Abstract
Study Objectives:
Reports on the association of polymorphisms in the gene encoding apolipoprotein E (APOE)—a vital macromolecule in cholesterol metabolism—with obstructive sleep apnea (OSA) have provided conflicting results. Our objective was to meta-analytically synthesize the existing evidence for the association of the APOE ε4 allele with the risk of OSA.
Design:
Random effects meta-analysis and meta-regression
Setting:
Genetic epidemiological studies reporting the association of APOE ε4 allele with OSA susceptibility.
Patients or Participants:
Synthesis of APOE ε4 allele data from 6,508 subjects including 1,901 cases of OSA and 4,607 controls.
Interventions:
None
Measurements and Results:
Eight studies were included in the random effects meta-analysis; the summary effect size measured as odds ratio (OR) for association of the APOE ε4 allele with the risk of OSA was found to be 1.13 (95% confidence interval 0.86–1.47). There was a statistically significant heterogeneity (I2 = 72%, P = 0.001) across study results that was not explained by the mean age, proportion of males, or the proportion possessing the APOE ε4 allele or when grouped based on the geographic location of the study.
Conclusions:
The hypothesis that the APOE ε4 allele may be causally associated with OSA cannot be supported on the basis of published literature.
Citation:
Thakre TP; Mamtani MR; Kulkarni H. Lack of association of the APOE ε4 allele with the risk of obstructive sleep apnea: meta-analysis and meta-regression. SLEEP 2009;32(11):1507-1511.
Keywords: Obstructive sleep apnea, epidemiology, apolipoprotein E, APOE, genetic
OBSTRUCTIVE SLEEP APNEA (OSA)—DEFINED AS EPISODIC AND REPETITIVE NARROWING OF UPPER AIRWAYS ACCOMPANIED BY LOSS OF TONE IN THE pharyngeal musculature leading to sleep disturbances such as snoring, frequent arousals, and daytime sleepiness—is widely prevalent at all ages and is associated with concomitant or consequential cardiovascular morbidities and obesity, disorders known to have a genetic basis.1–7 Since wide inter-individual variations exist in regards to the onset, severity, and the comorbidity mosaic of OSA, it is being increasingly argued that this condition may have a genetic component.8–10 However, the current understanding of the genetic basis of OSA is cursory.
Over the last decade, several investigators have evaluated the potential association between OSA and polymorphisms in the apolipoprotein E (APOE) gene.11–18 These genetic epidemiological studies attempted to build on the observations that the apoE molecule is implicated in the causation of Alzheimer disease,19,20 atherosclerosis,1,4 and HIV-associated dementia,21 owing to its role in cholesterol transport and metabolism.22 It is argued that the apoE macromolecule, consequent to its central nervous effects, may play a role in maintaining the muscle tone of pharyngeal muscle,17 and thus the APOE polymorphisms that can modulate gene expression may have bearing on OSA susceptibility. Of interest, the APOE ε4 allele has been the most intensely scrutinized one in the context of OSA. However, the exact biological mechanism and epidemiological contribution of APOE polymorphisms in OSA is still a matter of contention. Therefore, we report our results of a meta-analytic synthesis summarizing the strength of association of the APOE ε4 allele with susceptibility to OSA.
METHODS
Data Extraction
We conducted a search in the MEDLINE and CINAHL databases using the search query “sleep AND (apolipoprotein OR ApoE).” This strategy yielded 55 hits that included 22 articles which also contained the keyword “apnea.” Of these, there were 4 reviews, 8 studies related to outcomes other than OSA, and one non-English article. Thus, the association of APOE genotypes with OSA was the subject of 9 genetic association studies. The APOE ε2, ε3 and ε4 alleles are defined haplotypically by 2 single nucleotide polymorphisms (rs429358 and rs7412). One study23 genotyped only one of these 2 SNPs and therefore was excluded. We also conducted a web search using the same search strategy and found a completed case-control study. The results of this study are under preparation (Mignot E, Personal Communication) and were, at the time of this meta-analysis, unavailable. Thus, we had 8 studies11–18 available for our meta-analytic investigation. To further ensure that our search strategy had not inadvertently missed other published studies, using snowballing technique we also searched all the 249 references cited by the 8 included studies. Therefore our meta-analysis includes the aforementioned 8 published studies.
The included studies were independently reviewed by each of the study investigators with a special emphasis on extracting information for the following variables: number of OSA cases (defined as apnea/hypopnea index [AHI] > 15), number of controls (defined as those without OSA), mean age of the study subjects, proportion of males, proportion of study subjects possessing the APOE ε4 allele, and the geographic origin of the study. From each study, we extracted or derived a contingency table that summarized the association of possession of the APOE ε4 allele with risk of OSA. When such a contingency table was not directly presented in a study, we used the marginal frequencies and reported odds ratios (ORs) to derive a table. Only 5 studies11,12,16–18 reported the frequency of APOE haplotype pairs and, therefore, degree of association of other APOE alleles with OSA could not be ascertained.
Statistical Analysis
We conducted our analysis in 2 steps. First, we used the method of DerSimonian and Laird24 of random effects meta-analysis to combine the OR of OSA likelihood based on APOE ε4 allele possession. The random effects model considers both between-study and within-study variability in the estimates of OR. The assumption is that studies represent a random sample from the universe of all possible studies. The heterogeneity across the included studies was assessed using the I2 statistic, the statistical significance of which was tested using the Q statistic.25 Test for publication bias was assessed using Egger's method.26 This method relies on the assumption that generally larger studies tend to provide OR estimates that are closer to the true (and unknown) OR. Thus if one were to plot the inverse of standard error for log OR (referred to as precision) and the relative deviate of OR (referred to as standardized effect size) for each study, one should observe a negative sloping line. Publication bias is indicated if the points plotted in this standardized effect-precision space are not symmetric about the line with the negative slope. A statistical test (Egger's test26) is available for testing the significance of both the slope and bias deviate from a null hypothesis of zero. If significant heterogeneity was observed across studies, we conducted a meta-regression27,28 in which the log-OR for each study was treated as the outcome variable and continuous variables like mean age, proportion of males, proportion of subjects with European descent, and APOE ε4 allele frequency were used as explanatory variables after appropriate weighting based on the standard error of log-OR. All the analyses were conducted using Stata 10.2 (Stata Corp, College Station, TX). Statistical significance was assessed at a type I error rate of 0.05.
RESULTS
Our meta-analysis combined the genotype information from 8 studies representing 6,508 subjects (1,901 cases of OSA and 4,607 controls). Of 8 eight studies, 5 studies11,15–18 included subjects without any known comorbidities, one study12 recruited patients of Alzheimer disease, one study13 recruited patients at high risk of Alzheimer disease and the remaining study14 included patients who had hypertension (56%), coronary artery disease (18%), stroke (6%), and congestive heart failure (3%). Three studies11,12,18 were conducted exclusively on subjects of European descent, in 4 studies14–17 the proportion of subjects of European descent ranged from 55% to 97%, and one study13 was conducted in Japanese Asians. Thus, the 8 studies included 4,903 (75.3% of all subjects represented in this meta-analysis) subjects of European descent.
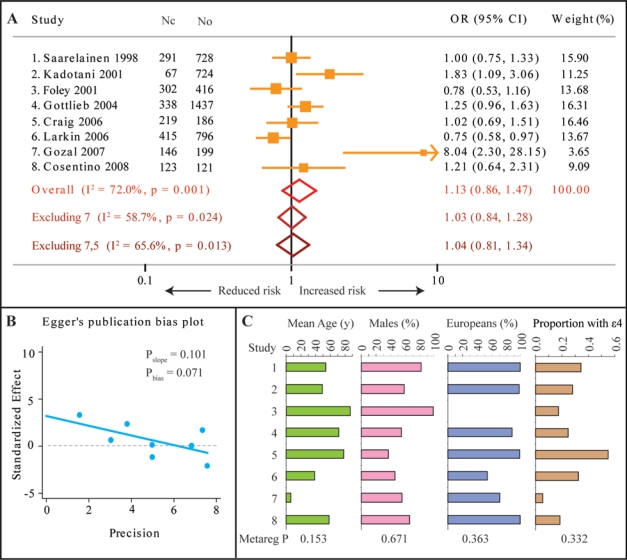
We found that (Figure 1A) the strength of association reported in the included studies varied widely, with 2 studies15,16 reporting a higher risk of OSA and one study17 reporting a reduced risk of OSA in the presence of the APOE ε4 allele, while the remaining 5 studies11–14,18 showed no association between OSA and the APOE ε4 allele. The summary OR was 1.13 (95% confidence interval [CI] 0.86–1.47), indicating that the current evidence fails to support a significant association between the APOE ε4 allele and the risk of OSA. However, there was a significantly high degree of heterogeneity across the studies (I2 = 72%, P = 0.001). The study of Gozal et al.15 reported very high OR for association, although the allele frequency for the APOE ε4 allele in this study was very small. Indeed, when we excluded this study from the meta-analysis, the I2 statistic shrunk by 13.3% and the summary OR became 1.03 (95% CI 0.84–1.28, Figure 1A). Also, the study of Craig et al.12 was conducted in patients with Alzheimer dementia, a comorbidity that is in itself influenced by the APOE ε4 allele. Therefore, we conducted meta-analysis by excluding both the Gozal study15 and the Craig study12 and observed that the summary odds ratio was 1.04 (95% CI 0.81–1.34, Figure 1A). There was still a significant heterogeneity across the remaining 6 studies (I2 = 65.6%, P = 0.013). The significance value for the slope of standardized effect size for a given study precision was 0.101, and there was no evidence for a significant publication bias (P = 0.071, Figure 1B).
Figure 1.
Results of meta-analysis and meta-regression for the association of the APOE ε4 allele with OSA susceptibility. (A) Forest plot summarizing the results of random-effects meta-analysis. Orange boxes and lines show the point and 95% confidence interval estimates of the odds ratios. Red hollow diamond is the overall summary effect with 95% confidence interval, light brown diamond is summary effect after excluding study number 7 (Gozal et al., 2007), and deep brown diamond is summary effect after excluding both study number 7 and study number 5 (Craig et al., 2006). Nc, number of cases of OSA; No, number of controls. (B) Egger's publication bias plot shown the precision (defined as 1/standard error of log OR) on the abscissa and standardized effect size (relative deviate for log OR) on the ordinate. Statistical significance is shown as Pslope and Pbias as explained in the text. A statistically nonsignificant bias (Pbias≥ 0.05) indicates no significant publication bias. (C) Continuous covariates used in meta-regression: mean age (in years, green bars), proportion of males (pink bars), proportion of subjects with European descent (blue bars) and proportion of subjects possessing the APOE ε4 allele (brown bars). The significance of the association of these covariates with study effect sizes is shown as Metareg P. The study number in panel C corresponds to that in panel A.
We therefore conducted a series of analyses to determine if some of the reported study characteristics accounted for the significant between-studies heterogeneity. For this, we meta-regressed the mean age, proportion of males, proportion of subjects with European descent, and proportion of subjects with the APOE ε4 allele onto log-OR. We observed (Figure 1C) that although these characteristics substantially varied across the included studies, they did not significantly explain the heterogeneity in log OR. We also conducted sub-group meta-analyses based on 2 potential confounders: the continent of study origin and the source of study subjects. With regard to the continent of origin, we observed that studies from Europe were homogeneous (I2 = 0%, P = 0.863) while the studies from North America were heterogeneous (I2 = 83.8%, P < 0.001). Still, both these subgroups had statistically insignificant summary ORs (Europe: OR = 1.03, 95% CI 0.83–1.28); North America: OR = 1.26, 95% CI 0.81–1.97). With regard to source of recruited subjects, we observed that within studies recruited from sleep centers11,12,15,18 or from community-based epidemiological studies13,14,16,17 the association was heterogeneous and statistically insignificant: Sleep center studies–summary OR 1.32 (95% CI 0.81–2.13), I2 78.8, P = 0.003; community-based studies–summary OR 1.05 (0.72–1.51), I2 71.2, P = 0.015). Lastly, in the 5 studies11,12,16–18 that reported the frequency of APOE genotypes, the odds ratios for the association of the ε4/ε4 genotype with OSA were 1.68, 3.9, 0.54, 0.99, and 0, respectively, again indicating large heterogeneity across studies
DISCUSSION
Our evidence-based results demonstrate that the hypothesized association between APOE polymorphisms and OSA is likely to be false. Indeed, research in this area seems to have come a full circle—Saarelainen et al.18 first suggested this lack of association over a decade ago, then came some human and animal studies16,29–31 that raised the expectation of this association; the more recent studies11,14,17 have generally negated the likelihood of such an association. Our observation of this lack of association could have resulted from a combination of one or more of the following possibilities: First, currently it is unclear how, if at all, APOE may mediate its biological impact upon OSA. Research appears to indicate that APOE polymorphisms can modulate the neuronal injury resulting from intermittent hypoxia thereby hastening the decline of cognitive function in patients of OSA29,32; however it is unlikely that these polymorphisms might increase the susceptibility to OSA itself. This likelihood is supported by the observation17 of a linkage peak in chromosome 19 that includes the APOE gene despite which there was no association with APOE polymorphisms.
Second, most of the studies included in this meta-analysis have used a definition of AHI > 15 as the definition of OSA. In reality, this definition reflects a spectrum of OSA severity33 and it is possible that the APOE polymorphisms may be associated with more severe forms of OSA (for example, AHI > 30). However, there was an insufficient number of studies using that diagnostic criterion for OSA to be included in this meta-analysis. Thus, larger studies that faithfully represent the spectrum of OSA severity are needed to answer this question. Third, we observed a statistically significant heterogeneity across the published studies—a finding that was not explained by variations in mean age, gender, ethnic background, APOE ε4 allele frequency and geographic location of the study. Thus, factors other than these may be playing important roles in the estimates of ORs obtained within studies. Specifically, it is conceivable that the phenotypic effects may be augmented by some thus far unidentified gene-gene interactions. Such interactions have been observed in the context of coronary artery disease, atherosclerosis, and primary open-angle glaucoma,34–38 but have not been determined in the context of OSA. Thus, future studies need to formally explore broader gene-gene interactions that might characterize OSA susceptibility.
Fourth, very few studies have evaluated the different genetic models that may show an association between APOE polymorphisms and OSA risk—most of the studies have conducted the analyses as presence or absence of the ε4 allele. In the context of human immunodeficiency associated dementia,21 the role of this allele is apparent only in a recessive state. Given the rarity of this genotype, small studies are unlikely to be sufficiently powered to statistically demonstrate a similar effect of this allele on OSA. In the same vein, another point that needs a mention while interpreting the observed lack of association is that, while statistically insignificant, the summary OR (1.13) across the 8 published studies exceeded unity. If that strength of association were to be considered as true, then it can be argued that studies attempting to tease out the association between the APOE ε4 allele and risk of OSA need to be based on large sample size. Assuming a type I error rate of 0.05, prevalence of the APOE ε4 allele in non-OSA population to be 27.8% (observed in the 8 studies included here) and power of 80%; to detect an OR of 1.13 studies with controls-to-case ratios of 1, 2, 3, and 4 will need to include 5,103, 3,795, 3,358, and 3,140 cases of OSA, respectively. These estimates translate to total sample sizes of 10,206, 11,384, 13,431 and 15,698, respectively. Clearly, all the published studies and, indeed, this meta-analysis itself may be underpowered to detect such weak association. Therefore, studies with much larger sample sizes will be needed before conclusively refuting the hypothesized association between the APOE ε4 allele and risk of OSA. Nevertheless, our study shows that the existing evidence points towards an overall lack of association between possession of the APOE ε4 allele and risk of OSA encouraging more robust approaches (e.g., genome-wide association studies) to uncover the genetic basis of OSA and to identify target genes for prevention and treatment of this condition.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
We sincerely thank the two anonymous reviewers for critical reviews and Dr. Emmanuel Mignot, Stanford University, USA and Dr. Daniel Gottlieb, Boston University, USA for help during the process of manuscript revision and refinement.
REFERENCES
- 1.Bradley TD, Floras JS. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 373:82–93. doi: 10.1016/S0140-6736(08)61622-0. [DOI] [PubMed] [Google Scholar]
- 2.Noda A, Okada T, Yasuma F, Sobue T, Nakashima N, Yokota M. Prognosis of the middle-aged and aged patients with obstructive sleep apnea syndrome. Psychiatry Clin Neurosci. 52:79–85. doi: 10.1111/j.1440-1819.1998.tb00977.x. [DOI] [PubMed] [Google Scholar]
- 3.Palomaki H, Partinen M, Erkinjuntti T, Kaste M. Snoring, sleep apnea syndrome, and stroke. Neurology. 42:75–81. discussion 82. [PubMed] [Google Scholar]
- 4.Parish JM, Somers VK. Obstructive sleep apnea and cardiovascular disease. Mayo Clin Proc. 79:1036–46. doi: 10.4065/79.8.1036. [DOI] [PubMed] [Google Scholar]
- 5.Redline S, Young T. Epidemiology and natural history of obstructive sleep apnea. Ear Nose Throat J. 72:20–1. 24–6. [PubMed] [Google Scholar]
- 6.Young T. Analytic epidemiology studies of sleep disordered breathing--what explains the gender difference in sleep disordered breathing? Sleep . 6:S1–2. doi: 10.1093/sleep/16.suppl_8.s1. [DOI] [PubMed] [Google Scholar]
- 7.Young T, Palta M, Dempsey J, Skatrud J, Weber S, Badr S. The occurrence of sleep-disordered breathing among middle-aged adults. N Engl J Med. 328:1230–5. doi: 10.1056/NEJM199304293281704. [DOI] [PubMed] [Google Scholar]
- 8.Carmelli D, Colrain IM, Swan GE, Bliwise DL. Genetic and environmental influences in sleep-disordered breathing in older male twins. Sleep. 27:917–22. doi: 10.1093/sleep/27.5.917. [DOI] [PubMed] [Google Scholar]
- 9.Ohayon MM. From wakefulness to excessive sleepiness: what we know and still need to know. Sleep Med Rev. 12:129–41. doi: 10.1016/j.smrv.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin AE, Polotsky VY, Balbir A, et al. Differences in sleep-induced hypoxia between A/J and DBA/2J mouse strains. Am J Respir Crit Care Med. 168:1520–7. doi: 10.1164/rccm.200304-462OC. [DOI] [PubMed] [Google Scholar]
- 11.Cosentino FI, Bosco P, Drago V, et al. The APOE epsilon4 allele increases the risk of impaired spatial working memory in obstructive sleep apnea. Sleep Med. 9:831–9. doi: 10.1016/j.sleep.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Craig D, Hart DJ, Passmore AP. Genetically increased risk of sleep disruption in Alzheimer's disease. Sleep. 29:1003–7. doi: 10.1093/sleep/29.8.1003. [DOI] [PubMed] [Google Scholar]
- 13.Foley DJ, Masaki K, White L, Redline S. Relationship between apolipoprotein E epsilon4 and sleep-disordered breathing at different ages. JAMA. 286:1447–8. doi: 10.1001/jama.286.12.1447. [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb DJ, DeStefano AL, Foley DJ, et al. APOE epsilon4 is associated with obstructive sleep apnea/hypopnea: the Sleep Heart Health Study. Neurology. 63:664–8. doi: 10.1212/01.wnl.0000134671.99649.32. [DOI] [PubMed] [Google Scholar]
- 15.Gozal D, Capdevila OS, Kheirandish-Gozal L, Crabtree VM. APOE epsilon 4 allele, cognitive dysfunction, and obstructive sleep apnea in children. Neurology. 69:243–9. doi: 10.1212/01.wnl.0000265818.88703.83. [DOI] [PubMed] [Google Scholar]
- 16.Kadotani H, Kadotani T, Young T, et al. Association between apolipoprotein E epsilon4 and sleep-disordered breathing in adults. JAMA. 285:2888–90. doi: 10.1001/jama.285.22.2888. [DOI] [PubMed] [Google Scholar]
- 17.Larkin EK, Patel SR, Redline S, Mignot E, Elston RC, Hallmayer J. Apolipoprotein E and obstructive sleep apnea: evaluating whether a candidate gene explains a linkage peak. Genet Epidemiol. 30:101–10. doi: 10.1002/gepi.20127. [DOI] [PubMed] [Google Scholar]
- 18.Saarelainen S, Lehtimaki T, Kallonen E, Laasonen K, Poussa T, Nieminen MM. No relation between apolipoprotein E alleles and obstructive sleep apnea. Clin Genet. 53:147–8. doi: 10.1111/j.1399-0004.1998.tb02663.x. [DOI] [PubMed] [Google Scholar]
- 19.Cosentino S, Scarmeas N, Helzner E, et al. APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology. 70:1842–9. doi: 10.1212/01.wnl.0000304038.37421.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahley RW, Huang Y. Apolipoprotein (apo) E4 and Alzheimer's disease: unique conformational and biophysical properties of apoE4 can modulate neuropathology. Acta Neurol Scand Suppl. 185:8–14. doi: 10.1111/j.1600-0404.2006.00679.x. [DOI] [PubMed] [Google Scholar]
- 21.Burt TD, Agan BK, Marconi VC, et al. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression. Proc Natl Acad Sci U S A. 105:8718–23. doi: 10.1073/pnas.0803526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function-from atherosclerosis to Alzheimer's disease to AIDS. [J Lipid Res 2008 Dec 22]; doi: 10.1194/jlr.R800069-JLR200. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalra M, Pal P, Kaushal R, et al. Association of ApoE genetic variants with obstructive sleep apnea in children. Sleep Med. 9:260–5. doi: 10.1016/j.sleep.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 26.Egger M, Smith GD. Bias in location and selection of studies. BMJ. 316:61–6. doi: 10.1136/bmj.316.7124.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Controlling the risk of spurious findings from meta-regression. Stat Med. 23:1663–82. doi: 10.1002/sim.1752. [DOI] [PubMed] [Google Scholar]
- 28.Thompson SG, Higgins JP. Treating individuals 4: can meta-analysis help target interventions at individuals most likely to benefit? Lancet. 365:341–6. doi: 10.1016/S0140-6736(05)17790-3. [DOI] [PubMed] [Google Scholar]
- 29.Kheirandish L, Row BW, Li RC, Brittian KR, Gozal D. Apolipoprotein E-deficient mice exhibit increased vulnerability to intermittent hypoxia-induced spatial learning deficits. Sleep. 28:1412–7. doi: 10.1093/sleep/28.11.1412. [DOI] [PubMed] [Google Scholar]
- 30.Robertson J, Curley J, Kaye J, Quinn J, Pfankuch T, Raber J. ApoE isoforms and measures of anxiety in probable AD patients and Apoe-/- mice. Neurobiol Aging. 26:637–43. doi: 10.1016/j.neurobiolaging.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Saarelainen S, Lehtimaki T, Nikkila M, Solakivi T, Nieminen MM, Jaakkola O. Association between apolipoprotein E alleles and autoantibodies against oxidised low-density lipoprotein. Clin Chem Lab Med. 38:477–8. doi: 10.1515/CCLM.2000.068. [DOI] [PubMed] [Google Scholar]
- 32.Spira AP, Blackwell T, Stone KL, et al. Sleep-disordered breathing and cognition in older women. J Am Geriatr Soc. 56:45–50. doi: 10.1111/j.1532-5415.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 33.Westbrook PR, Levendowski DJ, Cvetinovic M, et al. Description and validation of the apnea risk evaluation system: a novel method to diagnose sleep apnea-hypopnea in the home. Chest. 128:2166–75. doi: 10.1378/chest.128.4.2166. [DOI] [PubMed] [Google Scholar]
- 34.Infante J, Sanz C, Fernandez-Luna JL, Llorca J, Berciano J, Combarros O. Gene-gene interaction between interleukin-6 and interleukin-10 reduces AD risk. Neurology. 63:1135–6. doi: 10.1212/01.wnl.0000138570.96291.a8. [DOI] [PubMed] [Google Scholar]
- 35.Jia LY, Tam PO, Chiang SW, et al. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern Chinese. Mol Vis. 15:89–98. [PMC free article] [PubMed] [Google Scholar]
- 36.Kao WT, Yen YC, Lung FW. The effects of beta2 adrenergic receptor gene polymorphism in lipid profiles. Lipids Health Dis. 7:20. doi: 10.1186/1476-511X-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng DQ, Zhao SP, Nie S, Li J. Gene-gene interaction of PPARgamma and ApoE affects coronary heart disease risk. Int J Cardiol. 92:257–63. doi: 10.1016/s0167-5273(03)00101-3. [DOI] [PubMed] [Google Scholar]
- 38.Sorli JV, Corella D, Frances F, et al. The effect of the APOE polymorphism on HDL-C concentrations depends on the cholesterol ester transfer protein gene variation in a Southern European population. Clin Chim Acta. 366:196–203. doi: 10.1016/j.cca.2005.10.001. [DOI] [PubMed] [Google Scholar]