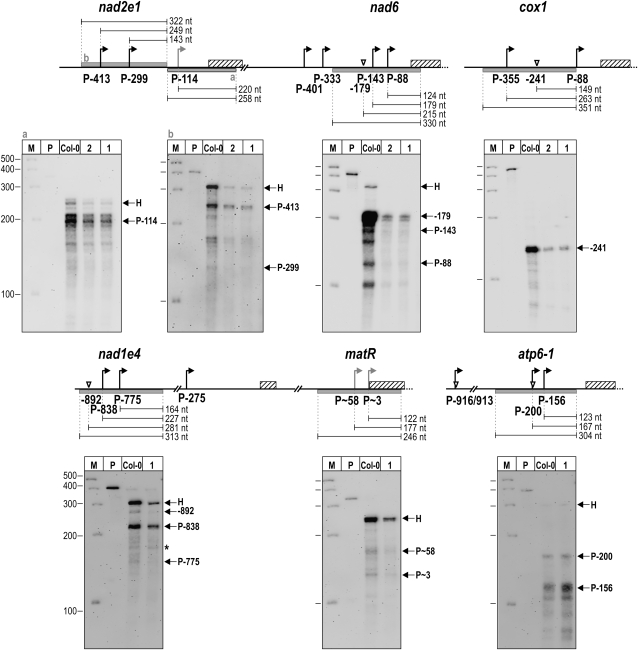
Transcript 5′ End Abundances in rpoTmp and Wild-Type Seedlings.
RPAs were performed on total seedling RNA prepared from rpoTmp-1 (lanes labeled 1), rpoTmp-2 (2), and the wild type (Col-0) using biotinylated riboprobes annealing to the nad2e1, nad6, cox1, nad1e4, matR, and atp6-1 5′ regions. Protected RNA fragments were separated on polyacrylamide gels alongside a molecular weight marker (lane M) and the probe (P) used in the respective assay; sizes are given in nucleotides. Specific protected fragments are indicated by arrows and labeled with the name of the promoter or processing site they correspond to. Signals marked H are derived from protected fragments corresponding to the complete homologous segment of a probe. An asterisk in the nad1e4 panel marks a signal that was considered nonspecific, as it was seen with nonspecific RNA in a control assay (data not shown). Diagrams of the nad2e1, nad6, cox1, nad1e4, matR, and atp6-1 5′-untranslated regions above each gel image illustrate positions of promoters (bent arrows) and processing sites (triangles); coding sequences are shown as hatched bars. Gray bars are drawn below sequences complementary to the riboprobes that were used in RPAs. The sizes of expected protected RNA fragments are given in nucleotides. The two protected fragments derived from the nad2e1 probe “a” migrated slightly faster than expected, possibly due to secondary structure formation. A fragment that could correspond to PmatR∼3 appears slightly larger than expected.
Altered Transcript Abundances in rpoTmp Coincide with Transcriptional Changes
The observed differences in mitochondrial transcript levels between rpoTmp and wild-type plants could be the result of changed rates of either transcript synthesis or/and transcript degradation. To pinpoint the molecular process(es) affected in the mutant, we performed mitochondrial run-on transcription assays with mitochondria prepared from rosettes of rpoTmp-2 and wild-type plants (Figure 4). We selected genes showing lowered transcript levels (nad2, nad6, rps4, and cox1) as well as genes with increased or unaltered transcript accumulation in rpoTmp (rrn26, atp6-1, ccmFc, and rps3) to be examined in these assays. Figure 4B shows the run-on transcription signals detected in two representative assays with mutant and wild-type mitochondria. We measured signals from three to four independent assays per genotype to quantify and compare the transcription of probed genes between rpoTmp and the wild type (Figure 4C). Transcription of rrn26, ccmFc, and atp6-1 in the mutant was significantly higher than in the wild type, while transcription of nad6, nad2, rps4, and cox1 was significantly reduced in the mutant (one-tailed t test, P ≤ 0.05). rps3 transcription did not differ significantly between mutant and wild-type plants.
The run-on transcription assays and the qRT-PCR assay on rpoTmp-2 and wild-type rosette leaves (Figure 2) were performed on the same set of plants, allowing the pairing of data from both experiments (Figure 4C). The transcriptional changes in rpoTmp generally coincided with the measured differences in transcript accumulation. The reduction in transcription of rps4 and nad2e1_e2 in the mutant was not as severe as the measured decrease in transcript accumulation. Although a contribution of altered transcript stabilities to changed transcript abundances in rpoTmp cannot be ruled out, it can be inferred from run-on transcription assays that transcriptional changes largely account for the changes in transcript abundances in the mutant.
rpoTmp Has Lower Levels of Respiratory Chain Complexes I and IV
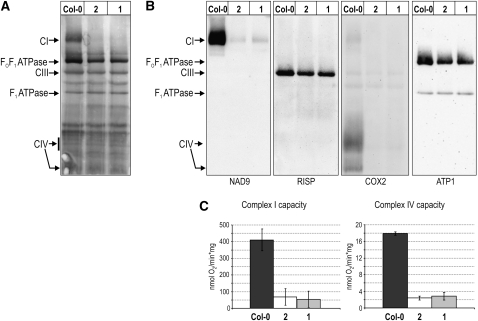
To examine if the observed reduction in mitochondrial transcripts in rpoTmp (particularly of the severely diminished nad6 and cox1 mRNAs) had consequences for the synthesis of proteins encoded by the mitochondrial genome, we analyzed the abundances of different respiratory chain complexes. Mitochondria were isolated from rpoTmp and wild-type rosette tissue, and mitochondrial membrane complexes were resolved using blue-native PAGE (BN-PAGE; Figure 5A). To validate the identification of bands in the BN-PAGE profiles, and to compare the abundances of membrane complexes between mutant and wild-type mitochondria, respiratory chain complexes I, III, and IV and complex V were immunolabeled (Figure 5B). Complex I (NADH:ubiquinone oxidoreductase) and complex IV were barely detectable in mutant mitochondria, whereas complex III (ubiquinol:cytochrome c oxidoreductase) and complex V (FOF1 ATP synthase) were clearly present (Figure 5A). While complex III and complex V appeared slightly more abundant in the wild type than in mutants, the F1 subcomplex of complex V was equally abundant in mutant and wild-type mitochondria. The unaltered levels of the F1 ATP synthase (which contains the ATP1 subunit encoded and synthesized in the mitochondrion) in rpoTmp indicate that mitochondrial protein synthesis is not impaired in mutants despite the reduced abundance of the rps4 mRNA (cf. Figure 3). We additionally compared protein synthesis in mutant and wild-type mitochondria by in organello protein synthesis assays and found no evidence for impeded translation in mutant mitochondria (see Supplemental Figure 4 online).
Figure 5.
Respiratory Chain Complexes I and IV Are Reduced in rpoTmp Mitochondria.
(A) Mitochondria were isolated from rpoTmp-1 (1), rpoTmp-2 (2), and wild-type (Col-0) rosettes, and mitochondrial membrane complexes were resolved by BN-PAGE, transferred onto a membrane, and Coomassie stained. Bands corresponding to respiratory chain complexes I (CI), III (CIII), and to the FOF1-ATPase and its F1 subcomplex are indicated. The gel areas in which complex IV (CIV) is migrating are marked.
(B) The membrane shown in (A) was probed with antibodies against NAD9, RISP (Rieske iron-sulfur protein), COX2, and ATP1 as marked to label CI, CIII, CIV, FOF1-ATPase, and the F1 subcomplex.
(C) The electron transport capacities of complex I and complex IV in mitochondrial membranes isolated from rpoTmp-1 (1), rpoTmp-2 (2), and wild-type (Col-0) rosettes were determined by measuring oxygen reduction in the presence of complex I–specific and complex IV–specific substrates. Three independent mitochondrial preparations per genotype were analyzed to calculate mean values and standard deviations.
In order to quantify the reduction in complex I and complex IV abundances in rpoTmp, we measured the electron transport capacities of complex I and complex IV in mitochondrial membranes prepared from mutant and wild-type plants (Figure 5C). Mutant membranes retained ∼15% of the complex I and complex IV capacities of the wild type.
The changes in the abundances of complex I and complex IV correlate with and are probably due to the highly reduced transcript levels seen for the genes nad6 and cox1 (Figure 3), which encode a complex I and a complex IV subunit, respectively.
Identical Promoters Are Active in rpoTmp and Wild-Type Plants
Considering the reported specificity of RPOTmp for selected promoters in plastids (Courtois et al., 2007; Swiatecka-Hagenbruch et al., 2008), we compared promoter utilization between rpoTmp and wild-type seedlings to test if the reduced transcript levels observed for several genes in mutants were due to the lack of transcription initiation from specific promoters. With the exception of cox1 (Kühn et al., 2005), none of the genes displaying reduced transcript abundance in rpoTmp had been characterized with regard to primary transcript 5′ ends and promoters. Two recent studies of mRNA termini in Arabidopsis have determined 5′ and 3′ ends of several mitochondrial transcripts, including transcripts such as nad2, nad6, ccmC, and ccmFc (Forner et al., 2007, 2008) that we observed to be changed in abundance in rpoTmp. However, those studies were not specifically directed at identifying transcription start sites (TSSs); they mapped major transcript 5′ ends and did not distinguish between primary and processed ends. We thus undertook a comprehensive analysis of TSSs and promoters in upstream regions of those genes displaying altered transcript accumulation in mutants.
In plant mitochondria, the promoter sequence comprises the TSS (Binder and Brennicke, 2003); promoters can therefore be identified by TSS mapping. We identified TSSs in rpoTmp mutant and wild-type plants using a 5′-rapid amplification of cDNA ends (5′-RACE) technique employed previously for determining primary and processed transcript 5′ ends and promoters in Arabidopsis mitochondria (Kühn et al., 2005). Figure 6 provides a graphical summary of TSSs and 5′ end processing sites determined in our study using total cellular RNA prepared from 7-d-old seedlings. Table 1 lists the mapped TSSs and processing sites and their surrounding sequences. Promoters (P) and their corresponding TSSs are specified with the gene name and the position of the initiating nucleotide with respect to the start of the coding sequence (e.g., Pcox1-355). Sites for which transcript 5′ end analysis did not support their functioning as TSS and that did not display sequence elements frequently associated with mitochondrial promoters are listed as processing sites. Nearly all of these sites have previously been identified as processing sites resulting from endonucleolytic 5′-terminal processing (Forner et al., 2007). For eight 5′ termini mapping to a promoter-like motif, such as Pnad2e1-114, 5′-RACE analysis did not distinguish a primary 5′ end (see Supplemental Figures 7, 9, 11, 13, and 14 online). Such sites are nonetheless listed as putative promoters because it is sometimes not possible to clearly identify primary transcript ends with the 5′-RACE technique applied here (e.g., when the triphosphates at primary 5′ ends are unstable) (Kühn et al., 2005; Forner et al., 2007). These putative promoters have been pointed out in Table 1. Images of all analyzed 5′-RACE products and detailed specifications of identified 5′ ends are provided in Supplemental Figures 5 to 14 online.
Figure 6.
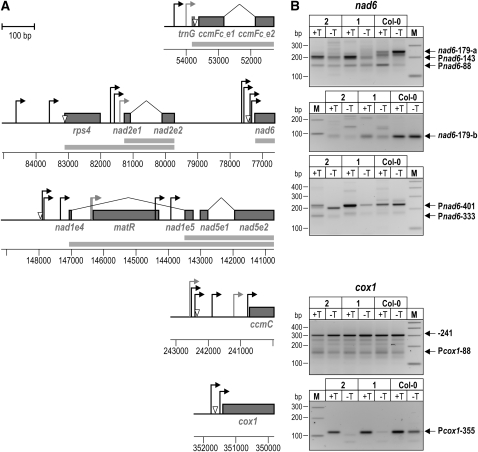
Analysis of TSSs in rpoTmp and Wild-Type Seedlings.
(A) Maps of mtDNA regions for which TSSs and transcript 5′ end processing sites were analyzed. Protein-coding sequences are displayed as gray boxes and labeled with the corresponding gene names. In the case of cis- and trans-spliced sequences, exons are labeled with the gene name, followed by the letter e and the exon number. Splicing events are indicated by a bent line between exons. TSSs mapped in 5′-RACE analyses are indicated by bent arrows; open triangles mark 5′ end processing sites. Putative promoters, for which primary 5′ termini could not be experimentally distinguished, are drawn in gray. Where indicated by experimental results, cotranscription of genes and exons is shown by gray bars below gene maps. Coordinates of the analyzed regions on the mtDNA are indicated.
(B) 5′-RACE analysis of nad6 and cox1 transcripts. 5′-RACE was performed on RNA extracted from 7-d-old rpoTmp-1 (lanes labeled 1), rpoTmp-2 (2), and wild-type (Col-0) seedlings using a 5′-RACE technique that exploits the difference in 5′ end phosphorylation between primary and processed transcript 5′ ends (Bensing et al., 1996). Products obtained from TAP-treated (+T) and untreated RNA (−T) were separated on agarose gels alongside a molecular weight marker (M). Products from primary transcript 5′ ends are labeled with the name of the corresponding promoter as listed in Table 1; products corresponding to processed ends are labeled accordingly. The product from the processed end nad6-179 was barely detectable in rpoTmp-1 and -2 (signal marked nad6-179-a) but could be amplified with a reverse primer not detecting any downstream 5′ ends (nad6-179-b).
Table 1.
Transcription Initiation Sites and Processing Sites Detected by 5′-RACE
| Col-0 |
rpoTmp |
|||||
|---|---|---|---|---|---|---|
| Gene | 5′ End | Sequence | +TAP | −TAP | +TAP | −TAP |
| cox1 | Pcox1-88 | CTCTCAAGAACTCGTAGACTATGGA | 2/8 | 0/7 | n.a. | n.a. |
| cox1 | cox1-241a,b | GAATCCAATGGTAGGCCTGGGCTCT | n.a. | n.a. | n.a. | n.a. |
| cox1 | Pcox1-355a | AATTTATTCAATTATATaATAATAA | n.a. | n.a. | 6/8# | n.a. |
| nad1 | Pnad1e4-275 | TACAAAAACATTCTAGGaAAGGGCT | 6/10 | 1/8 | 2/2 | n.a. |
| nad1 | Pnad1e4-775 | CGACTGCTATCTAAGAGAAgATAAG | 4, 3, 2/13 | 7, 0, 7/15 | 8/8 | n.a. |
| nad1 | Pnad1e4-838 | GGCAATTTATCGTATAGaAGAATAG | 3, 6/12 | 0, 0/13 | 14, 0/14 | n.a. |
| nad1 | nad1e4-892 | GACCAAGCAAATCTCCTCTTCTAGT | – | 2/2 | – | – |
| matR | PmatR∼3*,e | CAATATGAAGAAAGAAaGAAGGGTT | ||||
| matR | PmatR∼58*,e | GCCTTTGATTATAAAAAaGGGGAGC | ||||
| nad1 | Pnad1e5-427 | ACTAATTATATATATATaATAAGGT | 5/7 | 0/10 | 2/8 | n.a. |
| nad1 | Pnad1e5-927 | CAAATATTGTAAATCAAgAAGGTGG | 5/18 | 0/10 | 2/13 | n.a. |
| ccmC | PccmC-45 | CTTTGATTGCGTATTATaGATCCAT | 4/16 | 0/15 | 1/14 | n.a. |
| ccmC | PccmC-487ce | TATCGATCGGCGTAAGaAAAgATTC | 1, 2, 1/15 | 0, 2, 1/12 | 1, 1/8 | n.a. |
| ccmC | PccmC-1159 | GATTCGTCATGGTAGAGaAGAAAAA | 6/6 | – | 8/8 | – |
| ccmC | ccmC-1634 | CCTCGAATTCTGTAAGCTAGCTTGG | 1/1 | n.a. | 9/9 | n.a. |
| ccmC | PccmC-1677 | CTACCAAACGTGCTAATAAAAGAGC | 8/9 | 0/7 | – | – |
| ccmC | PccmC-1817 | AAGTAATTCATTATAAgATAAGTaA | 1, 3, 9/13 | 0, 0, 4/8 | 2, 2/9 | n.a. |
| ccmC | PccmC-1834e | ACACTACTATATAAGATAAGTAATT | 2, 0/2 | 1,4/6 | n.a. | n.a. |
| rps4 | rps4+2b | GAGTCCCAGGGACGCAAtGTGGCTG | 13/13 | n.d | 8/8# | n.a. |
| rps4 | Prps4-477 | AGAAAGCTCAAGTATGTaAACAACC | 6/7 | n.a. | 8/8 | n.a. |
| rps4 | Prps4-1509 | AGATAGGTCAGGTATATaTAGCGAT | 4/6 | 0/8 | 3/3# | – |
| nad2 | Pnad2e1-114be | CACTATCATTAtATTATATaTTCAT | 6,10, 4/32 | 5, 3, 2/18 | 3, 1/7 | n.a. |
| nad2 | Pnad2e1-299 | CAAGATATTGCGTATaAATATATAT | 2/28 | n.a. | 1/8 | n.a. |
| nad2 | Pnad2e1-413 | GAACAAAAGATGTAATGaAAAgATA | 2, 3/10 | n.a. | 4, 3/8 | n.a. |
| nad6 | Pnad6-88 | GAAGAAAATGAAATAGGAaCAACCG | 2,10/16 | 0,1/7 | 7/15 | n.a. |
| nad6 | Pnad6-143 | ATTGAGATTCCGTAAGTaACTCAGT | 3/8 | n.a. | 9/16 | n.a. |
| nad6 | nad6-179b | GGGTACAAGATCGAAAAgAATGCAT | 14/17 | 12/14 | 3/7 | n.a. |
| nad6 | Pnad6-333 | GATGATTATGTTGAAAAaGATCGAA | – | – | 8/8 | 0/7 |
| nad6 | Pnad6-401 | CGGGTACAGTAGTAGGTaAACTTGA | 18/19 | 2/8 | 8/8 | – |
| ccmFc | ccmFc-124b | AAGTCCCTCCTTCCGCTCCTGGTGT | n.a. | n.a. | n.a. | n.a. |
| ccmFc | PccmFc-379e | TTGGATTATCGTAGAATAAGAGGAG | 6/7 | 6/7 | n.a. | n.a. |
| ccmFc | PccmFc-743 | TTTATTATTCTATTATAaTAGTCTA | – | – | 8/8 | – |
| nad9 | nad9-211/210c | GGCGATCCACGAATTTGATCATTCG | 1, 2/8 | n.a. | n.a. | n.a. |
| nad9 | Pnad9-1241e | TACGAAATATAGTAAATATCGTAAA | 3/3 | n.a. | n.a. | n.a. |
| nad9 | Pnad9-1371 | TTCTCGTAATGTATTTAACCATAAA | 2/8 | n.a. | – | – |
| nad9 | nad9-1400 | CAAACTAAATGCTATAAGCATAGAA | 1/2 | n.a. | n.a. | n.a. |
|
nad9 |
Pnad9∼1730e |
GATTGATAAAGGTAATAGATATGAT |
n.a. |
n.a. |
n.a. |
n.a. |
TSSs and processing sites determined for the wild type (Col-0) are underlined in sequences; lowercase positions denote sites mapped for rpoTmp. CATA, CGTA, TCTA, ATTA, AGTA, and GGTA motifs frequently found associated with mitochondrial promoter sequences (Fey and Marechal-Drouard, 1999; Kühn et al., 2005) are in bold. The number of clones sequenced for each site is given together with the frequency of the respective transcript 5′ end as determined from TAP-treated (+TAP) or untreated (−TAP) seedling RNA. Where more than one nucleotide is marked in one sequence, 5′ end frequencies are given in 5′-to-3′ order. Mutant transcript 5′ ends were mostly analyzed from rpoTmp-2 5′-RACE products. In a few cases where products amplified for rpoTmp-2 were extremely faint but rpoTmp-1 products formed a clear band, the latter were cloned and sequenced (numbers marked #). As reported by Forner et al. (2007), matR 5′ ends were poorly defined (sites marked *; see Supplemental Figure 7 online for identified 5′ ends). The first codons of the annotated MATR and RPS4 coding sequences are italicized. –, Signal absent in 5′-RACE; n.a., not analyzed.
Determined previously (Kühn et al., 2005).
Determined previously (Forner et al., 2007).
Determined previously (Forner et al., 2008) with minor differences.
Putative promoters for which experimental data are insufficient to support their function as TSS.
The majority of 5′ ends detected in the wild-type plants were also detected in rpoTmp mutants (Figure 6, Table 1); 5′-RACE products amplified from cox1 transcript 5′ termini are shown as examples (Figure 6B). For some promoters, such as Pnad2e1-114 and Pnad2e1-297, 5′ ends did not map to exactly the same nucleotides in rpoTmp (lowercase positions in Table 1) and the wild type (underlined positions). This could be due to either technical inconsistency of the 5′-RACE technique or genotype-specific differences between initiating nucleotides in rpoTmp and the wild type. The first is more likely, as for some TSSs we also saw such differences between two independent 5′-RACE analyses performed on the same gene in the wild type (data not shown).
Selected transcript 5′ termini, such as the processed end nad6-179, were barely detectable in mutants (Figure 6B). On the other hand, a primary 5′ end mapping to Pnad6-143 downstream of this processing site was more easily detected in rpoTmp (Figure 6B). Products mapping to Pnad6-333 were not detected in the wild type or rpoTmp-1 but only in rpoTmp-2. The discrepancy between the two mutant lines, and also between rpoTmp-2 and the wild type, could be due to very low abundance of the Pnad6-333 5′ end. The apparent difference at Pnad6-333 between rpoTmp-2 and the wild type was therefore considered insignificant. These occasional inconsistencies are likely to be due to the fact that while the 5′-RACE technique employed here is a powerful tool for identifying primary transcript 5′ termini, it is inadequate for quantitative studies and may even fail to amplify selected 5′ ends. We therefore attempted to determine the abundances of these 5′ termini in mutants and the wild type using quantitative ribonuclease protection assays (RPAs; Figure 7).
Comparison of Transcript 5′ End Abundances between rpoTmp and Wild-Type Plants
For selected mitochondrial genes, we analyzed the abundance of their different transcript 5′ ends through RPAs. Because this technique uses the labeled probe in excess to saturate all target sequences present in one assay, it provides a quantitative detection method for the analysis of transcript 5′ ends. Using total cellular RNA prepared from 7-d-old rpoTmp-1, rpoTmp-2, and wild-type seedlings, we performed RPAs on 5′ termini of transcripts reduced in abundance in rpoTmp (nad1e4 and matR, nad2e1_e2, nad6, cox1, and ccmC) as well as a transcript accumulating more abundantly in mutants (atp6-1). atp6-1 was selected because it represents a gene with known multiple promoters that have been detected previously using the RPA technique (Kühn et al., 2005).
Figure 7 shows the PAGE analyses of protected fragments generated through RPAs performed on mitochondrial transcript 5′ ends. Linear diagrams in Figure 7 illustrate the upstream regions, promoters, and processing sites of genes examined by RPAs, as well as riboprobe target sequences. The fragments corresponding to Pnad6-143 and Pnad6-88 appeared to migrate in the PAGE as two major bands, with the larger of two transcripts seen for each Pnad6-143 and Pnad6-88 at the expected size. The two faster migrating transcripts show the same apparent reduction in size, which would agree with secondary structure formation. However, no equivalent lower signal is seen for the nad6-179 signal, which argues against secondary structure formation at the 3′ end. The additional nad6 signals could therefore not be assigned to known transcripts and have not been labeled in Figure 7.
Not only the 5′ end nad6-179, which had barely been detectable by 5′-RACE (Figure 6B) in rpoTmp mutants, but also the termini corresponding to Pnad6-143 and Pnad6-88 were greatly reduced in abundance in the mutants. We were unable to analyze the abundances of transcripts synthesized from nad6 promoters located upstream of the nad6-179 processing site because this region shows 97% sequence identity to the chloroplast rbcL transcript, which interfered strongly with the assay. From the signal corresponding to the complete protected probe (signal H in the nad6 panel of Figure 7) and from the abundance of the nad6-179 5′ end, the formation of which depends on transcription initiation from upstream sites, it can be inferred that transcripts starting at these sites were diminished in rpoTmp in comparison to the wild type. Signals corresponding to Pnad2e1-114, Pnad2e1-299, and Pnad2e1-413 were similarly reduced in mutants. Likewise, all analyzed nad1e4 and matR 5′ termini, primary or processed, were reduced in abundance in rpoTmp. By contrast, signals corresponding to Patp6-1-156 and Patp6-1-200 were enhanced in mutants.
The fairly prominent signal corresponding to the complete protected nad1e4 probe (signal H in the nad1e4 panel of Figure 7) indicates that significant transcription of nad1e4 and matR initiates from TSS upstream of the nad1e4-892 processing site. We were unable to consistently map these upstream TSSs in two independent 5′-RACE experiments and did not investigate them further; they are therefore not listed in Table 1. However, the signal intensities of the complete protected nad1e4 probe seen for rpoTmp mutants and the wild type indicate that nad1e4 and matR transcription from the undefined upstream TSS is also decreased in rpoTmp.
Primary 5′ ends corresponding to Pcox1-355 (Figure 7), PccmFc-379, and PccmFc-743 (data not shown) were not detectable in any genotype using RPAs, probably owing to rapid 5′ end processing of these transcripts.
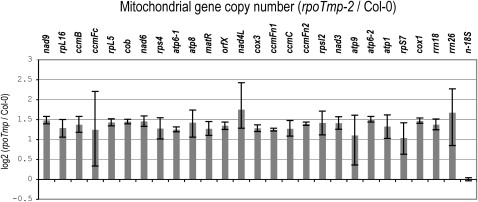
The quantitative analysis of transcript 5′ ends showed that all 5′ termini of a transcript were diminished in rpoTmp when the overall abundance of the transcript was reduced in the mutant. To ensure that this was not a consequence of lowered abundances of subgenomic DNA molecules in rpoTmp, we compared copy numbers of mitochondrial genes in mutant and wild-type plants using qPCR. Mitochondrial DNA levels in rpoTmp-2 were twofold to threefold higher than in the wild type uniformly across all genes tested; no gene-specific variations in gene copy numbers were detected (Figure 8). Nuclear DNA content in rpoTmp is unaltered (E. Cincu, J. Fuchs, and T. Börner, unpublished data; see Supplemental Figure 17 online), permitting the use of a nuclear reference gene in this analysis (Figure 8).
Figure 8.
Mitochondrial Gene Copy Numbers in rpoTmp-2 Seedlings.
Gene copy numbers are depicted as the log2 ratio of DNA levels in mutants compared with levels in wild-type plants (Col-0). Gene copy numbers of selected mitochondrial protein-encoding and rRNA genes were determined by qPCR in both mutant and wild-type seedlings employing the PCR protocol and primers used for qRT-PCR. Three technical replicates were averaged per genotype; standard errors are indicated. The nuclear 18S rRNA gene was used for data normalization.
Promoters of Genes with Reduced Transcription in rpoTmp Are Not Structurally Distinct
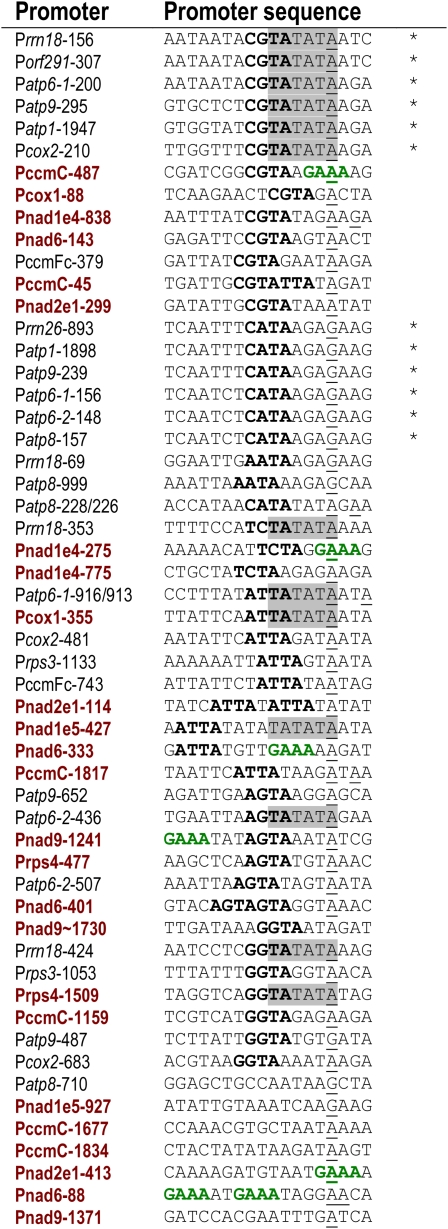
A list of all promoters and putative promoters identified in Arabidopsis mitochondria to date is displayed in Figure 9. Sequences spanning 18 nucleotides, from position −14 to +4 around the TSS, are displayed. This region has been defined by Dombrowski et al. (1999) to be required for promoter function in dicot plant mitochondria. The promoters and putative promoters of genes transcribed less actively in rpoTmp were computationally compared with all other known mitochondrial promoters in Arabidopsis to identify hexa-, penta-, or tetranucleotide elements statistically overrepresented in the subset of promoters affected in rpoTmp (see Supplemental Table 1 online). No RPOTmp-specific hexanucleotide or pentanucleotide elements were detected. Only the tetranucleotide GAAA was overrepresented among promoters of genes transcribed less actively in the mutant. Since the position of this element in the different promoter sequences is variable, with occurrences upstream of as well as comprising the TSS (Figure 9), this element is unlikely to be significant for RPOTmp-specific promoter function. Among the 25 tested promoters of genes affected in rpoTmp, only six displayed a GAAA element. We could find no criteria by which we could identify these promoters as a distinct subset of mitochondrial promoters, other than the fact that the transcripts initiated from them are less abundant in rpoTmp plants. A distance tree has been derived of all promoters as listed and aligned in Figure 9, in which rpoTmp-affected promoters are interspersed with unaffected promoters throughout most of the tree and do not cluster as an obvious subgroup (see Supplemental Figure 15 online).
Figure 9.
Promoter Sequences in Arabidopsis Mitochondria.
The list includes promoters identified previously (Kühn et al., 2005) and in this study; sequences have been sorted according to the presence of a CATA, CGTA, TCTA, ATTA, AGTA, or GGTA tetranucleotide (bold black) immediately upstream of the TSS (underlined). The motif TATATA, which is recurring in mitochondrial promoters, is highlighted. The names of promoters of genes showing reduced transcription in rpoTmp mutants are printed red. The tetranucleotide GAAA, which is overrepresented in this subset of promoters, is printed green. Promoters displaying a perfect CGTATATAA or CATAAGAA motif are marked with an asterisk.
DISCUSSION
Investigations of mitochondrial transcription in higher plants have found this process not to be stringently controlled (Holec et al., 2008). Transcription initiates from multiple sites that are not confined to the upstream regions of annotated mitochondrial genes, continuously giving rise to presumably nonfunctional RNAs and occasionally even detrimental transcripts that are usually efficiently removed by RNA-degrading mechanisms in the organelle (Finnegan and Brown, 1990; Holec et al., 2006). Posttranscriptional processes have been identified as a major determinant of mitochondrial RNA pools, as abundances of individual transcripts do not always reflect transcript synthesis rates (Giegé et al., 2000). Transcriptional mechanisms in mitochondria are thought to provide no significant means of regulating mitochondrial function. It is therefore surprising that in the mitochondria of eudicotyledonous plants, this basic process involves two RNA polymerases, RPOTm and RPOTmp, whose distinct roles have not yet been resolved. While indications of RPOTmp-dependent transcription in plastids have been reported (Courtois et al., 2007; Swiatecka-Hagenbruch et al., 2008), a transcriptional role for RPOTmp in mitochondria has been lacking experimental proof (Baba et al., 2004; Kühn et al., 2007). Our study substantiates and characterizes the function of RPOTmp in mitochondrial RNA synthesis and links this enzyme to a distinct, gene-specific transcriptional mechanism. We demonstrate that while RPOTm is vital for plant development, RPOTmp is required to optimally transcribe a subset of mitochondrial genes that, unexpectedly, are not defined by common promoter sequences.
RPOTmp Is Required for the Transcription of Distinct Mitochondrial Genes
Mitochondrial transcription and steady state transcript abundances were found to be severely altered in the absence of RPOTmp (Figures 2 and 4), establishing a fundamental role for this enzyme in RNA synthesis in mitochondria. The altered transcript levels are unlikely to be the secondary result of an unknown primary defect leading to reduced Complex I and Complex IV levels because the mitochondrial transcriptome profile of rpoTmp is very different from that of the otp43 mutant lacking Complex I due to defective nad1 splicing (comparison to unpublished data by A. Falcon de Longevialle; see Supplemental Figure 18 online). In particular, nad6, rps4, and cox1 transcript levels are not reduced in otp43. As the transcript changes observed in rpoTmp are gene specific, with distinct transcripts showing reduced levels but others exhibiting unaltered or elevated accumulation, RPOTmp function must be considered gene specific. The differences between rpoTmp mutant and wild-type mitochondrial transcript profiles are stable from the seedling to the rosette stage. The requirement for this enzyme corresponds with its expression throughout the Arabidopsis life cycle, primarily in tissues with high mitochondrial activity (Emanuel et al., 2006). Remarkably, transcriptional defects incurred by the absence of RPOTmp cannot be compensated for by subsequent mechanisms of gene expression, such as transcript stabilization or enhanced translation. This is manifested by the reduced levels of respiratory chain complexes I and IV in rpoTmp mutants (Figure 5) that are probably the result of diminished amounts of the nad6 and cox1 transcripts. Complex I function is not essential in Arabidopsis, but its absence leads to developmental defects (Falcon de Longevialle et al., 2007). Complex IV, on the other hand, is vital for plant development (Gu et al., 1994). The importance of RPOTmp for the expression of principal mitochondrial components adds substantially to present views of the contribution of transcriptional processes to steady state RNA pools and protein synthesis in plant mitochondria.
Our data are compatible with either a role of RPOTmp in transcribing several mitochondrial genes or, alternatively, a role in stimulating the transcription of these genes by RPOTm. Mitochondrial promoter specificity of recombinant RPOTm but not RPOTmp has been detected in a minimal in vitro transcription assay; however, RNA polymerase activity has been demonstrated for both (Kühn et al., 2007). Because of the latter finding, we are in favor of RPOTmp being an RNA polymerase transcribing specific mitochondrial genes, including nad6 and cox1 and thus supplementing the transcriptional role performed by RPOTm.
The accumulation of numerous mitochondrial transcripts was enhanced in the absence of RPOTmp. Although stimulation of RPOTm expression and/or activity in rpoTmp mutants could account for this, we would mostly attribute it to elevated cellular mitochondrial (mt) DNA levels in mutants. A two- to threefold increase in gene copy number was seen for all mitochondrial genes tested (Figure 8). The increase in abundance of a transcript usually did not exceed the increase in gene copy number per cell (cf. Figures 2 and 8). Interestingly, enhanced mitochondrial transcript levels as a consequence of elevated mitochondrial gene copy numbers have been observed in the barley (Hordeum vulgare) mutant albostrians with impaired chloroplast function (Hedtke et al., 1999a). Therefore, plants may respond in general to energy constraints caused by defects in the energy-converting organelles by increasing cellular mtDNA levels. Elevated mtDNA levels per nuclear genome could be due to either increased mtDNA copy numbers per mitochondrion or an increased number of mitochondria per cell.
Mitochondrial transcript profiles in rpoTmp appear to be the combined result of elevated cellular mtDNA levels and reduced transcription of specific mitochondrial genes, with the extent of reduction in RNA synthesis varying between those genes. Therefore, transcripts with unchanged or mildly elevated abundance in mutants (rps3 and atp6-1) might still be made at lower levels per gene copy in the absence of RPOTmp. The synthesis of those RNAs would mainly be performed by RPOTm and depend on RPOTmp to a minor extent. In the cases of nad6 and cox1, the transcription per gene copy is likely to be even more reduced than measured, while transcription of ccmFc appears to not require RPOTmp.
The rps4 gene coding for a ribosomal protein is among the genes showing reduced transcription and steady state transcript levels in the absence of RPOTmp. Mitochondrial protein analyses showed no evidence for mitochondrial translation being inhibited due to the reduced availability of the rps4 mRNA (Figure 5). Consequently, rps4 mRNA abundance appears not to be limiting for activity of mitochondrial ribosomes. The functionality of the mitochondrial rps4 gene in Arabidopsis has recently been questioned because the 5′-processed form of the rps4 mRNA lacks the first nucleotide of the annotated start codon (Forner et al., 2007). The absence of a detectable effect on mitochondrial translation in rpoTmp contrasts with the reduced levels of respiratory chain complexes I and IV, which we attribute to the limited availability of nad6 and cox1 mRNAs in the mutants. The nad6 and cox1 mRNAs were significantly more affected in mutants than the rps4 mRNA (Figure 3).
An earlier study of RPOTmp-deficient plants found all analyzed mitochondrial transcripts, which included matR and cox1, unchanged (Baba et al., 2004). Baba et al. studied the effect of RPOTmp loss on the light-induced accumulation of organelle transcripts and conducted their analysis of mitochondrial RNAs on seedlings grown entirely in the dark or seedlings transferred into continuous light for 6 d following the dark growth period. The discrepancies between our mitochondrial transcript analysis in rpoTmp mutants and the earlier report could be due to the different growth conditions and/or the analysis of transcripts in seedlings of different ages. RPOTmp may not be detectably active in wild-type mitochondria in the conditions applied in the earlier study. Alternatively, the different technical approach taken by Baba et al., who studied mitochondrial transcript abundances by hybridizing labeled cDNAs to macroarrays, could account for the different findings.
RPOTmp Activity Is Gene Specific Rather Than Promoter Specific
Although RPOTmp, unlike RPOTm, was not shown to specifically initiate transcription from mitochondrial promoters in vitro (Kühn et al., 2007), our data suggest that RPOTm and RPOTmp transcribe from overlapping sets of promoters. Mitochondrial transcription in RPOTmp-deficient plants initiates essentially from the same promoters as in wild-type plants. All 5′ ends of a transcript reduced in abundance in mutants appear diminished in comparison to the wild type; hence, transcription from all promoters of a gene transcribed less actively in rpoTmp appears to be reduced. RPOTmp activity is therefore gene specific rather than promoter specific.
For eight 5′ ends considered in this study as primary 5′ ends because they map to promoter-like sequences, experimental evidence for the termini being primary rather than processed is insufficient (see Supplemental Figures 7, 9, 11, 13, and 14 online). The experimental identification of a primary transcript 5′ ends depends on the presence of its 5′-triphosphate. Considering the poor stability of triphosphates at several known primary transcript 5′ ends in mitochondria (Kühn et al., 2005), the lack of distinguishable 5′ triphosphates at 5′ termini does not preclude them from mapping to promoters. For 19 out of 27 promoters characterized in this study, the analysis of 5′-RACE products was nevertheless sufficient to validate them as TSSs.
The neighboring genomic positions of genes showing reduced transcription in rpoTmp raise the question if some of these genes, such as rps4, nade1_e2, and nad6, could be transcriptionally linked. According to the results of our transcript analyses, cotranscription can only partly account for reduced transcript levels of adjacent genes. While nad2e1_e2 appears to be partly cotranscribed with rps4, promoters are active in the rps4-nad2e1_e2 intergenic region from which transcription also seemed reduced in rpoTmp (Figure 7). We have found no indication of cotranscription of nad2e1_e2 and nad6, either because 5′ end processing of the nad6 mRNA is extremely efficient or because there is no cotranscription. Transcription from the promoters Pnad6-88 and Pnad6-143 immediately upstream of the nad6 coding region is also reduced (Figure 7). While matR is mostly, if not exclusively, cotranscribed with nad1e4, nad1e5 is at least partly transcribed from promoters immediately upstream of nad1e5; a transcript whose 5′ end maps to one of these promoters is also reduced (Figure 6; see Supplemental Figures 3 and 8 online). Altogether, we found no evidence that reduced transcript levels of clustered genes in mutants were entirely due to the reduction in a polycistronic message spanning all genes; rather, transcription from all promoters in the examined region appeared to be reduced.
The analysis of promoters and putative promoters of genes transcribed less actively in rpoTmp and their comparison to promoters of other mitochondrial genes did not identify RPOTmp-specific promoter sequence motifs. We noticed that none of the genes possessing in their upstream region a perfect CATAGAGAA or CGTATATAA promoter motif showed reduced mitochondrial transcript levels in rpoTmp, which can be interpreted as a link between these promoters and RPOTm function. However, because several genes exhibiting unaltered or elevated RNA levels in rpoTmp have none of the cited motifs in their upstream region, these promoters could only represent a minor portion of promoters used by RPOTm. ccmFc does not seem to require RPOTmp for its transcription, yet its promoter sequences are not noticeably different from those of genes with decreased RNA abundances in rpoTmp.
Promoter utilization in rpoTmp mutants indicates that there are no RPOTmp-specific promoter sequences in Arabidopsis mitochondria. Instead, additional gene-specific elements distinct from the known promoters are likely to direct the transcription of specific mitochondrial genes by RPOTmp. These elements may act through RPOTmp-specific trans-factors. The identification of the proposed factors may be aided by the knowledge of genes transcribed by RPOTmp. The known promoters, rather than functioning as elements controlling transcription through recruitment of a specific RNA polymerase, may simply function as sites at which the double helix is easily opened to initiate transcription. Although mitochondrial promoter selection has been reported to be controlled by nucleus-encoded factors in alloplasmic lines of Nicotiana tabacum and maize (Zea mays; Newton et al., 1995; Edqvist and Bergman, 2002), these factors and their possible roles in regulating mitochondrial function remain to be identified. Due to frequent rearrangements of mitochondrial sequences, the mtDNAs of dicotyledonous plant species show occasional substitutions of promoters between different mitochondrial genes (Unseld et al., 1997; Kubo et al., 2000; Handa, 2003; Forner et al., 2008). Given that promoters seem to be interchangeable between genes and that RPOTmp function is gene specific rather than promoter specific, we speculate that the transcription of mitochondrial genes may partly be controlled through elements within the transcribed regions rather than upstream of the TSS.
From comparing analyses of mitochondrial transcripts (this study) and of plastidial transcripts (Baba et al., 2004; Courtois et al., 2007) in mutants lacking RPOTmp, the dual-targeted RNA polymerase is likely to have dissimilar roles in the two organelles. The presence of three transcriptases in plastids of eudicotyledonous plants (Liere and Börner, 2007) complicates individual functional studies of RPOTmp in this organelle. In Arabidopsis plastids, the rrn16 Pc promoter is highly RPOTmp specific and nearly inactive in the absence of RPOTmp (Courtois et al., 2007). In addition to the Pc promoter, RPOTmp is capable of initiating transcription from several other plastidial promoters (Swiatecka-Hagenbruch et al., 2008). RPOTmp in plastids has been reported to be immobilized and inactivated following germination (Azevedo et al., 2008). This is in agreement with the developmental variation of the Pc promoter activity in Arabidopsis and with our plastidial transcript screen, which showed no differences between wild-type and mutant transcript levels in plastids of 7-d-old plantlets (see Supplemental Figure 2 online). The apparent developmentally limited importance of RPOTmp in plastids contrasts with the requirement of this enzyme for the expression of mitochondrial genes in seedlings as well as in adult plants.
Roles of RPOTmp- and RPOTm-Dependent Transcriptional Mechanisms
Our study confirms that RPOTm functions as the basic RNA polymerase in mitochondria. It is unconditionally required for the transcription of most, if not all, mitochondrial genes. RPOTmp fine-tunes this basic level of transcription by enhancing the levels of specific mitochondrial RNAs. RPOTmp-dependent RNA synthesis may present a transcriptional mechanism allowing mitochondria to independently control the expression of a subset of mitochondrial genes. A challenge for future research into RPOTmp function lies in exploring the conditions where such a mechanism might operate. It could have evolved as a control on the abundances of respiratory chain complexes I and IV, thereby enabling mitochondria to fine-tune the electron flow through these complexes in response to the developmental or metabolic requirements of the organelle and the cell.
METHODS
Plant Material and Culture Conditions
The T-DNA insertion lines SALK_132842 (rpoTmp-2) and SALK_086115 (rpoTmp-3) were obtained from the ABRC seed stock center; line GABI_286E07 (rpoTmp-1) was acquired from GABI-Kat (Rosso et al., 2003). The Arabidopsis thaliana (ecotype Col) T-DNA mutants and wild-type plants were germinated on half-strength Gamborg B5 agar containing 1% sucrose at 22°C and a light intensity of 80 μmol quanta m−2 s−1 in a 16-h photoperiod. Seedlings for DNA and RNA isolation were harvested after 7 d; plants were further grown on soil (two parts Scotts Shamrock growing medium and one part vermiculite) under identical conditions. For the isolation of RNA and mitochondria from rosettes, mutant and wild-type plants were germinated on soil at 22°C and a light intensity of 80 μmol quanta m−2 s−1 in a 16-h photoperiod. After 10 d, plants were grown further in an 8-h photoperiod; plant tissue was harvested from 7-week-old (wild type) and 9-week-old plants (mutants).
DNA and RNA Isolation and RT-PCR Analysis
Genomic DNA isolated from seedlings as described by Edwards et al. (1991) was used for PCR analysis of rpoTmp alleles with Taq DNA Polymerase (Qiagen; see Supplemental Figure 16 online for primer information and PCR results). RNA was isolated from rpoTmp-1, rpoTmp-2, rpoTmp-3, and Col-0 seedlings grown on agar in a 16-h photoperiod for 7 d and from rosette leaves of plants grown on soil in an 8-h photoperiod for 9 weeks (rpoTmp-1 and rpoTmp-2) or 7 weeks (Col-0); this difference in age was necessary to obtain plants at a similar developmental stage. The plant RNeasy extraction kit (Qiagen) was used for RNA isolation according to the manufacturer's instructions. The RNA was then subjected to two consecutive treatments with TURBO DNase (Ambion) and confirmed by PCR to be free of detectable amounts of DNA. Three micrograms of RNA were reverse-transcribed with Superscript III (Invitrogen) according to the manufacturer's protocol using random hexamers (Invitrogen). One microliter of the 25-fold diluted reverse transcription reaction was used for PCR with Taq DNA Polymerase (Qiagen; see Supplemental Figure 3 online for primer information and RT-PCR results).
Quantitative RT-PCR Analysis
RNA was isolated from seedlings and rosette leaves and reverse-transcribed as detailed above. Quantitative RT-PCR was performed in 384-well plates using the LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics) and a LightCycler 480 Roche real-time PCR system as described previously (Falcon de Longevialle et al., 2008); primer sequences are listed in Supplemental Table 2 online. Primer pairs specific for the nuclear 18S rRNA gene and the nuclear ACT2 and ACT8 genes were used to measure the accumulation of nucleus-encoded transcripts. The specificity of each primer pair was tested by sequencing the PCR product and subsequently by melting curve analysis. For each primer pair and each comparison between mutant and wild type, a standard curve based on serial dilutions of the wild-type cDNA was included. The accumulation of each transcript was analyzed in triplicate. Values displayed in Figure 2 represent means of triplicates; the transcript analysis was done on mutant lines rpoTmp-1, rpoTmp-2, and rpoTmp-3 representing three independent biological replicates. The raw data were analyzed using the LightCycler 480 software release 1.5 (Roche Diagnostics) and crossing points determined by second derivative maximum analysis. The accumulation of mitochondrial transcripts was normalized by setting the average ratio of nuclear transcripts to 1.
RNA Gel Blot Analysis
Total RNA extracted from Arabidopsis seedlings was resolved on 1.2% agarose-formaldehyde gels and transferred onto Hybond N+ nylon membranes (GE Healthcare). Hybridizations with biotinylated oligonucleotides or in vitro–biotinylated complementary RNA (cRNA) probes were performed as detailed by Falcon de Longevialle et al. (2008). The Pierce chemiluminescent nucleic acid detection module kit (Thermo Scientific) was used for cRNA probe detection; an ImageQuant-RT ECL system (GE Healthcare) was employed for chemiluminescence imaging.
To generate cRNA probe templates for nad2, nad6, cox1, and ccmFc, fragments of mitochondrial genes were amplified from total Arabidopsis DNA using Phusion Hot Start high-fidelity DNA polymerase (Finnzymes) with primer pairs rps4-t7-R (5′-taatacgactcactatagggATTCTCTTGTGCTCAGCGAAGGA-3′) and rps4-t7-F (5′-CAAGCAAGGCAGCCGATAAGTC-3′), nad2-t7-R (5′-taatacgactcactatagggCTTCCGTGGAAAATTCAGACT-3′) and nad2-t7-F (5′-TGGCGCACCTCTCCTAACTATTG-3′), nad6-t7-R (5′-taatacgactcactatagggAGCATTTCGTCGGAATACATCCTG-3′) and nad6-t7-F (5′-CGTTTTGTTTCCCATCCCAGTCT-'), cox1-t7-R (5′-taatacgactcactatagggATACCGAATCCAGGCAGAATGAG-3′) and cox1-t7-F (5′-GTAGGTAGCGGCACTGGGTG-3′), ccmC-t7-R (5′-taatacgactcactatagggAAATAGAAGCCGGTTCGACAGG-3′) and ccmC-t7-F (5′-TTGGTTGTTCTTAACAGCGATGG), nad3-t7-R (5′-taatacgactcactatagggACAGATCAATCTTGTTGGGAGGTAC-3′) and nad3-t7-F (5′-CGCATAAGGTCTGCAAGCCT-3′), and ccmFc-t7-R (5′-taatacgactcactatagggCGCTTCGCTGACCTATCGC-3′) and ccmFc-t7-F (5′-ACTATTGAAATGGTTCGTCAGTAGAGATG-3′). Lowercase nucleotides in primer sequences represent the T7 promoter sequence. The template for transcribing the rrn18 probe was amplified with primers rrn18-F (5′-CAGCTCGTGTCGTGAGATGT-3′) and rrn18-R (5′-CCCACCTTCCTCCAGTATATCA-3′), ligated in the appropriate orientation into pDrive (Qiagen), and reamplified with rrn18-R and the M13R universal primer to generate a template with an attached T7 promoter. The PCR products were used as templates for synthesizing biotinylated cRNA probes using biotin-CTP (Invitrogen) and the Ambion MAXIscript kit (Applied Biosystems) according to the manufacturer's instructions.
5′-RACE Analysis of Mitochondrial Transcripts
Primary ends of mitochondrial transcripts carry 5′ triphosphates that have to be converted to monophosphates by tobacco acid pyrophosphatase (TAP) in order to be detected by 5′-RACE; amplification of processed ends requires no such modification. 5′-RACE will yield products from TAP-treated RNA for both primary and processed transcripts, whereas without exposure to TAP, products resulting from primary transcript termini will be significantly reduced or absent. The comparison of 5′-RACE products obtained from TAP-treated and untreated RNA will thus identify primary transcripts after sequencing of cloned 5′-RACE products (Table 1).
RNA was subjected to TAP treatment and 5′-RACE as described previously (Kühn et al., 2005). Five micrograms of RNA were incubated with 10 units of TAP (Epicentre Technologies), extracted, and subsequently ligated to an RNA oligonucleotide 5′-adapter using T4 RNA ligase (Epicentre Technologies). Transcripts were then reverse transcribed using gene-specific primers (see Supplemental Table 3 online) and Superscript III (Invitrogen). Adapter primer P1a (5′-CGAATTCCTGTAGAACGAACACTAGAAG-3′) served as forward primer in PCR reactions; a list of all gene-specific reverse primers is provided in Supplemental Table 3 online. Sequencing of cloned 5′-RACE products was performed by Macrogen.
Ribonuclease Protection Assay
To generate cRNA probe templates, fragments of mitochondrial genes were amplified from total Arabidopsis DNA using Taq DNA polymerase (Qiagen) with primer pairs RPA-nad1e4-F (5′-AGATTCCGCACAGAAGACCAAG-3′) and RPA-nad1e4-R (5′-AGCTCGCAGAGCATAAGTCAGC-3′), RPA-nad2e1-F1 (5′-CATTTTTTTATTGAGCCGAATCACT-3′) and RPA-nad2e1-R1 (5′-CATGAATGAGCAAAATGGAGGT-3′), RPA-nad2e1-F (5′-CCTAGTCCGGGTGGGTGG-3′) and P5-nad2e1 (5′-GAAAAGAAAGGGCGGAATAGCA-3′), RPA-nad6-F (5′-CTCGCTACCATACCACAAGGC-3′) and P4-nad6 (5′-GACCAAAGCAGGGCTCGAC-3′), RPA-cox1-F1 (5′-CGAGGAAGAAAGTTTTGATCTGTG-3′) and P4-cox1-b (5′-GAGTTCTTGAGAGTCCCGTGG-3′), RPA-matR-F (5′- CCAAAAAGACTGAACTGAGGGAAG-3′) and P3-matR (5′-CGGAAATGCGATGTGTCTGG-3′), and RPA-atp6-1-F (5′-CACCGCACGAGTCAGACCT-3′) and RPA-atp6-1-R (5′-TCCAGACAGCTTCACTCCGTC-3′) and ligated into pDrive (Qiagen) in the appropriate orientation. Biotinylated cRNA probes were made from these plasmids through in vitro transcription in the presence of biotin-CTP (Invitrogen) using the Ambion MAXIscript kit (Applied Biosystems) according to the manufacturer's instructions. Transcripts were then extracted with phenol/chloroform/isoamyl alcohol (25:24:1), ethanol-precipitated from the aqueous phase, and dissolved in ultrapure water.
Between 20 and 40 μg of seedling RNA and 5 ng of biotinylated cRNA probe were subjected to ribonuclease protection using the Ambion RPA III ribonuclease protection assay kit (Applied Biosystems) according to the manufacturer's protocol, except that hybridizations were performed overnight at 55 to 65°C. Protected transcripts were separated in 5% urea-polyacrylamide gels and transferred onto Hybond N+ nylon membranes (GE Healthcare). Signal detection was performed as described for RNA gel blots.
Isolation of Mitochondria from Arabidopsis Plants
The aerial parts of plants were ground at 4°C in extraction buffer (0.3 M sucrose, 5 mM tetrasodium pyrophosphate, 10 mM KH2PO4, pH 7.5, 2 mM EDTA, 1% [w/v] PVP40, 1% [w/v] BSA, 5 mM cysteine, and 1 mM DTT) using a Polytron (30 mL of buffer for 10 g of plants, speed 4). The homogenate was centrifuged 5 min at 3000g and the supernatant centrifuged 10 min at 20,000g. The pellet was resuspended in wash buffer (0.3 M sucrose, 1 mM EGTA, and 10 mM MOPS/KOH, pH 7.2) and subjected to the same low-speed (3000g) and high-speed (20,000g) centrifugations. The pellet was then resuspended in a small volume of wash buffer and loaded on top of a Percoll step gradient (from bottom to top: 1 volume 50%, 5 volumes 25%, and 1 volume 18% Percoll in wash buffer, equivalent to 20 g of plant loaded per gradient). The gradient was centrifuged at 40,000g for 45 min. The mitochondrial fraction located at the interface between the 50 and 25% layers was collected and washed three times in wash buffer before being used in run-on transcription assays or stored as a mitochondrial suspension at −80°C.
Run-On Transcription Assay
Previously described protocols (Giegé et al., 2000; Zoschke et al., 2007) were adapted for use with biotin-CTP. Intact mitochondria (the equivalent of 400 μg of mitochondrial protein) were disrupted in 100 μL of reaction mix containing 20 mM Tris, pH 7.6, 40 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.5 mM of each ATP, GTP, and UTP, 0.3 mM CTP, 1 mM biotin-CTP [Invitrogen], and 40 units ribonuclease inhibitor [Invitrogen]) and incubated at 25°C for 10 min. The reactions were terminated by adding 100 μL of stop solution (50 μg/mL sodium lauroylsarcosine, 50 mM Tris, pH 8.0, and 25 mM EDTA), and transcripts were extracted twice with phenol/chloroform 5:1, pH ∼4.5 (Sigma-Aldrich), once with choroform/isoamyl alcohol 24:1, ethanol-precipitated, and resuspended in 100 μL of ultrapure water. Transcripts were hybridized overnight in 250 mM NaH2PO4, pH 7.2, 7% (w/v) SDS, and 2.5 mM EDTA at 58°C to DNA probes (500 ng) dot-blotted in duplicates onto Hybond N+ nylon membranes (GE Healthcare). A slot blotting apparatus (MilliBlot-S; Millipore) was used for filter preparation. The experiment was repeated four times per genotype with independently labeled RNA obtained from two separate mitochondrial preparations per genotype. Signal detection was performed as described for RNA gel blots. Signals were analyzed using the ImageQuantTL 1D gel analysis software (Amersham Biosciences). The background signal measured for the dot-blotted nonspecific pDrive plasmid sequence was subtracted from each gene-specific signal.
To generate DNA probes, mitochondrial DNA fragments were amplified with Taq DNA polymerase (Qiagen) using gene-specific primers (see Supplemental Table 4 online) and ligated into pDrive (Qiagen). From these plasmids, DNA probes were amplified using M13 primers; the vector fragment used as nonspecific probe was amplified from self-ligated pDrive.
BN-PAGE and Immunodetections
BN-PAGE was performed according to the method described by Jänsch et al. (1996). Mitochondrial proteins (100 μg) were solubilized with dodecylmaltoside (1% [w/v] final) in ACA buffer (750 mM amino caproic acid, 0.5 mM EDTA, and 50 mM Tris-HCl, pH 7.0) and incubated 20 min at 4°C. The samples were centrifuged 10 min at 20,000g, and Serva Blue G (0.2% [v/v] final) was added to the supernatant. The samples were loaded onto a 4.5 to 16% gradient gel. After migration, the gel was transferred on a polyvinylidene difluoride membrane (ImmobilonP; Millipore) in Cathode buffer (50 mM Tricine and 15 mM Bis-Tris-HCl, pH 7.0) for 16 h at 4°C. The membrane was stained with a Coomassie Brilliant Blue solution (40% [v/v] methanol, 7% [v/v] acetic acid, and 0.02% [w/v] Coomasie R250) and scanned. After destaining in 100% (v/v) methanol, the membrane was probed with antibodies against ATP1 (gift from T. Elthon, University of Nebraska, Lincoln, NE), COXII (AS04 053A; Agrisera), the Rieske iron-sulfur protein (D. Price, Australian National University, Canberra, Australia), or NAD9 (G. Bonnard, Institut de Biologie Moléculaire des Plantes, Strasbourg, France). A secondary antibody linked to horseradish peroxidase (Sigma-Aldrich) was used, and signals were detected by chemiluminescence (ECL plus; GE Healthcare) and recorded using an ImageQuant-RT ECL system (GE Healthcare). Between immunodetections, antibodies were stripped off the membrane by incubating the membrane at 50°C for 30 min in a solution containing 2% (w/v) SDS and 1% (v/v) β-mercaptoethanol.
Activity Measurements
The activity of complex I and complex IV were measured using a Clark-type oxygen electrode (Hansatech Instrument). A membrane suspension (100 μg proteins) was incubated in 1 mL of respiration buffer (0.3 M sucrose, 5 mM KH2PO4, 10 mM TES, 10 mM NaCl, 2 mM MgSO4, and 0.1% [w/v] BSA, pH 6.8), and specific substrates were added: 0.5 mM deaminoNADH for complex I or ascorbate (10 mM) and cytochrome c (25 μM) in the presence of Triton X-100 (0.05% [w/v]) for complex IV. Oxygen consumption rate was monitored and used to calculated specific activities. The specificity of the reactions was controlled by addition of the specific inhibitors rotenone (5 mM) or KCN (1 mM) for complex I and complex IV, respectively.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RPOTmp, At5g15700 (RPOT2); RPOTm, At1g68990 (RPOMT); ArthMr003 (18S, rrn18), ArthMp035 (atp6-1), ArthMp079 (ccmC, ccb256), ArthMp017 (ccmFc, ccb452), ArthMp109 (cox1), ArthMp045 (matR), ArthMp044 (nad1), ArthMp026 (nad2), ArthMp086 (nad3), ArthMp006 (nad5), ArthMp024 (nad6), ArthMp007 (nad9), ArthMp085 (rps12), and ArthMp027 (rps4). T-DNA insertion mutants used were SALK_005875 (allele rpoTm-1), GK-350F01 (rpoTm-2), GK-286E07 (rpoTmp-1), SALK_132842 (rpoTmp-2), and SALK_086115, (rpoTmp-3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Lethality of Loss of RPOTm.
Supplemental Figure 2. Plastidial Steady State Transcript Levels in rpoTmp Mutants.
Supplemental Figure 3. RNA Gel Blot Hybridizations with Probes Specific to Exons nad1e4, nad1e5, and nad5e2.
Supplemental Figure 4. In Organello Protein Synthesis by Mitochondria Isolated from rpoTmp and Wild-Type Plants.
Supplemental Figure 5. 5′-RACE Analysis of cox1 Transcripts.
Supplemental Figure 6. 5′-RACE Analysis of nad1e4 Transcripts.
Supplemental Figure 7. 5′-RACE Analysis of matR Transcripts.
Supplemental Figure 8. 5′-RACE Analysis of nad1e5 Transcripts.
Supplemental Figure 9. 5′-RACE Analysis of ccmC Transcripts.
Supplemental Figure 10. 5′-RACE Analysis of rps4 Transcripts.
Supplemental Figure 11. 5′-RACE Analysis of nad2e1_e2 Transcripts.
Supplemental Figure 12. 5′-RACE Analysis of nad6 Transcripts.
Supplemental Figure 13. 5′-RACE Analysis of ccmFc Transcripts.
Supplemental Figure 14. 5′-RACE Analysis of nad9 Transcripts.
Supplemental Figure 15. Neighbor Joining Distance Tree of Mitochondrial Promoter Sequences in Arabidopsis.
Supplemental Figure 16. Identification of rpoTmp Mutant Lines.
Supplemental Figure 17. Nuclear Genome Ploidy Levels.
Supplemental Figure 18. Mitochondrial Steady State Transcript Levels in the Arabidopsis otp43 Mutant.
Supplemental Table 1. Sequence Motifs Significantly Overrepresented Among Promoters of Genes Showing Reduced Transcription in rpoTmp.
Supplemental Table 2. Primers Used for Quantitative RT-PCR Analysis.
Supplemental Table 3. Primers Used for 5′-RACE.
Supplemental Table 4. Gene-Specific Primers Used to Amplify DNA Probes for the Detection of Mitochondrial Run-On Transcripts.
Supplemental Data Set. Promoter Sequence Alignment File Used to Calculate the Distance Tree of Mitochondrial Promoters Shown in Supplemental Figure 15.
Supplementary Material
Acknowledgments
This work was supported by an Australian Research Council Centre of Excellence Grant CEO561495, a Research Grant by the University of Western Australia Research Grants Scheme to K.K., an ARC Australian Professorial Fellowship to A.H.M., a WA State Premier's Fellowship to I.D.S., and a Deutsche Forschungsgemeinschaft grant (SFB 429) to T.B. We thank the ABRC for providing seed stocks.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kristina Kühn (kristina@cyllene.uwa.edu.au).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Azevedo, J., Courtois, F., Hakimi, M.A., Demarsy, E., Lagrange, T., Alcaraz, J.P., Jaiswal, P., Marechal-Drouard, L., and Lerbs-Mache, S. (2008). Intraplastidial trafficking of a phage-type RNA polymerase is mediated by a thylakoid RING-H2 protein. Proc. Natl. Acad. Sci. USA 105 9123–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, K., Schmidt, J., Espinosa-Ruiz, A., Villarejo, A., Shiina, T., Gardestrom, P., Sane, A.P., and Bhalerao, R.P. (2004). Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J. 38 38–48. [DOI] [PubMed] [Google Scholar]
- Bensing, B.A., Meyer, B.J., and Dunny, G.M. (1996). Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 93 7794–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, S., and Brennicke, A. (2003). Gene expression in plant mitochondria: Transcriptional and post-transcriptional control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 181–188, discussion 188–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caoile, A.G., and Stern, D.B. (1997). A conserved core element is functionally important for maize mitochondrial promoter activity in vitro. Nucleic Acids Res. 25 4055–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.C., Sheen, J., Bligny, M., Niwa, Y., Lerbs-Mache, S., and Stern, D.B. (1999). Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11 911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois, F., Merendino, L., Demarsy, E., Mache, R., and Lerbs-Mache, S. (2007). Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis. Plant Physiol. 145 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski, S., Hoffmann, M., Guha, C., and Binder, S. (1999). Continuous primary sequence requirements in the 18-nucleotide promoter of dicot plant mitochondria. J. Biol. Chem. 274 10094–10099. [DOI] [PubMed] [Google Scholar]
- Edqvist, J., and Bergman, P. (2002). Nuclear identity specifies transcriptional initiation in plant mitochondria. Plant Mol. Biol. 49 59–68. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel, C., von Groll, U., Muller, M., Borner, T., and Weihe, A. (2006). Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta 223 998–1009. [DOI] [PubMed] [Google Scholar]
- Emanuel, C., Weihe, A., Graner, A., Hess, W.R., and Börner, T. (2004). Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J. 38 460–472. [DOI] [PubMed] [Google Scholar]
- Falcon de Longevialle, A., Hendrickson, L., Taylor, N.L., Delannoy, E., Lurin, C., Badger, M., Millar, A.H., and Small, I. (2008). The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 56 157–168. [DOI] [PubMed] [Google Scholar]
- Falcon de Longevialle, A., Meyer, E.H., Andres, C., Taylor, N.L., Lurin, C., Millar, A.H., and Small, I.D. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg, M., Gaspari, M., Rantanen, A., Trifunovic, A., Larsson, N.G., and Gustafsson, C.M. (2002). Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31 289–294. [DOI] [PubMed] [Google Scholar]
- Fey, J., and Marechal-Drouard, L. (1999). Compilation and analysis of plant mitochondrial promoter sequences: An illustration of a divergent evolution between monocot and dicot mitochondria. Biochem. Biophys. Res. Commun. 256 409–414. [DOI] [PubMed] [Google Scholar]
- Finnegan, P.M., and Brown, G.G. (1990). Transcriptional and posttranscriptional regulation of RNA levels in maize mitochondria. Plant Cell 2 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner, J., Holzle, A., Jonietz, C., Thuss, S., Schwarzlander, M., Weber, B., Meyer, R.C., and Binder, S. (2008). Mitochondrial mRNA polymorphisms in different Arabidopsis accessions. Plant Physiol. 148 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner, J., Weber, B., Thuss, S., Wildum, S., and Binder, S. (2007). Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: T-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 35 3676–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, P., Hoffmann, M., Binder, S., and Brennicke, A. (2000). RNA degradation buffers asymmetries of transcription in Arabidopsis mitochondria. EMBO Rep. 1 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.W., and Lang, B.F. (1998). Transcription in chloroplasts and mitochondria: A tale of two polymerases. Trends Microbiol. 6 1–3. [DOI] [PubMed] [Google Scholar]
- Greenleaf, A.L., Kelly, J.L., and Lehman, I.R. (1986). Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc. Natl. Acad. Sci. USA 83 3391–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J., Dempsey, S., and Newton, K.J. (1994). Rescue of a maize mitochondrial cytochrome oxidase mutant by tissue culture. Plant J. 6 787–794. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa, H. (2003). The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): Comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31 5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277 809–811. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Legen, J., Weihe, A., Herrmann, R.G., and Börner, T. (2002). Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J. 30 625–637. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Meixner, M., Gillandt, S., Richter, E., Börner, T., and Weihe, A. (1999. b). Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo. Plant J. 17 557–561. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Wagner, I., Börner, T., and Hess, W.R. (1999. a). Inter-organellar crosstalk in higher plants: Impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 19 635–643. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., and Börner, T. (1999). Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 190 1–59. [DOI] [PubMed] [Google Scholar]
- Holec, S., Lange, H., Canaday, J., and Gagliardi, D. (2008). Coping with cryptic and defective transcripts in plant mitochondria. Biochim. Biophys. Acta 1779 566–573. [DOI] [PubMed] [Google Scholar]
- Holec, S., Lange, H., Kuhn, K., Alioua, M., Börner, T., and Gagliardi, D. (2006). Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 26 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricova, A., Quesada, V., and Micol, J.L. (2006). The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 141 942–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, T.M., and Gray, M.W. (1999). Identification and characterization of T3/T7 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol. Biol. 40 567–578. [DOI] [PubMed] [Google Scholar]
- Jänsch, L., Kruft, V., Schmitz, U.K., and Braun, H.P. (1996). New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9 357–368. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Dokiya, Y., and Sugita, M. (2001). Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem. Biophys. Res. Commun. 289 1106–1113. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Nishizawa, S., Sugawara, A., Itchoda, N., Estiati, A., and Mikami, T. (2000). The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 28 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, K., Bohne, A.V., Liere, K., Weihe, A., and Börner, T. (2007). Arabidopsis phage-type RNA polymerases: Accurate in vitro transcription of organellar genes. Plant Cell 19 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, K., Weihe, A., and Börner, T. (2005). Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 33 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi, K., Yara, A., Mitsui, N., Tozawa, Y., and Iba, K. (2004). Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol. 45 1194–1201. [DOI] [PubMed] [Google Scholar]
- Liere, K., and Börner, T. (2007). Transcription and transcriptional regulation in plastids. In Topics in Current Genetics: Cell and Molecular Biology of Plastids, R. Bock, ed (Berlin/Heidelberg, Germany: Springer), pp. 121–174.
- Liere, K., Kaden, D., Maliga, P., and Börner, T. (2004). Overexpression of phage-type RNA polymerase RpoTp in tobacco demonstrates its role in chloroplast transcription by recognizing a distinct promoter type. Nucleic Acids Res. 32 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupold, D.S., Caoile, A.G., and Stern, D.B. (1999). The maize mitochondrial cox2 gene has five promoters in two genomic regions, including a complex promoter consisting of seven overlapping units. J. Biol. Chem. 274 3897–3903. [DOI] [PubMed] [Google Scholar]
- Masters, B.S., Stohl, L.L., and Clayton, D.A. (1987). Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51 89–99. [DOI] [PubMed] [Google Scholar]
- Newton, K.J., Winberg, B., Yamato, K., Lupold, S., and Stern, D.B. (1995). Evidence for a novel mitochondrial promoter preceding the cox2 gene of perennial teosintes. EMBO J. 14 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp, W.D., Lupold, D.S., Mack, S., and Stern, D.B. (1993). Architecture of the maize mitochondrial atp1 promoter as determined by linker-scanning and point mutagenesis. Mol. Cell. Biol. 13 7232–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp, W.D., and Stern, D.B. (1992). A conserved 11 nucleotide sequence contains an essential promoter element of the maize mitochondrial atp1 gene. EMBO J. 11 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P., Khafif, M., Riondet, C., Droux, M., Bonnard, G., and Meyer, Y. (2007). Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19 1851–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch, M., Emanuel, C., Hedtke, B., Liere, K., and Börner, T. (2008). Impaired function of the phage-type RNA polymerase RpoTp in transcription of chloroplast genes is compensated by a second phage-type RNA polymerase. Nucleic Acids Res. 36 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch, M., Liere, K., and Börner, T. (2007). High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Genet. Genomics 277 725–734. [DOI] [PubMed] [Google Scholar]
- Tiranti, V., Savoia, A., Forti, F., D'Apolito, M.F., Centra, M., Rocchi, M., and Zeviani, M. (1997). Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the expressed sequence tags database. Hum. Mol. Genet. 6 615–625. [DOI] [PubMed] [Google Scholar]
- Tracy, R.L., and Stern, D.B. (1995). Mitochondrial transcription initiation: Promoter structures and RNA polymerases. Curr. Genet. 28 205–216. [DOI] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15 57–61. [DOI] [PubMed] [Google Scholar]
- Van Aken, O., Pecenkova, T., van de Cotte, B., De Rycke, R., Eeckhout, D., Fromm, H., De Jaeger, G., Witters, E., Beemster, G.T., Inze, D., and Van Breusegem, F. (2007). Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 52 850–864. [DOI] [PubMed] [Google Scholar]
- Zoschke, R., Liere, K., and Börner, T. (2007). From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50 710–722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.